Abstract
High fructose consumption in the Western diet correlates with disease states such as obesity and metabolic syndrome complications, including type II diabetes, chronic kidney disease, and nonalcoholic fatty acid liver disease. Liver and kidneys are responsible for metabolism of 40–60% of ingested fructose, while the physiological fate of the remaining fructose remains poorly understood. The primary metabolic pathway for fructose includes the fructose-transporting solute-like carrier transport proteins 2a (SLC2a or GLUT), including GLUT5 and GLUT9, ketohexokinase (KHK), and aldolase. Bioinformatic analysis of gene expression encoding these proteins (glut5, glut9, khk, and aldoC, respectively) identifies other organs capable of this fructose metabolism. This analysis predicts brain, lymphoreticular tissue, placenta, and reproductive tissues as possible additional organs for fructose metabolism. While expression of these genes is highest in liver, the brain is predicted to have expression levels of these genes similar to kidney. RNA in situ hybridization of coronal slices of adult mouse brains validate the in silico expression of glut5, glut9, khk, and aldoC, and show expression across many regions of the brain, with the most notable expression in the cerebellum, hippocampus, cortex, and olfactory bulb. Dissected samples of these brain regions show KHK and aldolase enzyme activity 5–10 times the concentration of that in liver. Furthermore, rates of fructose oxidation in these brain regions are 15–150 times that of liver slices, confirming the bioinformatics prediction and in situ hybridization data. This suggests that previously unappreciated regions across the brain can use fructose, in addition to glucose, for energy production.
Keywords: fructose toxicity, brain energy metabolism, fructose transporters
1. Introduction
The amount of fructose in the Western diet has increased by 26% since 1977, and it is a major sweetener in processed foods (Gaby). Currently, fructose may constitute 5–15% of daily caloric intake (Ishimoto et al., 2012; Ouyang et al., 2008). In humans, chronic fructose ingestion correlates with the development of a number of disease states including the development of nonalcoholic fatty liver disease (NAFLD), inflammation of liver and kidneys resulting in steatophepatitis and chronic kidney disease, insulin resistance and type II diabetes, atherosclerosis, hyperuricemia, lactic acidosis, obesity, dyslipidemia, and cataracts (Basaranoglu et al., 2015; Bergheim et al., 2008; Douard and Ferraris, 2008; Gersch et al., 2007; Mayes, 1993; Nakayama et al., 2010; Ouyang et al., 2008; Thuy et al., 2008). The liver and kidneys absorb approximately 40–60% of fructose (Holdsworth and Dawson, 1964; Wolfe et al., 1975), but the fate of the remaining fructose remains poorly understood. These disease states that correlate with excessive fructose consumption highlight the need for a better understanding of fructose metabolism throughout the body.
In order for a tissue to participate in fructose metabolism, cells must express the genes for fructose-specific transporters, such as glut51 (expressed in the kidneys and small intestine) and glut2 (expressed in the liver and small intestine) (Burant and Saxena, 1994; Burant et al., 1992; Douard and Ferraris, 2008; Helliwell et al., 2000; Litherland et al., 2004). Recently, the number of transporters in the solute-carrier 2A family (SLC2A, protein symbol GLUT) that have been characterized as being fructose transporters has expanded. GLUTs 2, 5, 7, 9, and 11 are all localized to the plasma membrane and have fructose-transporting activity. GLUTs 2, 7, 9, and 11 transport other small molecules in addition to fructose (Anzai et al., 2008; Gaster et al., 2004; Scheepers et al., 2005; Thorens and Mueckler, 2010), whereas GLUT5 is specific for fructose transport (Rand et al., 1993).
Once fructose enters the cell, metabolism proceeds through either the fructose-1-phosphate (Fru 1-P) or the fructose-6-phosphate (Fru 6-P) pathways, depending on the activity of ketohexokinase (KHK) or hexokinase (HK), respectively. The KHK-dependent Fru-1-P pathway starts with KHK phosphorylation of fructose at the C1-hydroxyl group, yielding Fru 1-P, which is then cleaved by aldolase into dihydroxyacetone phosphate (DHAP) and glyceraldehyde. Fru-1-P metabolism via KHK, which is not product-inhibited (Adelman et al., 1967), bypasses two of the major regulatory enzymes in glycolysis, HK, and phosphofructokinase I. The importance of KHK in disease due to this unregulated metabolism was recently demonstrated in mice genetically lacking the ubiquitously expressed isoform, KHK-A, but still expressing the liver and kidney isoform, KHK-C (Ishimoto et al., 2012). In the khkA−/− mouse, fructose metabolism can only occur in the liver and kidneys, and high dietary fructose more readily induces liver and kidney pathology compared with wild-type (WT) mice expressing both KHK-A and -C exposed to the same dietary fructose loads. This demonstrates extra-hepatic fructose metabolism via KHK-A activity is physiologically relevant, and highlights the importance of understanding whole-body fructose metabolism.
In the hexokinase (HK)-dependent Fru-6-P pathway, fructose competes with glucose for phosphorylation by HK at the C6-hydroxyl, creating the glycolytic intermediate Fru 6-P. Glucose is the primary substrate for HK; the ubiquitously expressed HK-I has a Km value <1 mM for glucose (Schimke and Grossbard, 1968; Sols and Crane, 1954). The Km value of HK for fructose is 10-fold higher (Stocchi et al., 1982) than that for glucose. While physiological concentrations of fructose realized by different organs are currently unknown, it is clear that physiological concentrations of glucose would provide stiff competition for fructose utilization of HK in vivo. The competing Fru-1-P and Fru-6-P pathways, along with the participating enzymes, are depicted in Fig 1.
Fig 1. Fru-1-P and Fru-6-P pathways for fructose catabolism.
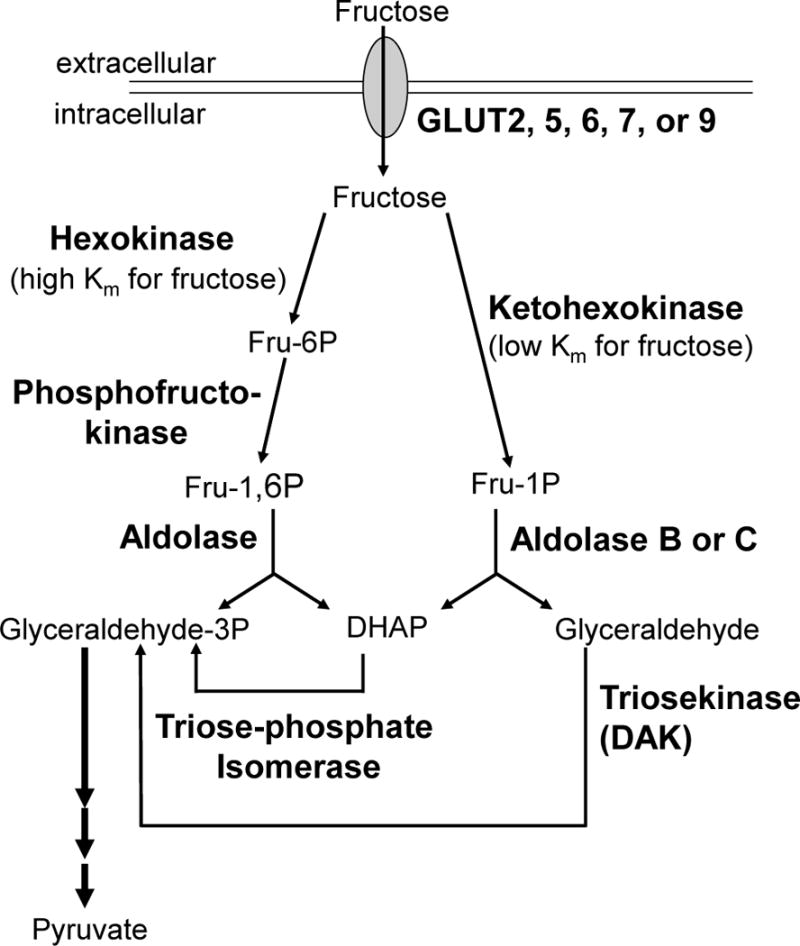
Arrows depict pathways from fructose to pyruvate. Plasma membrane is indicated by double line. Transporters are all capital bold and critical enzymes are bold. Intermediates Fru 6-P, Fru 1-P, Fru 1,6-P2, DHAP, Glyceraldehyde-3-P, Pyruvate, and Glyceraldehyde are indicated.
After phosphorylation by KHK to Fru 1-P, or by HK and phosphofructokinase to Fru 1,6-P2, fructose metabolism continues through cleavage by aldolase. The three aldolase isozymes (A, B, and C) differ in their tissue distribution and their enzymatic efficiency toward Fru 1-P (Penhoet et al., 1969). Aldolase B is expressed in the liver and kidneys, and has the smallest difference in values for the substrate specificity constants (kcat/Km) toward cleavage of either Fru 1-P or Fru 1,6-P2 (Penhoet and Rutter, 1971; Pezza, et al., 2003; Kusakabe, et al., 1994) (see Supplemental Material, Table S4). Aldolase A is expressed ubiquitously, including in cells where aldolase C is concurrently being expressed, and has the largest difference in values for the substrate specificity constants for these two phosphorylated hexoses. Aldolase C is expressed in nervous tissue and smooth muscle (Baron et al., 1999). The role of aldolase C has been an enigma since its discovery due to its properties intermediate between A and B, including its substrate specificities toward these two hexoses (Penhoet and Rutter, 1971). Interestingly, human aldolase C has ratios of kcat/Km for Fru 1,6-P2 to Fru-1-P that are closer to aldolase B (3–5 fold) than to aldolase A (15–26 fold)(Kusakabe et al., 1994; Penhoet and Rutter, 1971; Pezza et al., 2003)(see Supplemental Material, Table S4). This indicates that among the human isozymes aldolase C is more likely participating in fructose metabolism than aldolase A given that its substrate preferences are closer to aldolase B.
Accordingly, any tissue expressing a GLUT gene capable of constitutive fructose transport from the blood stream (gluts 2, 5, 7, 9, or 11), the KHK gene (khk), and aldolase B or C genes (aldoB or aldoC) should be capable of metabolizing fructose via the Fru-1-P pathway. Tissues capable of fructose metabolism were identified by bioinformatic analysis using the Virtual Northern Blot (VNB) (Funari et al., 2010). In addition to expression in the liver and kidney, this analysis revealed significant expression of genes necessary for fructose metabolism in the brain. In situ hybridization (ISH) of brain slices, along with Western Blots of GLUTs 5 and 9, and measurements of specific activity of the enzymes KHK and aldolase, confirmed the expression of these proteins. Measurement of hexose oxidation rates of dissected brain tissue was used to investigate the rate of fructose metabolism, and the contribution of KHK versus HK, to this metabolism in the brain. It was clear from these data that several regions of the brain, including the cerebellum, hippocampus, cerebral cortex, and olfactory bulb, were capable of significant fructose metabolism. Additionally, this fructose metabolism was enhanced in brains after exposure to high-fructose in the diet.
2. Results
2.1 Bioinformatic prediction of the brain as a site for fructose metabolism
Prediction of tissues other than the liver and kidneys that express genes required for fructose metabolism via the Fru-1-P pathway was performed using the virtual northern blot program (VNB) (Funari et al., 2010). This tool compiled qualitative and quantitative expression profiles for the genes in the Fru-1-P pathway (fructose transporters gluts 2, 5, 7, 9, and 11; khk, aldoB, and aldoC), and revealed brain and nervous tissue as potentially major sites capable of fructose metabolism (see Supplemental Material, Tables S1–S3). The VNB correctly identified the liver and kidney as sites for expression of all the genes necessary for this pathway, but notably, spleen/lymphoreticular tissue and brain/nervous tissue were also in the top four tissues predicted to have high expression of khk, with expression levels higher than or comparable to kidney for the other genes (aldos and gluts). By mass, the brain is the largest of these organs, and was the focus of this study.
2.2 In situ hybridization (ISH) shows certain brain regions express all the genes in the Fru-1-P pathway
The predicted expression of genes necessary for fructose metabolism in the brain/nervous tissue (Table 1) was verified by ISH. Both sense (non-complementary) and antisense (complementary) DIG-labeled RNA probes were generated for glut5, glut9, khk, and aldoC. Each probe was used in succession on adjacent 14 μm coronal slices of brains from adult male BL6 mice. All sense probes hybridized to tissue at background levels for all of the genes tested (see Supplemental Material, Fig S1).
Table 1.
Activities of enzymes in Fru-1-P pathway in fructose-metabolizing brain regions
| Enzyme | Specific Activity (U/μg)a | |
|---|---|---|
| Brain | Liver | |
| KHK | 300 ± 15 | 65 ± 10 |
| Aldolase | 760 ± 60 | 71 ± 5 |
| HK | 460 ± 40 | – |
Specific activity of KHK, aldolase, and hexokinase were measured in the supernatant fractions of mixtures of sonicated dissected cerebellum, cerebral cortex, hippocampus, and olfactory bulb regions of the brains or liver from mice (n = 3–9) as described in Experimental Procedures. Error expressed as standard error of the means for 3–9 assays.
Using the antisense probes, sections of the cerebellum (Fig 2) showed strong khk expression in the Purkinje cell layer (PCL), while the molecular cell layer (MCL) and granular cell layer (GCL) had little to no expression (Fig 2, top left). Expression of aldoC in adjacent slices showed the same pattern as khk in the PCL (Fig 2, top right), while missing from the MCL and GCL. This heterogeneous labeling was expected for aldolase C, which is also known as zebrin II and shows this pattern in immunostaining (Abbott et al., 1996; Ahn et al., 1994; Chung et al., 2008; Hawkes and Herrup, 1995; Leclerc et al., 1992; Sarna et al., 2006). Probes for glut5 and glut9 showed strong expression in the PCL as well (Fig 2, bottom panels).
Fig 2. Genes required in the Fru-1-P pathway are expressed in Purkinje cells.
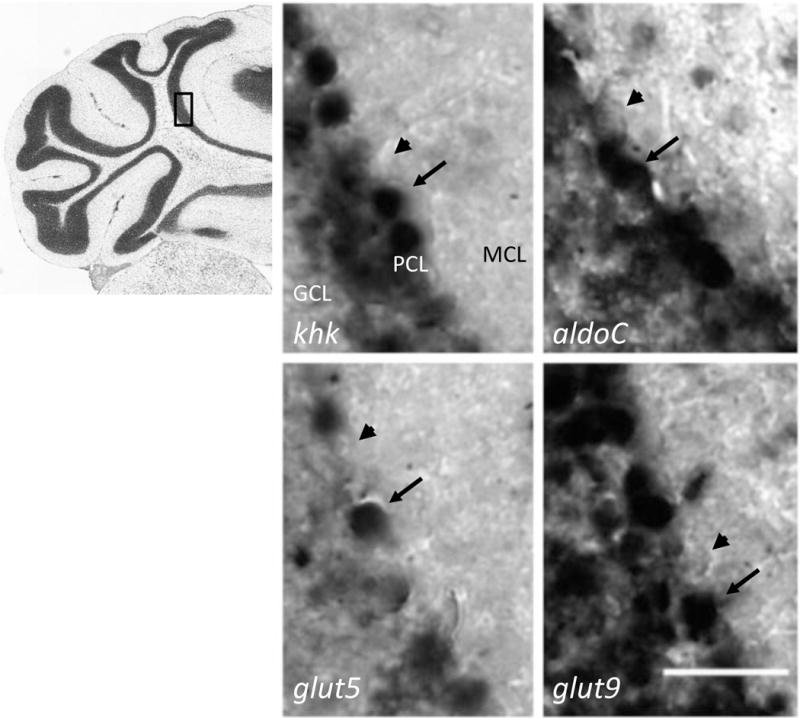
Coronal slices (14 μm) of adult mouse brain (n = 6) were probed with DIG-conjugated antisense mRNA complementary to the genes for ketohexokinase (khk), aldolase C (aldoC), GLUT5 (glut5), and GLUT9 (glut9) at the same concentration. Expression was visualized by color development from horseradish peroxidase-conjugated anti-DIG antibody. Scale bar represents 50 μm. An index micrograph from the Allen Brain Atlas (ABA) for the same layer of these coronal sections is shown to the left with a box indicating the field of observation for the micrographs. The cell layers are labeled in the upper left micrograph; PCL, Purkinje cell layer; MCL, molecular cell layer; GCL, granule cell layer. Arrow notes positive expression and arrowheads indicate negative expression.
Another brain region that showed expression of Fru-1-P-pathway genes was the hippocampus (Fig 3). In all of the cell bodies in the hippocampal formation there was strong expression of khk. In addition, expression of aldoC was clearly apparent in these same regions of the hippocampus. Among the fructose transporters, it appeared that glut5 was not expressed in the hippocampus, while glut9 was expressed. If fructose were metabolized in the hippocampus, it likely would be transported into cells via GLUT9.
Fig 3. Cells of the hippocampus express genes necessary for fructose metabolism.
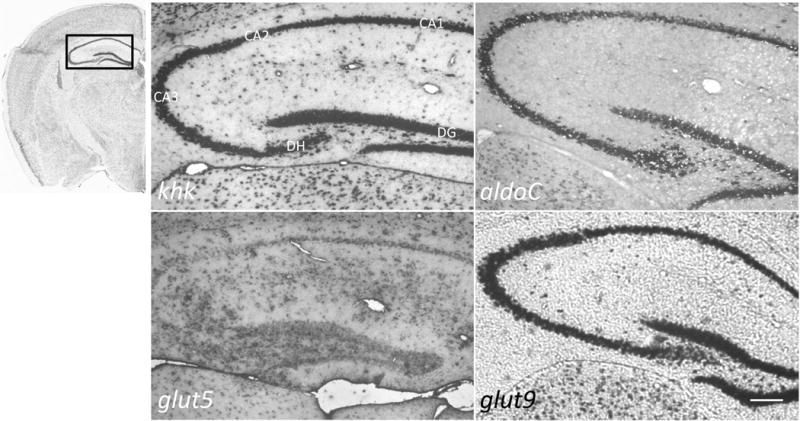
Antisense probes for the genes listed in the legend for Fig 2 and noted in the bottom left of each panel were used for ISH of coronal slices (14 μm) of adult mouse brain (n = 6) with ABA index micrograph and field of observation boxed scale bar represents 100 μm. The cell layers are labeled in the upper left micrograph; CA1-3, dorsal hippocampus (DH), and dentate gyrus (DG).
In slices of the cerebral cortex, at least some cells in all layers, with possible exception of layer I, were expressing khk, aldoC, glut5, and glut9 (Fig 4). The expression of genes in the Fru-1-P pathway was not ubiquitous to all cells in the cortex, and it was ambiguous from the in situ data alone which types of neurons and/or glial cells were expressing these genes.
Fig 4. Select cells in the cortex express genes necessary for fructose metabolism.
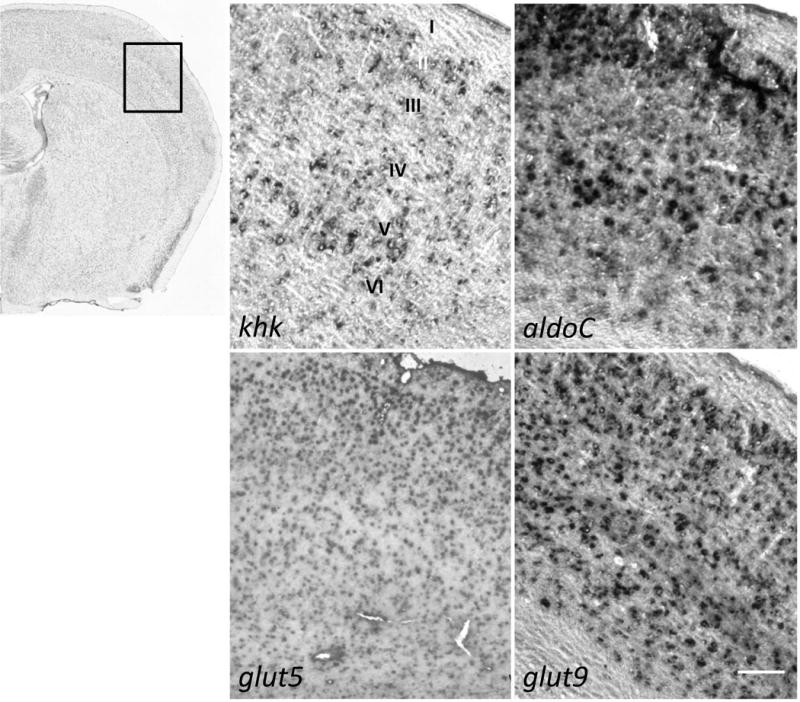
Antisense probes for the genes listed in the legend for Fig 2 and noted in the bottom left of each panel were used for in situ hybridization of coronal slices (14 μm) of adult mouse brain (n = 6) with ABA index micrograph and field of observation boxed. Scale bar represents 100 μm. The cell layers (I–VI) are labeled in the upper left micrograph.
Lastly, the ISH staining in the olfactory bulb indicated strong expression of the genes in the Fru-1-P pathway (Fig 5). Neurons in the granule cell layer (GCL), the mitral cell layer (MCL), and the periglomerular cells in the glomerulus layer (GL) were positive for expression of khk and aldoC. These cells expressed both glut5 and glut9, with glut9 expression being stronger in these layers of the olfactory bulb using similar concentrations of probes for each of these transcripts.
Fig 5. Cells in the olfactory bulb express genes necessary for fructose metabolism.
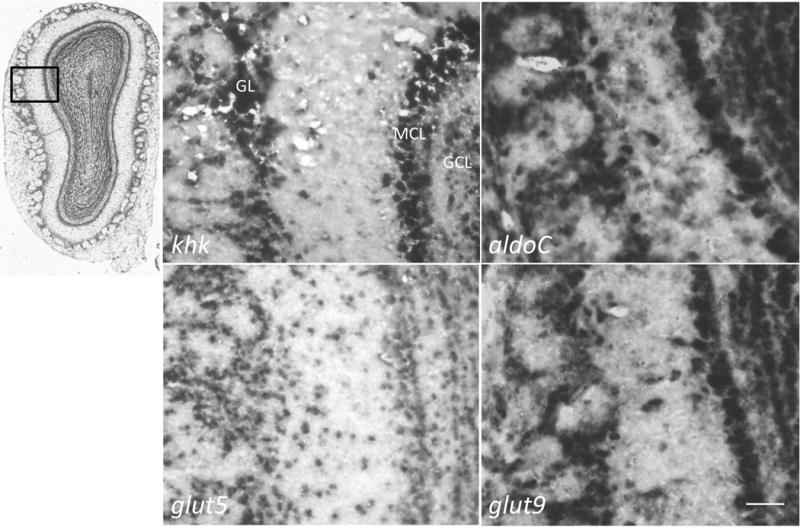
Antisense probes for the genes listed in the legend for Fig 2 and noted in the bottom left of each panel were used for ISH of coronal slices (14 μm) of adult mouse brain (n = 6) with ABA index micrograph and field of observation boxed. Scale bar represents 50 μm. The cell layers are labeled in the upper left micrograph; glomerular layer (GL), mitral cell layer (MCL), and granule cell layer (GCL).
2.3 Regions of the brain positive for gene expression of the Fru-1-P-pathway genes express functional proteins
Finding further evidence for gene expression beyond observing mRNA products in various cells of the brain was verified by measuring the presence of the protein products. Protein expression of transporters was measured by immunoblotting and protein expression of enzymes was measured by enzyme-activity assays. Both these measurements have limited sensitivity due to the amount of tissue obtainable from individual brain regions and necessitated combining those regions that were ISH-positive in the experiments shown above. Cerebellum, hippocampus, cortex, and olfactory bulbs were dissected and combined to a “Fru-1-P positive” sample. Protein samples from Fru-1-P positive brain tissue were prepared by sonication and centrifugation. Immunoblotting for GLUT3, 5, or 9 analyzed the membrane portion of these samples. These blots revealed that the positive control of GLUT3, which is highly expressed in the brain, was present in the combined Fru-1-P positive regions of the brain (Fig 6). Moreover, both of the fructose-transporting GLUTs (5 and 9), which were predicted by VNB and in situ hybridization, were present. A control of similarly prepared membrane fractions from liver correctly showed that GLUT9 was expressed there while GLUT5 was not. While macrophages of the brain and cells in the blood brain barrier express glut5 (Funari et al., 2007; Horikoshi et al., 2003), the presence of either GLUT5 or 9 in the regions identified here had never been shown.
Fig 6. Immunoblotting for detecting GLUT Expression.
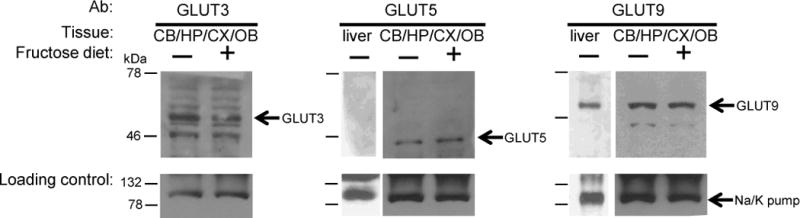
Membrane proteins were extracted from pooled samples of Fru-1-P positive brain regions. Protein (50 μg) extracted from membrane fractions was separated by SDS-PAGE and blotted with GLUT3, GLUT5, or GLUT9 antibodies as described in Experimental Procedures. The loading control was an antibody to the A1 subunit of the ubiquitous membrane protein Na+/K+ ATPase pump, used to probe the same blots. Representative blots of combined membrane preparations from the cerebellum (CB), hippocampus (HP), cortex (CX), and olfactory bulbs (OB) (the Fru-1-P-positive brain regions) or liver, from adult mice fed standard chow and water (−) or from mice fed standard chow and a 40% (w/v) fructose drinking solution (+) for 3 weeks. Arrows indicate the migration distance of GLUT3, 5, 9, and the loading control, which migrated as proteins of 50, 45, 54, and 100 kDa, respectively, based on known molecular weight marker proteins from the Kaleidoscope pre-stained markers (BioRad) as indicated to the left with dashed on the left of each blot.
The amount of functional KHK and aldolase enzymes in the Fru-1-P-positive brain tissue was quantified by measuring enzyme specific activity (Units/μg total protein; activity of an enzyme relative to total protein in a sample) in supernatant fractions of the sonicated Fru-1-P-positive brain samples. HK was measured in these fractions as a baseline for enzyme activity in the brain (Table 1). The specific activity of KHK was nearly five times higher in Fru-1-P-positive brain tissue than in liver despite the use of a different isozyme (KHK-A in brain, and KHK-C in liver) (Ishimoto et al., 2012). The specific activity of aldolase was about 10-fold higher in these brain regions than in liver, but this was consistent with the use of isozymes with different kcat values towards Fru 1,6-P2 (aldolase A and C in brain, and aldolase B in liver) (Kusakabe et al., 1994; Penhoet et al., 1969; Pezza et al., 2003).
2.4 Dissected brain slices from various regions of the brain oxidize fructose faster than liver slices
Observing functional protein products from the genes involved in fructose metabolism using immunoblots and enzyme activity assays indicated that various regions in the brain could utilize this carbon source. However, demonstration of actual functional metabolism of fructose is needed. The rate of fructose oxidation in brain samples assessed the physiological relevance of the expression of genes in the Fru-1-P pathway. Since expression of the Fru-1-P pathway genes was qualitatively different in some of the Fru-1-P positive-brain regions identified by ISH (cerebellum, hippocampus, cortex, and olfactory bulb), these regions were dissected separately, along with liver as a control, from adult mice. All tissues were left intact when used for assay of fructose oxidation to carbon dioxide. By not homogenizing the tissue, the integrity of the cell membrane was maintained. This method allowed for indirectly assessing the transport of fructose into cells, since tissue not transporting fructose into its cells will not oxidize it. The rate of carbon dioxide production by the tissue samples was measured after the addition of radioactive fructose to the media containing the slices (Table 2). As a control, the rate of glucose oxidation was measured for all tissue samples. The rate of oxidation of glucose and fructose by the liver was in the same range as reported previously (Funari et al., 2005). The glucose oxidation rates by the various brain regions were 2–20 fold higher than that of liver, which was consistent with the higher glycolytic capacity of brain relative to the liver (Gaitonde and Richter, 1966). Importantly, all four regions of the brain oxidized fructose, which corroborated with the ISH, immunoblots, and enzyme-activity data. Moreover, the rates of fructose oxidation by these dissected brain regions were 15–150 fold greater than that of the liver. While it appears that there were differences in oxidative capacity for both hexoses among the different brain regions, ANOVA analysis failed to show any significance (p>0.12), with the exception of glucose metabolism by the cerebellum (p<0.5). Comparing the ratio of glucose oxidation to fructose oxidation for each tissue (brain and liver), revealed that the rate glucose oxidation was consistently about 5.9 fold (±1.2) times higher than fructose oxidation throughout all tissues.
Table 2.
Rates of Oxidation of Hexoses by Different Regions of the Brain as Compared to the Liver
| Tissue | Glucose | Fructose |
|---|---|---|
| (μmol of CO2/mg wet weight•hour)a | ||
| Liver | 2.8 ±0.6 | 0.09 ±0.02 |
| Cerebellum | 9.5 ±2.0 | 1.9 ±0.2 |
| Hippocampus | 41 ±10 | 6.2 ±1 |
| Olfactory Bulb | 36 ±5 | 7.7 ±3 |
| Cortex | 24 ±7 | 3.9 ±0.4 |
Oxidation rates of glucose and fructose were determined after dissection of brains from wild type mice (n = 3–5) and error expressed as standard error of the means for n ≥3 assays. A one-way ANOVA analysis of the fructose metabolism among the brain regions showed no significant differences (p>0.12).
2.5 Both KHK and hexokinase participate in fructose phosphorylation in the brain
The determination that these particular regions of the brain (cerebellum, hippocampus, cortex, and olfactory bulb) are capable of fructose metabolism lead to the question whether the brain indeed utilize the same pathway (through Fru 1-P) as the liver (see Fig 1). Given there was no significant difference in oxidative capacity of fructose between the regions, and to increase total oxidative signal (see Table 2), these brain regions were pooled to afford as large a difference between uninhibited and inhibited treatments. To answer the question whether the Fru-1-P pathway has an important role, Fru-1-P-positive-brain regions were used for oxidation assays using 14C-fructose (1 mM) in four ways: 1) using 14C-fructose (1 mM) alone to measure overall metabolism, 2) including 2.5 mM glucose to measure any change due to completion with HK using the Fru-6-P pathway, 3) including 1 μM of a pyrimindinopyrimidine inhibitor of KHK to measure any change if the Fru-1-P pathway is blocked (this inhibitor concentration is >80-fold excess of the 0.012 μM IC50 value determined in vitro and 3-fold in excess of its in vivo IC50 value in HepG2 cells (Maryanoff et al., 2011)), or 4) using both 2.5 mM glucose and 1 μM the KHK inhibitor, which would measure any metabolism of fructose not blocked by these additions (Fig 7). The [fructose] of 1 mM is at the high range of physiological concentration in blood immediately after ingestion of fructose (Topping and Mayes, 1971) and at sufficient concentration to get a signal as well as allow competition with 2.5 mM glucose. The specificity of the pyrimindinopyrimidine KHK inhibitor is high; with a panel of 31 kinases the IC50 values were less than 800-fold higher than for KHK (Maryanoff et al., 2011). In the reactions using fructose alone, these brain regions oxidized fructose at similar rates as the individually dissected brain regions (see Table 2). In reactions where fructose was either competing with glucose for phosphorylation by HK, or its phosphorylation was blocked with the pyrimindinopyrimidine inhibitor of KHK, there was a decreasing trend, although not significant, when compared to reactions with fructose alone. However, only when both modes of blocking fructose metabolism were employed was there a significant decrease in oxidation rates, which showed 14% the amount of fructose oxidation compared to fructose alone. This demonstrated that both pathways are affected by these additions, and are indeed responsible for metabolism of 80–90% of fructose. Clearly, both kinases play a major role in the metabolism of fructose and the Fru-1-P pathway is a major contributing pathway in these brain regions.
Fig 7. Inhibiting fructose phosphorylation by both KHK and HK is required to reduce total fructose oxidation.
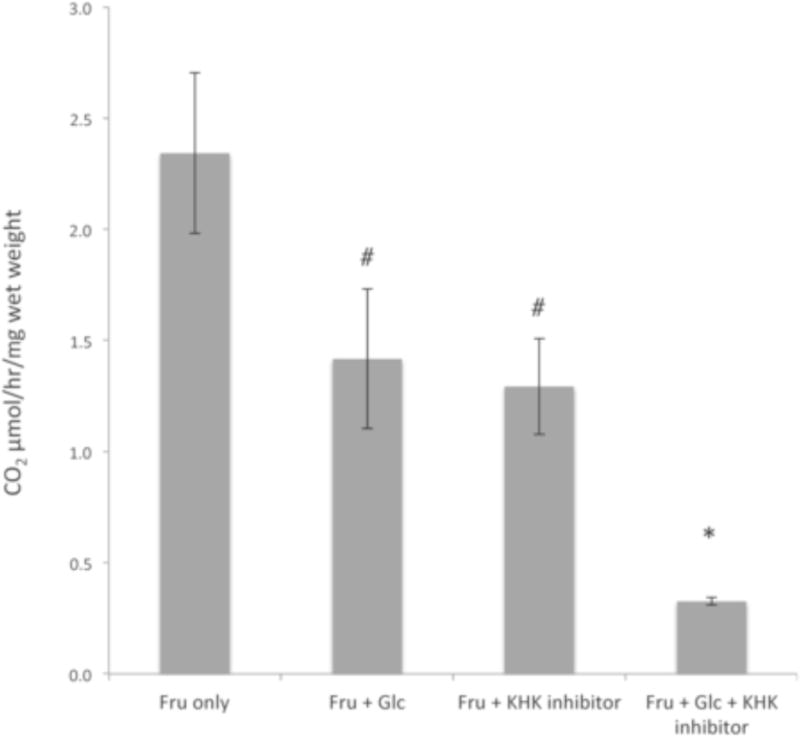
Oxidation of radioactive fructose (1 mM)(U-14C at 0.065 – 0.073 μCi/mmole) was measured in Fru-1-P positive brain samples (n = 4–6) with fructose alone (Fru only), in the presence of 2.5 mM cold glucose (to compete for HK phosphorylation; Fru + Glc), 1 μM of a pyrimindinopyrimidine inhibitor of KHK (Fru + KHK inhibitor), or both cold glucose and the KHK inhibitor (Fru + Glc + KHK inhibitor), for 1 hr. Protein content, measured by dye-binding assays (Bradford, 1976), was used to normalize samples. The measured oxidation rates were the average of ≥3 assays and statistical significance among the groups analyzed by ANOVA. Error bars indicate 1σ SEM with an asterisk indicating p < 0.05 and the # indicating no significance (p>0.10).
2.6 KHK activity increases in brains of fructose-fed mice
In the liver, high amounts of dietary fructose induce up-regulation of glut5, and increases in KHK and aldolase B specific activities (Burant and Saxena, 1994; Koo et al., 2008; Korieh and Crouzoulon, 1991; Shu et al., 1998). Similar increases in the brain would indicate that dietary fructose can induce such changes in brain metabolism, which could lead to possible detrimental metabolic dysregulation in the brain of individuals with a high fructose diet, as is seen in other organs. To assess if dietary fructose can increase GLUT5 and/or 9, KHK, and/or aldolase expression in brain, mice were give a 40% (w/v) drinking solution, or water, ad libitum for three weeks after weaning, and then sacrificed. Immunoblots of membrane fractions from Fru-1-P-positive brain regions did not show a difference in protein levels of either GLUT5 or 9 from the Fru-1-P positive brain regions with or without a fructose-rich diet (Fig 6). A more sensitive quantitative measurement of changes in gene expression, at least for genes encoding enzymes, can be attained using enzyme assays. The specific activity of KHK from these same brain regions of mice on a high-fructose diet increased 3-fold over that of mice not exposed to fructose (Table 3). HK specific activity, whose induction by a fructose diet is not known, was measured as a negative control. As a positive control, aldolase specificity activity was measured in livers of mice on high fructose diets, whose induction by a fructose diet is well known (Koo et al., 2008; Korieh and Crouzoulon, 1991). Indeed, in liver there was a 2.5-fold increase in aldolase specific activity. Likewise, KHK specific activity was induced about 2-fold, which was similar to another report (Ouyang et al., 2008). However, in the brain KHK specific activity was increased 3-fold. This increased activity was not accompanied by a significant increase in aldolase specific activity, as it was in the liver.
Table 3.
Increase of KHK activity in brain and liver after high-fructose feeding
| Tissue | Enzyme | Specific Activity (U/μg total protein) | Fold Change | |
|---|---|---|---|---|
| Normal diet | High Fructose Diet | |||
| Brain | KHK | 300 ±15 | 900 ±60* | 3 ±0.25 |
| HK | 460 ±40 | 515 ±38 | 1.1 ±0.13 | |
| Aldolase | 757 ±58 | 909 ±54 | 1.2 ±0.12 | |
| Liver | KHK | 68 ±11 | 139 ±13* | 2 ±0.38 |
| Aldolase | 71 ±5 | 173 ±19* | 2.5 ±0.31 | |
Specific activity of KHK, aldolase, and hexokinase were measured in the supernatant fractions of sonicated dissected brain (combination of cerebellum, cerebral cortex, hippocampus, and olfactory bulb regions) or liver, from three week old mice with access to either normal chow and water, or chow and a 40% (w/v) fructose drinking solution since weaning. The same supernatant fractions were measured for protein concentration as described in Experimental Procedures. Enzyme specific activities were calculated as the mean and standard error of the means for the error of n ≥ 5 trials; asterisks represent p ≤ 0.002 in pairwise t tests between the different diets for individual enzymes (n = 4–6).
3. Discussion
The data presented here support the idea that fructose is metabolized by tissues other than liver and kidney, specifically, the brain. Transcriptional profiling highlighted the importance of the brain as a site for fructose metabolism by predicting expression levels of the enzymes in the Fru-1-P pathway as being comparable to those in the liver. In situ hybridization of adult mouse brain slices showed that the genes necessary for this fructose metabolism were expressed across several brain regions, although expression was not uniform in all cells. Previously, we have reported the expression of KHK and aldolase C in Purkinje cells in the cerebellum (Funari et al., 2005). Here, it is reported that in addition to the cerebellum, the hippocampus, cortex, and olfactory bulb contained high populations of neurons and/or glial cells expressing the genes in the Fru-1-P pathway. Together, these regions of the brain express GLUT5 and/or 9, and had specific activities of KHK nearly 5-fold higher than KHK activity found in liver. Oxidation assays of dissected brain regions from adult mice confirmed that these regions could actively metabolize fructose. Interestingly, khk expression was not ubiquitously expressed in the PCL, with heterogeneity of khk-positive and khk-negative Purkinje cells, similar to that of aldoC expression (see Fig 2). This pattern would be consistent with a number of other differentially expressed proteins in the PCL (Chung et al., 2008; Sarna et al., 2006; Slemmer et al., 2007), including aldolse C, which was first identified as such and termed zebrin II (Abbott et al., 1996; Ahn et al., 1994; Chung et al., 2008; Hawkes and Herrup, 1995; Leclerc et al., 1992; Sarna et al., 2006). KHK had not previously been reported to show this pattern.
The issue of fructose metabolism by the brain in vivo has been controversial and elusive, despite knowing for over 50 years that fructose can be metabolized by brain tissue from experiments conducted on cultured brain slices (Chain et al., 1969; Izumi and Zorumski, 2009; Stein and Cohen, 1976; Wada et al., 1998). One reason for this confusion is that accessibility of fructose to brain cells has been difficult to quantify. The expression of GLUT5 in the blood-brain barrier (BBB) (Mantych et al., 1993) and microglial (Maher et al., 1994) allow fructose to enter the brain, but provides little insight into the concentrations attained physiologically. The ability of fructose to cross into cerebral spinal fluid of dogs has been investigated, though this study did not investigate other neural tissues, nor was metabolism of fructose measured (Fishman, 1963). Given the results presented here, it is clear that dissected brain regions can metabolize fructose much faster than the liver. Still, the liver is responsible for clearing at least half of all ingested fructose from the body (Craig et al., 1951; Mendeloff and Weichselbaum, 1953). It may be that the BBB presents the rate-limiting step in fructose metabolism by the brain, and could be why this organ is not responsible for the majority of fructose metabolism in the body despite its potential for rapid oxidation of fructose.
While recent studies have reported fructose metabolism in some regions of the brain that express khk (including the eye, cortex, and the rostral ventrolateral medulla) (Hassel et al., 2015; Meakin et al., 2007; Wu et al., 2014), this study reveals a more pervasive capacity for multiple brain regions for fructose metabolism. Furthermore, these studies revealed for the first time that KHK activity in the mouse brain increased on a high fructose diet. Interestingly, the rise in KHK activity was not accompanied by a rise in aldolase activity, as is known for aldolase B in the liver and small intestine (Koo et al., 2008; Korieh and Crouzoulon, 1991). It could be that mouse aldo3 expression (aldoC in humans) does not respond to differences in concentration of dietary fructose the way aldo2 (aldoB in humans) does in the liver. The up-regulation of KHK, but not HK or aldolase, has the potential to increase Fru-1-P levels, and shift the flux of fructose metabolism to favor the KHK-mediated Fru-1-P pathway over the HK-mediated Fru-6-P pathway since KHK activity in not product-inhibited as is HK activity. An increase in KHK activity without an increase in aldolase activity has the potential to create an inorganic phosphate sink if there is a lag between Fru-1-P phosphorylation and its cleavage to glyceraldehyde and DHAP. Such inorganic phosphate sinks in the liver potentially lead to liver damage (Lanaspa et al., 2012; Ludwig et al., 2001). Future studies will need to investigate what effects this increase in KHK activity has on the brain. Regardless, it is clear that dietary fructose has the ability to increase KHK activity, at least in the mouse brain. KHK is an enzyme specific to fructose metabolism, indicating that not only is the brain exposed to dietary fructose, but that it is also responsive to it.
The impact that high fructose exposure has on the human brain, which continues developing from birth through puberty (Blakemore and Choudhury, 2006; Levitt, 2003), is currently unknown. Since fructose is produced endogenously by the body only through the sorbitol/polyol pathway under hyperglycemic conditions in diabetic patients (Chung et al., 2003), the exposure of the brain to fructose is almost entirely diet dependent in non-diabetic individuals. Additionally, given that exposure to high concentrations of fructose has toxic effects such as hypophosphatemia and hyperuricemia, along with other metabolic consequences including lipid dysregulation, and activation of pro-inflammatory pathways, in liver and kidneys (Bergheim et al., 2008; Gersch et al., 2007; Mayes, 1993; Nakayama et al., 2010; Ouyang et al., 2008; Rayssiguier et al., 2006; Thuy et al., 2008), it is important to quantify the amount of fructose absorbed by the brain from the diet, and the intermediate metabolites of the Fru-1-P pathway.
4. Experimental Procedures
4.1 Care and use of animals
Animal care was performed in accordance with institutional guidelines. The Boston University, Charles River Campus, Institutional Animal Care and Use Committee (IACUC) approved all protocols. Mice were kept on normal chow diets (LabDiet, St. Louis, MO; formula 5P75), unless otherwise stated (Protocol 12-030).
4.2 Fructose exposure of mice
Mice were maintained under normal conditions, and fed either fructose-free chow (Bio-Serv, Flemington, NJ; formula #F6700 with 200 g/kg casein, 3 g/kg L-cystine, 129.5 g/kg corn starch, 500 g/kg dextrose, 50 g/kg cellulose, 70 g/kg soybean oil, 35 g/kg AIN-93G mineral mix, 10 g/kg AIN-93 vitamin mix, 2.5 g/kg choline bitartrate, 0.0014 tert-butylhydroquinone) with access to either water, or a 40% (w/v) fructose drinking solution.
4.3 In silico expression profiling using a Virtual Northern Blot program
The VNB program was used as described previously (Funari et al., 2010). Briefly, ClustalW alignments of human cDNA sequences from RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq/) for each gene in the Fru-1-P pathway, and its known homologs, were input into the VNB to generate gene-specific probes. GLUT homologs included the genes encoding human GLUT1 through GLUT13 (NM 006516, NM 00340, NM 006931; NM 001042, NM 001135585, NM 017585, NM 207420, NM 014580, NM 001001290, NM 030777.3, NM 030807.3, NM 145176.2, and NM 052885.3, respectively); aldolase homologs included all three isozymes, A, B, and C (NM 000034, NM 000035, and NM 005165, respectively). KHK (NM 000221) isozymes are generated by alternative splicing from one gene (Hayward and Bonthron, 1998); DAK (NM 015533) does not have isozymes. VNB generated alignments for these sequences using parameters of 20-base probe size and 8-base window size and created a set of highly specific probes for each gene. The VNB program then collected expressed sequence tags (ESTs) from a BLAST search of dbEST (http://www.ncbi.nlm.nih.gov/dbEST) that exactly matched the gene-specific probes generated from the alignment, which were subsequently mapped to an EST library using annotation from Entrez. Finally, these identities were used to search library construction information in the Cancer Genome Anatomy Project (CGAP) database (http://cgap.nci.nih.gov/). Summing ESTs from similar tissues tabulated results; ESTs from cell lines, tumor, or disease-derived tissues were not included in this analysis. Summing all EST “hits” for similar tissues generated a qualitative profile of gene expression. Summing all EST “hits” from un-manipulated or otherwise not “normalized” libraries for similar tissues generated a quantitative profile of gene expression. The denominators for the total ESTs from all libraries for each tissue were derived from information in CGAP.
4.4 Deoxygenin (DIG)-labelling of RNA probes
All work was done using RNAse-free conditions. RNA probes were generated using the DIG nucleic acid detection kit from Boehringer Mannheim Biochemical (Indianapolis, IN) according to manufacturer’s instructions. The appropriate RNA polymerase (SP6, T7, or T3) (Ambion) was used to generate probes from template cDNA plasmids containing the mouse glut5 (GenBank ID: AI786845), glut9 (BC018897), khk (AI256253), aldolase C (AI180787), or dak (triosekinase, BC021917). Probe concentration was determined by dot blot quantification using the Boerhinger Genius System and DIG-labeled control RNA (Roche).
4.5 Tissue collection and dissections
All experiments were carried out in accordance with the EC Directive 86/609/EEC for animal experiments. Mice were sacrificed by CO2 asphyxiation with cervical dislocation as a secondary procedure to confirm death. Livers and brains were dissected immediately and put on ice. Once the skull was removed from over the top of the brain, care was taken to gently remove the brain as an entire structure, including the olfactory bulb and cerebellum, out of the bottom of the skull and onto a glass plate, and kept on ice. The cerebellum, hippocampus, olfactory bulb, and front one half of the cortex were dissected away from the rest of the brain.
4.6 Tissue sectioning
The brains of adult mice perfused with 10% (v/v) paraformaldehyde were dissected on ice and stored at 4 °C in phosphate-buffered saline (PBS) containing 30% (w/v) sucrose and 0.1% (v/v) diethylpyrocarbonate. Brain tissues were mounted using Tissue-Tek O.C.T. compound (Sakura) at −20 °C and coronal sections (14 μm) were prepared using a Cryocut 1800 cryostat. Tissue cryosections were collected on Gold Seal Micro slides, placed inside a slide box, desiccated for 30 min under a vacuum, and stored at −80 °C.
4.7 RNA In situ hybridization (ISH) of tissue slices
ISH of brain tissue slices were performed essentially as described previously (Funari et al., 2005). Sections were probed with both anti-sense probes to detect for gene expression, and sense probes as a negative control. Briefly, cryosections were rehydrated by washing in decreasing concentrations of ethanol (100%, 95%, 70%, and 50%) and finally in 2X SSC. Rehydrated tissues were digested with proteinase K (10 μg/ml) in 0.1 M TEA, pH 8.0, at 37 °C for 30 min, then transferred to 28 mM acetic anhydride in the same buffer and incubated at 37 °C for 10 min. After rinsing with 2X SSC, the sections were dehydrated in increasing concentrations of ethanol (50%, 75%, 90%, 100%). DIG-labeled RNA probes (2 ng/μL) were incubated at 68 °C for 10 min and cooled on ice before diluting (1:25) in hybridization buffer with a final concentrations of 1.4 mM Tris-HCl, pH 7.5, 42 mM NaCl, 28 μM EDTA, 53% (v/v) deionized formamide, 10% (v/v) dextran sulfate, and 1.3% (v/v) Blocking Reagent (Roche). This hybridization solution was sealed over the sections with DPX Mountant (Fluka), and incubated at 55 °C for 18–36 hrs. Slides were unsealed and washed in 2X SSC at 55 °C for 30 min, in 2X SSC, 50% (v/v) formamide at 55 °C for 30 min, and finally twice in 2X SSC at 37 °C. Slides were treated with RNase A (20 μg/mL in 0.5 M NaCl, 1 mM EDTA, 10 mM Tris-HCl, pH 7.5) at 37 °C for 30 min, and washed with buffer only at 55 °C for 30 min. Visualization using DIG-antibody (1:1000)(Roche) was performed according to the manufacturers instructions. Following color development, glycerol was applied to the tissue sections and they were mounted with coverslips and sealed with nail polish. Dried slides were stored at 4 °C in the dark, and pictures were taken on an Olympus 1×70 microscope using an attached Olympus camera (Model BH2-RFL-T3) and Pictureframe software.
4.8 Preparation of cell lysis and membrane fractions of dissected tissues
Fresh liver and brains were either dissected and used immediately for assays, or were stored at −80 °C. If needed to attain sufficient material, the cerebellum, hippocampus, cortex, and olfactory bulbs were dissected away from the rest of the brain, and combined to create a “Fru-1-P positive region.” Fragments (60–120 μg) of dissected liver, or brain samples, were weighed, and 1 volume of native lysis buffer (150 mM NaCl, 20 mM HEPES, pH 7.4, 1 mM EDTA, 1% (v/v) Triton X100) was added to samples. Tissues were subjected to sonication in a Bioruptor Twin bath sonicator set to high for 12 30-s cycles. The sonicate was centrifuged for 60 min at 20,000g at 4 °C. Supernatant fractions were separated from the pellet fractions. The pellet fractions were solubilization by boiling for 5 min in 1% (w/v) sodium dodecyl sulfate (SDS) to create a membrane fraction for detection of membrane-bound proteins. Membrane fractions were clarified by centrifugation at 20,000g for 10 min at 4 °C. Final volumes varied depending on the original pellet size, and ranged from 80–150 μL.
4.9 Enzymatic activity assays
Enzymatic activity of ketohexokinase was measured using coupled assay that measures the conversion of ATP to ADP during the production of fructose 1-phosphate. The ADP was converted back to ATP using phosphoenolpyruvate catalyzed by pyruvate kinase during which pyruvate was produced. The pyruvate was converted to lactate, toward which the equilibrium lies, by catalysis with lactate dehydrogenase. During this conversion to lactate, NADH was oxidized to NAD+ and the loss of absorbance at 340 nm was measured (Donaldson et al., 1993).
The enzymatic activity of aldolase was measured using a coupled assay that measures the converstion of the two triose phosphates (interconverted by triosephosphate isomerase, see Fig 1) to glycerol 3-phosphate through the catalytic reduction of dihydroxyacetone phosphate by glycerol-3-phosphate dehydrogenase. During this conversion to glycerol 3-phosphate, NADH was oxidized to NAD+ and the loss of absorbance at 340 nm was measured (Blostein and Rutter, 1963).
The enzymatic activity of hexokinase was measured by a coupled assay that measures the glucose 6-phosphate after its subsequent oxidation to 6-phosphogluconate catalyzed by glucose-6-phosphate dehydrogenase. This conversion was accompanied by the the reduction of NADP+ to NADPH and the gain in absorbance at 340 nm was measured (Parry and Pedersen, 1983).
All enzymatic activity assays combined 200 μL of the appropriate cocktail containing excess coupling enzymes, cofactors, substrates, and buffers as previously as appropriate for each assay described above with 50 μL of protein sample from control enzymes or tissue extract supernatant fractions that were appropriately diluted to yield a rate of ΔA340/min of 0.005 to 0.025. Identical assays were preformed, except without substrate, and these background rates were subtracted from rates of assays that included substrate. Absorbance was measured in triplicate for 30 min. Protein concentration of all extracts was measured using a dye-binding assay (Bradford, 1976).
4.10 Western blotting for membrane fraction proteins
Protein samples (30 – 50 μg) from membrane fractions of sonicated tissue were electrophoresed on SDS-PAGE gels. Gels and Protean Extra Thick Blotting Paper (Biorad) were incubated with in 250 mM Tris base, 192 mM glycine, and 20% (v/v) methanol for 15 min. PVDF membranes (Biorad) were submerged and incubated in 100% methanol for 3 min. Protein from the SDS-PAGE gel was transferred to the PVDF membrane between two saturated pieces of blotting paper using a semi-dry transfer cell (BioRad) at 18–20 mA for 40–45 min. Nonspecific protein binding was blocked by incubating membranes in a solution of TBST (50 mM Tris, 150 mM NaCl, pH 7.4, 0.1 % v/v Tween 20/20) with 5% (w/v) powdered milk (Carnation) and 2% (w/v) BSA (Gibco BRL) for 3 h at 25 °C, or overnight at 4 °C. Membranes were then incubated in a solution containing primary antibodies against GLUT3 (1:1000, Santa Cruz 1–14), which is ubiquitously expressed in the brain (Choeiri et al., 2002; Maher et al., 1993), GLUT5, (1:5000, Santa Cruz H-200), which is not expressed in the liver (Kayano et al., 1990), GLUT9 (1:5000, Novus NBP1-05054), which is expressed mostly in kidney and liver, but at low levels in many tissues(Augustin et al., 2004; Phay et al., 2000), or the A1 subunit of the Na+/K+ ATPase (1:200, Abcam AB7671), which is a ubiquitous integral membrane protein in adult brain (Herrera et al., 1994). The antibodies were diluted in a solution of TBST with 1% (v/v) normal goat serum (NGS) and 0.02% (w/v) sodium azide, for 3 h at 25 °C, or overnight at 4 °C, then rinsed 3 times for 5 min each in TBST. All antibodies were specific for the isoforms tested according to the manufacturer’s quality-control tests. After rinsing with TBST, membranes were incubated for 1 hr at 25 °C with appropriately diluted secondary antibodies in TBST, which were all conjugated to horseradish peroxidase. Membranes were rinsed with TBST for 10 min, repeated twice more, and rinsed twice for 20 min with the same buffer without detergent. Proteins recognized by the appropriate primary antibodies were visualized using SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) according to manufacturer’s instructions. Briefly, a 1:1 mixture of the two SuperSignal West substrate solutions was applied evenly to membranes. Blots were visualized by autoradiography by exposing to x-ray film for 1 to 180 s and developed using KODAK M-35A X-Omat Processor.
4.11 Oxidation rates of hexoses by tissues
Tissues were dissected from BL6 adult mice following cervical dislocation, immediately chilled on ice, and weighed. Weighed tissues (30–140 mg) were placed into airtight flasks containing phosphate-buffered saline with [U-14C]-hexose (0.06–0.15 mCi/mmol) at 4 °C. The flasks had a glass-blown center well (G. Finkenbeiner Inc. Waltham, MA), into which a 2 N KOH-saturated fluted Whatman 1 filter paper was placed. The flasks were incubated in a shaking incubator for 60 min at 37 °C after which HCl was added to a final concentration of 0.6 N, and tissues were incubated for 15 min to drive out any remaining CO2 trapped in the tissue. The contents of the center well were collected for liquid scintillation counting.
4.12 Statistics
A one-way ANOVA (independent samples, weighted) test was used to compare statistical significance among groups of three or more measurements. A student t-test was used to compare significance between pairs of measurements. Assays performed in triplicate at the same time used the mean and standard deviation reported at one sigma. Assays that used multiple trials at different times used the mean and the standard error of the means at one sigma.
Supplementary Material
Highlights.
Bioinformatics predicts that brain expresses genes specific for fructose metabolism.
ISH confirms bioinformatic predictions and visualizes gene expression in mouse brain.
Select cells in specific brain regions express genes in fructose-metabolism pathway.
Fructose oxidation by brain regions confirms brain’s ability to metabolize fructose.
Increased dietary fructose up-regulates KHK activity in specific brain regions.
Acknowledgments
Authors wish to thank Dr. Hans Kornberg for many helpful discussions and help with the oxidation experiments, and Dr. Angela Ho for help with brain dissections and review of manuscript. We would also like to thank Pfizer for their generous gift of the pyrimindinopyrimidine inhibitor of KHK. This work was supported by a Public Health Service grant (DK065089).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations used: solute carrier 2A, glucose transporter (GLUT); fructose 1-phosphate (Fru 1-P), fructose-6-phosphate (Fru 6-P), Fructose 1,6-bisphosphate (Fru-1,6-P2), ketohexokinase (KHK), hexokinase (HK), dihydroxyacetone phosphate (DHAP), triosekinase or dihydroxyacetone kinase (DAK), Virtual Northern Blot (VNB), Cancer Genome Anatomy Project (CGAP), In situ hybridization (ISH), sodium dodecyl sulfate (SDS), Purkinje cell layer (PCL), granule cell layer (GCL), mitral cell layer (MCL), glomerulus layer (GL), dentate gyrus (DG), dorsal hippocampus (DH), wild-type (WT), nonalcoholic fatty liver disease (NAFLD)
References
- Abbott LC, Isaacs KR, Heckroth JA. Co-localization of tyrosine hydroxylase and zebrin II immunoreactivities in Purkinje cells of the mutant mice, tottering and tottering/leaner. Neurosci. 1996;71:461–75. doi: 10.1016/0306-4522(95)00444-0. [DOI] [PubMed] [Google Scholar]
- Adelman RC, Ballard FJ, Weinhouse S. Purification and properties of rat liver fructokinase. J Biol Chem. 1967;242:3360–5. [PubMed] [Google Scholar]
- Ahn AH, Dziennis S, Hawkes R, Herrup K. The cloning of zebrin II reveals its identity with aldolase C. Development. 1994;120:2081–90. doi: 10.1242/dev.120.8.2081. [DOI] [PubMed] [Google Scholar]
- Anzai N, Ichida K, Jutabha P, Kimura T, Babu E, Jin CJ, Srivastava S, Kitamura K, Hisatome I, Endou H, Sakurai H. Plasma urate level is directly regulated by a voltage-driven urate efflux transporter URATV1 (SLC2a9) in humans. J Biol Chem. 2008;283:26834–8. doi: 10.1074/jbc.C800156200. [DOI] [PubMed] [Google Scholar]
- Augustin R, Carayannopoulos MO, Dowd LO, Phay JE, Moley JF, Moley KH. Identification and characterization of human glucose transporter-like protein-9 (GLUT9): Alternative splicing alters trafficking. J Biol Chem. 2004;279:16229–36. doi: 10.1074/jbc.M312226200. [DOI] [PubMed] [Google Scholar]
- Baron CB, Tolan DR, Choi KH, Coburn RF. Aldolase A ins(1,4,5)P3-binding domains as determined by site-directed mutagenesis. Biochem J. 1999;341:805–12. [PMC free article] [PubMed] [Google Scholar]
- Basaranoglu M, Basaranoglu G, Bugianesi E. Carbohydrate intake and nonalcoholic fatty liver disease: Fructose as a weapon of mass destruction. Hepatobiliary Surg Nutr. 2015;4:109–16. doi: 10.3978/j.issn.2304-3881.2014.11.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergheim I, Weber S, Vos M, Kramer S, Volynets V, Kaserouni S, McClain CJ, Bischoff SC. Antibiotics protect against fructose-induced hepatic lipid accumulation in mice: Role of endotoxin. J Hepatol. 2008;48:983–92. doi: 10.1016/j.jhep.2008.01.035. [DOI] [PubMed] [Google Scholar]
- Blakemore SJ, Choudhury S. Development of the adolescent brain: Implications for executive function and social cognition. J Child Psychol Psychiatry. 2006;47:296–312. doi: 10.1111/j.1469-7610.2006.01611.x. [DOI] [PubMed] [Google Scholar]
- Blostein R, Rutter WJ. Comparative studies of liver and muscle aldolase. J Biol Chem. 1963;238:3280–5. [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burant CF, Saxena M. Rapid reversible substrate regulation of fructose transporter expression in rat small intestine and kidney. Am J Physiol. 1994;267:G71–9. doi: 10.1152/ajpgi.1994.267.1.G71. [DOI] [PubMed] [Google Scholar]
- Burant CF, Takeda J, Brot-Laroche E, Bell GI, Davidson NO. Fructose transporter in human spermatozoa and small intestine is GLUT5. J Biol Chem. 1992;267:14523–6. [PubMed] [Google Scholar]
- Chain EB, Rose SP, Masi I, Pocchiari F. Metabolism of hexoses in rat cerebral cortex slices. J Neurochem. 1969;16:93–100. doi: 10.1111/j.1471-4159.1969.tb10346.x. [DOI] [PubMed] [Google Scholar]
- Choeiri C, Staines W, Messier C. Immunohistochemical localization and quantification of glucose transporters in the mouse brain. Neurosci. 2002;111:19–34. doi: 10.1016/s0306-4522(01)00619-4. [DOI] [PubMed] [Google Scholar]
- Chung SH, Marzban H, Croci L, Consalez GG, Hawkes R. Purkinje cell subtype specification in the cerebellar cortex: Early B-cell factor 2 acts to repress the zebrin II-positive Purkinje cell phenotype. Neurosci. 2008;153:721–32. doi: 10.1016/j.neuroscience.2008.01.090. [DOI] [PubMed] [Google Scholar]
- Chung SS, Ho EC, Lam KS, Chung SK. Contribution of polyol pathway to diabetes-induced oxidative stress. J Am Soc Nephrol. 2003;14:S233–6. doi: 10.1097/01.asn.0000077408.15865.06. [DOI] [PubMed] [Google Scholar]
- Craig JW, Drucker WR, Miller M, Owens JE, Woodward H, Jr, Brofman B, Pritchard WH. Metabolism of fructose by the liver of diabetic and nondiabetic subjects. Proc Soc Exp Biol Med. 1951;78:698–702. doi: 10.3181/00379727-78-19186. [DOI] [PubMed] [Google Scholar]
- Donaldson IA, Doyle TC, Matas N. Expression of rat liver ketohexokinase in yeast results in fructose intolerance. Biochem J. 1993;291:179–86. doi: 10.1042/bj2910179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douard V, Ferraris RP. Regulation of the fructose transporter GLUT5 in health and disease. Am J Physiol Endocrinol Metab. 2008;295:E227–37. doi: 10.1152/ajpendo.90245.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman RA. Studies of the transport of sugars between blood and cerebrospinal fluid in normal states and in meningeal carcinomatosis. Trans Am Neurol Assoc. 1963;88:114–8. [PubMed] [Google Scholar]
- Funari VA, Crandall JE, Tolan DR. Fructose metabolism in the cerebellum. Cerebellum. 2007;6:130–40. doi: 10.1080/14734220601064759. [DOI] [PubMed] [Google Scholar]
- Funari VA, Herrera VLM, Freeman D, Tolan DR. Genes required for fructose metabolism are expressed in Purkinje cells in the cerebellum. Molec Brain Res. 2005;142:115–22. doi: 10.1016/j.molbrainres.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Funari VA, Voevodski K, Leyfer D, Yerkes K, Cramer D, Tolan DR. Quantitative gene-expression profiles in real time from expressed sequence tag database. Gene Expr. 2010;14:321–36. doi: 10.3727/105221610x12717040569820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaby AR. Adverse effects of dietary fructose. Altern Med Rev. 2005;10:294–306. [PubMed] [Google Scholar]
- Gaitonde MK, Richter D. Changes with age in the utilization of glucose carbon in liver and brain. J Neurochem. 1966;13:1309–16. doi: 10.1111/j.1471-4159.1966.tb04293.x. [DOI] [PubMed] [Google Scholar]
- Gaster M, Handberg A, Schurmann A, Joost HG, Beck-Nielsen H, Schroder HD. GLUT11, but not GLUT8 or GLUT12, is expressed in human skeletal muscle in a fibre type-specific pattern. Pflugers Arch. 2004;448:105–13. doi: 10.1007/s00424-003-1219-4. [DOI] [PubMed] [Google Scholar]
- Gersch MS, Mu W, Cirillo P, Reungjui S, Zhang L, Roncal C, Sautin YY, Johnson RJ, Nakagawa T. Fructose, but not dextrose, accelerates the progression of chronic kidney disease. Am J Physiol Renal Physiol. 2007;293:F1256–61. doi: 10.1152/ajprenal.00181.2007. [DOI] [PubMed] [Google Scholar]
- Hassel B, Elsais A, Froland AS, Tauboll E, Gjerstad L, Quan Y, Dingledine R, Rise F. Uptake and metabolism of fructose by rat neocortical cells in vivo and by isolated nerve terminals in vitro. J Neurochem. 2015;133:572–81. doi: 10.1111/jnc.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes R, Herrup K. Aldolase c/zebrin ii and the regionalization of the cerebellum. J Mol Neurosci. 1995;6:147–58. doi: 10.1007/BF02736761. [DOI] [PubMed] [Google Scholar]
- Hayward BE, Bonthron DT. Structure and alternative splicing of the ketohexokinase gene. Eur J Biochem. 1998;257:85–91. doi: 10.1046/j.1432-1327.1998.2570085.x. [DOI] [PubMed] [Google Scholar]
- Helliwell PA, Richardson M, Affleck J, Kellett GL. Stimulation of fructose transport across the intestinal brush-border membrane by pma is mediated by GLUT2 and dynamically regulated by protein kinase C. Biochem J. 2000;350(Pt 1):149–54. [PMC free article] [PubMed] [Google Scholar]
- Herrera VL, Cova T, Sassoon D, Ruiz-Opazo N. Developmental cell-specific regulation of Na(+)-K(+)-ATPase alpha 1-, alpha 2-, and alpha 3-isoform gene expression. Am J Physiol. 1994;266:C1301–12. doi: 10.1152/ajpcell.1994.266.5.C1301. [DOI] [PubMed] [Google Scholar]
- Holdsworth CD, Dawson AM. The absorption of monosaccharides in man. Clin Sci. 1964;27:371–9. [PubMed] [Google Scholar]
- Horikoshi Y, Sasaki A, Taguchi N, Maeda M, Tsukagoshi H, Sato K, Yamaguchi H. Human GLUT5 immunolabeling is useful for evaluating microglial status in neuropathological study using paraffin sections. Acta Neuropathol (Berl) 2003;105:157–62. doi: 10.1007/s00401-002-0627-4. [DOI] [PubMed] [Google Scholar]
- Ishimoto T, Lanaspa MA, Le MT, Garcia GE, Diggle CP, Maclean PS, Jackman MR, Asipu A, Roncal-Jimenez CA, Kosugi T, Rivard CJ, Maruyama S, Rodriguez-Iturbe B, Sanchez-Lozada LG, Bonthron DT, Sautin YY, Johnson RJ. Opposing effects of fructokinase C and A isoforms on fructose-induced metabolic syndrome in mice. Proc Natl Acad Sci USA. 2012;109:4320–5. doi: 10.1073/pnas.1119908109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Zorumski CF. Glial-neuronal interactions underlying fructose utilization in rat hippocampal slices. Neurosci. 2009;161:847–54. doi: 10.1016/j.neuroscience.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayano T, Burant CF, Fukumoto H, Gould GW, Fan YS, Eddy RL, Byers MG, Shows TB, Seino S, Bell GI. Human facilitative glucose transporters. Isolation, functional characterization, and gene localization of cdnas encoding an isoform (GLUT5) expressed in small intestine, kidney, muscle, and adipose tissue and an unusual glucose transporter pseudogene-like sequence (GLUT6) J Biol Chem. 1990;265:13276–82. [PubMed] [Google Scholar]
- Koo HY, Wallig MA, Chung BH, Nara TY, Cho BH, Nakamura MT. Dietary fructose induces a wide range of genes with distinct shift in carbohydrate and lipid metabolism in fed and fasted rat liver. Biochim Biophys Acta. 2008;1782:341–8. doi: 10.1016/j.bbadis.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Korieh A, Crouzoulon G. Dietary regulation of fructose metabolism in the intestine and in the liver of the rat. Duration of the effects of a high fructose diet after the return to the standard diet. Arch Int Physiol Biochim Biophys. 1991;99:455–60. [PubMed] [Google Scholar]
- Kusakabe T, Motoki K, Hori K. Human aldolase C: Characterization of the recombinant enzyme expressed in Escherichia coli. J Biochem (Tokyo) 1994;115:1172–7. doi: 10.1093/oxfordjournals.jbchem.a124475. [DOI] [PubMed] [Google Scholar]
- Lanaspa MA, Sanchez-Lozada LG, Cicerchi C, Li N, Roncal-Jimenez CA, Ishimoto T, Le M, Garcia GE, Thomas JB, Rivard CJ, Andres-Hernando A, Hunter B, Schreiner G, Rodriguez-Iturbe B, Sautin YY, Johnson RJ. Uric acid stimulates fructokinase and accelerates fructose metabolism in the development of fatty liver. PLoS One. 2012;7:e47948. doi: 10.1371/journal.pone.0047948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc N, Schwarting GA, Herrup K, Hawkes R, Yamamoto M. Compartmentation in mammalian cerebellum: Zebrin II and P-path antibodies define three classes of sagittally organized bands of Purkinje cells. Proc Natl Acad Sci USA. 1992;89:5006–10. doi: 10.1073/pnas.89.11.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P. Structural and functional maturation of the developing primate brain. J Ped. 2003;143:S35–45. doi: 10.1067/s0022-3476(03)00400-1. [DOI] [PubMed] [Google Scholar]
- Litherland GJ, Hajduch E, Gould GW, Hundal HS. Fructose transport and metabolism in adipose tissue of zucker rats: Diminished GLUT5 activity during obesity and insulin resistance. Mol Cell Biochem. 2004;261:23–33. doi: 10.1023/b:mcbi.0000028734.77867.d2. [DOI] [PubMed] [Google Scholar]
- Ludwig DS, Peterson KE, Gortmaker SL. Relation between consumption of sugar-sweetened drinks and childhood obesity: A prospective, observational analysis. Lancet. 2001;357:505–8. doi: 10.1016/S0140-6736(00)04041-1. [DOI] [PubMed] [Google Scholar]
- Maher F, Vannucci SJ, Simpson IA. Glucose transporter isoforms in brain: Absence of GLUT3 from the blood-brain barrier. J Cereb Blood Flow Metab. 1993;13:342–5. doi: 10.1038/jcbfm.1993.43. [DOI] [PubMed] [Google Scholar]
- Maher F, Vannucci SJ, Simpson IA. Glucose transport proteins in brain. FASEB J. 1994;8:1003–11. doi: 10.1096/fasebj.8.13.7926364. [DOI] [PubMed] [Google Scholar]
- Mantych GJ, James DE, Devaskar SU. Jejunal/kidney glucose transporter isoform (GLUT-5) is expressed in the human blood-brain barrier. Endocrinol. 1993;132:35–40. doi: 10.1210/endo.132.1.8419132. [DOI] [PubMed] [Google Scholar]
- Maryanoff B, O’Neill JC, McComsey DF, Yabut SC, Luci DK, Jordan A, Jr, Masucci J, Jones WJ, Abad MC, Gibbs A, Petrounia I. Inhibitors of ketohexokinase: Discovery of pyrimidinopyrimidines with specific substitution that complements the atp-binding site. ACS Med Chem Lett. 2011;2:538–43. doi: 10.1021/ml200070g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes PA. Intermediary metabolism of fructose. Am J Clin Nutr. 1993;58:754S–65S. doi: 10.1093/ajcn/58.5.754S. [DOI] [PubMed] [Google Scholar]
- Meakin PJ, Fowler MJ, Rathbone AJ, Allen LM, Ransom BR, Ray DE, Brown AM. Fructose metabolism in the adult mouse optic nerve, a central white matter tract. J Cereb Blood Flow Metab. 2007;27:86–99. doi: 10.1038/sj.jcbfm.9600322. [DOI] [PubMed] [Google Scholar]
- Mendeloff AI, Weichselbaum TE. Role of the human liver in the assimilation of intravenously administered fructose. Metabol. 1953;2:450–8. [PubMed] [Google Scholar]
- Nakayama T, Kosugi T, Gersch M, Connor T, Sanchez-Lozada LG, Lanaspa MA, Roncal C, Perez-Pozo SE, Johnson RJ, Nakagawa T. Dietary fructose causes tubulointerstitial injury in the normal rat kidney. Am J Physiol Renal Physiol. 2010;298:F712–20. doi: 10.1152/ajprenal.00433.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang X, Cirillo P, Sautin Y, McCall S, Bruchette JL, Diehl AM, Johnson RJ, Abdelmalek MF. Fructose consumption as a risk factor for non-alcoholic fatty liver disease. J Hepatol. 2008;48:993–9. doi: 10.1016/j.jhep.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry DM, Pedersen PL. Intracellular localization and properties of particulate hexokinase in the novikoff ascites tumor. Evidence for an outer mitochondrial membrane location. J Biol Chem. 1983;258:10904–12. [PubMed] [Google Scholar]
- Penhoet EE, Kochman M, Rutter WJ. Isolation of fructose diphosphate aldolases A, B, and C. Biochemistry. 1969;8:4391–5. doi: 10.1021/bi00839a025. [DOI] [PubMed] [Google Scholar]
- Penhoet EE, Rutter WJ. Catalytic and immunochemical properties of homomeric and heteromeric combinations of aldolase subunits. J Biol Chem. 1971;246:318–23. [PubMed] [Google Scholar]
- Pezza JA, Choi KH, Berardini TZ, Beernink PT, Allen KN, Tolan DR. Spatial clustering of isozyme-specific residues reveals unlikely determinants of isozyme specificity in fructose 1,6-bisphosphate aldolase. J Biol Chem. 2003;278:17307–13. doi: 10.1074/jbc.M209185200. [DOI] [PubMed] [Google Scholar]
- Phay JE, Hussain HB, Moley JF. Cloning and expression analysis of a novel member of the facilitative glucose transporter family, SLC2a9 (GLUT9) Genomics. 2000;66:217–20. doi: 10.1006/geno.2000.6195. [DOI] [PubMed] [Google Scholar]
- Rand EB, Depaoli AM, Davidson NO, Bell GI, Burant CF. Sequence, tissue distribution, and functional characterization of the rat fructose transporter GLUT5. Am J Physiol. 1993;264:G1169–G76. doi: 10.1152/ajpgi.1993.264.6.G1169. [DOI] [PubMed] [Google Scholar]
- Rayssiguier Y, Gueux E, Nowacki W, Rock E, Mazur A. High fructose consumption combined with low dietary magnesium intake may increase the incidence of the metabolic syndrome by inducing inflammation. Magnes Res. 2006;19:237–43. [PubMed] [Google Scholar]
- Sarna JR, Marzban H, Watanabe M, Hawkes R. Complementary stripes of phospholipase Cbeta3 and Cbeta4 expression by Purkinje cell subsets in the mouse cerebellum. J Comp Neurol. 2006;496:303–13. doi: 10.1002/cne.20912. [DOI] [PubMed] [Google Scholar]
- Scheepers A, Schmidt S, Manolescu A, Cheeseman CI, Bell A, Zahn C, Joost HG, Schurmann A. Characterization of the human SLC2a11 (GLUT11) gene: Alternative promoter usage, function, expression, and subcellular distribution of three isoforms, and lack of mouse orthologue. Mol Membr Biol. 2005;22:339–51. doi: 10.1080/09687860500166143. [DOI] [PubMed] [Google Scholar]
- Schimke RT, Grossbard L. Studies on isozymes of hexokinase in animal tissues. Ann N Y Acad Sci. 1968;151:332–50. doi: 10.1111/j.1749-6632.1968.tb11899.x. [DOI] [PubMed] [Google Scholar]
- Shu R, David ES, Ferraris RP. Luminal fructose modulates fructose transport and GLUT-5 expression in small intestine of weaning rats. Am J Physiol. 1998;274:G232–9. doi: 10.1152/ajpgi.1998.274.2.G232. [DOI] [PubMed] [Google Scholar]
- Slemmer JE, Haasdijk ED, Engel DC, Plesnila N, Weber JT. Aldolase C-positive cerebellar Purkinje cells are resistant to delayed death after cerebral trauma and ampa-mediated excitotoxicity. Eur J Neurosci. 2007;26:649–56. doi: 10.1111/j.1460-9568.2007.05708.x. [DOI] [PubMed] [Google Scholar]
- Sols A, Crane RK. Substrate specificity of brain hexokinase. J Biol Chem. 1954;210:581–95. [PubMed] [Google Scholar]
- Stein HH, Cohen J. The measurement by a microtitration technique of carbon dioxide production in rat brain slices. Studis with glucose, fructose, and xylitol. Anal Biochem. 1976;71:444–51. doi: 10.1016/s0003-2697(76)80010-3. [DOI] [PubMed] [Google Scholar]
- Stocchi V, Magnani M, Canestrari F, Dacha M, Fornaini G. Multiple forms of human red blood cell hexokinase. Preparation, characterization, and age dependence. J Biol Chem. 1982;257:2357–64. [PubMed] [Google Scholar]
- Thorens B, Mueckler M. Glucose transporters in the 21st century. Am J Physiol Endocrinol Metab. 2010;298:E141–5. doi: 10.1152/ajpendo.00712.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuy S, Ladurner R, Volynets V, Wagner S, Strahl S, Konigsrainer A, Maier KP, Bischoff SC, Bergheim I. Nonalcoholic fatty liver disease in humans is associated with increased plasma endotoxin and plasminogen activator inhibitor 1 concentrations and with fructose intake. J Nutr. 2008;138:1452–5. doi: 10.1093/jn/138.8.1452. [DOI] [PubMed] [Google Scholar]
- Topping DL, Mayes PA. The concentration of fructose, glucose and lactate in the splanchnic blood vessels of rats absorbing fructose. Nutr Metab. 1971;13:331–8. doi: 10.1159/000175352. [DOI] [PubMed] [Google Scholar]
- Wada H, Okada Y, Uzuo T, Nakamura H. The effects of glucose, mannose, fructose and lactate on the preservation of neural activity in the hippocampal slices from the guinea pig. Brain Res. 1998;788:144–50. doi: 10.1016/s0006-8993(97)01532-1. [DOI] [PubMed] [Google Scholar]
- Wolfe BM, Ahuja SP, Marliss EB. Effects of intravenously administered fructose and glucose on splanchnic amino acid and carbohydrate metabolism in hypertriglyceridemic men. J Clin Invest. 1975;56:970–7. doi: 10.1172/JCI108177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Hung CY, Chan JY, Wu CW. An increase in adenosine-5′-triphosphate (ATP) content in rostral ventrolateral medulla is engaged in the high fructose diet-induced hypertension. J Biomed Sci. 2014;21:8. doi: 10.1186/1423-0127-21-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


