Abstract
Low temperature is one of the major environmental stresses that affects plant growth and development, and leads to decrease in crop yield and quality. Thellungiella salsuginea (salt cress) exhibits high tolerance to chilling, is an appropriate model to investigate the molecular mechanisms of cold tolerance. Here, we compared transcription changes in the roots and leaves of T. salsuginea under cold stress using RNA-seq. We identified 2,782 and 1,430 differentially expressed genes (DEGs) in leaves and roots upon cold treatment, respectively. The expression levels of some genes were validated by quantitative real-time-PCR (qRT-PCR). Among these DEGs, 159 (11.1%) genes in roots and 232 (8.3%) genes in leaves were annotated as various types of transcription factors. We found that five aquaporin genes (three TIPs, one PIPs, and one NIPs) responded to cold treatment. In addition, the expression of COR47, ICE1, and CBF1 genes of DREB1/CBF-dependent cold signaling pathway genes altered in response to low temperature. KEGG pathway analysis indicated that these cold regulated genes were enriched in metabolism, photosynthesis, circadian rhythm, and transcriptional regulation. Our findings provided a complete picture of the regulatory network of cold stress response in T. salsuginea. These cold-responsive genes could be targeted for detail functional study and utilization in crop cold tolerance improvement.
Keywords: Thellungiella salsuginea, salt cress, cold stress, gene expression
Introduction
Plants generally are rooted in one place, and have to face drought, salinity, high temperature, cold, and other adverse stresses which may cause significant loss of crop yield (Boyer, 1982; Kawasaki and Bohnert, 2001). Low temperature is one of the major environmental stresses that affect plant growth and development, crop yield and quality. In plant tissues, the intercellular fluid generally has a higher freezing point than the intracellular fluid. When temperature decreased below freezing point, intercellular spaces of plant tissues form ice prior to intracellular region. So, the water potential decreases rapidly outside the cells, and causes the movement of water from inside the cell to the intercellular spaces. Consequently, cold stress could lead to severe cellular dehydration (Thomashow, 1998). In addition, low temperature can lead to the formation of adhesion between the intercellular ice and the cell walls and membranes (Levitt, 1980). Low temperature damage could lead to growth inhibition, wilting, and weak seedling.
During the evolution history, most plants developed the capacity to tolerate cold. Cold acclimation is a strategy for the plants to acquire freezing tolerance by a prior exposure to low nonfreezing temperature (Guy, 1990). To adapt to low temperature environment, many physiological and molecular changes occur during cold acclimation (Thomashow, 1999). Exposure to low temperature, linolenic acid and membrane lipid unsaturation increased, and the plasma membrane H+-ATPase activity increased and these changes are essential for the plants to withstand low temperature (Shi et al., 2008). In addition, calcium-dependent protein kinase confers cold tolerance via the regulation of calcium channel in plasma membrane (Xiong et al., 2002; Komatsu et al., 2007). Early studies identified a number of genes in plants which were response to cold treatment, and these genes were known as cold regulated (COR) genes (Thomashow, 1999; Lee et al., 2005). For example, a total of 939 cold regulated genes were identified in Arabidopsis thaliana (Lee et al., 2005). The cold regulated genes were involved in a variety of functions such as metabolism, protein synthesis, signal transduction, transcription regulation, and hormone biosynthesis and signaling (Thomashow, 1999; Lee et al., 2005).
Among these COR genes, a family of transcription factor known as C-repeat/dehydration-responsive element-binding (CBF) was identified as the key factor to regulate response to cold stress in many plants (Gao et al., 2002; Xiong and Fei, 2006; Nakamura et al., 2011; Wisniewski et al., 2011). In Arabidopsis, three members of CBFs were identified, including CBF1, CBF2, and CBF3 (also name as DREB1b, DREB1c, and DREB1a, respectively). Overexpression of CBF1 induced COR genes and increased freeze tolerance of the transgenic plants (Jaglo-Ottosen et al., 1998; Yamaguchi-Shinozaki and Shinozaki, 2001). Deletion of all three CBF genes the transgenic plants are extremely sensitive to freezing after cold acclimation, suggesting that the three CBF genes together are essential for cold acclimation (Zhao et al., 2016). Recently, ICE1 (inducer of CBF expression 1) was identified as an upstream transcription factor regulating the transcription of CBF, and its overexpression activated the expression of CBF regulon under cold condition and improved freeze tolerance of the transgenic plants (Chinnusamy et al., 2003). However, transcriptome profiling experiments showed that the number of CBF regulon gene accounts only 6.5 % of the total number of COR genes, suggesting that other transcription factors are also involved in the regulation of COR genes and the low-temperature regulatory network beyond the CBF pathway is complex and highly interconnected (Park et al., 2015). Recently, many other COR genes were identified from CBF-independent pathways. For example, SCOF-1 encodes a cold-inducible zinc finger protein from soybean, and Osmyb4 encodes a member of MYB transcription factor from rice, these genes also contributed to cold tolerance in plants (Kim et al., 2001; Vannini et al., 2004).
Thellungiella salsuginea also named as T. halophile or salt cress, is a close relative of Arabidopsis. Compare to Arabidopsis, T. salsuginea exhibits higher tolerance to cold, and it could complete its life cycles at 5°C, and could survive at extreme low temperature of –21°C after cold acclimation (Griffith et al., 2007). Thus, Thellungiella was proposed as an appropriate model to investigate the molecular mechanisms of plant adapted to cold stress (Griffith et al., 2007; Amtmann, 2009). To illustrate how Thellungiella adapts to low-temperature, cold regulated genes were identified from both the mRNA and protein levels. In a survey of 3,628 Thellungiella cDNAs, 76 cold induced transcripts including COR47, ERD10, and COR15b were identified using microarray methods (Wong et al., 2006). Northern blot analysis demonstrated that some cold response genes (CBF1, COR15a, and COR47) from Arabidopsis were also induced in Thellungiella (Griffith et al., 2007). Two-dimensional electrophoresis (2-DE) approach was used in Thellungiella, and found 66 protein spots were significantly affected by cold in Thellungiella rosette leaves (Gao et al., 2009). These studies provided useful clues for understanding the mechanism of cold tolerance in Thellungiella. However, due to the limited genomic sequences, these studies fail to provide a comprehensive interpretation of the transcriptomic changes of Thellungiella in response to cold. To gain insight into the molecular networks underlying Thellungiella cold tolerance, more comprehensive genome-wide gene expression profiling studies are required.
Recently, the whole genome sequence of Thellungiella was completed, which provides new opportunity to understand the cold tolerance mechanism in Thellungiella (Dassanayake et al., 2011; Wu et al., 2012). In this study, we carried out genome-wide analysis of gene expression in roots and leaves of Thellungiella under cold treatment using RNA-seq technology. The aim of the study is to identify cold responsive genes and biological pathways that may contribute to cold tolerance in Thellungiella. We identified thousands of cold-responsive genes and provided an overall picture of the regulatory network in response to cold stress in Thellungiella. These cold-responsive genes could be targeted as potential candidates for further functional validation, and have potential application value for increasing cold tolerance in crops.
Results and Discussion
High Throughput Sequencing and Gene Expression Profiles
To gain the profiles of gene expression in Thellungiella under cold condition, eight cDNA libraries were constructed using roots and leaves under normal (control) and low temperature (cold) for 24 h, respectively. The cDNA libraries were sequenced by Illumina Hiseq2000 platform using the paired end method. After removing low quality, N-containing and adaptor-contaminated reads, a total of 96,305,447 clean reads were generated, with an average of ∼12 million reads per libraries (Table 1). Approximately 90% reads from leaves and 85% reads from roots were mapped to the Thellungiella reference genome, and about 7% reads from leaves and 3.5% reads from roots were mapped to multiple regions, respectively (Table 1). The RNA-seq raw sequencing data from this study have been submitted in the SRA database1 under BioProjects: PRJNA377594 (SRA: SRP101369).
Table 1.
Summary of read numbers from leaves and roots under cold treatment.
| Samples | Reads in leaf (%) |
Reads in root (%) |
||||||
|---|---|---|---|---|---|---|---|---|
| Control 1 | Control 2 | Cold 1 | Cold 2 | Control 1 | Control 2 | Cold 1 | Cold 2 | |
| Total clean reads | 11,839,336 | 12,464,847 | 12,606,715 | 11,696,922 | 12,135,076 | 11,814,795 | 11,896,522 | 11,851,234 |
| Mapped reads | 10,682,216 (90.23%) | 11,286,644 (90.55%) | 11,306,780 (89.69%) | 10,432,315 (89.19%) | 10,299,132 (84.87%) | 10,110,549 (85.58%) | 10,284,125 (86.45%) | 10,185,035 (85.94%) |
| Unique match | 9,750,932 (82.36%) | 10,387,629 (83.34%) | 10,457,647 (82.95%) | 9,659,258 (82.58%) | 9,852,257 (81.19%) | 9,659,737 (81.76%) | 9,859,595 (82.88%) | 9,775,273 (82.48%) |
| Multi-position Match | 931,284 (7.87%) | 899,015 (7.21%) | 849,133 (6.74%) | 773,057 (6.61%) | 446,875 (3.68%) | 450,812 (3.82%) | 424,530 (3.57%) | 409,762 (3.46%) |
| Unmapped reads | 1,157,120 (9.77%) | 1,178,203 (9.45%) | 1,299,935 (10.31%) | 1,264,607 (10.81%) | 1,835,944 (15.13%) | 1,704,246 (14.42%) | 1,612,397 (13.55%) | 1,666,199 (14.06%) |
Among the 29,284 genes deposited in Thellungiella genome database, 22,414 (76.5%) genes were detected in the control and cold-treated libraries. A total of 2,782 and 1,430 differentially expressed genes (DEGs) were identified from leaves and roots, respectively (Figure 1 and Supplementary Tables S1, S2). Under cold treatment, 579 and 1,691 genes were up-regulated, 851 and 1,091 genes were down-regulated in roots and leaves, respectively (Figure 1). Our results demonstrated that the expression patterns of the majority of DEGs were different in roots and leaves. For example, among the 1,691 up-regulated genes in leaves, only 269 (15.9%) were also induced in roots (Figure 2). Interestingly, some genes showed the opposite expression trend in roots and leaves upon cold treatment. For example, fifteen DEGs were up-regulated in root, but down-regulated in leaves. There were 68 DEGs down-regulated in root, but up-regulated in leaves (Figure 2). Moreover, 463 DEGs were with the same expression trend in roots and leaves, including 269 up-regulated and 194 down-regulated DEGs (Figure 2 and Supplementary Table S1).
FIGURE 1.
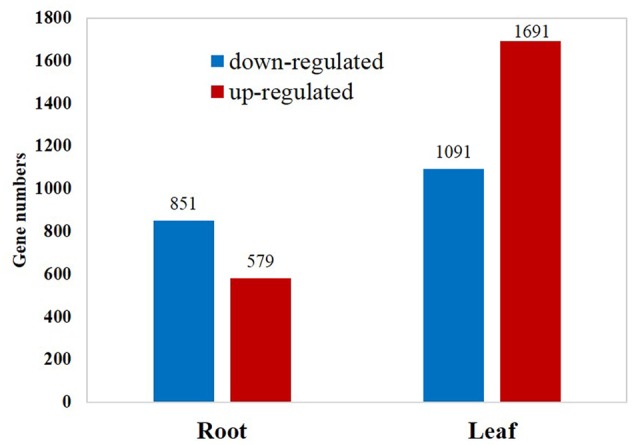
Numbers of differentially expressed genes in response to cold treatment.
FIGURE 2.
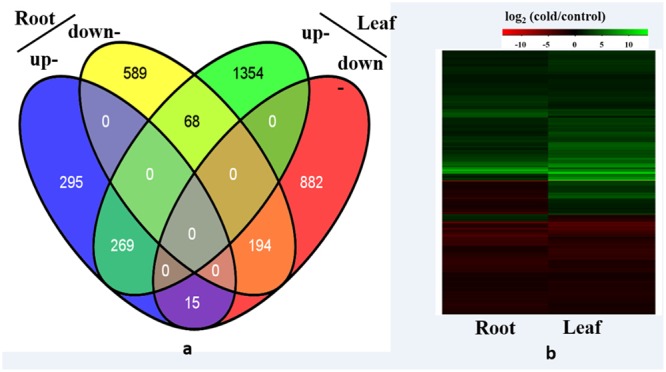
Differentially expressed genes analysis. (a) Venn diagram demonstrated the common and specific differentially expressed genes (DEGs) in roots and leaves, (b) Heat map demonstrated the expression profile DEGs in roots and leaves.
Functional Analysis of DEGs
The DEGs were characterized by the assignment of gene ontology (GO) terms using Blast2GO (Conesa et al., 2005) program. GO enrichment analysis showed 241 and 374 GO terms were significant enriched in root and leaf, respectively (Supplementary Table S3). Then, we employed WEGO web-based tool to visualize the biological process, molecular function and cellular component main categories (Figure 3). In the cellular component category, the terms of “cell”, “cell part”, “organelle”, and “membrane” were enriched, implying the potential contribution of cell and cell structure in the process of Thellungiella response to cold condition. For the category of molecular function, “binding”, “catalytic activity” and “nucleic acid binding transcription factor activity” were the top terms. The most abundant terms of biological process were “cellular process”, “metabolic process”, “response to stimulus” and “single-organism process”, suggesting a high degree of metabolic activity changes upon cold treatment (Figure 3).
FIGURE 3.
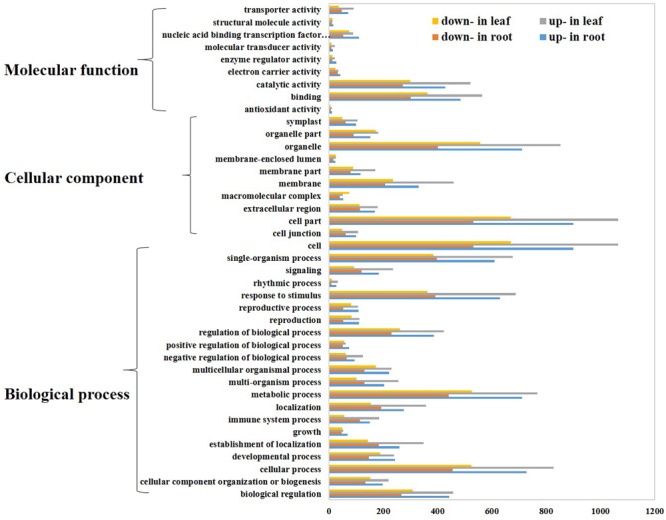
Gene Ontology (GO) analysis of differentially expressed genes. Genes were classified into three main categories: biological process, cellular component, and molecular function. The x-axis indicates the number of genes in a category, and the y-axis means the GO terms.
In order to obtain more biological information for understanding the molecular mechanism and regulatory network of Thellungiella cold tolerance, KEGG enrichment pathway analysis was performed. By applying a cut-off criterion of Q-value < 0.05 and P-value < 1E-05, the result showed that ten and fifteen pathways were significantly enriched from roots and leaves, respectively (Table 2). Previous studies demonstrated that many metabolic changes for enhancing freeze tolerance in Arabidopsis, such as increasing accumulation of soluble sugars and other compatible osmolytes (Wanner and Junttila, 1999). We found six pathways, “biosynthesis of secondary metabolites”, “metabolic pathways”, “nitrogen metabolism”, “tryptophan metabolism”, “cysteine and methionine metabolism”, and “sulfur metabolism” were enriched both in roots and leaves in response to cold. Interestingly, all these pathways were involved in a particular metabolic process, suggesting that the metabolic process was activated via cold treatment (Table 2). Proline is one of the most effective organic osmolytes in plants, and there is a positive correlation between proline accumulation and plant stress tolerance. A number of studies showed that proline played beneficial roles in plants when exposed to cold condition (Hayat et al., 2012). Previous studies demonstrated that Thellungiella contained higher levels of proline than Arabidopsis under non-stressed condition (Kant et al., 2006). Twenty-six genes involved in “Arginine and proline metabolism” pathway were found to be regulated by cold in our study. Most of these DEGs (18 of 26) were up-regulated, suggesting that these cold induced genes might promote the accumulation of proline in Thellungiella to enhance cold tolerance.
Table 2.
The top enriched pathways of DEGs in roots and leaves.
| Pathway | DEGs (%) | All genes | P-value | Q-value | Pathway ID |
|---|---|---|---|---|---|
| Root: 1043 DEGs with KEGG pathway annotation | |||||
| Biosynthesis of secondary metabolites | 217 (20.81%) | 1727 (11.11%) | 2.99E-21 | 3.52E-19 | ko01110 |
| Metabolic pathways | 326 (31.26%) | 3484 (22.41%) | 4.96E-12 | 2.93E-10 | ko01100 |
| Nitrogen metabolism | 22 (2.11%) | 76 (0.49%) | 2.73E-09 | 1.07E-07 | ko00910 |
| Phenylpropanoid biosynthesis | 49 (4.70%) | 306 (1.97%) | 1.04E-08 | 3.08E-07 | ko00940 |
| Flavonoid biosynthesis | 33 (3.16%) | 172 (1.11%) | 3.47E-08 | 8.20E-07 | ko00941 |
| Phenylalanine metabolism | 28 (2.68%) | 151 (0.97%) | 7.72E-07 | 1.52E-05 | ko00360 |
| Tryptophan metabolism | 22 (2.11%) | 127 (0.82%) | 3.48E-05 | 5.87E-04 | ko00380 |
| Cysteine and methionine metabolism | 21 (2.01%) | 123 (0.79%) | 6.48E-05 | 8.08E-04 | ko00270 |
| Sulfur metabolism | 13 (1.25%) | 56 (0.36%) | 6.51E-05 | 8.08E-04 | ko00920 |
| Galactose metabolism | 16 (1.53%) | 80 (0.51%) | 6.85E-05 | 8.08E-04 | ko00052 |
| Leaf: 2419 DEGs with KEGG pathway annotation | |||||
| Biosynthesis of secondary metabolites | 408 (16.87%) | 1727 (11.11%) | 7.46E-21 | 9.55E-19 | ko01110 |
| Metabolic pathways | 687 (28.40%) | 3484 (22.41%) | 3.31E-14 | 2.12E-12 | ko01100 |
| Photosynthesis - antenna proteins | 16 (0.66%) | 27 (0.17%) | 2.62E-07 | 1.12E-05 | ko00196 |
| Nitrogen metabolism | 29 (1.20%) | 76 (0.49%) | 1.40E-06 | 4.47E-05 | ko00910 |
| Photosynthesis | 30 (1.24%) | 81 (0.52%) | 1.92E-06 | 4.92E-05 | ko00195 |
| Pyruvate metabolism | 40 (1.65%) | 125 (0.80%) | 3.22E-06 | 6.87E-05 | ko00620 |
| Biosynthesis of unsaturated fatty acids | 22 (0.91%) | 53 (0.34%) | 5.16E-06 | 9.44E-05 | ko01040 |
| Valine, leucine and isoleucine biosynthesis | 17 (0.70%) | 37 (0.24%) | 1.20E-05 | 1.92E-04 | ko00290 |
| Sulfur metabolism | 22 (0.91%) | 56 (0.36%) | 1.49E-05 | 2.12E-04 | ko00920 |
| Amino sugar and nucleotide sugar metabolism | 47 (1.94%) | 167 (1.07%) | 2.30E-05 | 2.94E-04 | ko00520 |
| Glyoxylate and dicarboxylate metabolism | 26 (1.07%) | 74 (0.48%) | 2.70E-05 | 3.14E-04 | ko00630 |
| Cysteine and methionine metabolism | 37 (1.53%) | 123 (0.79%) | 3.48E-05 | 3.71E-04 | ko00270 |
| Circadian rhythm - plant | 45 (1.86%) | 162 (1.04%) | 4.81E-05 | 4.74E-04 | ko04712 |
| Tryptophan metabolism | 37 (1.53%) | 127 (0.82%) | 7.41E-05 | 6.44E-04 | ko00380 |
| Glycine, serine and threonine metabolism | 29 (1.20%) | 91 (0.59%) | 7.55E-05 | 6.44E-04 | ko00260 |
The “biosynthesis of unsaturated fatty acids” pathway was also enriched in Thellungiella leaves. A total of 22 genes involved in this pathway were affected by low temperature (Table 2). Previous study suggested that membrane lipid composition, especially the concentration of unsaturated fatty acid, is highly correlated with plant freezing tolerance (Thomas et al., 2012). The expression regulation of genes for unsaturated fatty acid synthesis might be a key factor contributing to cold tolerance in Thellungiella.
We observed that the pathways related to biosynthesis of phenylpropanoid and flavonoid were enriched in roots of Thellungiella. In addition, “Photosynthesis - antenna proteins” and “Photosynthesis” were enriched. Most of the DEGs in “Photosynthesis-antenna proteins” and “Photosynthesis” pathways were down-regulated in leaves, suggesting the adverse effect of low temperature on photosynthetic carbohydrate metabolism and photochemical reaction (Supplementary Table S2).
In Arabidopsis, environmental temperature affected the expression of clock component related genes, such as timing of cab expression 1 (TOC1), GIGANTEA (GI), circadian clock associated 1 (CCA1), and late elongated hypocotyl (LHY). Here, our results demonstrated that the pathway of “circadian rhythm - plant” was enriched in Thellungiella after cold treatment. The expression of many genes involved in circadian rhythm including LHY, CCA1, TOC1 and GI were also induced or inhibited in Thellungiella upon cold treatment (Table 2 and Supplementary Table S2).
Validation of RNA-Seq Results Using Quantitative Real-Time PCR
To validate the RNA-seq data, quantitative real-time PCR (qRT-PCR) was performed for 25 genes with different expression levels and functional assignments (Supplementary Table S4). Among them, seven genes were significantly induced upon cold treatment both in roots and leaves. Two of these genes encoded salt and low temperature response protein, one encoded dehydrin. A putative phosphatidylethanolamine-binding protein gene, and three genes with unknown function were also selected for qRT-PCR validation. Other ten selected genes were down-regulated both in roots and leaves. These genes included one aquaporin TIP2-1, one NAC domain protein, one stress-induced sti1-like protein coding genes and several genes with unknown function. In addition, eight genes that down-regulated in roots but up-regulated in leaves were also selected for qRT-PCR analysis. These genes included O-methyltransferase family protein, peroxidase, cytochrome P450, leucine-rich repeat receptor-like protein kinase, MYB and zine finger AN1 domain-contain protein coding genes (Supplementary Table S4). The qRT-PCR results of these 25 genes in leaves were in agreement with the RNA-seq data. The relative expression level (log2 cold/control) estimated by RNA-seq and qRT-PCR were strongly correlated (R2=0.9695) in leaves (Supplementary Table S4 and Figures 4, 5). In roots, the majority of genes (23 of 25) showed similar expression patterns except Thhalv10004977m and Thhalv10002333m, which were down-regulated in RNA-seq, but up-regulated in qRT-PCR. In roots, the Pearson’s coefficient was 0.7664 which was lower than that in leaves (Figure 4 and Supplementary Figure S2).
FIGURE 4.
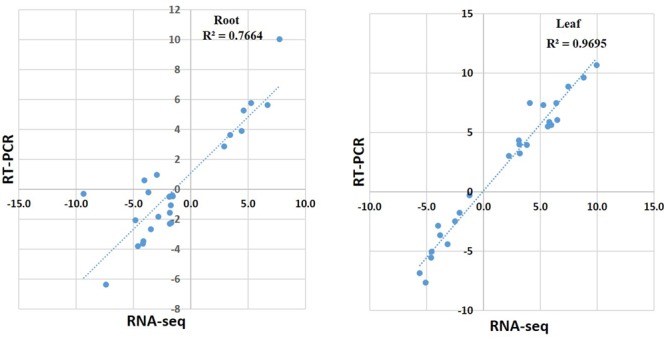
Pearson’s correlation of RNA-seq and qRT-PCR results. qRT-PCR validation of DEGs in roots and leaves under cold condition. The correlation of the fold change analyzed by RNA-Seq (x-axis) with data obtained using qRT-PCR (y-axis).
FIGURE 5.
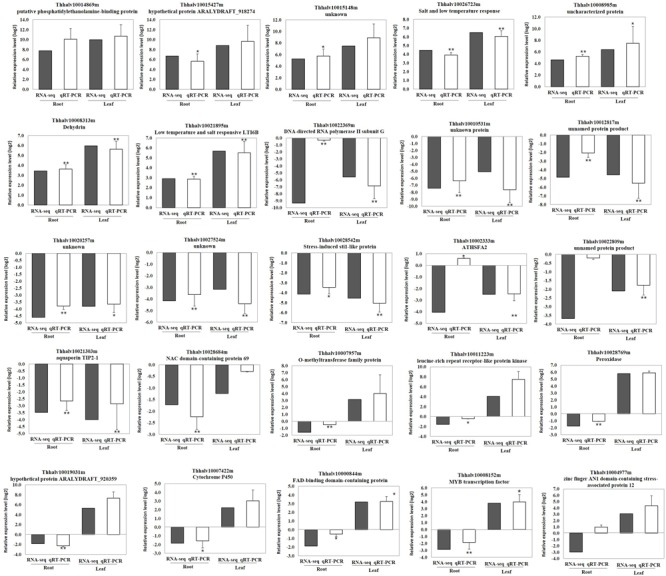
Validation of gene expression by qRT-PCR. Error bars indicate ± SE obtained from three biological repeats. Student’s t-test was performed to analyze the changes in the gene expression after treated with cold. ∗∗denotes the p-value <0.01 and ∗denotes the p-value <0.05.
Cold Related Transcription Factors
Studies showed that transcription factors played important roles in plant response to low temperature and other adverse stresses (Singh et al., 2002; Chen and Zhu, 2004; Agarwal P.K. et al., 2006). We observed that many transcript factors were response to cold in Thellungiella. Among the 1,430 DEGs in roots, 159 genes (11.1%) were annotated as different types of transcription factors (Table 3). About half of these transcription factor genes (74 of 159) were up-regulated, the rest 85 genes were down-regulated. According to functional annotation, these transcription factors were classified into 18 categories, such as abscisic acid responsive, NAC, Zinc finger domain, AP2, MYB, bHLH and WRKY etc. The transcription factors Zinc finger domains (27 genes, 17.0%), ethylene responsive (11 genes, 6.9%) and calcium ion binding (10 genes, 6.3%) were the three major families of the cold-regulated transcription factors in roots.
Table 3.
Cold-regulated transcription factors.
| Category | Number of transcription factors in root |
Number of transcription factors in leaf |
||||
|---|---|---|---|---|---|---|
| Total | Up-regulated | Down-regulated | Total | Up-regulated | Down-regulated | |
| Abscisic acid responsive | 4 | 3 | 1 | 1 | 1 | 0 |
| AP2 | 1 | 0 | 1 | 2 | 1 | 1 |
| Auxin-responsive | 4 | 2 | 2 | 5 | 2 | 3 |
| Basic leucine zipper | 3 | 1 | 2 | 1 | 0 | 1 |
| bHLH | 4 | 0 | 4 | 4 | 1 | 3 |
| BTB and TAZ domain | 1 | 1 | 0 | 1 | 0 | 1 |
| Calcium ion binding | 10 | 0 | 10 | 6 | 5 | 1 |
| Ethylene responsive | 11 | 4 | 7 | 5 | 3 | 2 |
| F-box | 5 | 1 | 4 | 5 | 0 | 5 |
| Heat shock protein | 1 | 0 | 1 | 9 | 5 | 4 |
| Homeobox-leucine zipper | 1 | 1 | 0 | 4 | 0 | 4 |
| Leucine-rich repeat | 6 | 1 | 5 | 7 | 2 | 5 |
| MADS-box | 2 | 2 | 0 | 1 | 0 | 1 |
| MYB | 6 | 3 | 3 | 10 | 3 | 7 |
| NAC-domain | 2 | 1 | 1 | 9 | 7 | 2 |
| WRKY | 3 | 0 | 3 | 12 | 11 | 1 |
| Zinc finger domain | 27 | 15 | 12 | 43 | 33 | 10 |
| Ulassification | 68 | 39 | 29 | 107 | 57 | 50 |
| Total | 159 | 74 | 85 | 232 | 131 | 101 |
Among the 2,782 DEGs in leaves, 232 (8.3%) were transcription factors, including 131 up-regulated and 101 down-regulated genes. These transcription factors were classified into 18 categories (Table 3). Among these differential expressed transcription factors in leaves, Zinc finger domain, WRKY and MYB are enriched. Heat shock proteins (HSPs) were implicated in plant heat stress tolerance (Nover et al., 2002). Here, we found five HSPs were up-regulated and four HSPs were down-regulated upon cold treatment. WRKY transcription factors played important roles in plant responses to biotic and abiotic stress (Eulgem et al., 2000). Twelve differentially expressed WRKY transcription factor genes were identified in leaves, and eleven of them were up-regulated. NAC transcription factors were involved in many aspects of plant growth and development, and response to abiotic stress (Nuruzzaman et al., 2013). In our RNA-seq data, nine and three differentially expressed NAC transcription factor genes were identified in leaves and roots, respectively. These results indicated that HSF, WRKY and NAC transcription factors were involved in plant responses to various stresses, and suggested that cold stress might share common molecular mechanism with other abiotic stresses. In addition, we noticed that the expression of ten calcium ion binding transcription factors was all suppressed in roots upon cold treatment, while most of them were induced in leaves, implying that these genes might be functioning in different ways in roots and leaves (Table 3 and Supplementary Table S1). These results suggested the existence of differences in cold responsive regulatory networks between roots and leaves in Thellungiella.
Previous studies showed that some MYB transcription factors were involved in cold stress tolerance, such as AtMYB15 in Arabidopsis (Agarwal M. et al., 2006), MYB55 and OsMYBS3 in rice (Su et al., 2010; Elkereamy et al., 2012). In our data, the expression of six and ten MYB genes was altered after cold treatment in roots and leaves, respectively (Table 3 and Supplementary Tables S1, S2). Interestingly, Thhalv10008152m encoding a MYB transcription factor displayed opposite expression trend in roots and leaves. In roots, the expression of this gene was inhibited by cold treatment, while it was induced in leaves (Figure 5).
Basic helix-loop-helix (bHLH)-type transcription factors played important regulatory roles in diverse biological processes in plants (Toledoortiz et al., 2003). The latest evidences showed that bHLH transcription factor acted as positive regulators of CBF-pathway and conferred cold tolerance in plants (Feng et al., 2012; Peng et al., 2013; Xu et al., 2014). We found that the expression of four and four bHLH genes was altered after cold treatment in Thellungiella roots and leaves, respectively (Table 3 and Supplementary Tables S1, S2). However, only one of these bHLH gene (Thhalv10025656m) was induced by cold, and the others were down-regulated in response to cold treatment. Functional annotation showed that Thhalv10025656m was homologous of bHLH69 of Arabidopsis, and named as TsbHLH69. In Arabidopsis, bHLH69 contributed to the regulation of circadian periodicity by reducing the expression of LHY and TOC1 (Hanano et al., 2008; Harmer, 2009). These data suggested that the cold-induced TsbHLH69 might participate in the regulation of rhythm of Thellungiella under cold condition.
Cold Regulation of Genes Related to Plant Hormone Biosynthesis and Signaling
In this study, we found that the expression of many genes related to plant hormone biosynthesis and signaling were altered upon cold treatment (Supplementary Tables S1, S2). A total of 134 DEGs were related to plant hormone biosynthesis and signaling in leaves, 71 of which were up-regulated and 73 of which were down-regulated. DELLA, an key factor in gibberellins (GA) signaling, was also involved in the signal transduction of other hormones suggesting that DELLA functions as a modulator of plant development and response to stresses (Achard et al., 2003; Willige et al., 2007). Studies showed that DELLA contributed to CBF1-induced cold acclimation and was considered as components of CBF1-mediated cold stress response (Achard et al., 2008). We found the expression of a DELLA encoding gene (Thhalv10015535m) was slightly induced in both roots (1.40 fold) and leaves (1.39 fold) upon cold treatment. Meanwhile, gibberellin 2-oxidase (Thhalv10008179m), an enzyme inactivating the bioactive gibberellins (GAs), was significantly induced (2.6-fold) (Supplementary Table S2). These results implied that GA metabolism and signaling might contribute to cold stress tolerance in Thellungiella.
Other Cold-Regulated (COR) Genes
Studies demonstrated that the expression of COR genes was strongly induced after plants were shifted to cold temperature (Hajela et al., 1990; Griffith et al., 2007). RNA-seq results revealed five COR genes including COR27, COR47, and three COR15 were dramatically induced in both roots and leaves under cold treatment (Table 4). Interestingly, we found that the expression levels of all three COR15 increased by 1,000-fold in leaves after cold treatment. Among all these COR genes, COR15 represented the most induced genes, whose expression level increased more than 8,000-fold to compare with the control. Sequence alignment results showed that the DNA sequences of these three COR15 genes were highly similar, implying these genes might have the same functions in cold tolerance (Supplementary Figure S1). The COR47 gene encoding a member of dehydrin has been isolated in several plant species. Previous studies showed that the promoter region of COR47 contained C-repeat, dehydration-responsive element, low temperature-responsive element (CRT/DRE/LTREs) and ABA regulatory element (ABRE). COR47 was the downstream gene of CBF/DREB1, and CBF/DREB1 binds to the promoter of COR47 to induce its expression (Jaglo-Ottosen et al., 1998). Functional annotation and sequence alignment showed that Thhalv10027405m, a TsCBF/DREB1 gene, was a homolog of Arabidopsis CBF1/2/3. RNA-seq data revealed that Thhalv10027405m was up-regulated in both roots and leaves under cold treatment (Table 4 and Supplementary Table S2). The expression profiles of TsCBF/DREB1 and COR47 in Thellungiella was consistent with their counterparts in Arabidopsis, suggesting that CBF/DREB1 and COR47 genes might play important roles in DREB1/CBF-dependent cold signaling pathway in Thellungiella.
Table 4.
The expression profile of cold-regulated genes.
| Gene | Annotation | Relative expression level (cold/control) |
Expression trend | |||
|---|---|---|---|---|---|---|
| log2 (leaf) | Q-value (leaf) | log2 (root) | Q-value (root) | |||
| Thhalv10017395m | Cold response protein (COR15) | 13.40 | 1.000 | 3.36 | 0.830 | Up |
| Thhalv10017393m | Cold response protein (COR15) | 10.73 | 0.961 | 7.44 | 0.540 | Up |
| Thhalv10017394m | Cold response protein (COR15) | 10.22 | 0.943 | 8.11 | 0.932 | Up |
| Thhalv10003264m | COR27 | 9.26 | 0.979 | 1.18 | 0.809 | Up |
| Thhalv10008706m | COR47 | 6.80 | 0.999 | 6.54 | 0.998 | Up |
| Thhalv10027405m | CBF/DREB1 | 2.85 | 0.89 | 1.61 | 0.90 | Up |
In addition, our RNA-seq data indicated that there might be more genes involved in DREB1/CBF pathway in Thellungiella. For example, we found DREB2B gene (Thhalv10021161m) was up-regulated in leaves under cold treatment (Supplementary Table S2). In addition to COR47 gene, we observed other two dehydrin genes, Thhalv10010821m and Thhalv10008313m were also induced by cold treatment (Figure 5 and Supplementary Table S2).
In Arabidopsis, INDUCER OF CBF EXPRESSION 1 (ICE1) could induce CBF3 expression by binding to its promoter. It was an important upstream regulatory factor in DREB1/CBF signaling pathway (Chinnusamy et al., 2003). In Thellungiella, we observed that the expression of ICE1 (Thhalv10004059m) were up-regulated in roots upon cold treatment (1.57-fold). However, the expression level of ICE1 was similar in cold treated and control samples in leaves. These results indicated that the function of ICE1 might be different in Arabidopsis and Thellungiella.
Aquaporins (or water channel proteins) played a crucial role in plant water relations. According to their distinct sub-cellular localization, aquaporins could be divided into four subgroups, the tonoplast intrinsic proteins (TIPs), plasma membrane intrinsic proteins (PIPs), Nod26-like intrinsic membrane proteins (NIPs) and small basic intrinsic proteins (SIPs). TIPs were abundantly expressed in vacuole (Maurel, 2007) and played an important role in maintenance of the intracellular space by controlling the water influx in vacuole (Leitão et al., 2014). A member of tonoplast intrinsic proteins GhTIP1;1 was responsive to cold stress and contributed to freezing-tolerance in cotton (Li D.D. et al., 2009). In the current study, five cold regulated aquaporin genes including three TIPs, one PIPs and one NIPs were identified. RNA-seq and qRT-PCR results demonstrated that TIP2-1 (Thhalv10021303m) was significantly down-regulated in both roots and leaves when the plants were under cold treatment. Another two TIP genes, Thhalv10011730m (also annotated as TIP2-1) and Thhalv10002084m (TIP4-1), were also significantly down-regulated upon cold treatment. These results suggested that the decreased expression of TIP genes might be beneficial to reduce water in/out of vacuole which is important for maintaining the stability of the cells in cold condition (Figure 5 and Supplementary Tables S1, S2). Moreover, we observed that the expression of PIP2-7 (Thhalv10025940m) was significantly decreased, suggesting that the water in/out of the cell might be reduced.
Conclusion
In this study, we compared the transcriptome of Thellungiella roots and leaves in response to cold treatment using RNA-seq. We identified a number of cold-responsive genes which were involved in different pathways closely related to environmental adaptation and other biological processes, suggesting the complex responses of Thellungiella toward cold stress (Figure 6). Our findings provided an overall picture of the regulatory network in response to cold stress in Thellungiella. These cold-responsive genes could be targeted as potential candidates for further functional validation, and have potential application value for increasing cold tolerance in crops.
FIGURE 6.
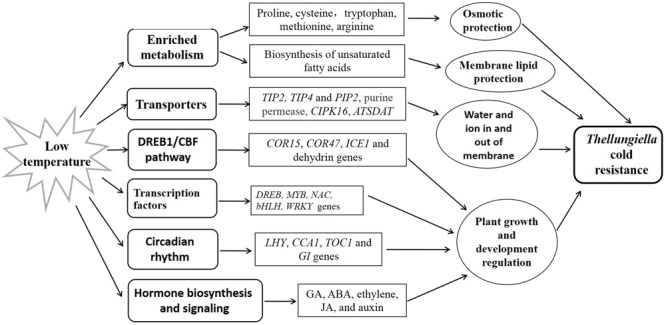
Simple model deduced based on the transcriptome data.
Materials and Methods
Plant Materials
Seeds of T. salsuginea (Shandong ecotype) were surface-sterilized and plated on 1/2 MS-agar plates for synchronize germination at 4°C for a week. The plates were moved to growth chamber with 16 h light at 26°C with light intensity of 3000 lx and 8 h dark at 22°C. Seven-day-old seedlings were transferred to soil for 5 weeks. For cold treatment, seedlings were exposed to a growth chamber under a 16/8 h light/dark regime at 8/4°C for 24 h. For the control, the seedlings were grown at 22/16°C. Leaves and roots of control and cold treated seedlings were collected, frozen immediately in liquid nitrogen, and stored at -80°C. To minimize the plant to plant variation, nine individuals were used as one biological replicate, the tissue from nine individuals were pooled into one independent biological replicate. For both leaves and roots, three biological replicates were prepared.
RNA Isolation, cDNA Library Construction, and Sequencing
Total RNA was isolated from leaves and roots using RNAiso Reagent (Takara, China), and treated with DNase I (Takara, China) to remove the contaminated genomic DNA according to the manufacturer’s protocols. RNA quality was detected by electrophoresis on 1.0% agarose gels and NanoDrop. The mRNA was enriched and cleaved into short fragments (about 200 nt). The mRNA fragments were used as templates to synthesize the first strand cDNA using random hexamer-primer. The first strand cDNA was further incubated with DNA polymerase I, buffer, dNTPs and RNase H to synthesize the second strand. Following end repair, a single nucleotide (adenine) was added, and then sequencing adaptors were ligated to the fragments. Finally, the fragments were purified and enriched with PCR amplification to construct cDNA library. Two biological replicates of each sample were used for RNA-Seq via Illumina HiSeqTM2000 platform by Beijing Genomics Institute (BGI).
Bioinformatics Analysis of RNA-Seq Data
To acquire clean reads, the low-quality reads, adaptor sequences, and empty reads were removed. All clean reads were mapped with the genome sequences of T. salsuginea2 using SOAP2 program under the criterion of no more than two mismatches in the alignment (Li R. et al., 2009). The gene expression level was calculated using RPKM (Reads per Kb per Million reads) method according to previous studies (Morrissy et al., 2009; Qu et al., 2016). The relative gene expression level between different samples was calculated by log2 ratio. Differentially expressed genes (DEGs) were identified using NOIseq under the criteria of probability ≥0.8 and the absolute value of log2Ratio ≥ 1. The probability (P-value) was calculated according to the manufacturer’s protocol with the default parameters (Tarazona et al., 2011).
Gene Ontology annotation was conducted using Blast2GO (Conesa et al., 2005) by comparing all DEGs with GO terms in the database, which covered three domains: cellular component, molecular function and biological process3. The significantly enriched GO terms in DEGs were identified using hypergeometric test comparing to the genome background under the standard of p-value ≤ 0.05. Then GO picture was generated using WEGO4 (Web Gene Ontology Annotation Plot) (Ye et al., 2006). KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis was performed by mapping the DEGs to specific biochemical pathways in KEGG database5. Significantly enriched metabolic pathways or signal transduction pathways were identified using enrichment analysis by comparing DEGs with the whole genome background.
qRT-PCR Validation of RNA-Seq Results
The primers used for qRT-PCR validation were designed using primer premier 5.0 software6 and were listed in Supplementary Table S5. The primers were designed according to the transcript sequences of T. salsuginea downloaded from database 7. qRT-PCR was performed in ABI7500 Real Time System (Applied Biosystems) using SYBR Green I (Roche) with the following reaction: 94°C 10 min; 94°C 15 s, 60°C 10 s and 72°C 25 s for 40 cycles. All reactions were performed in biological triplicates, ubiquitin, and actin were used as internal reference genes. The relative expression of genes was calculated by the software of ABI7500 Real-Time PCR System using the 2-△△ Ct method.
Author Contributions
Manuscript draft: JW, CZ, and XW; Analyzing data: CZ and FC; Experiment: JW, QZ, LH, SZ, HX, JQ, TL, and YZ; Conception and supervision of the research: CZ and XW.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31500217, 31401034), Natural Science Foundation of Shandong province (ZR2014CQ046, ZR2014CM041), Agricultural Science and Technology Innovation Project (CXGC2016C08) and Young Talents Training Program of Shandong Academy of Agricultural Sciences.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00713/full#supplementary-material
Sequence alignment of three COR15 genes.
Hierarchical cluster and correlation analysis between qRT-PCR and RNA-seq.
References
- Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20 2117–2129. 10.1105/tpc.108.058941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Vriezen W. H., Van Der Straeten D., Harberd N. P. (2003). Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15 2816–2825. 10.1105/tpc.015685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M., Hao Y., Kapoor A., Dong C. H., Fujii H., Zheng X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281 37636–37645. 10.1074/jbc.M605895200 [DOI] [PubMed] [Google Scholar]
- Agarwal P. K., Agarwal P., Reddy M. K., Sopory S. K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25 1263–1274. 10.1007/s00299-006-0204-8 [DOI] [PubMed] [Google Scholar]
- Amtmann A. (2009). Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant 2 3–12. 10.1093/mp/ssn094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J. S. (1982). Plant productivity and environment. Science 218 443–448. 10.1126/science.218.4571.443 [DOI] [PubMed] [Google Scholar]
- Chen W. J., Zhu T. (2004). Networks of transcription factors with roles in environmental stress response. Trends Plant Sci. 9 591–596. 10.1016/j.tplants.2004.10.007 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B. H., Hong X., Agarwal M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17 1043–1054. 10.1101/gad.1077503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. (2005). Blast2go: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Dassanayake M., Oh D. H., Haas J. S., Hernandez A., Hong H., Ali S., et al. (2011). The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43 913–918. 10.1038/ng.889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkereamy A., Bi Y. M., Ranathunge K., Beatty P. H., Good A. G., Rothstein S. J. (2012). The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 7:e52030 10.1371/journal.pone.0052030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 199–206. 10.1016/S1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- Feng X. M., Zhao Q., Zhao L. L., Qiao Y., Xie X. B., Li H. F., et al. (2012). The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1encodes an ICE-like protein in apple. BMC Plant Biol. 12:22 10.1186/1471-2229-12-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Zhou Y., Zhu W., Li X., Fan L., Zhang G. (2009). Proteomic analysis of cold stress-responsive proteins in Thellungiella rosette leaves. Planta 230 1033–1046. 10.1007/s00425-009-1003-6 [DOI] [PubMed] [Google Scholar]
- Gao M. J., Allard G., Byass L., Flanagan A. M., Singh J. (2002). Regulation and characterization of four CBF transcription factors from Brassica napus. Plant Mol. Biol. 49 459–471. 10.1023/A:1015570308704 [DOI] [PubMed] [Google Scholar]
- Griffith M., Timonin M., Wong A. C. E., Gray G. R., Akhter S. R., Saldanha M., et al. (2007). Thellungiella: an Arabidopsis-related model plant adapted to cold temperatures. Plant Cell Environ. 30 529–538. 10.1111/j.1365-3040.2007.01653.x [DOI] [PubMed] [Google Scholar]
- Guy C. L. (1990). Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu. Rev. Plant Physiol. 41 187–223. 10.1146/annurev.pp.41.060190.001155 [DOI] [Google Scholar]
- Hajela R. K., Horvath D. P., Gilmour S. J., Thomashow M. F. (1990). Molecular cloning and expression of cor (cold-regulated) genes in Arabidopsis thaliana. Plant Physiol. 93 1246–1252. 10.1104/pp.93.3.1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano S., Stracke R., Jakoby M., Merkle T., Domagalska M. A., Weisshaar B., et al. (2008). A systematic survey in Arabidopsis thaliana of transcription factors that modulate circadian parameters. BMC Genomics 9:182 10.1186/1471-2164-9-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S. L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60 357–377. 10.1146/annurev.arplant.043008.092054 [DOI] [PubMed] [Google Scholar]
- Hayat S., Hayat Q., Alyemeni M. N., Wani A. S., Pichtel J., Ahmad A. (2012). Role of proline under changing environments: a review. Plant Signal. Behav. 7 1456–1466. 10.4161/psb.21949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen K. R., Gilmour S. J., Zarka D. G., Schabenberger O., Thomashow M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106. 10.1126/science.280.5360.104 [DOI] [PubMed] [Google Scholar]
- Kant S., Kant P., Raveh E., Barak S. (2006). Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na + uptake in T. halophila. Plant Cell Environ. 29 1220–1234. 10.1111/j.1365-3040.2006.01502.x [DOI] [PubMed] [Google Scholar]
- Kawasaki S., Bohnert H. J. (2001). Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13 889–905. 10.1105/tpc.13.4.889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. C., Lee S. H., Cheong Y. H., Yoo C. M., Lee S. I., Chun H. J., et al. (2001). A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. Cell Mol. Biol. 25 247–259. 10.1046/j.1365-313x.2001.00947.x [DOI] [PubMed] [Google Scholar]
- Komatsu S., Yang G., Khan M., Onodera H., Toki S., Yamaguchi M. (2007). Over-expression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Mol. Genet. Genomics 277 713–723. 10.1007/s00438-007-0220-6 [DOI] [PubMed] [Google Scholar]
- Lee B., Henderson D. A., Zhu J. K. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17 3155–3175. 10.1105/tpc.105.035568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitão L., Prista C., Loureiro-Dias M. C., Moura T. F., Soveral G. (2014). The grapevine tonoplast aquaporin TIP2;1 is a pressure gated water channel. Biochem. Biophys. Res. Commun. 450 289–294. 10.1016/j.bbrc.2014.05.121 [DOI] [PubMed] [Google Scholar]
- Levitt J. (1980). Responses of Plants to Environmental Stresses. Cambridge, MA: Academic Press. [Google Scholar]
- Li D. D., Tai F. J., Zhang Z. T., Li Y., Zheng Y., Wu Y. F., et al. (2009). A cotton gene encodes a tonoplast aquaporin that is involved in cell tolerance to cold stress. Gene 438 26–32. 10.1016/j.gene.2009.02.023 [DOI] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y., Lam T. W., Yiu S. M., Kristiansen K., et al. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25 1966–1967. 10.1093/bioinformatics/btp336 [DOI] [PubMed] [Google Scholar]
- Maurel C. (2007). Plant aquaporins: novel functions and regulation properties. FEBS Lett. 581 2227–2236. 10.1016/j.febslet.2007.03.021 [DOI] [PubMed] [Google Scholar]
- Morrissy A. S., Morin R. D., Delaney A., Zeng T., Mcdonald H., Jones S., et al. (2009). Next-generation tag sequencing for cancer gene expression profiling. Genome Res. 19 211–241. 10.1101/gr.094482.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura J., Yuasa T., Thi H. T., Harano K., Tanaka S., Iwata T., et al. (2011). Rice homologs of inducer of CBF expression (OsICE) are involved in cold acclimation. Plant Biotechnol. 28 303–309. 10.5511/plantbiotechnology.11.0421a [DOI] [Google Scholar]
- Nover L., Bharti K., Döring P., Mishra S. K., Ganguli A., Scharf K. D. (2002). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M., Sharoni A. M., Kikuchi S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248 10.3389/fmicb.2013.00248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Lee C. M., Doherty C. J., Gilmour S. J., Kim Y., Thomashow M. F. (2015). Regulation of the Arabidopsis cbf regulon by a complex low-temperature regulatory network. Plant J. 82 193–207. 10.1111/tpj.12796 [DOI] [PubMed] [Google Scholar]
- Peng H. H., Shan W., Kuang J. F., Lu W. J., Chen J. Y. (2013). Molecular characterization of cold-responsive basic helix-loop-helix transcription factors MabHLHs that interact with MaICE1 in banana fruit. Planta 238 937–953. 10.1007/s00425-013-1944-7 [DOI] [PubMed] [Google Scholar]
- Qu C. P., Xu Z. R., Hu Y. B., Lu Y., Yang C. J., Sun G. Y., et al. (2016). RNA-SEQ reveals transcriptional level changes of poplar roots in different forms of nitrogen treatments. Front. Plant Sci. 7:51 10.3389/fpls.2016.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., An L., Zhang M., Huang C., Zhang H., Xu S. (2008). Regulation of the plasma membrane during exposure to low temperatures in suspension-cultured cells from a cryophyte (Chorispora bungeana). Protoplasma 232 173–181. 10.1007/s00709-008-0291-1 [DOI] [PubMed] [Google Scholar]
- Singh K. B., Foley R. C., Oñate-Sánchez L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5 430–436. 10.1016/S1369-5266(02)00289-3 [DOI] [PubMed] [Google Scholar]
- Su C. F., Wang Y. C., Tsaihung H., Lu C. A., Tunghai T., Yu S. M. (2010). A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 153 145–158. 10.1104/pp.110.153015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona S., Garcíaalcalde F., Dopazo J., Ferrer A., Conesa A. (2011). Differential expression in RNA-seq: a matter of depth. Genome Res. 21 2213–2223. 10.1101/gr.124321.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D., Patrick G., Zuther E., Bettina S., Hincha D. K., Willmitzer L. (2012). Differential remodeling of the lipidome during cold acclimation in natural accessions of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 72 972–982. 10.1111/tpj.12007 [DOI] [PubMed] [Google Scholar]
- Thomashow M. F. (1998). Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 118 1–8. 10.1104/pp.118.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow M. F. (1999). PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 50 571–599. 10.1146/annurev.arplant.50.1.571 [DOI] [PubMed] [Google Scholar]
- Toledoortiz G., Huq E., Quail P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770. 10.1105/tpc.013839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C., Locatelli F., Bracale M., Magnani E., Marsoni M., Osnato M., et al. (2004). Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 37 115–127. 10.1046/j.1365-313X.2003.01938.x [DOI] [PubMed] [Google Scholar]
- Wanner L. A., Junttila O. (1999). Cold-induced freezing tolerance in Arabidopsis. Plant Physiol. 120 391–400. 10.1104/pp.120.2.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige B. C., Ghosh S., Nill C., Zourelidou M., Dohmann E. M., Maier A., et al. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19 1209–1220. 10.1105/tpc.107.051441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski M., Norelli J., Bassett C., Artlip T., Macarisin D. (2011). Ectopic expression of a novel peach (Prunus persica) CBF transcription factor in apple (Malus × domestica) results in short-day induced dormancy and increased cold hardiness. Planta 233 971–983. 10.1007/s00425-011-1358-3 [DOI] [PubMed] [Google Scholar]
- Wong C. E., Li Y., Labbe A., Guevara D., Nuin P., Whitty B., et al. (2006). Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 140 1437–1450. 10.1104/pp.105.070508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. J., Zhang Z., Wang J. Y., Oh D. H., Dassanayake M., Liu B., et al. (2012). Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. U.S.A. 109 12219–12224. 10.1073/pnas.1209954109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Schumaker K. S., Zhu J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Fei S. Z. (2006). Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.). Planta 224 878–888. 10.1007/s00425-006-0273-5 [DOI] [PubMed] [Google Scholar]
- Xu W., Zhang N., Jiao Y., Li R., Xiao D., Wang Z. (2014). The grapevine basic helix-loop-helix (bHLH) transcription factor positively modulates CBF-pathway and confers tolerance to cold-stress in Arabidopsis. Mol. Biol. Rep. 41 5329–5342. 10.1007/s11033-014-3404-2 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2001). Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Novartis Found. Symp. 236 176–186. [PubMed] [Google Scholar]
- Ye J., Fang L., Zheng H., Zhang Y., Chen J., Zhang Z., et al. (2006). WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34 293–297. 10.1093/nar/gkl031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Zhang Z., Xie S., Si T., Li Y., Zhu J. K. (2016). Mutational evidence for the critical role of cbf transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171 2744–2759. 10.1104/pp.16.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of three COR15 genes.
Hierarchical cluster and correlation analysis between qRT-PCR and RNA-seq.


