Abstract
Background and Objectives:
Bacteria need iron for growth and most of them can actively acquire Fe ions using especial iron-chelating proteins which named siderophores. We aimed to determine the frequencies of iucA, iroN and irp2 genes in the uropathogenic Escherichia coli (UPEC) isolates. We also analyzed the effects of siderophore genes beside iron supplements on growth rate of the isolates.
Materials and Methods:
Totally, 170 E. coli strains were isolated from urinary tract infections and the presence of 3 siderophore genes were analyzed using PCR among them. Three final concentrations of 0.1, 0.5 and 1 mMFe(II) and Fe(III) ions were made in M9 broth medium. Inoculated cultures were incubated at 37°C for 33 hours and bacterial density in the suspension was measured with 1 hour intervals using spectrophotometer.
Results:
The frequency of iucA, iroN and irp2 genes among 170 UPEC isolates were 29 (17.1%), 52 (30.6%) and 116 (68.2%), respectively. In addition, Our findings showed that Fe(II) supplements had significantly higher promoting effects on UPEC growth rate almost in all of the three applied concentrations (0.1, 0.5 and 1 mM) compared to the control group (P<0.0001). Differences between Fe(III) supplemented groups and the controls were statistically significant when 1 mM concentration was added into the medium (p<0.05).
Conclusion:
irp2 gene probably plays a major role in the pathogenesis of UPEC strains. Promoting or inhibitory effects of iron on bacterial growth mainly depend on the iron concentration in the culture medium however different siderophores have different potentials for capturing and assimilation of Fe ions by the bacteria, especially inside the host cell.
Keywords: Escherichia coli, Siderophore, Aerobactin, Salmochelin, Yersiniabactin, Fe (II) supplements
INTRODUCTION
Iron is an essential element for supporting growth and development of any living microorganism including E. coli. Iron, as a unique catalyzer, contributes to the initiation and progression of many biological pathways including tricarboxylic acid (TCA) cycle, electron transport chain (ETC), oxidative phosphorylation, nitrogen fixation and biosynthesis of many aromatic compounds (1). Bacterial growth and development need at least 10−6 M iron (2). On the other hand, the higher iron concentrations may be cytotoxic and show bactericidal or antimicrobial effects on the microorganisms. Association between iron concentration and microbial infection has been demonstrated by many scientists through empirical studies in humans or laboratory animals (3). Mammals usually employ an efficient defense strategy against bacterial pathogens such as high affinity iron-chelating molecules (like transferrin and lactoferrin). To scavenge free iron ions, these molecules will be released which lead to deprivation of invader microorganism from accessing the ions (4). In contrast, during urinary tract infections, uropathogenic E. coli (UPEC) strains employ different strategies including synthesis of several high affinity and low molecular weight proteins which are named siderophores (1). Siderophores belong to a chemically diverse family of molecules produced by many pathogenic and non-pathogenic bacteria to chelate iron ions (5). Siderophores are produced and secreted into the surrounding environment, have higher affinity to ferric than ferrous ions. Ferric-bound siderophore is captured and internalized by the bacteria using specific outer membrane receptors (1). E. coli strains may synthesize 4 types of siderophores named enterobactin, salmochelin, yersiniabactin and aerobactin. Many non-pathogenic E. coli strains were only capable for synthesizing enterobactin siderophore which is encoded by ent-fes-fep gene cluster (6). However, UPEC strains are also able to synthesize siderophores other than enterobactin which help bacteria for better survival in the iron-limited conditions like urinary tract. Bacterial aerobactin is encoded by iuc DBAC gene cluster whereas the salmochelin is encoded by iroA gene cluster and synthesis of yersiniabactin is encoded by high pathogenicity island (HPI) and irp2, irp1, ybtSETUXPQA gene clusters (7–9). Several studies have demonstrated the significant associations between aerobactin synthesis and certain infectious conditions like pyelonephritis and cystitis (10). Further surveys of female patients with recurrent urinary tract infections have revealed significant higher frequency of salmochelin and aerobactinsiderophore expression in isolated bacterial strains and these siderophores were considered as the main factors highly associated with recurrent infections and drug resistance (11). Current study was designed to survey the effect of two or three valent iron ions concentrations on the growth rate of UPEC strains isolated from urinary tract infections. We also determined the frequency of genes encoding aerobactin, salmochelin and yersiniabactin siderophores among isolated strains.
MATERIALS AND METHODS
Sample collection and bacterial identification
A cross-sectional descriptive analytic study has been done. During the study period, mid-stream urine samples were collected from suspected urinary tract infections from patients who admitted to university dependent healthcare centers in Gorgan, center of Gholestan province (North of Iran) or samples from outpatients with the same condition referred to the private sector medical laboratories. Standard conventional procedures were applied for isolation and identification of enterobacteriaceae including IMViC biochemical tests and finally a total of 170 E. coli strains were isolated in M9 broth media after 24 h incubation at 37°C.
Genomic DNA extraction and PCR amplification
Boiling method was used genomic DNA extraction according to a previously published study (12). To examine the presence of IucA, IroN and Irp2 genes among E. coli isolates, we used 3 specific pairs of primers (Table 1). PCR conditions were set according to the study conducted by Karimian et al (13) and the products were visualized on 1.5% agarose gel.
Table 1.
Primer pairs for PCR amplification of IucA, IroN and Irp2 genes in uropathogenic E. coli isolates
Culture of bacteria in media supplemented with Fe(II) or Fe(III) ions:
Selection of representative strains
Based on PCR results, e.g., presence or absence of irp2, iroN, and iucA genes in the bacteria, 170 UPEC isolates were divided into 8 groups and one representative strain from each group were selected for evaluating iron element effects on bacterial growth rate and correlating the rate with the types of siderophores produced by the bacteria. Eight representative strains and details of their genotypes were presented in Table 2.
Table 2.
Based on PCR results, eight representative strains were selected to study iron effect on microbial growth rate.
| Strain name | iucA | iroN | irp2 |
|---|---|---|---|
| E-132 | + | − | − |
| E-331 | − | − | + |
| E-131 | + | + | − |
| E-462 | − | + | − |
| E-335 | − | + | + |
| E-477 | + | − | + |
| E-66 | + | + | + |
| E-501 | − | − | − |
Preparation of Fe2+ and Fe3+ stock solutions
For preparation of 20 mM Fe2+ stock solution, 556 mg ferrous sulfate (FeSO4, 7H2O, Merk, Germany) was weighed and dissolved in distilled water (final volume: 100ml). For preparation of 20 mM Fe3+ stock solution, 541 mg ferric chloride (FeCl3, 6H2O, Merk) was weighed and dissolved in DW (final volume 100 ml). Both stock solutions were filter sterilized (using a 0.4 micron pore size filter).
Culture media
M9 broth medium (45-63011-500G-F, Fluka, Germany) supplemented with 0.5% glucose was used for supporting bacterial growth. The medium was prepared according to the manufacturer recommendations and autoclaved at 121°C for 15 min for sterilization.
Plotting growth curve in the absence and presence of iron supplements
Representative strains were cultured in M9 broth medium and incubated at 37°C for 24 hours. One ml of the suspension was inoculated into 4 ml freshly prepared M9 broth media and incubated again at 37°C until the turbidity of the suspension was reached to 0.5 McFarland standard turbidity (1.5 × 108 cells/ml). A 2.5 ml sample from each standardized bacterial suspension was added to 50 ml of M9 broth media and serial dilution were prepared. Finally a working suspension containing 106 bacterial cells per ml was resulted. Then 250, 1250 and 2500 μl from the two stock iron solutions were added to the bacterial suspensions (final iron concentrations equal to 0.1, 0.5 and 1 mM). Turbidity of these tubes was measured and considered as the zero point for bacterial growth curve prediction. A series of control tubes containing media plus bacterial suspension, but omitting iron addition step was included. Blank tube series were also included in the experiment. The blanks had M9 broth media and iron but not bacterial strains. All tests, control, and blank tubes were incubated at 37°C with shaking. Optical density (OD) of tubes was read at 600 nm with one hour intervals using a spectrophotometer and the resulting data was used for plotting growth curve of the each bacterial isolate (17–19). To achieve the highest accuracy, all experiments were repeated two or more times. For plotting growth curves log10 values of bacterial densities were presented on Y axis and the time points, as hours, went on X.
Statistical analysis
SPSS software (v. 16.0) was used for statistical analysis. ANOVA was used for comparing the mean values between all studied groups. Tukey test was used for testing equality of variances. Two independent-samples t-test was used for binary comparisons. P value less than 0.05 was considered as statistically significant.
RESULTS
Distribution of siderophore genes
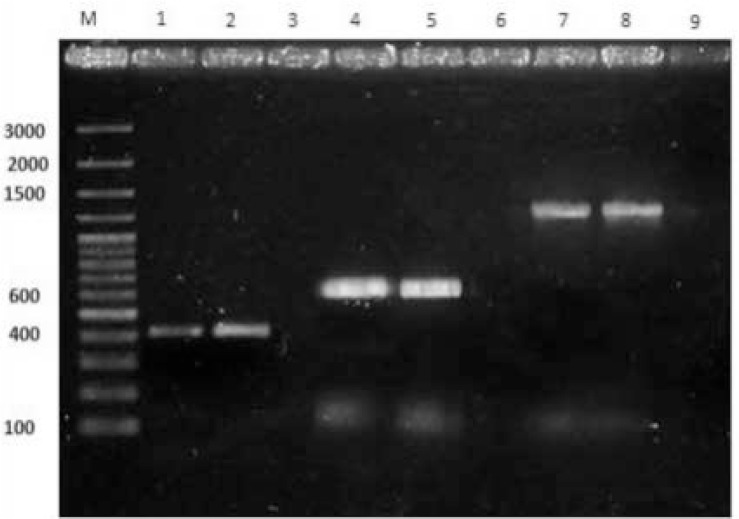
PCR amplification of three siderophore genes among 170 UPEC isolates revealed that iucA, iroN and irp2 genes were present among 29 (17.1), 52 (30.8%) and 116 (68.2%) strains, respectively (Fig. 1). In addition, 38 isolates (22.4%) did not possess any of the studied genes and 7 isolates (4.1%) had all of the 3 genes. Four isolates (2.4%) had only aerobactin gene (iucA), 60 isolates (35.3%) were only positive for yersiniabactin gene (irp2) and 10 isolates (5.9%) only represented salmochelin gene (iroN). Two isolates (1.2%) carried both aerobactin and salmochelin genes (iucA/iroN). Sixteen isolates (9.4%) showed positivity for both aerobactin and yersiniabactin genes (iucA/irp2). Finally, 33 isolates (19.4%) had both of salmochelin and yersiniabactin genes (iroN/irp2).
Fig. 1.
PCR amplification of irp2, iroN and iucA. M: DNA molecular weight marker, lanes 1, 2(Irp2), 4, 5(IroN), 7, 8(IucA): PCR aproducts from positive strains relating to the three siderophore genes. lanes of 3,6,9: Negative strains lacking any of the three detected siderophore genes.
Effects of supplemental Fe2+/Fe3+ ions on bacterial growth
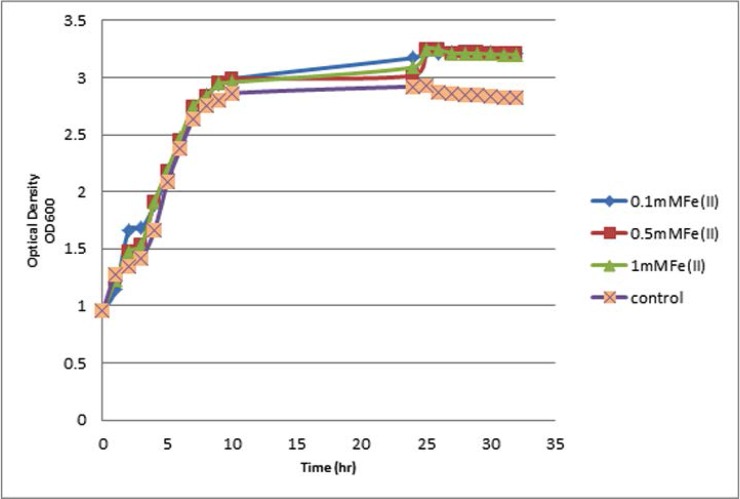
Incubation of selected strains in broth media supplemented with 0.5% glucose and three distinct concentrations of Fe2+ or Fe3+ ions (0.1, 0.5 and 1 mM) and continuous monitoring of cellular density in both test and control groups revealed that strains harboring siderophores were better survived and regenerated in all three concentrations of iron ions. In addition comparing the Fe2+ ion supplementation effects on the bacterial growth rate using ANOVA statistical test clearly showed that there were obvious differences between test and control cultures regarding cell growth rate. Supplementing the media with Fe2+ represented growth promoting effects on tested bacteria nearly in all three concentrations which represented statistically significant differences (P<0.0001) (Fig. 2).
Fig. 2.
Growth curve of E. coli E-331 strain harboring yersiniabactinsiderophore in the presence or absence of Fe(II) ions
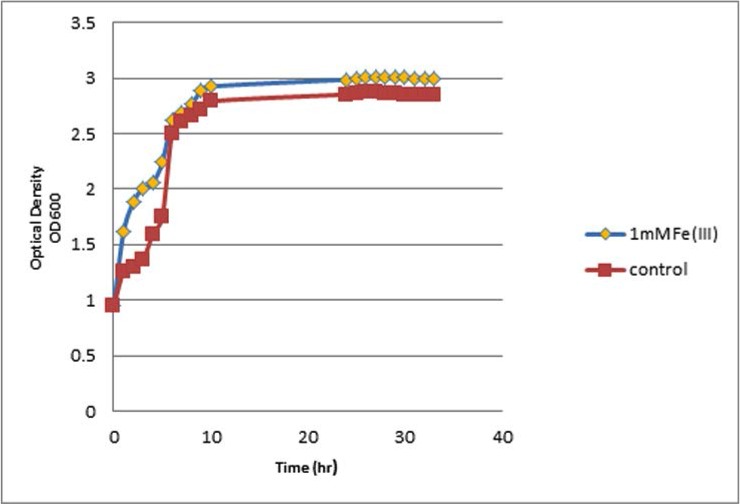
The effects of different Fe3+ ion concentrations revealed that Fe3+ ions at concentrations of 0.1, 0.5, and 1 mM have increased growth effects on the bacterial strains; however, bacteria cultured in media supplemented with 1 mM Fe3+ ions exhibited a statistically significant increase in growth rates, when compared with the control cultured with no iron supplements (Tukey test, P<0.05) (Fig. 3). More importantly, our findings showed that bacteria cultured with Fe2+ supplementation had higher growth rates as compared with those cultured with Fe3+ supplementation (P<0.0001, Table 3).
Fig. 3.
Growth curve of E. coli E-131 strain harboring both aerobactin and salmochelinsiderophores in the presence or absence of Fe(III) ions
Table 3.
Compare statistical effect Fe2+/Fe3+ ions on productivity growth of strains according to the type of siderophore
| Strian | Fe3+ | Fe2+ | ||||
|---|---|---|---|---|---|---|
| 1mM | 0.5mM | 0.1mM | 1mM | 0.5mM | 0.1mM | |
| E-132 (IucA) | ||||||
| E-131 (IroN-IucA) | ||||||
| E-331 (Irp2) | ||||||
| E-462 (IroN) | ||||||
| E-335 (Irp2-IroN) | P value=0.0001 | P value=0.05 | ||||
| E-477 (IucA-Irp2) | ||||||
| E-66 (IroN-IucA-Irp2) | ||||||
| E-501 (Non siderophor) | ||||||
The effect of siderophore types on bacterial growth in the presence of Fe2+/Fe3+ ions
Among selected strains cultured in the presence of Fe2+ ion, E-331 strain containing irp2 gene had the highest growth rate in the presence of 0.1, 0.5 and 1 mM Fe2+ followed by E-66 strain containing all three identified siderophore genes (iucA, iroN and irp2). E-131 strain containing salmochelin and aerobactin genes also showed higher growth rate in the presence of 1 mM concentration of Fe2+.
Among selected strains cultured and supplemented with Fe3+ ions, E-335 strain harboring yersiniabactin and salmochelin genes had the highest growth rate in the presence of 0.1, 0.5 and 1 mM Fe3+. In the presence of 1 mM Fe3+, the highest bacterial density belonged to E-132 strain represented aerobactin gene (iucA) followed by E-131 strain containing aerobactin and salmochelin genes (iucA/iroN). In general, all bacterial control samples lacking iron supplementation showed obvious delay in growth compared to the supplemented media.
DISCUSSION
Access to iron is of crucial importance for bacterial infections. The invading microbes often have to overcome the iron-limiting conditions imposed by the presence of high affinity iron-binding proteins in the body fluids of vertebrate hosts. UPEC strains, especially during urinary tract infections, implement different strategies for efficient iron acquisition from the iron-limited surrounding environment. One of the important strategies for survival and growth of bacteria in the environment is the synthesis of low molecular weight, high affinity iron-chelating agents named siderophores. Hitherto et al. has shown that UPEC strains may express aerobactin, salmochelin or yersiniabactin siderophores during the infection. Siderophores are usually bound to free Fe(III) ions and are also capable to dissociate ions from host-related iron-chelating molecules like transferrin, lactoferrin or haptoglobin. Finally, Fe(III)-siderophore molecules are recaptured from the environment and internalized by the bacteria using receptor-mediated mechanisms (1–4). Our findings showed that among E. coli strains isolated from urinary tract infections, the most frequently detected siderophore gene is irp2 (68.2%) which encodes for yersiniabactin. This finding is in agreement with the results published by Abdi et al. in which 89% of isolated UPEC strains originated from urinary tract infections contained irp2 gene positive (20). In the study conducted by Karimian et al. different frequencies were observed for these three siderophore genes among UPEC strains isolated from patients admitted to the emergency section of a university hospital in Tehran. The reported frequencies for iucA, iroN and irp2 genes in this study were 10.6, 42.3 and 11.4%, respectively (13). Which are clearly different from our findings. The differences may be attributable to the different geographical locations. Momtaz et al. investigated frequency of irp2 gene among 123 UPEC strains isolated from urinary tract infections of Iranian patients and showed that 14 out of 123 strains (11.4%) had the gene sequence in their genomic DNA (21). Nateghi et al. also analyzed the frequency of irp2 and iucD genes among 100 UPEC isolates from Iranian patients and the frequencies were 33 and 0%, respectively (22). In 2014, Ali-Abdi et al. found that 40 and 83.6% of UPEC strains isolated from urinary tract infections of Iranian patients had iroN and iucA genes, respectively (23), which is clearly higher compared with our results. In 2011, Usein et al. studied the frequency of irp2 and iucC genes among 93 E. coli strains isolated from vaginal fluid samples of Romanian women and found that the frequency of these genes was 85 and 51%, respectively (24). Inconsistent with our results, frequency of irp2 gene was high (85 vs. 68.2% in our study). In a comparative study, Bauer et al. analyzed the frequency of iroN gene among E. coli strains isolated from urinary tract infections as well as feces of American patients and the results showed that 28 out of 74 UTI isolates (38%) and 34 out of 184 feces isolates (18.5%) had iroN gene (25). iroN gene frequency in this study was in close consistency with our results (38 vs. 30.8% in our study). In another study from Brazil, the frequency of iucD gene among 162 UPEC strains isolated from cystitis patients was 27.8percnt; (26) which is very close to our finding.
Several studies have been conducted to explore the effect of iron supplementation on bacterial growth. Kalantari et al. evaluated the influence of iron on growth rate of E. coli in nutrient broth as the culture medium (17). Obvious reduction in bacterial growth rate was reported in the presence of 1 mM Fe+3 or 0.5 mM Fe+2 compared with the control cultures without any supplemented iron. In addition, bacterial growth was totally inhibited in the presence of 1 mM Fe+2 (17). In the present study which was carried out using M9 broth medium, 8 UPEC strains containing different combinations of siderophore genes, iron addition not only had no toxic (inhibitory) effect but also showed promoting effect on bacterial growth even in the presence of up to 1 mM Fe+2 or Fe+3 ions. Although, both Fe+2 or Fe+3 ions had positive effects on the bacterial growth, however, the effect is further prominent in cultures supplemented with Fe+2 ions in comparison with matched cultures supplemented with Fe+3 and the differences were statistically significant. A study conducted by Chatterjee et al. the effect of iron oxide nano particles on the growth kinetics of E. coli was investigated in India. LB broth media supplemented with 50, 100, 150 and 200 μg/ml of the nano particles (Fe3O4) were used for supporting bacterial growth. In this experiment, iron nano-particles showed notable inhibitory effects on the growth of the exposed strains and the time needed for reaching to logarithmic growth phase of the bacteria was gradually decreased when the particle concentration in the medium was harmonically increased. In addition, bacterial growth was totally inhibited in higher concentrations of the nano particles (27). Another experiment Kalantari et al. compared the effect of ferric and ferrous ions on the growth of two bacterial species, Bacillus cereous and E. coli. The study showed that 11.2 mg/l concentration of ferrous or ferric ions in the culture medium could completely cease the growth of E. coli and B. cereous. In addition, 5.6 mg/l concentration of these two ions led to 48.4% reduction in the growth of B. cereous after 3.5 hours of incubation but did not show any inhibitory effect on the growth of E. coli strains. This study emphasized on the toxic effect of high concentrations of iron ions for these two bacteria (18). During the last decade, studies have demonstrated that pathogenic E. coli strains had higher iron requirements than environmental isolates. It has also been demonstrated that pathogenic strains isolated from human infections are able to express higher levels of siderophores in both iron-limited and iron-enriched conditions (28, 29).
Our study showed all the eight isolated UPEC can grow at higher rate in Fe2+ and Fe3+ concentrations including 0. 1, 0.5 and 1mM, regardless of siderophore genes. Nonetheless isolated UPEC bearing yersiniabactin and aerobactin siderophore genes has the highest growth in compare with other siderophore. This finding suggests that the higher growth rate of this to UPEC may be result in higher affinity of this siderophore to iron in compare with other siderophore. a hypothesis that should be checked in the future experiments.
CONCLUSION
As for the highest frequency of irp2 gene, encoding for yersiniabactin siderophore, observed among 170 UPEC isolates in the current study, it seems that irp2 gene plays a major role in the pathogenesis of the bacteria and as a potential virulence factor which may assure the bacterial survival in the iron-limited conditions. Iron is an important element in the culture media which can promote or inhibit bacterial growth based on its concentrations. We also found that different siderophores have different potentials in capturing and assimilation of Fe ions by the bacteria.
ACKNOWLEDGEMENT
We thank the staff of Golestan University of Medical Sciences, Maya Babai and Hanie Bagheri for providing isolates and Masood Bazori and Naemeh Javid for their logistical supports. This study was supported by a grant awarded to the Infectious Disease Research Center and Technology and Research Deputy of Golestan University of Medical Sciences (contract no.930611110).
REFERENCES
- 1.Messenger AJ, Barclay R. Bacteria, iron and pathogenicity. Biochem Educ 1983;11:54–63. [Google Scholar]
- 2.Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A 2003;100:3584–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelvan D. Enhancement of adriamycin toxicity by iron chelates is not a free radical mechanism. Biol Trace Elem Res 1997;56:295–309. [DOI] [PubMed] [Google Scholar]
- 4.Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol 2004;2(12):946–953. [DOI] [PubMed] [Google Scholar]
- 5.Bagg A, Neilands J. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev 1987;51:509–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol 2000;54:881–941. [DOI] [PubMed] [Google Scholar]
- 7.Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins, siderophores of Salmonella enterica and Uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor Iron. Proc Natl Acad Sci U S A 2003;100:3677–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carniel E. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect 2001;3:561–569. [DOI] [PubMed] [Google Scholar]
- 9.Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev 1991;4:80–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slavchev G, Pisareva E, Markova N. Virulence of Uropathogenic Escherichia coli. J Cult Collect 2013;6:3–9. [Google Scholar]
- 11.Ejrnæs K. Bacterial characteristics of importance for recurrent urinary tract infections caused by Escherichia coli. Dan Med Bull 2011;58:B4187. [PubMed] [Google Scholar]
- 12.Salehi TZ, Madani SA, Karimi V, Khazaeli FA. Molecular genetic differentiation of Avian Escherichia coli by RAPD-PCR. Braz J Microbiol 2008;39:494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karimian A, Momtaz H, Mahbobe Madani M. Detection of Uropathogenic Escherichia coli virulence factors in patients with urinary tract infections in Iran. Afr J Microbiol Res 2012;6:6811–6816. [Google Scholar]
- 14.Tivendale KA, Allen JL, Ginns CA, Crabb BS, Browning GF. Association of iss and iucA, but not tsh, with plasmid-mediated virulence of Avian pathogenic Escherichia coli. Infect Immun 2004;72:6554–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 2000;181:261–272. [DOI] [PubMed] [Google Scholar]
- 16.Ewers C, Janben T, Kiebling S, Philipp H-C, Wieler LH. Rapid detection of virulence-associated genes in Avian pathogenic Escherichia coli by multiplex polymerase chain reaction. Avian Dis 2005;49:269–273. [DOI] [PubMed] [Google Scholar]
- 17.Kalantari N, Ghaffari S. Evaluation of toxicity of heavy metals for Escherichia coli growth. IJEHSE 2008;5:173–178. [Google Scholar]
- 18.kalantari N, Moshtaghi AS. Comparison of the effects of ferric and ferrous iron on the growth of B. cereus and E. coli (persian). J Gorgan Uni Med Sci 2001;3:37–40. [Google Scholar]
- 19.Murugappan R, Aravinth A, Rajaroobia R, Karthikeyan M, Alamelu M. Optimization of MM9 medium constituents for enhancement of siderophoregenesis in marine Pseudomonas putida using response surface methodology. Indian J Microbiol 2012;52:433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rashki A. The phylogenetic study of Uropathogenic Escherichia coli strains in Sistan of Iran. J Birjand Uni Med Sci 2014;21:385–393. [Google Scholar]
- 21.Momtaz H, Karimian A, Madani M, Safarpoor Dehkordi F, Ranjbar R, Sarshar M, et al. Uropathogenic Escherichia coli in Iran: serogroup distributions, virulence factors and antimicrobial resistance properties. Ann Clin Microbiol Antimicrob 2013;12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nateghi F, Jafarpour M, Nazemi A. A survey for detection of eight correlated genes of Avian pathogenic Escherichia coli in human Uropathogenic Escherichia coli. JMW 2010;3:169–176. [Google Scholar]
- 23.Abdi HA, Rashki A. Comparison of Virulence Factors Distribution in Uropathogenic E. coli Isolates From Phylogenetic Groups B2 and D. Int J Enteric Pathog 2014;2:e21725. [Google Scholar]
- 24.Usein C-R, Grigore LA, Georgescu RM, Bãltoiu MC, Condei M, Teleman MD. Phylogenetic background and extraintestinal virulence genotypes of Escherichia coli vaginal strains isolated from adult women. Rev Romana Med Lab 2011; 19: 37. [Google Scholar]
- 25.Bauer RJ, Zhang L, Foxman B, Siitonen A, Jantunen ME, Saxen H, et al. Molecular epidemiology of 3 putative virulence genes for Escherichia coli urinary tract infection–usp, iha, and iroN E. coli. J Infect Dis 2002;185:1521–1524. [DOI] [PubMed] [Google Scholar]
- 26.Tiba MR, Yano T, Leite DdS. Genotypic characterization of virulence factors in Escherichia coli strains from patients with cystitis. Rev Inst Med Trop São Paulo 2008;50:255–260. [DOI] [PubMed] [Google Scholar]
- 27.Chatterjee S, Bandyopadhyay A, Sarkar K. Effect of iron oxide and gold nanoparticles on bacterial growth leading towards biological application. J Nanobiotechnology 2011;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinberg ED. Iron and infection. Microbiol Rev 1978;42:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinberg ED. Iron withholding: a defense against infection and neoplasia. Physiol Rev 1984;64:65–102. [DOI] [PubMed] [Google Scholar]