Abstract
The occurrence of nonulosonic acids in bacteria is wide-spread and linked to pathogenicity. However, the knowledge of cognate nonulosonic acid transferases is scarce. In the periodontopathogen Tannerella forsythia, several proposed virulence factors carry strain-specifically either a pseudaminic or a legionaminic acid derivative as terminal sugar on an otherwise structurally identical, protein-bound oligosaccharide. This study aims to shed light on the transfer of either nonulosonic acid derivative on a proximal N-acetylmannosaminuronic acid residue within the O-glycan structure, exemplified with the bacterium's abundant S-layer glycoproteins. Bioinformatic analyses provided the candidate genes Tanf_01245 (strain ATCC 43037) and TFUB4_00887 (strain UB4), encoding a putative pseudaminic and a legionaminic acid derivative transferase, respectively. These transferases have identical C-termini and contain motifs typical of glycosyltransferases (DXD) and bacterial sialyltransferases (D/E-D/E-G and HP). They share homology to type B glycosyltransferases and TagB, an enzyme catalyzing glycerol transfer to an N-acetylmannosamine residue in teichoic acid biosynthesis. Analysis of a cellular pool of nucleotide-activated sugars confirmed the presence of the CMP-activated nonulosonic acid derivatives, which are most likely serving as substrates for the corresponding transferase. Single gene knock-out mutants targeted at either transferase were analyzed for S-layer O-glycan composition by ESI-MS, confirming the loss of the nonulosonic acid derivative. Cross-complementation of the mutants with the nonnative nonulosonic acid transferase was not successful indicating high stringency of the enzymes. This study identified plausible candidates for a pseudaminic and a legionaminic acid derivative transferase; these may serve as valuable tools for engineering of novel sialoglycoconjugates.
Keywords: Bacteroidetes, glycoengineering, glycosyltransferase, nonulosonic acids, periodontitis
Introduction
Glycosylation as the most frequent modification of proteins (Messner 1997; Ohtsubo and Marth 2006; Varki 2006; Faridmoayer and Feldman 2010; Nothaft and Szymanski 2010; Messner et al. 2013; Tytgat and Lebeer 2014; Schäffer and Messner 2017) is well known to modify protein properties, endowing them with a wide repertoire of glycan-mediated functions. These can be as diverse as enabling site-directed delivery of glycoconjugates as well as mediating signaling events and modulating cell adhesion processes (Varki 1993; Taylor and Drickamer 2003). In the context of bacterial physiology, glycosylation can trigger the colonization of a specific host (region). Due to the increasing discovery of glycoconjugates in pathogenic bacteria, their investigation in a biomedical context is of great relevance. Especially, bacterial cell surface glycans, which represent the immediate contact zone of bacteria with the environment/host, seem to be prone to act as specific ligands for cell–cell or cell–bacterium interactions, or to serve as virulence factors based on molecular mimicry of host glycans (Varki et al. 2009).
Nonulosonic acids are particularly important at the interface of bacterial pathogenicity and human physiology (Varki 2008; Morrison and Imperiali 2014). They share a nine-carbon carbohydrate monomer as a common core structure, with additional structural variations mostly occurring at the C-5 and C-7 leading to over 50 derivatives identified so far across archaea, bacteria and eukaryotes (Angata and Varki 2002; Knirel et al. 2003). Sialic acid (Sia; neuraminic acid-Neu5Ac) carrying an N-acetyl group at the C-5 are the most abundant naturally occurring and best studied nonulosonic acids. Two exclusively bacterial derivatives, present in pathogens, are pseudaminic acid (5,7-diacetamido-3,5,7,9-tetradeoxy-l-glycero-α-l-manno-non-2-ulopyranosonic acid; Pse) and legionaminic acid (5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-non-2-ulopyranosonic acid; Leg) (Zunk and Kiefel 2014). The exact biological roles of pseudaminic acid and legionaminic acid and their derivatives are not yet fully understood. The modification of flagellins with these nonulosonic acids in Campylobacter and Helicobacter species confers bacterial virulence in facilitating bacterial host interactions (Thibault et al. 2001; Schirm et al. 2003). In two Campylobacter jejuni strains, the modification of flagellin subunits with Pse5Ac7Ac was found to be necessary for the assembly of a functional flagellum and, consequently, bacterial motility (Goon et al. 2003). Further, the structural similarity of pseudaminic acid and legionaminic acid to eukaryotic sialic acid indicates molecular mimicry as a basic strategy these pathogens may employ to evade the host immune response (Knirel et al. 2003; Vimr et al. 2004). However, it should be noted here that Pse and Leg have different stereochemistry, making Leg a potentially better mimic of host neuraminic acids than Pse.
The complete five-step biosynthesis pathway of cytidine monophosphate-activated pseudaminic acid (CMP-Pse) representing the biologically active form of pseudaminic acid for its incorporation into glycoconjugates has been characterized in detail in Helicobacter pylori (Schoenhofen et al. 2006), the pathway leading to CMP-Leg has been elucidated in C. jejuni (Schoenhofen et al. 2009). However, the subsequent and necessary transfer step of the nonulosonic acid from its nucleotide activator onto the acceptor—which might be the glycosylation site on a target polypeptide or a sugar residue within an oligosaccharide—by a dedicated nonulosonic acid transferase remains elusive. So far, the only report of a candidate pseudaminic acid transferase found in the literature concerns the motility-associated factor Maf1 predicted to be involved in the transfer of pseudaminic acid onto the flagellin of Aeromonas caviae (Parker et al. 2012). This was concluded from pull-down experiments between Maf1 and the flagellin, without provision of direct evidence of enzymatic function (Parker et al. 2014). With regard to legionaminic acid transferases, no predictions of such enzymes are presently available, neither in the literature nor in databases. Interestingly, selected sialyltransferases, i.e., porcine ST3Gal1, Pasteurella multocida sialyltransferase, Photobacterium α2,6-sialyltransferase and Neisseria meningitidis MC58 α2,3-sialyltransferase, were shown to accept CMP-Leg5Ac7Ac as a donor substrate to replace Sia as terminal sugar (Watson et al. 2011, 2015).
Tannerella forsythia provides the unique situation of a bacterium that strain-specifically displays either a pseudaminic or a legionaminic acid derivative as terminal sugar on an otherwise structurally very similar, protein-bound oligosaccharide (Posch et al. 2011; Friedrich et al. 2017). Tannerella forsythia is a Gram-negative bacterium that is recognized as a key periodontal pathogen (Socransky et al. 1998; Holt and Ebersole 2005) following the polymicrobial synergy and dysbiosis model of periodontal disease etiology (Hajishengallis and Lamont 2012; Hajishengallis 2014). The molecular basis of its pathogenicity is only slowly unraveling. Among several identified virulence factors (Veith et al. 2009; Sharma 2010), such as the outer membrane protein BspA (Onishi et al. 2008), KLIKK-proteases (Ksiazek et al. 2015) and outer membrane vesicles (Friedrich et al. 2015; Veith et al. 2015), are the two glycosylated cell surface (S-) layer proteins TfsA and TfsB which self-assemble into a 2D crystalline array on the bacterial cell surface, completely covering the outer membrane (Sabet et al. 2003; Sekot et al. 2011; Posch et al. 2012). We have shown for the T. forsythia ATCC 43037 type strain that the S-layer proteins as well as several other cell surface and outer membrane proteins of T. forsythia are modified at multiple sites at the conserved D(S/T)(A/I/L/V/M/T) motif with the same O-linked dekasaccharide (Fletcher et al. 2009; Posch et al. 2011), which displays a pseudaminic acid with an acetamidino (Am) group at C-5 and a glyceric acid at C-7 (Gra)—Pse5Am7Gra—as terminal, nonreducing end residue.
The biosynthesis of CMP-Pse in T. forsythia ATCC 43037 was only recently shown to be encoded by a dedicated gene locus (Friedrich et al. 2017) present in immediate vicinity to the general protein O-glycosylation gene cluster of T. forsythia ATCC 43037 (Posch et al. 2011). In fact, we expressed and confirmed the activity of all five necessary enzymes from the CMP-Pse biosynthetic pathway in that strain (Friedrich et al. 2017), which proceeds in analogy to what has been described for H. pylori (Schoenhofen et al. 2006). Since candidates for the enzymes modifying the pseudaminic acid in T. forsythia ATCC 43037 have not been identified so far, a crucial point still to be answered concerns the biosynthetic stage at which the modifications are transferred onto the nonulosonic acid.
On the genome of T. forsythia UB4 (Genbank accession number FMMN00000000; Stafford et al. 2016), the genes encoding the biosynthetic enzymes for CMP-Leg are replacing those for CMP-Pse in strain ATCC 43037. This was confirmed in step-wise in vitro assays using the recombinant enzymes (Friedrich et al. 2017), based on the knowledge of the CMP-Leg biosynthetic pathway in C. jejuni (Schoenhofen et al. 2009). In terms of basic sugar composition and glycan structure, the O-glycan of strain UB4 was found to be identical to that of strain ATCC 43037 (Posch et al. 2011) apart from a mass defect of 29 Da on the terminal Leg residue, likely reflecting different noncarbohydrate substituents as compared to the Gra and Am modifications of the ATCC 43037 pseudaminic acid residue, as well as a missing methyl group on the proximal N-acetylmannosaminuronic acid residue (−14 Da) (Friedrich et al. 2017).
Considering that the T. forsythia S-layer is classified as a virulence factor (Sharma 2010), it might well be that the involvement of the S-layer in the cell adhesion and invasion capability of the bacterium (Sakakibara et al. 2007) as well as in the delay of the host immune response against the bacterium (Honma et al. 2007; Sekot et al. 2011) is impacted by the bacterium's nonulosonic acids.
In this study, we investigated a putative pseudaminic acid derivative transferase (Tanf_01245) from the oral pathogen T. forsythia ATCC 43037 and a putative legionaminic acid derivative transferase (TFUB4_00887) from the clinical isolate T. forsythia UB4, using the abundant T. forsythia S-layer glycoproteins as a model system (Posch et al. 2011). Specifically, this included (i) construction of T. forsythia deletion mutants targeted at the candidate nonulosonic acid derivative transferases and subsequent determination of the effect of the gene deletion on S-layer O-glycan composition by mass spectrometry; (ii) analysis of the mutants for their cellular pool of nucleotide-activated sugars in order to exclude interference of the candidate transferases with the biosynthetic pathways of the nonulosonic acid precursor and to unravel if the nonulosonic acid modifications are present already at the CMP-bound state and (iii) a cross-complementation experiment of the nonulosonic acid derivative transferase mutants with the nonnative T. forsythia enzyme to learn about the specificity of the enzymes, considering that the core structures of pseudaminic acid and legionaminic acid are stereoisomers and that these residues would be transferred onto the same sugar acceptor residue within the S-layer O-glycan structure.
This is the first report on plausible enzyme candidates for the transfer of a pseudaminic acid (Pse5Am7Gra) and a legionaminic acid derivative as a terminal residue onto a glycoprotein glycan.
Results
Bioinformatic analyses of candidate genes for the transfer of nonulosonic acids in T. forsythia
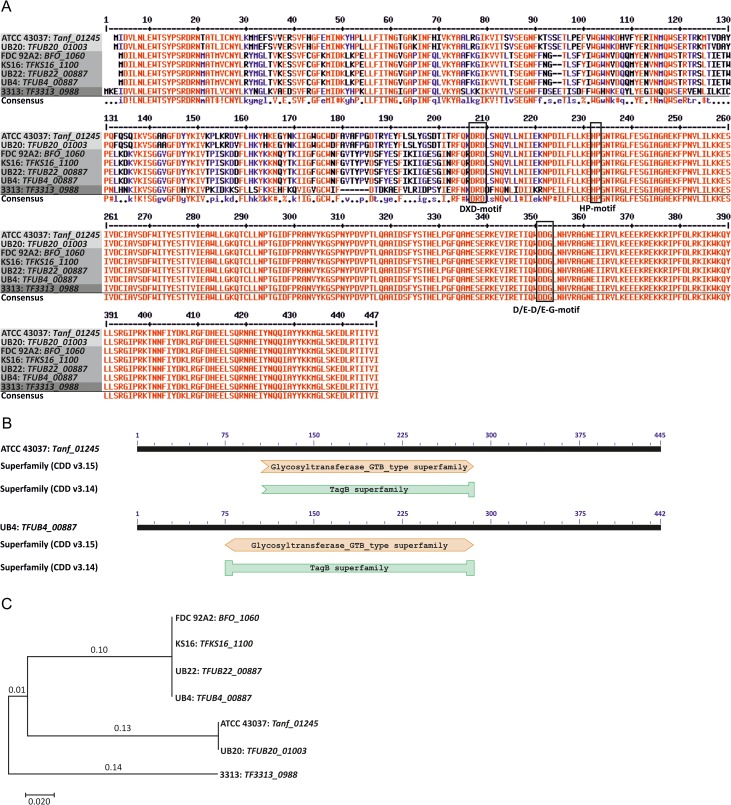
Recent findings from our laboratory revealed the presence of a functional biosynthetic pathway for either CMP-Pse or CMP-Leg in different T. forsythia strains (ATCC 43037, UB4 and 92A2, respectively) (Friedrich et al. 2017), elaborated in the course of the general T. forsythia protein O-glycosylation system (Markus Tomek, Valentin Friedrich, Christina Schäffer, unpublished data). To investigate the subsequent transfer step of the activated nonulosonic acid derivative on a proximal N-acetylmannosaminuronic acid residue as present in T. forsythia protein O-glycans, we were focusing here on two genes encoding putative nonulosonic acid derivative transferases. The candidate Pse derivative transferase gene Tanf_01245 is located immediately down-stream of the CMP-Pse biosynthesis locus on the T. forsythia ATCC 43037 genome and the candidate Legderivative transferase gene TFUB4_00887 is found on the T. forsythia UB4 genome down-stream of the CMP-Leg biosynthesis gene locus, separated only by a putative methyltransferase (Friedrich et al. 2017). The predicted transferases Tanf_01245 (445 amino acids; calculated molecular weight, 51.9 kDa) and TFUB4_00887 (442 amino acids; calculated molecular weight, 51.3 kDa) share 81% amino acid sequence identity. Precisely, the sequences of the 241-amino acid long C-terminal domains are identical in both proteins (Figure 1A). A DXD motif starting at position D205 (strain ATCC 43037) and D202 (strain UB4), respectively, is present at the beginning of the conserved C-terminal domain (Figure 1A). This short motif is found in many families of glycosyltransferases (GTs), which add a range of different sugars to other sugars, phosphates and proteins. All DXD-containing GTs use nucleoside diphosphate sugars as donors and require divalent cations, typically manganese (Breton et al. 2006). Usually, however, DXD-motifs are absent in sialyltransferases (Brockhausen 2014), which do not require divalent metal ions for enzymatic activity. Two recently identified functional motifs (D/E-D/E-G and HP), found in N. meningitidis (NmB-polyST) and P. multocida (PmST1), are highly conserved in bacterial sialyltransferases and important to enzyme catalysis and CMP-Neu5Ac binding (Freiberger et al. 2007). Both motifs are present in the putative nonulosonic acid transferases studied here (Figure 1A). Further bioinformatic analyses (Basic Local Alignment Search Tool—BLAST at http://blast.ncbi.nlm.nih.gov/Blast.cgi) suggested a GT B-type superfamily domain using NCBI's conserved domain database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, version CDD v3.15) or a putative conserved TagB superfamily domain (version CDD v3.14) (Marchler-Bauer et al. 2015), spanning the same amino acid residues, to be present in both proteins (Figure 1B). The TagB protein has been implicated in the priming step of poly(glycerol phosphate) wall teichoic acid synthesis in Bacillus subtilis (Swoboda et al. 2010). There, TagB adds a single glycerol phosphate residue to the nonreducing end of undekaprenyl-phosphate-linked N-acetylmannosamine-β(1,4)-N-acetylglucosamine-1-phosphate, which reveals analogy in basic sugar structure with the N-acetylmannosaminuronic acid residue present as acceptor in the T. forsythia O-glycan. Secondary structure predictions performed at http://bioinf.sce.carleton.ca/PCISS/start.php indicate for T. forsythia ATCC 43037 Tanf_01245 and T. forsythia UB4 TFUB4_00887, an α-helix content of 34.38% and 32.58%, a β-sheet content of 20.90% and 22.40%, respectively, and 45% turns, each. No transmembrane regions are predicted for the nonulosonic acid derivative transferases using the prediction server TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
Fig. 1.
Bioinformatic and molecular phylogenetic analyses of putative nonulosonic acid derivative transferases from different T. forsythia strains. (A) Amino acid sequence alignment of Tanf_01245 (strain ATCC 43037), TFUB4_00887 (strain UB4), TFUB20_01003 (strain UB20), BFO_1060 (strain FDC 92A2), TFKS16_1100 (strain KS16), TFUB22_00887 (strain UB22) and TF3313_0988 (strain 3313) illustrates a high sequence identity (81%) for all compared sequences except for that from strain 3313, and conservation of the C-terminal domain. Conserved motifs (DXD, D/E-D/E-G and HP) are indicated within boxes (alignment was done with the software Multalin at http://multalin.toulouse.inra.fr). (B) A conserved TagB superfamily domain (version CDD v3.14) or a GT B-type (version CDD v3.15) superfamily domain was identified in all candidate nonulosonic acid transferases using the NCBI's CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Marchler-Bauer et al. 2015). (C) The evolutionary history of nonulosonic acid transferases from different T. forsythia strains was inferred by using the Maximum Likelihood method based on the JTT matrix-based model (Jones et al. 1992). Four investigated strains (FDC 92A2, KS16, UB22 and UB4) have identical amino acid sequences and group together in the phylogenetic tree, thus representing strains with legionaminic acid transferases, while two strains, including the ATCC type strain (ATCC 43037) and strain UB20 are representing strains with pseudaminic acid transferases. Interestingly, strain 3313 does not group in neither of the nonulosonic acid transferases and thus has an unknown GT activity. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016). This figure is available in black and white in print and in color at Glycobiology online.
To obtain insight in the prevalence of the predicted nonulosonic acid derivative transferases in T. forsythia strains, we included in the multiple sequence alignment further sequences of homologous proteins from other publically available genomes of T. forsythia strains, including Tanf_01245 (strain ATCC 43037), TFUB4_00887 (strain UB4), TFUB20_01003 (strain UB20), BFO_1060 (strain FDC 92A2), TFKS16_1100 (strain KS16), TFUB22_00887 (strain UB22) and TF3313_0988 (strain 3313), revealing 246 identical C-terminal amino acid residues in all compared sequences. Based on differences at the amino acid level in the N-terminal regions, all aligned proteins except TF3313_0988 matched either the Tanf_01245 sequence from strain ATCC 43037 or the TFUB4_00887 sequence from strain UB4 (Figure 1A). This result is also reflected by analyzing the phylogenetic relationship of nonulosonic acid transferases (Jones et al. 1992; Kumar et al. 2016) (Figure 1C).
The migration behavior of S-layer glycoproteins on sodium dodecyl sulfate-polyacrylamide gel electrophoresis is affected by knocking-out the putative Pse and Leg derivative transferases
The glycosylated S-layer proteins TfsA and TfsB are the most abundant and best characterized glycoproteins in T. forsythia strains (Posch et al. 2011; Sekot et al. 2012) and, thus, were chosen as model glycoproteins for investigating the putative nonulosonic acid derivative transferases.
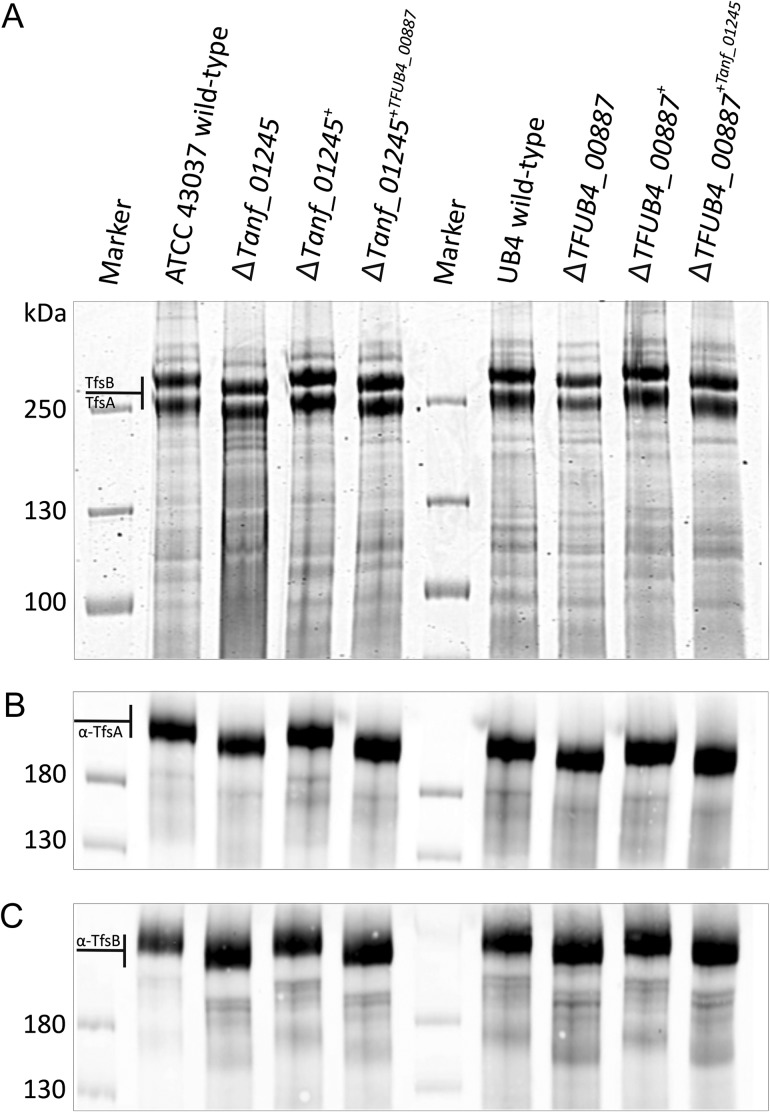
To assess an overall involvement of Tanf_01245 (candidate Pse derivative transferase) and TFUB4_00887 (candidate Leg derivative transferase) in S-layer protein glycosylation, the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) pattern of the respective gene deletion mutants T. forsythia ATCC 43037 ΔTanf_01245 and T. forsythia UB4 ΔTFUB4_00887 was compared to that of the corresponding parent strains and the reconstituted strains (T. forsythia ATCC 43037 ΔTanf_01245+ and T. forsythia UB4 TFUB4_00887+). Upon protein staining of crude cell extracts with Coomassie Brilliant Blue (CBB) (Figure 2A), the S-layer glycoproteins of both strain ATCC 43037 and UB4 appeared as prominent high-molecular mass bands, with TfsA migrating at ~230 kDa (calculated MW, 135 kDa) and TfsB at ~270 kDa (calculated MW, 152 kDa), as expected (Posch et al. 2011). A clear down-shift of both S-layer glycoproteins (~20 kDa, each) was visible in ATCC 43037 ΔTanf_01245, and the native migration behavior could be restored in the reconstituted strain ATCC 43037 ΔTanf_01245+ (Figure 2A). The same result was obtained in an analogous experiment performed with T. forsythia UB4, where the parent strain, UB4 ΔTFUB4_00887 and the reconstituted strain were compared (Figure 2A). Western immunoblots using polyclonal antibodies directed against the S-layer proteins TfsA (Figure 2B) and TfsB (Figure 2C) confirmed the identity of the said proteins in either T. forsythia strain, and simultaneously pinpointed a role of Tanf_01245 and TFUB4_00887 in the general protein O-glycosylation pathway of ATCC 43037 and UB4, respectively.
Fig. 2.
SDS-PAGE and Western immunoblot analyses of T. forsythia ATCC 43037 and T. forsythia UB4 wild-type and mutants. (A) CBB staining of crude cell extracts from T. forsythia ATCC 43037 wild-type, ΔTanf_01245 mutant, reconstituted mutant ΔTanf_01245+ and cross-complemented mutant ΔTanf_01245+TFUB4_00887 after separation on a 7.5% SDS-PA gel. The S-layer glycoproteins (labeled TfsA and TfsB) are indicated and the down-shift resulting from the loss of the Pse5Am7Gra residue can be observed in the deletion mutant and in the cross-complemented mutant, while in the reconstituted strain the bands are up-shifted again to wild-type level. The same migration profiles could be observed for T. forsythia UB4 wild-type, ΔTFUB4_00887 mutant, reconstituted mutant ΔTFUB4_00887+ and cross-complemented mutant ΔTFUB4_00887+Tanf_01245. PageRuler Plus Prestained Protein Ladder (Thermo Fisher Scientific) was used as a protein molecular weight marker. The S-layer glycoprotein bands were further processed for MS analyses. Western immunoblots probed with anti-TfsA antiserum (B) and anti-TfsB antiserum (C) confirmed the identity of the S-layer glycoproteins in all analyzed T. forsythia species. PageRuler Prestained Protein Ladder (Thermo Fisher Scientific) was used as a molecular weight marker.
O-Glycans of T. forsythia Pse and Leg derivative transferase knock-out mutants are devoid of the terminal nonulosonic acid
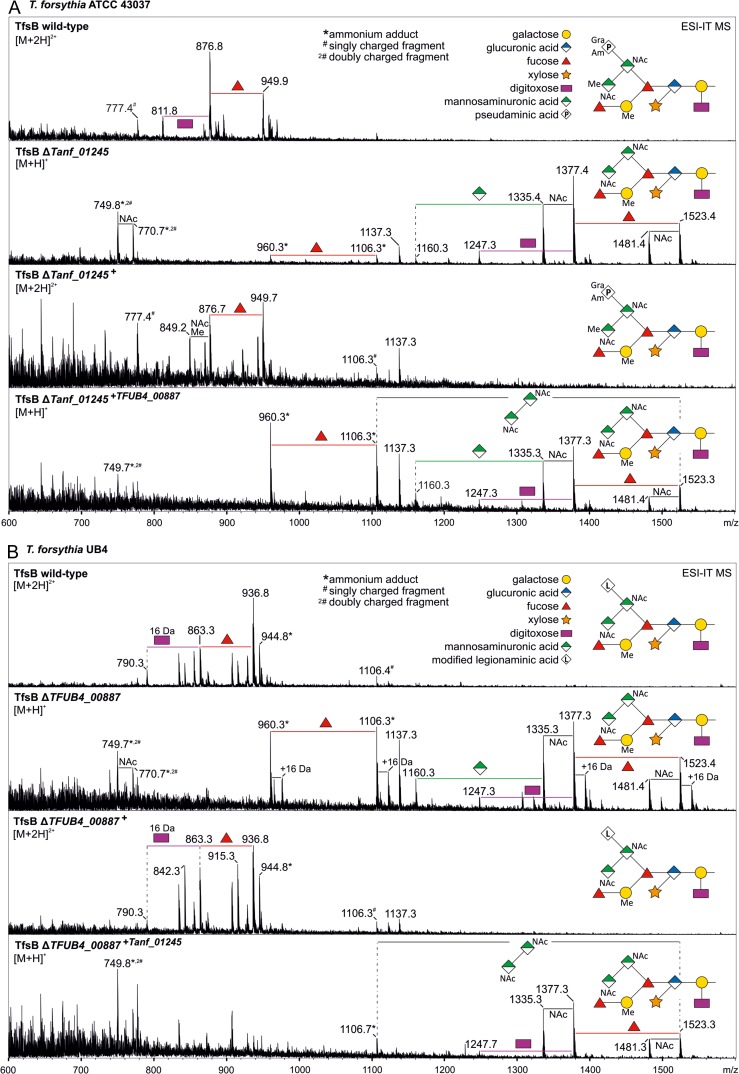
Knocking-out Tanf_01245 resulted in a loss of the terminal Pse5Am7Gra residue on the O-glycans of T. forsythia ATCC 43037 as revealed by electrospray ionization ion-trap mass spectrometry (ESI-IT-MS) analyses of β-eliminated S-layer glycans. Deconvoluted mass spectra showed the highest mass peak at m/z = 1523.4 [M + H]+, which conforms with the m/z value of the T. forsythia O-glycan lacking the Pse5Am7Gra residue (361.2 Da) and, additionally, one methyl group (14.0 Da); the latter was determined to be missing at the branching N-acetylmannosaminuronic acid residue (Figure 3A). Additional peaks with a typical fragmentation pattern of a glycan moiety were assigned and showed the subsequent loss of sugar residues and side chain modifications of the mutant O-glycan. The doubly charged complete wild-type O-glycan exhibited an m/z signal of 949.9 [M + 2 H]2+, which corresponds to m/z = 1898.8 when calculating a singly charged form thereof. In the reconstituted strain ATCC 43037 ΔTanf_01245+, Pse5Am7Gra transfer could be fully restored, as evidenced by the detection of a glycan with m/z = 949.7 [M + 2 H]2+, which corresponds to the mass of the wild-type glycan (Figure 3A).
Fig. 3.
Deconvoluted ESI-IT-MS sum spectra of β-eliminated TfsB O-glycans from T. forsythia parent and mutant strains. (A) Comparison of the spectra from T. forsythia ATCC 43037 wild-type, ΔTanf_01245 mutant, reconstituted mutant ΔTanf_01245+ and cross-complemented mutant ΔTanf_01245+TFUB4_00887. (B) Comparison of the spectra from T. forsythia UB4 wild-type, ΔTFUB4_00887 mutant, reconstituted mutant ΔTFUB4_00887+ and cross-complemented mutant ΔTFUB4_00887+Tanf_01245. Another glycan species with additional +16 Da at the position of the digitoxose was observed, indicative of the presence of a deoxyhexose instead of a dideoxyhexose in some forms of the glycan. The glycan structures of the highest mass peaks are shown as symbolic representations. Mass peaks from the subsequent fragmentation pattern were assigned according to the loss of carbohydrate units and modifications. Relative peak intensities of occurring peaks are given on the y axis. This figure is available in black and white in print and in color at Glycobiology online.
The wild-type O-glycan of T. forsythia UB4, where the Pse derivative is replaced by a Leg residue with calculated acetyl (Ac) and glycolyl (Gc) modifications (Friedrich et al. 2017), exhibits m/z = 936.8 [M + 2 H]2+, which corresponds to an m/z = 1872.6 of the [M + H]+ ion. In the UB4 ΔTFUB4_00887 mutant, the Leg derivative (350.2 Da) as well as one methyl group (−14.0 Da) modifying the N-acetylmannosaminuronic acid residue was no longer present as evidenced by the prominent peak with m/z = 1523.4 [M + H]+ (Figure 3B). As reported recently (Friedrich et al. 2017), also in this analysis, both in the UB4 wild-type and the UB4 ΔTFUB4_00887 mutant another glycan species with additional +16 Da at the position of the digitoxose could be observed, indicating the presence of a deoxyhexose instead of a dideoxyhexose in some forms of the glycan. Still, in the reconstituted strain T. forsythia UB4 ΔTFUB4_00887+, the production of the Leg derivative was fully restored, resulting in m/z = 936.8 [M + 2 H]2+, which conforms with that of the T. forsythia UB4 wild-type glycan (Figure 3B).
These data corroborated the involvement of Tanf_01245 and TFUB4_00887 in the transfer of the Pse and Leg derivative, respectively, during T. forsythia O-glycan assembly. Furthermore, it is indicated that methylation of the branching N-acetylmannosaminuronic acid residue occurs only after transfer of the respective nonulosonic acid derivative to the other, terminal N-acetylmannosaminuronic acid residue of T. forsythia ATCC 43037 O-glycan.
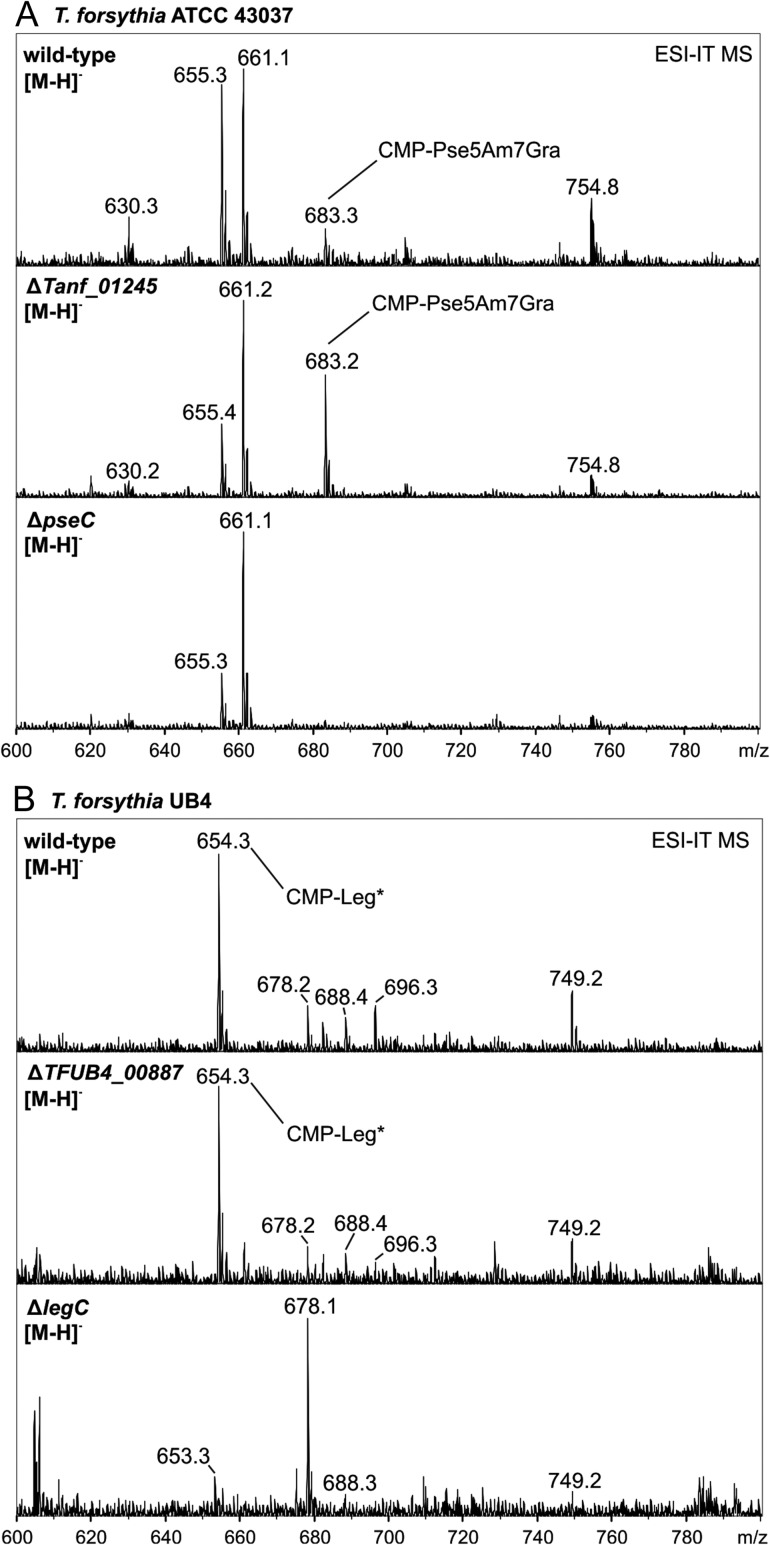
CMP activation and modification of nonulosonic acids occur prior to their transfer onto the O-glycan
Considering that the nonulosonic acids present in the T. forsythia O-glycans carry modifications, i.e., the proven Am (at C-5) and Gra (at C-7) modifications of the Pse residue in ATCC 43037 and the calculated Ac and Gc modifications (based on mass spectrometry (MS) data) of the Leg residue in UB4 (Posch et al. 2011; Friedrich et al. 2017) and that the modifying enzymes are still unknown, we wanted to rule out that the candidate transferases (Tanf_01245 and TFUB4_00887) would be involved in the modification instead of the predicted transfer process, which would also result in a loss of the nonulosonic acid on the O-glycans if the modifications were a prerequisite for the transfer to occur. Thus, the cellular pool of nucleotide-activated sugars was analyzed for the T. forsythia ATCC 43037 ΔTanf_01245 and T. forsythia UB4 ΔTFUB4_00887 mutants and compared to that of the parent strains. Besides known uridine 5′-diphosphate (UDP)- and guanosine-5′-diphosphate (GDP)-activated sugars, CMP-Pse5Am7Gra with m/z = 683.2 (Posch et al. 2011) representing the fully modified Pse precursor was clearly visible in the pool of nucleotide-activated sugars prepared from cell extracts of both the ATCC 43037 wild-type and the ATCC 43037 ΔTanf_01245 mutant (Figure 4A). As a negative control, the ATCC 43037 ΔpseC mutant available from a recent study (Friedrich et al. 2017), where the m/z = 683.2 compound is missing as a result of abolishing Pse biosynthesis, was used (Figure 4A), confirming the validity of the analysis.
Fig. 4.
ESI-IT-MS analysis of cellular nucleotide sugar pools from T. forsythia strains. (A) CMP-activated Pse5Am7Gra (m/z 683.3) was detected in the T. forsythia ATCC 43037 wild-type and in the ΔTanf_01245 mutant, whereas this mass was absent in a Pse biosynthesis deficient strain (ΔpseC) which served as a negative control. (B) In T. forsythia UB4 wild-type and in the ΔTFUB4_00887 mutant, a m/z 654.3 peak was identified, which was attributed to a CMP-activated Leg derivative (CMP-Leg*). This mass is consistent with having Ac and Gc modifications on Leg, based on calculation. Notably, this peak was absent in the Legbiosynthesis deficient strain (ΔlegC) which served as a negative control. Relative peak intensities are given on the y axis.
Legionaminic acid modification prior to transfer to the O-glycan was confirmed for T. forsythia UB4 as well, where a unique peak at m/z = 654.3 could be detected in the cellular pool of nucleotide-activated sugars of both the T. forsythia UB4 wild-type and the UB4 ΔTFUB4_00887 mutant (Figure 4B). This compound eluted just after CMP-Pse5Am7Gra on a porous graphitized carbon (PGC) column and showed the typical m/z = 322.0 fragment peak of CMP upon collision induced decay (Supplementary Figure S1), supporting its identity as a CMP-activated Leg derivative. In analogy to the data presented for T. forsythia ATCC 43037 (Figure 4A), the said m/z = 654.3 peak was absent in a UB4 ΔlegC mutant which is deficient in CMP-Leg biosynthesis (Friedrich et al. 2017).
Tannerella forsythia strains cross-complemented with the nonnative putative nonulosonic acid derivative transferase remain without nonulosonic acid derivative attachment to the protein O-glycan
Considering the high amino acid sequence similarity (81%) of the two candidate transferases under investigation and the stereoisomeric character of the Pse and Leg backbones, cross-complementation experiments of the T. forsythia mutants with proven deficiency in the respective nonulosonic acid derivative in the O-glycan structure (Figure 3A and B) with the nonnative putative nonulosonic derivative transferase were performed. Specifically, the T. forsythia ATCC 43037 ΔTanf_01245 mutant was complemented with the TFUB4_00887 gene, and vice versa, i.e., the UB4 ΔTFUB4_00887 mutant was complemented with the ATCC derived Tanf_01245 gene (Supplementary Figures S2A and S3A).
The cross-complemented strains T. forsythia ATCC 43037 ΔTanf_01245+TFUB4_00887 and T. forsythia UB4 ΔTFUB4_00887+Tanf_01245 were first analyzed on CBB-stained SDS-PAGE gels and Western immunoblots targeted at the TfsA and TfsB S-layer glycoproteins. However, neither of the cross-complemented strains showed the up-shift of the S-layer glycoproteins to the wild-type level (Figure 2). This result was confirmed by mass spectrometric analyses, in which both cross-complemented strains exhibited an m/z signal of 1523.3 [M + H]+ as maximum glycan mass. This mass was clearly indicative of the presence of T. forsythia O-glycans without the terminal nonulosonic derivative and without a methyl group (Figure 3).
In conclusion, these data suggest that the candidate transferases Tanf_01245 and TFUB4_00887 have high stringency for their CMP-substrate.
Discussion
Pathogenic bacteria frequently decorate their cell envelope with nonulosonic acids (Lewis et al. 2009; Morrison and Imperiali 2014). Major roles have been attributed to these sugar acids in biology and disease, including involvement in bacterial biofilm formation and motility. For instance, Leg glycan modification in C. jejuni was shown to be important for colonization of chickens (Howard et al. 2009), nonulosonic acid modification of the LPS of Vibrio vulnificus was shown to promote survival in the host bloodstream (Lubin et al. 2015), and there are various reports on the requirement of Pse modification for flagellar filament assembly in Campylobacter strains (Goon et al. 2003) and in Treponema denticola (Kurniyati et al. 2017). Further, it is speculated that pathogens may use nonulosonic acids other than sialic acids in molecular mimicry (Varki and Gagneux 2012), with a direct interaction between Pse on C. jejuni flagella and Siglec-10 receptor having been experimentally demonstrated (Stephenson et al. 2014).
Biosynthetic pathways for Sia, Pse and Leg can be predicted with a wide distribution among archaea and bacteria, including the bacterial phylum Bacteroidetes to which T. forsythia is affiliated (Lewis et al. 2009). While the biosynthetic pathways leading from UDP-GlcNAc to CMP-Pse and that from GDP-GlcNAc to CMP-Leg have been elucidated in detail in different bacteria (Schoenhofen et al. 2006, 2009), and, recently, also in T. forsythia (Friedrich et al. 2017), it is still unknown how the indispensable subsequent transfer step of Pse and Leg or derivatives thereof from the activated state onto an acceptor saccharide or polypeptide is elaborated. Considering the increasing documentation of the occurrence of Pse and Leg in pathogens, it is surprising that reports on putative Pse- or Leg-transferases in the literature are scarce (Parker et al. 2012; Watson et al. 2015).
This study was designed to shed light on putative nonulosonic acid derivative transferases encoded in the genome of different strains of T. forsythia, a periodontal pathogen which was shown to decorate its cell surface with either a Pse or a Leg derivative present as a terminal residue on otherwise structurally identical and abundant S-layer glycoprotein O-glycans (Posch et al. 2011; Friedrich et al. 2017). Specifically, we investigated the candidate CMP-Pse5Am7Gra:ManNAcA transferase Tanf_01245 from T. forsythia ATCC 43037 and the candidate CMP-Leg derivative:ManNAcA transferase TFUB4_00887 from T. forsythia UB4 (which is also present in T. forsythia FDC 92A2) (for glycan structure, see Friedrich et al. 2017). The identification of biosynthetic pathway genes for either CMP-Pse or CMP-Leg in conjunction with a gene encoding a putative nonulosonic derivative transferase in immediate vicinity on the genome of different strains of the same bacterium is interesting from an evolutionary point of view, since it raises the question about what has shaped the genome content of different T. forsythia strains and how this may contribute to the pathogenesis of periodontal diseases.
The high similarity of the two candidate transferases at primary sequence level was not unexpected, since their nonulosonic acid substrates are stereoisomers, albeit with different noncarbohydrate modifications (5Am7Gra in Pse vs. calculated AcGc in Leg). A multiple sequence alignment revealed an identical, 245-amino acid comprising, C-terminal protein region of Tanf_01245 and TFUB4_00887, and a GT-B or TagB superfamily domain (vide infra) is predicted (Figure 1). The prediction of two domains on the amino acid sequence level is in agreement with the two-domain presentation of known 3D structures from crystallized GTs with both GT-A and GT-B folds (Breton et al. 2006; Lairson et al. 2008). A membrane associated third-fold family has been identified based on the crystal structure and kinetic data from an α-2,3/2,8-sialyltransferase (CstII) in C. jejuni (Chiu et al. 2004). In agreement with the experimental evidence provided in the present study, bioinformatic analysis predicts a GT-fold for the putative T. forsythia nonulosonic derivative transferases. However, it cannot be ruled out that a novel GT-fold might be involved in nonulosonic acid transfer, which makes these enzymes exciting objects to study.
Interestingly, a DXD motif as well as a D/E-D/E-G and HP motif, common to bacterial sialyltransferases, were identified in all investigated nonulosonic acid transferases (Figure 1A). Glycosyltransferases from the A-type family share this common DXD motif, which interacts primarily with the phosphate group of the respective nucleotide donor (Breton et al. 2006; Lairson et al. 2008). Yet, a DXD motif is only a decent indication of a GT, since it is not necessarily present in this class of enzymes as exemplified by a CMP-Sia utilizing enzyme from C. jejuni (Chiu et al. 2004). A significant support for a transferase function of both enzymes, Tanf_01245 and TFUB4_00887, is derived from homology searches predicting a TagB superfamily domain or a GT B-type superfamily domain located at the rather N-terminal part of the proteins (Figure 1B). TagB is a glycerophosphate transferase from the wall teichoic acid biosynthesis in B. subtilis 168 where it catalyzes the transfer of a single phosphoglycerol unit from CDP-glycerol onto the C-4 hydroxyl of a ManNAc residue (Swoboda et al. 2010). Even though glycerol and nonulosonic acids are clearly different substrates, two remarkable analogies are evident; first, the ManNAc residue from the teichoic acid backbone has the same basic structure as the ManNAcA residue, which is the acceptor saccharide for the nonulosonic acids in T. forsythia O-glycan biosynthesis, and secondly, an 1,4-linkage as formed upon TagB catalysis is also implemented in the terminal transfer of the Pse and Leg derivative onto the ManNAcA residue within the known O-glycan structure (for glycan structure, see Friedrich et al. 2017).
Thus, it is conceivable to assume that the conserved C-terminal protein region of Tanf_01245 and TFUB4_00887, encompassing the DXD, the D/E-D/E-G and HP motifs, is involved in the binding of the CMP activator of both the Pse and Leg derivative, while the TagB domain might specifically bind the nonulosonic acid portion of CMP-Pse5Am7Gra and the CMP-Leg derivative, respectively (Figure 1A).
Experimentally, we have successfully proven the requirement of Tanf_01245 from T. forsythia ATCC 43037 and TFUB4_00887 from T. forsythia UB4 for the full saccharide assembly of the T. forsythia protein O-glycans. Deletion of Tanf_01245 and TFUB4_00887, respectively, resulted in a Pse5Am7Gra and Leg derivative deficient phenotype as revealed by LC-ESI-MS of β-eliminated O-glycans from TfsB (Figure 3) and TfsA (not shown). To exclude that polar effects would cause the loss of the respective nonulosonic acid, we showed that reconstituted strains regained the Pse5Am7Gra and Leg derivative transfer activity yielding the native dekasaccharide form of the glycan (Figure 3).
Since the biosynthetic enzymes for the noncarbohydrate modifications on the T. forsythia nonulosonic acids are currently unknown, it was important to demonstrate that these have not been unintentionally targeted in the deletion mutants and that, consequently, indeed the fully modified sugar acid would be available in CMP-activated form. For instance, deletion of genes involved in the biosynthesis of the acetamidino group in Methanococcus maripaludis resulted in flagellins with a truncated glycan that did not only miss the acetamidino group present on an inner sugar (a modified ManNAc residue) but also the terminal sugar of the flagellin glycan (Jones et al. 2012). This indicates that the loss of noncarbohydrate substituents within a glycan might influence down-stream enzymes. ESI-IT-MS analyses of the cellular pools of nucleotide-activated sugars from the ΔTanf_01245 and ΔTFUB4_00887 mutant strains in comparison to the ATCC 43037 and UB4 parent strains demonstrated the presence of the mature Pse and Leg derivative, respectively, at m/z = 683.2 in both ATCC 43037 samples and m/z = 654.3 in both UB4 samples (Figure 4). Further, the presence of CMP was proven by the appearance of a typical m/z = 322.0 fragment in the MS/MS spectrum (Supplementary, Figure S1), which was especially important for the T. forsythia UB4 strain, as the structure of its Leg derivative has not been fully elucidated so far.
To learn about the substrate specificity of the predicted T. forsythia Pse5Am7Gra and Leg derivative transferase, cross-complementation experiments were performed. Complementation of T. forsythia ATCC 43037 ΔTanf_01245 and T. forsythia UB4 ΔTFUB4_00887 with the nonnative enzyme could not restore the native O-glycan phenotype (Figure 3), despite identity of the underlying O-glycan structure. Considering that even cross-glycosylation experiments between different species of the Bacteroidetes, namely between T. forsythia ATCC 43037 and Bacteroides fragilis have been successful (Posch et al. 2013), it is conceivable to assume that Tanf_01245 from T. forsythia ATCC 43037 and TFUB4_00887 from T. forsythia UB4 possess high stringency for the CMP-activated nonulosonic acid substrate. Whether stringency relates to the stereoisomery or the modifications of the nonulosonic acids or both remains to be investigated.
While we have provided strong evidence of the discovery of two new nonulosonic acid transferases in different strains of the periodontal pathogen T. forsythia, the development of a dedicated in vitro assay to unequivocally prove the enzymatic activity of the investigated T. forsythia enzymes is currently under way. Given the power of current glycoengineering approaches the present study may contribute to the design of novel nonulosonic acid-based glycoconjugates of potential therapeutic relevance in the future.
Materials and methods
Bacterial strains and growth conditions
Tannerella forsythia ATCC 43037 type-strain (American Type Culture Collection—ATCC, Manassas, VA) (Friedrich et al. 2015) and T. forsythia UB4 (clinical isolate kindly provided by Dr. Ashu Sharma, State University of New York at Buffalo, NY) and defined mutants (Table I) were grown anaerobically as described previously (Tomek et al. 2014). Briefly, brain heart infusion (BHI) liquid medium (Oxoid, Basingstoke, UK) with a concentration of 37.0 g/L was supplemented with 10.0 g/L of yeast extract (Oxoid), 1.0 g/L l-cysteine (Sigma, Vienna, Austria), 5.0 µg/mL hemin (Sigma), 2.0 µg/mL menadione (Sigma), 20 µg/mL N-acetylmuramic acid (Carbosynth, Compton, UK) and 5% (v/v) horse serum (Thermo Fisher Scientific, Vienna, Austria). For cultivation of T. forsythia wild-type and mutant strains on BHI agar plates (0.8%, w/v), incubation was performed in anaerobic jars (AnaeroJar; Oxoid) at 37°C. Media were supplemented with 50 µg/mL gentamycin, 5 µg/mL erythromycin or 10 µg/mL chloramphenicol (Cat), when appropriate.
Table I.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and use or description | Source or reference |
|---|---|---|
| Escherichia coli strain | ||
| DH5α | F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1; cloning strain | Invitrogen, Austria |
| Tannerella forsythia strains | ||
| ATCC 43037 | Type strain, wild-type | ATCC; Friedrich et al. (2015) |
| ATCC 43037 ΔTanf_01245 | ΔTanf_01245::ermF; knock-out strain of Tanf_01245 | This work |
| ATCC 43037 ΔTanf_01245+ | ΔTanf_01245::Tanf_01245 cat; reconstituted knock-out strain | This work |
| ATCC 43037 ΔTanf_01245+TFUB4_00887 | ΔTanf_01245:: TFUB4_00887cat; cross-complemented knock-out strain | This work |
| ATCC 43037 ΔpseC | ΔpseC::ermF; knock-out strain of Tanf_01190 | Friedrich et al. (2017) |
| UB4 | Clinical isolate; wild-type strain | Stafford et al. (2016) |
| UB4 ΔTFUB4_00887 | ΔTFUB4_00887::ermF; knock-out strain of TFUB4_00887 | This work |
| UB4 ΔTFUB4_00887+ | ΔTFUB4_00887::cat; reconstituted knock-out strain | This work |
| UB4 ΔTFUB4_00887+Tanf_01245 | ΔTFUB4_00887::Tanf_01245 cat; cross-complemented knock-out strain | This work |
| UB4 ΔlegC | ΔlegC::ermF; knock-out strain of TFUB4_00900 | Friedrich et al. (2017) |
| FDC 92A2 | Wild-type strain | ATCC; Tanner et al. (1986) |
| Plasmids | ||
| pJET1.2/blunt | Cloning vector; ampR | Thermo Scientific, Austria |
| pJET/ΔTF0955ko | Vector for amplification of the erythromycin resistance gene | Tomek et al. (2014) |
| pEXALV | Vector for amplification of the Cat resistance gene | Zarschler et al. (2009) |
| pJET1.2/ΔTanf_01245 | Tanf_01245 knock-out cassette; ampRermFR | This work |
| pJET1.2/ΔTanf_01245+ | Cassette for reconstitution of ΔTanf_01245; ampRcatR | This work |
| pJET1.2/ΔTanf_01245+TFUB4_00887 | Cassette for cross-complementation of ΔTanf_01245 with TFUB4_00887; ampRcatR | This work |
| pJET1.2/ΔTFUB4_00887 | TFUB4_00887 knock-out cassette; ampRermFR | This work |
| pJET1.2/ΔTFUB4_00887+ | Cassette for reconstitution of ΔTFUB4_00887; ampRcatR | This work |
| pJET1.2/ΔTFUB4_00887+Tanf_01245 | Cassette for cross-complementation of ΔTFUB4_00887 with Tanf_01245; ampRcatR | This work |
Escherichia coli strains (Table I) were grown under standard conditions in lysogeny broth (LB) medium supplemented with 100 µg/mL ampicillin, when appropriate.
Construction of knock-out strains deficient in pseudaminic or legionaminic acid transferase activity
Vectors were constructed to knock-out candidate genes for a Pse5Am7Gra transferase in T. forsythia ATCC 43037 (gene Tanf_01245) and for a Leg derivative in T. forsythia UB4 (gene TFUB4_00887) (Table I). A detailed description of the cloning procedure and the transformation of knock-out cassettes into T. forsythia is published elsewhere (Tomek et al. 2014). For PCR amplifications, Phusion High-Fidelity DNA polymerase (Thermo Fisher Scientific) was used according to the manufacturer's instructions. Oligonucleotides (Thermo Fisher Scientific) used in this study are listed in Table II. Extraction of genomic DNA was performed according to a published protocol (Cheng and Jiang 2006).
Table II.
Oligonucleotide primers used for PCR amplification reactions
| Primers | Sequence (5′–3′) |
|---|---|
| 1a | GGTACCCCCGATAGCTTCCGCTATTGC |
| 2 | CTACGAAGGATGAAATTTTTCAGGG |
| 3a | GCAATAGCGGAAGCTATCGGGGGTACC |
| 4 | CCCTGAAAAATTTCATCCTTCGTAG |
| 48 | GTCAGATAGGCCTAATGACTGGC |
| 76 | TTATAAAAGCCAGTCATTAGGCCTATCTGAC |
| 77b | aatcaGCATGC GGTACCTTATAAAAGCCAGTCATTAGGCCTATCTGAC |
| 118 | ATGGCTACAATGGTCTGTAATTATCTTC |
| 119 | TTATATTACTGTTATTGTTCGTAGATCC |
| 120 | CCATGATAATCTCGACTTCGG |
| 121 | GCAATAGCGGAAGCTATCGGGGGTACCATTCCTATCTCTTGAAGGATAGG |
| 122 | CCCTGAAAAATTTCATCCTTCGTAGGTATAGAGGTACAATGGATATAGGGC |
| 123 | GCACCCATTTATCTAAATAATCTTC |
| 124 | GGCCCTCAACCTTTTCTGGC |
| 125 | CCTATCCTTTAGGTATCTATATG |
| 126b | CGAACAATAACAGTAATATAATGTACAATGAACTTTAATAAAATTGATTTAGAC |
| 127 | aatcaCATATGGGTACCTTATAAAAGCCAGTCATTAGGCCTATCTGAC |
| 128 | aatcaGGTACCGTATAGAGGTACAATGGATATAGGGC |
| 130 | CCAATTGTCTAAATCAATTTTATTAAAGTTCATTGTACATTATATTACTGTTATTGTTCGTAGATCC |
| 133 | aatcaTGTACATTATATTACTGTTATTGTTCGTAGATCCTC |
| 134 | aatcaCTCGAGCCATGATAATCTCGACTTCGG |
| 140 | aatcaCATATGGCACCCATTTATCTAAATAATCTTC |
| 152 | CTAAATTTCTTTCATAATAATTCTTTGTATAATGATTGATGTTTTAAATTTAGAATGGACC |
| 153 | GGTCCATTCTAAATTTAAAACATCAATCATTATACAAAGAATTATTATGAAAGAAATTTAG |
| 460c | ATGACAAAAAAGAAATTGCCCGTTCGTTTTAC |
| 461c | CTACGAAGGATGAAATTTTTCAGGGACAAC |
| 474d | CGGGCAATTTCTTTTTTGTCATTTCTTAATGTAATCTAAGTCCAACCG |
| 475d | TTGTAGCAGAACTATCAGCCAATCAC |
| 476d | GTTGTCCCTGAAAAATTTCATCCTTCGTAGGTATAGAGGTACAATGGATATAG |
| 477d | CCCAGACTCTTCTTTAACAAGAAACC |
| 512 | TGCAGGCTGCAATTGATTCC |
| 513 | GATCCCACGTGAAAGCAAATA |
| 524 | GTAAAACGAACGGGCAATTTCTTTTTTGTCAT |
| 525 | CCCTGAAAAATTTCATCCTTCGTAG |
| 530 | CGTATGATATTTGCAGTCTTG |
| 531 | GTAATAACCATATCTGCCTCTGGAAC |
| 540 | CAACAATTGTAGCAGAACTATCAGCC |
| 541 | aatcaGGTACCTATAAGTATAGAGGTACAATGGATATAGG |
| 542 | aatcaGCATGCGGTATCTATATGAAGTACGCACC |
| 543 | ctgaCTCGAGCAACAATTGTAGCAGAACTATC |
| 565 | CTAAATCAATTTTATTAAAGTTCAT |
| 575 | TCTAAATTAAGAATATCCATTTCTTAATGTAATCTAAGTCCAACCGCATTCC |
| 576 | GACTTAGATTACATTAAGAAATGGATATTCTTAATTTAGAATGGACTTCC |
Nucleotides used for overlap-extension PCRs are written in bold, artificial restriction sites are underscored. Lowercase letters indicate artificially introduced bases to improve restriction enzyme digestion.
dSequence (3′–5′).
The knock-out vectors for the construction of a T. forsythia ATCC 43037 ΔTanf_01245 and a T. forsythia UB4 ΔTFUB4_00887 mutant consisted of approximately 1-kbp up- and down-stream homology regions and an erythromycin resistance marker (ermF) cloned in between. Primer pairs 474/475 and 476/477, respectively, were used to amplify the up- and down-stream homology regions from genomic DNA of T. forsythia ATCC 43037, and primer pairs 120/121 and 122/123 were used to amplify those from genomic DNA of T. forsythia UB4. The antibiotic resistance gene ermF (805 bp) was amplified from pJET/TF0955ko (Tomek et al. 2014), either without the promotor region using primers 460 and 461 or including the native promotor using primers 1 and 2. Subsequently, each knock-out cassette was blunt-end cloned into the cloning vector pJET1.2, creating the final knock-out vectors pJET1.2/ΔTanf_01245 and pJET1.2/ΔTFUB4_00887. Transformed and viable clones on selective plates containing erythromycin were further tested for correct integration of the knock-out cassette by screening PCR (Supplementary Figures S2 and S3).
To reconstitute the transferase function in the T. forsythia ATCC 43037 ΔTanf_01245 and T. forsythia UB4 ΔTFUB4_00887mutants, the genes of interest were re-integrated by homologous recombination including cat as an alternate resistance gene for selection purposes (Supplementary Figures S2 and S3). The up-stream region, including the approximately 1-kbp up-stream homology region and the 1338-bp Tanf_01245 gene, was amplified with primer pair 540/130 from genomic DNA of T. forsythia ATCC 43037. The 650-bp cat gene (without the native promotor) was amplified using pEXALV as a template (Zarschler et al. 2009) and primers 126/77 prior to addition to the up-stream region by way of overlap-extension PCR. The construct was blunt-end ligated into the cloning vector pJET1.2. The approximately 1-kbp down-stream homology region (541/542) was cloned into the above mentioned pJET1.2 construct using the restriction sites KpnI and SphI, assembling the final construct pJET1.2/ΔTanf_01245+. After control digestion (not shown), electrocompetent T. forsythia ΔTanf_01245 cells were transformed with the final construct.
Analogously, up-stream and TFUB4_00887 gene primers 120/130, cat primers 126/127 and down-stream primers 128/140 (with KpnI and NdeI restriction sites) were used to construct the reconstitution cassette pJET1.2/ΔTFUB4_00887+. Subsequent steps were performed as described for the Tanf_01245-based construct, yielding the final construct pJET1.2/ΔTFUB4_00887+.
Cross-complementation of nonulosonic acid transferase deficient mutants
Based on the reconstitution cassette pJET1.2/ΔTanf_01245+, a cross-complementation cassette was constructed by replacing the native gene Tanf_01245 with TFUB4_00887. The native up-stream region was amplified with primers 543 (including an XhoI restriction site) and 575. The UB4 TFUB4_00887 gene was amplified using primer pair 576/133(BsrGI) and cloned to the up-stream region by overlap-extension-PCR. Via the restriction sites XhoI and BsrGI, the native gene was replaced by the nonnative gene, assembling the final cross-complementation cassette pJET1.2/ΔTanf_01245+TFUB4_00887.
Analogously, the cross-complementation cassette pJET1.2/ΔTFUB4_00887+Tanf_01245 for the UB4 strain was constructed. Briefly, primers 134(XhoI)/153 were used for the amplification of the up-stream homology region and 152/133(BsrGI) for the amplification of the Tanf_01245 gene. Clones were tested for correct integration on genomic level after transformation and selection (Supplementary Figures S2 and S3).
SDS-PAGE and Western immunoblotting
SDS-PAGE gels (7.5%) were prepared according to a standard protocol (Laemmli 1970). Crude cell extracts of T. forsythia ATCC 43037, UB4 and defined mutants thereof were run in a Mini Protean electrophoresis apparatus (Bio-Rad, Vienna, Austria) and proteins were visualized with colloidal CBB R-250. For Western immunoblot analyses, proteins were transferred onto a polyvinylidene difluoride membrane (Bio-Rad) using a Mini Trans-Blot Cell (Bio-Rad). Polyclonal antisera raised in rabbits against the recombinant S-layer proteins TfsA (α-TfsA) and TfsB (α-TfsB) (Sekot et al. 2012) were used as primary antibodies followed by a monoclonal goat anti-rabbit secondary antibody labeled with IRDye 800CW (LI-COR Biosciences, Lincoln, NE). S-layer protein bands were visualized at 800 nm using an Odyssey Infrared Imaging System (LI-COR Biosciences).
S-layer O-glycan preparation and liquid chromatography ESI-IT-MS
The preparation and release of O-glycans from the glycosylated S-layer proteins TfsA and TfsB by in-gel reductive β-elimination was performed as described previously (Posch et al. 2011; Tomek et al. 2014). Removal of excess salt via PGC cartridges was performed according to published protocols (Pabst and Altmann 2008; Stadlmann et al. 2008) and the final purification of borohydride reduced O-glycans prior to ESI-IT-MS analyses was completed using preparative PGC-HPLC (Posch et al. 2011). The glycan mixture was analyzed using a Dionex Ultimate 3000 system directly linked to an ion trap instrument (amaZon speed EDT, Bruker, Germany) equipped with the standard ESI source in the positive ion, DDA mode (= switching to MSMS mode for eluting peaks). MS-scans were recorded (range, 450-1650 m/z; icc target was set to 100,000; maximum accumulation time, 200 ms) and the highest peaks were selected for fragmentation. Instrument calibration was performed using ESIcalibration mixture (Agilent, Vienna, Austria). For separation of the glycans, a Thermo Hypercarb separation column (5 µm particle size; 100 × 0.360 mm) was used. A gradient from 99% solvent A and 1% solvent B (solvent A, 65 mM ammoniumformate buffer (pH 3.0); B, 100% acetonitrile) to 21% B in 20 min was applied, followed by a 10-min gradient from 21% B to 50% B, at a flow rate of 6 µl/min. Data were evaluated using the DataAnalysis 4.0 software (Bruker).
Extraction, purification and analysis of nucleotide-activated sugars
Cellular pools of nucleotide-activated sugars were extracted and purified as described before (Posch et al. 2011). Briefly, 2 mL of bacterial solution was harvested by centrifugation, washed in phosphate-buffered saline and lysed by ultrasonication in 62 mM sodium fluoride buffer before loading onto a Hypersep Hypercarb 10 mg column (Thermo Scientific, Vienna, Austria). Elution of GDP-, UDP- and CMP-activated sugars was done with 50% acetonitrile in 50 mM ammoniumformate buffer pH 9.0. The glycan mixture was analyzed using a Dionex Ultimate 3000 system directly linked to an ion trap instrument (amaZon speed EDT) equipped with the standard ESI source in the negative ion, DDA mode (= switching to MSMS mode for eluting peaks). MS-scans were recorded (range, 200–900; m/z; icc target was set to 15,000; maximum accumulation time, 200 ms; target mass was set to 600 m/z) and the five highest peaks were selected for fragmentation. Instrument calibration was performed using ESIcalibration mixture (Agilent). For separation of the analytes, a Thermo Hypercarb separation column (5 µm particle size, 100 × 0.360 mm) was used. A gradient from 99% solvent A and 1% solvent B (solvent A, 0.3% formic acid adjusted to pH 9.0 with ammonia solution, B, 100% acetonitrile) to 25% B in 20 min was applied, followed by a 10-min gradient from 25% B to 50% B, at a flow rate of 6 µL/min. Data were evaluated using the DataAnalysis 4.0 software (Bruker).
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Ashu Sharma (State University of New York at Buffalo, NY) and Dr. Graham Stafford (University of Sheffield, UK) for kindly providing the clinical isolate T. forsythia UB4.
Supplementary data
Funding
The Austrian Science Fund FWF, projects P24317-B22 (to C.S.), P24305-B20 (to P.M.) and the FWF Doctoral Program “Biomolecular Technology of Proteins” W1224.
Abbreviations
Am, N-acetyl or acetamido; Am, N-acetimidoyl or acetamidino; BHI, brain heart infusion; Cat, chloramphenicol; CBB, Coomassie Brilliant Blue; CMP, cytidine-5′-monophosphate; ESI-IT-MS, electrospray ionization ion-trap mass spectrometry; Gc, N-glycolyl; GDP, guanosine-5′-diphosphate; GT, glycosyltransferase; Gra, N-glyceroyl or N-2,3-dihydroxypropionyl or glycerate group; LB, lysogeny broth; LC, liquid chromatography; Leg, legionaminic acid (Leg5,7Ac2), 5,7-diacetamido-3,5,7,9-tetradeoxy-d-glycero-d-galacto-non-2-ulopyranosonic acid; ManNAc, N-acetylmannosamine; ManNAcA, N-acetylmannosaminuronic acid; ManNAcCONH2, N-acetylmannosaminuronamide; MS, mass spectrometry; NeuAc, N-acetylneuraminic acid; PGC, porous graphitized carbon; Pse, pseudaminic acid (Pse5,7Ac2), 5,7-bis(acetylamino)-3,5,7,9-tetradeoxy-l-glycero-α-l-manno-non-2-ulopyranosonic acid; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; Sia, sialic acid (Neu5Ac), 5-acetamido-3, 5-dideoxy-d-glycero-d-galacto-nonulosonic acid; UDP, uridine 5′-diphosphate
References
- Angata T, Varki A. 2002. Chemical diversity in the sialic acids and related α-keto acids: An evolutionary perspective. Chem Rev. 102:439–470. [DOI] [PubMed] [Google Scholar]
- Breton C, Šnajdrová L, Jeanneau C, Koča J, Imberty A. 2006. Structures and mechanisms of glycosyltransferases. Glycobiology. 16:29R–37R. [DOI] [PubMed] [Google Scholar]
- Brockhausen I. 2014. Crossroads between bacterial and mammalian glycosyltransferases. Front Immunol. 5:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H-R, Jiang N. 2006. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol Lett. 28:55–59. [DOI] [PubMed] [Google Scholar]
- Chiu CPC, Watts AG, Lairson LL, Gilbert M, Lim D, Wakarchuk WW, Withers SG, Strynadka NCJ. 2004. Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat Struct Mol Biol. 11:163–170. [DOI] [PubMed] [Google Scholar]
- Faridmoayer A, Feldman MF. 2010. Bacterial protein glycosylation In: Mander L, Liu H-W, editors. Comprehensive natural products II: Chemistry and biology. Kidlington (UK): Elsevier Ltd; p. 351–380. [Google Scholar]
- Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. 2009. A general O-glycosylation system important to the physiology of a major human intestinal symbiont Bacteroides fragilis. Cell. 137:321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiberger F, Claus H, Günzel A, Oltmann-Norden I, Vionnet J, Mühlenhoff M, Vogel U, Vann WF, Gerardy-Schahn R, Stummeyer K. 2007. Biochemical characterization of a Neisseria meningitidis polysialyltransferase reveals novel functional motifs in bacterial sialyltransferases. Mol Microbiol. 65:1258–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich V, Gruber C, Nimeth I, Pabinger S, Sekot G, Posch G, Altmann F, Messner P, Andrukhov O, Schäffer C. 2015. Outer membrane vesicles of Tannerella forsythia: Biogenesis, composition, and virulence. Mol Oral Microbiol. 30:451–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich V, Janesch B, Windwarder M, Maresch D, Braun ML, Megson ZA, Vinogradov E, Goneau MF, Sharma A, Altmann F, et al. 2017. Tannerella forsythia strains display different cell-surface nonulosonic acids: biosynthetic pathway characterization and first insight into biological implications. Glycobiology, 27:342–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich V, Pabinger S, Chen T, Messner P, Dewhirst FE, Schäffer C. 2015. Draft genome sequence of Tannerella forsythia type strain ATCC 43037. Genome Announc. 3: e00660–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goon S, Kelly JF, Logan SM, Ewing CP, Guerry P. 2003. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol Microbiol. 50:659–671. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. 2014. Immunomicrobial pathogenesis of periodontitis: Keystones, pathobionts, and host response. Trends Immunol. 35:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 27:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 38:72–122. [DOI] [PubMed] [Google Scholar]
- Honma K, Inagaki S, Okuda K, Kuramitsu HK, Sharma A. 2007. Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb Pathog. 42:156–166. [DOI] [PubMed] [Google Scholar]
- Howard SL, Jagannathan A, Soo EC, Hui JP, Aubry AJ, Ahmed I, Karlyshev A, Kelly JF, Jones MA, Stevens MP, et al. 2009. Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infect Immun. 77:2544–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 8:275–282. [DOI] [PubMed] [Google Scholar]
- Jones GM, Wu J, Ding Y, Uchida K, Aizawa S, Robotham A, Logan SM, Kelly J, Jarrell KF. 2012. Identification of genes involved in the acetamidino group modification of the flagellin N-linked glycan of Methanococcus maripaludis. J Bacteriol. 194:2693–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirel YA, Shashkov AS, Tsvetkov YE, Jansson PE, Zähringer U. 2003. 5,7-Diamino-3,5,7,9-tetradeoxynon-2-ulosonic acids in bacterial glycopolymers: Chemistry and biochemistry. Adv Carbohydr Chem Biochem. 58:371–417. [DOI] [PubMed] [Google Scholar]
- Ksiazek M, Mizgalska D, Eick S, Thøgersen IB, Enghild JJ, Potempa J. 2015. KLIKK proteases of Tannerella forsythia: putative virulence factors with a unique domain structure. Front Microbiol. 6:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurniyati K, Kelly JF, Vinogradov E, Robotham A, Tu Y, Wang J, Liu J, Logan SM, Li C. 2017. A novel glycan modifies the flagellar filament proteins of the oral bacterium Treponema denticola. Mol Microbiol. 103:67–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. 2008. Glycosyltransferases: Structures, functions, and mechanisms. Annu Rev Biochem. 77:521–555. [DOI] [PubMed] [Google Scholar]
- Lewis AL, Desa N, Hansen EE, Knirel YA, Gordon JI, Gagneux P, Nizet V, Varki A. 2009. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc Natl Acad Sci USA. 106:13552–13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin JB, Lewis WG, Gilbert NM, Weimer CM, Almagro-Moreno S, Boyd EF, Lewis AL. 2015. Host-like carbohydrates promote bloodstream survival of Vibrio vulnificus in vivo. Infect Immun. 83:3126–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu SN, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res. 43:D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner P. 1997. Bacterial glycoproteins. Glycoconj J. 14:3–11. [DOI] [PubMed] [Google Scholar]
- Messner P, Schäffer C, Kosma P. 2013. Bacterial cell-envelope glycoconjugates. Adv Carbohydr Chem Biochem. 69:209–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison MJ, Imperiali B. 2014. The renaissance of bacillosamine and its derivatives: Pathway characterization and implications in pathogenicity. Biochemistry. 53:624–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. 2010. Protein glycosylation in bacteria: Sweeter than ever. Nat Rev Microbiol. 8:765–778. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K, Marth JD. 2006. Glycosylation in cellular mechanisms of health and disease. Cell. 126:855–867. [DOI] [PubMed] [Google Scholar]
- Onishi S, Honma K, Liang S, Stathopoulou P, Kinane D, Hajishengallis G, Sharma A. 2008. Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythia leucine-rich repeat protein BspA. Infect Immun. 76:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst M, Altmann F. 2008. Influence of electrosorption, solvent, temperature, and ion polarity on the performance of LC-ESI-MS using graphitic carbon for acidic oligosaccharides. Anal Chem. 80:7534–7542. [DOI] [PubMed] [Google Scholar]
- Parker JL, Day-Williams MJ, Tomas JM, Stafford GP, Shaw JG. 2012. Identification of a putative glycosyltransferase responsible for the transfer of pseudaminic acid onto the polar flagellin of Aeromonas caviae Sch3N. Microbiologyopen. 1:149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Lowry RC, Couto NAS, Wright PC, Stafford GP, Shaw JG. 2014. Maf-dependent bacterial flagellin glycosylation occurs before chaperone binding and flagellar T3SS export. Mol Microbiol. 92:258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G, Pabst M, Brecker L, Altmann F, Messner P, Schäffer C. 2011. Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia. J Biol Chem. 286:38714–38724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G, Pabst M, Neumann L, Coyne MJ, Altmann F, Messner P, Comstock LE, Schäffer C. 2013. “Cross-glycosylation” of proteins in Bacteroidales species. Glycobiology. 23:568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G, Sekot G, Friedrich V, Megson ZA, Koerdt A, Messner P, Schäffer C. 2012. Glycobiology aspects of the periodontal pathogen Tannerella forsythia. Biomolecules. 2:467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabet M, Lee S-W, Nauman RK, Sims T, Um H-S. 2003. The surface (S-) layer is a virulence factor of Bacteroides forsythus. Microbiology. 149:3617–3627. [DOI] [PubMed] [Google Scholar]
- Sakakibara J, Nagano K, Murakami Y, Higuchi N, Nakamura H, Shimozato K, Yoshimura F. 2007. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology. 153:866–876. [DOI] [PubMed] [Google Scholar]
- Schäffer C, Messner P. 2017. Emerging facets of prokaryotic glycosylation. FEMS Microbiol Rev. 41:49–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm M, Soo EC, Aubry AJ, Austin J, Thibault P, Logan SM. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol Microbiol. 48:1579–1592. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, McNally DJ, Brisson JR, Logan SM. 2006. Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology. 16:8C–14C. [DOI] [PubMed] [Google Scholar]
- Schoenhofen IC, Vinogradov E, Whitfield DM, Brisson JR, Logan SM. 2009. The CMP-legionaminic acid pathway in Campylobacter: Biosynthesis involving novel GDP-linked precursors. Glycobiology. 19:715–725. [DOI] [PubMed] [Google Scholar]
- Sekot G, Posch G, Messner P, Matejka M, Rausch-Fan X, Andrukhov O, Schäffer C. 2011. Potential of the Tannerella forsythia S-layer to delay the immune response. J Dent Res. 90:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekot G, Posch G, Oh YJ, Zayni S, Mayer HF, Pum D, Messner P, Hinterdorfer P, Schäffer C. 2012. Analysis of the cell surface layer ultrastructure of the oral pathogen Tannerella forsythia. Arch Microbiol. 194:525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. 2010. Virulence mechanisms of Tannerella forsythia. Periodontol 2000. 54:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol. 25:134–144. [DOI] [PubMed] [Google Scholar]
- Stadlmann J, Pabst M, Kolarich D, Kunert R, Altmann F. 2008. Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics. 8:2858–2871. [DOI] [PubMed] [Google Scholar]
- Stafford GP, Chaudhuri RR, Haraszthy V, Friedrich V, Schäffer C, Ruscitto A, Honma K, Sharma A. 2016. Draft genome sequences of three clinical isolates of Tannerella forsythia isolated from subgingival plaque from periodontitis patients in the United States. Genome Announc. 4: e01286–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson HN, Mills DC, Jones H, Milioris E, Copland A, Dorrell N, Wren BW, Crocker PR, Escors D, Bajaj-Elliott M. 2014. Pseudaminic acid on Campylobacter jejuni flagella modulates dendritic cell IL-10 expression via Siglec-10 receptor: A novel flagellin-host interaction. J Infect Dis. 210:148714–148798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda JG, Campbell J, Meredith TC, Walker S. 2010. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem. 11:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner ACR, Listgarten MA, Ebersole JL, Strezempko MN. 1986. Bacteroides forsythus sp. nov., a slow-growing, fusiform Bacteroides sp. from the human oral cavity. Int J Syst Bacteriol. 36:213–221. [Google Scholar]
- Taylor ME, Drickamer K. 2003. Introduction to glycobiology. Oxford and New York: Oxford University Press. [Google Scholar]
- Thibault P, Logan SM, Kelly JF, Brisson JR, Ewing CP, Trust TJ, Guerry P. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J Biol Chem. 276:34862–34870. [DOI] [PubMed] [Google Scholar]
- Tomek MB, Neumann L, Nimeth I, Koerdt A, Andesner P, Messner P, Mach L, Potempa JS, Schäffer C. 2014. The S-layer proteins of Tannerella forsythia are secreted via a type IX secretion system that is decoupled from protein O-glycosylation. Mol Oral Microbiol. 29:307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytgat HLP, Lebeer S. 2014. The sweet tooth of bacteria: Common themes in bacterial glycoconjugates. Microbiol Mol Biol Rev. 78:372–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. 1993. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 3:97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. 2006. Nothing in glycobiology makes sense, except in the light of evolution. Cell. 126:841–845. [DOI] [PubMed] [Google Scholar]
- Varki A. 2008. Sialic acids in human health and disease. Trends Mol Med. 14:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Hudson FH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. 2009. Essentials of glycobiology, 2nd ed Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Varki A, Gagneux P. 2012. Multifarious roles of sialic acids in immunity. Ann NY Acad Sci. 1253:16–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith PD, Chen Y-Y, Chen D, O'Brien-Simpson NM, Cecil JD, Holden JA, Lenzo JC, Reynolds EC. 2015. Tannerella forsythia outer membrane vesicles are enriched with substrates of the type IX secretion system and TonB-dependent receptors. J Proteome Res. 14:5355–5366. [DOI] [PubMed] [Google Scholar]
- Veith PD, O'Brien-Simpson NM, Tan Y, Djatmiko DC, Dashper SG, Reynolds EC. 2009. Outer membrane proteome and antigens of Tannerella forsythia. J Proteome Res. 8:4279–4292. [DOI] [PubMed] [Google Scholar]
- Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. 2004. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 68:132–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson DC, Leclerc S, Wakarchuk WW, Young NM. 2011. Enzymatic synthesis and properties of glycoconjugates with legionaminic acid as a replacement for neuraminic acid. Glycobiology. 21:99–108. [DOI] [PubMed] [Google Scholar]
- Watson DC, Wakarchuk WW, Leclerc S, Schur MJ, Schoenhofen IC, Young NM, Gilbert M. 2015. Sialyltransferases with enhanced legionaminic acid transferase activity for the preparation of analogs of sialoglycoconjugates. Glycobiology. 25:767–773. [DOI] [PubMed] [Google Scholar]
- Zarschler K, Janesch B, Zayni S, Schäffer C, Messner P. 2009. Construction of a gene knockout system for application in Paenibacillus alvei CCM 2051T, exemplified by the S-layer glycan biosynthesis initiation enzyme WsfP. Appl Environ Microbiol. 75:3077–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunk M, Kiefel MJ. 2014. The occurrence and biological significance of the α-keto-sugars pseudaminic acid and legionaminic acid within pathogenic bacteria. RSC Adv. 4:3413–3421. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.