Abstract
We sought to identify drugs that could counteract cytarabine resistance in acute myeloid leukemia (AML) by generating eight resistant variants from MOLM-13 and SHI-1 AML cell lines by long-term drug treatment. These cells were compared with 66 ex vivo chemorefractory samples from cytarabine-treated AML patients. The models and patient cells were subjected to genomic and transcriptomic profiling and high-throughput testing with 250 emerging and clinical oncology compounds. Genomic profiling uncovered deletion of the deoxycytidine kinase (DCK) gene in both MOLM-13- and SHI-1-derived cytarabine-resistant variants and in an AML patient sample. Cytarabine-resistant SHI-1 variants and a subset of chemorefractory AML patient samples showed increased sensitivity to glucocorticoids that are often used in treatment of lymphoid leukemia but not AML. Paired samples taken from AML patients before treatment and at relapse also showed acquisition of glucocorticoid sensitivity. Enhanced glucocorticoid sensitivity was only seen in AML patient samples that were negative for the FLT3 mutation (P=0.0006). Our study shows that development of cytarabine resistance is associated with increased sensitivity to glucocorticoids in a subset of AML, suggesting a new therapeutic strategy that should be explored in a clinical trial of chemorefractory AML patients carrying wild-type FLT3.
Introduction
Cytarabine (Ara-C or 1-beta-D-arabinofuranosylcytosine) has remained the cornerstone of therapy for adult acute myeloid leukemia (AML) patients for decades.1, 2 Induction therapy comprised of cytarabine in combination with anthracyclines, leads to responses in 60–70% of adult AML patients.3, 4 However, relapse due to acquired resistance is common and the overall long-term survival of adult AML patients is <40%.5, 6 Therefore, better insights are needed about potential therapeutic regimens to either prevent resistance from arising or to treat patients resistant to cytarabine.2
There are several mechanisms playing a role in cytarabine resistance in different types of leukemia. Mutation, deletion and reduced gene and protein expression of deoxycytidine kinase (DCK) have been reported in AML and ALL cell lines and clinical samples.7, 8, 9, 10, 11 In addition, genetic aberrations in the SLC29A1, CDA and NT5C2 genes have been associated with clinical resistance as well as acquired and/or intrinsic in vitro resistance in AML.12, 13, 14 Deregulation of apoptosis, such as BCL-2 overexpression, has also shown to be responsible for cytarabine resistance in both AML cell lines and clinical samples.15, 16 Upregulation of NK cell-activating receptor NKG2D gene was reported to be associated with cytarabine resistance in T-cell lymphoid leukemia cell lines.17 Variation in the function of multi-drug resistance (MDR) genes often causes resistance to nucleoside analog drugs.18 Negoro et al.19 characterized five cytarabine-resistant cell lines from different blood cell lineages and reported that differential expression of eight genes associated with cytarabine resistance. These studies of resistance mechanisms have not yet led to the development of therapeutic strategies to prevent or overcome resistance.
It is important to identify drugs able to overcome cytarabine resistance. A clinical trial showed that clofarabine induces cytarabine-mediated cytotoxicity by chemical inhibition of ribonucleotide reductase in chemorefractory AML patients.20 Other approaches reported to overcome cytarabine resistance include (i) targeting sonic hedgehog pathway gene GLI1,21, 22 (ii) treatment with BH3 mimetics23 or (iii) BCL-2 inhibitor YC137 in combination with guanine arabinoside.24 These studies have highlighted the strategies to counter cytarabine resistance but their clinical translation has not yet been achieved.
Here we hypothesized that generation of drug resistance to a chemotherapeutic agent is likely to increase vulnerability to other drugs. We first generated a series of cytarabine-resistant variants from the MOLM-13 and SHI-1 AML cell lines. To identify drugs effective against the resistance, the cytarabine-resistant variants were subjected to high-throughput drug sensitivity and resistance testing (DSRT) with 250 oncology drugs. The drug-resistant cells were also characterized for gene expression, copy number variation and mutations. The molecular and functional data from these in vitro models were compared with data from ex vivo analysis of 66 relapsed and refractory AML patient samples, including paired samples from patients taken before and after chemotherapy, including cytarabine.
Materials and methods
Development of cytarabine-resistant AML cell lines
Cytosine β-D-arabinofuranoside (cytarabine, Ara-C, Sigma-Aldrich, St. Louise, MO, USA) was dissolved in DMSO. MOLM-13 and SHI-1 AML cell lines were purchased from the DSMZ (Braunschweig, Germany). MOLM-13 and SHI-1 cells were cultured DSMZ-specified media. Both AML cell lines were treated with cytarabine and doses were doubled when the AML cells started to proliferate at an equal rate as the untreated parental cells. Cell line variants resistant to 160, 320, 640 and 1280 nM concentrations were named as M 160 Ara-C, M 320 Ara-C, M 640 Ara-C, M 1280 Ara-C for MOLM-13 and S 160 Ara-C, S 320 Ara-C, S 640 Ara-C, S 1280 Ara-C for SHI-1. The authenticity of each cell type was tested StemElite ID (Promega, Madison, WI, USA) kit.
Patient samples
Peripheral blood or bone marrow aspirates (n=66) were collected from 48 individual AML patients and 15 healthy donors. Skin biopsies were used as a germline control. Informed consent was obtained from all the patients and the samples were collected using approved study protocols (Helsinki Ethical Committee 239/13/03/00/2010 and 303/13/03/01/2011). Paired samples were taken from AML patients before and after relapse under cytarabine therapy. Clinical details of AML patients are given in Table 1.
Table 1. Clinical characteristics of adult AML patients.
| Patient identifier | Diagnosis | Sample number | Sample type | Age | Disease status | Time from dignosis (months) | Cytogenetics | Previous therapies for AML |
|---|---|---|---|---|---|---|---|---|
| 1145 | Therapy-related AML | 1145_2 | Bone marrow | 41 | Diagnosis | 0 | Hyperdipo 43, t(5; 6), t(7; 9), −19, −20, −Y | – |
| 1145_3 | Bone marrow | 43 | Relapse | 15 | Cytarabine, azacitidine, allogenic HSCT | |||
| 1064 | Therapy-related AML | 1064_1 | Bone marrow | 37 | Diagnosis | 0 | abn(3) | – |
| 1064_3 | Bone marrow | 40 | First relapse | 30 | abn(3) | Cytarabine-antracycline, HSCT (MUD) | ||
| 3443 | AML without maturation | 3443_3 | Bone marrow | 21 | Resistant disease | 1 | del 17p, −2 (both), −17 (both), −5, −6, −7, −11, −12, −18, −22, +13,+21, 6–8 marker chromosomes | Cytarabine-antracycline |
| 3443_6 | Bone marrow | 22 | Resistant disease | 4 | As above, 8–12 marker chromosomes | Cytarabine-antracycline, ruxolitinib-everolimus, clofarabine-cytarabine |
Abbreviations: abn(3), abnormal chromosome 3; AML, acute myeloid leukemia; del, deletion; HSCT, hematopoietic stem cell transplantation; MUD, matched unrelated donor.
Drug sensitivity and resistance testing (DSRT)
Briefly, DSRT was performed with MOLM-13 and SHI-1 parental and respective cytarabine-resistant cells with 250 active chemical compounds (Supplementary Table 1). DSRT assay details are given in Supplementary methods. Drug efficacy was quantified with a drug sensitivity score (DSS), which is modified area-under-the-curve measurement.25 The DSS of cytarabine-resistant cell lines and patient samples 3443_3 and 3443_6 are provided in Supplementary Table 1. Selective DSS was calculated by subtracting from the patient DSS, the average of healthy control DSS. The selective DSS of glucocorticoids—dexamethasone, methylprednisolone and prednisolone are listed for all patient samples in Supplementary Table 2.
Molecular profiling and western blot analysis
We analyzed gene expression, mutation and copy number changes in AML cell lines. The methodology and data analysis pipeline details are described in Supplementary methods. Briefly, copy number and mutation data were analyzed as described previously.26, 27, 28 Exome sequencing were performed for 29 samples and the data were analyzed as described earlier.26 Western blot analysis is described in Supplementary Methods.
Target addiction scoring
Target addiction score (TAS) is a quantitative measure of the functional sensitivity of cell line variants to the therapeutic targets, calculated on the basis of estimated level of addiction of cells to a target protein.29 The TAS algorithm integrates DSS profiles with global compound–target interaction networks. All the on and off targets of the 202 targeted compounds were collected and integrated using the Kinase inhibitor BioActivity (KiBA) model.30 To select targets of the compounds, we applied the KiBA cutoff ⩽3 for 181 compounds and the KiBA cutoff ⩽4 for the rest of the 21 compounds. For a given target t, TAS was computed as an average of the DSS over those n inhibitors that target the protein t:
TAS calculation generated target addiction profiles of the individual cell types including both parental and cytarabine-resistant cells.
Statistical analyses
The dose response percent inhibition values were fitted with non-linear regression and four parameter logistic curve. DSS and TAS were calculated as previously described.25, 29 TAS and gene expression were compared using Spearman rank test. The Student's t-test, Mann–Whitney U-test and correlation analyses were performed using Prism software version 6 (GraphPad Prism, San Diego, CA, USA). Statistical significance was considered at two-tailed P< 0.05.
Results
Development of cytarabine-resistant MOLM-13 and SHI-1 variants
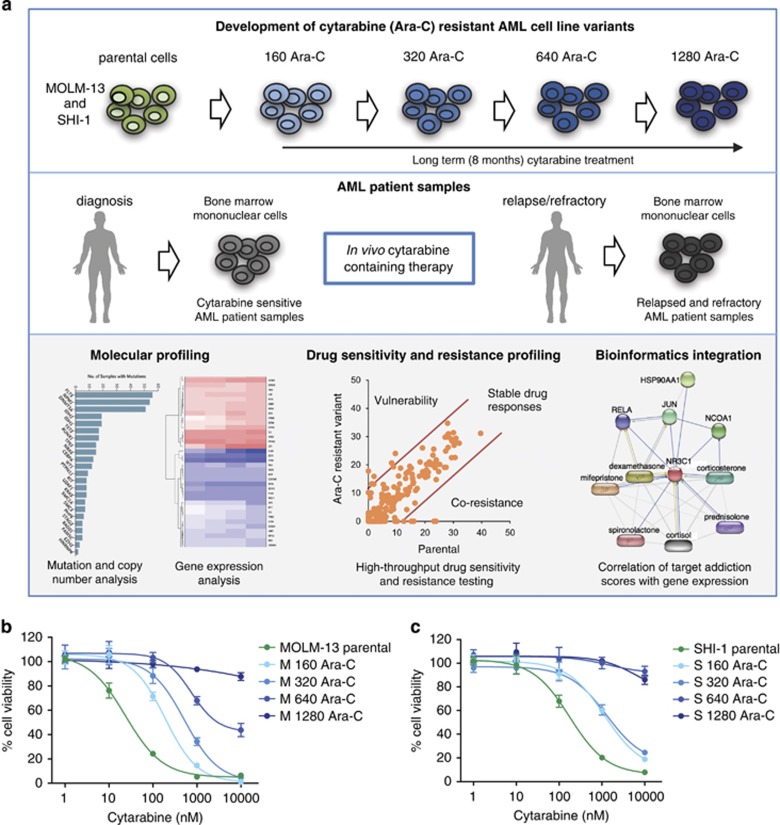
A schematic overview of this study is presented in Figure 1a. First, we generated cytarabine-resistant AML cell line models from SHI-1 and MOLM-13 followed by systematic molecular profiling and drug testing. High-throughput drug testing data showed drugs with co-resistance and novel vulnerability patterns in cytarabine-resistant cells compared with parental cells. These in vitro findings were compared with ex vivo cytarabine-treated relapsed and refractory AML patient samples to identify the clinical relevance of results observed in cytarabine resistance cells.
Figure 1.
Overview of the study design. (a) Schematic diagram illustrating generation of cytarabine-resistant cell line variants (upper panel) and collection of bone marrow mononuclear cells before and after cytarabine treatment in AML patients (middle panel). Subsequently, molecular profiling, high-throughput drug testing and bioinformatics data integration from cytarabine-resistant cell line variants and patient samples were studied (lower panel) to clinically validate the results. (b, c) The sensitivity of MOLM-13 and SHI-1 parental and cytarabine-resistant variants to cytarabine was tested with cell viability assay after 72 h incubation with drug concentrations ranging from 1 to 10 000 nM. The bars represent mean ±s.d.
The MOLM-13 cell line was selected as cells harbor an internal tandem duplication (ITD) in FLT3, a most common aberration in adult AML patients.31 The SHI-1 cell line carries a hot spot mutation in the KRAS and MLL-AF6 rearrangement, representing another established subset of AML patients.32 MOLM-13 and SHI-1 cytarabine variants, 160 Ara-C, 320 Ara-C, 640 Ara-C and 1280 Ara-C, generated in 8 months depicted stepwise development of resistance (Figure 1b and c). The drug-resistant variants were confirmed to represent the parental MOLM-13 or SHI-1 cell line by genetic authentication. The growth rates of the cytarabine-resistant variants were measured and found to be similar to the parental cells in MOLM-13 and SHI-1 cells (Supplementary Figure 1A and B). In addition, we cultured M 1280 Ara-C cells without cytarabine for 3 weeks, which did not result in the loss of cytarabine resistance (Supplementary Figure 1C). This indicates that cytarabine resistance was not reversible and thereby unlikely to be due to transient cell signaling changes or epigenetic modifications.
Copy number and gene expression profiling showed deletion and downregulation of DCK in cytarabine-resistant cells
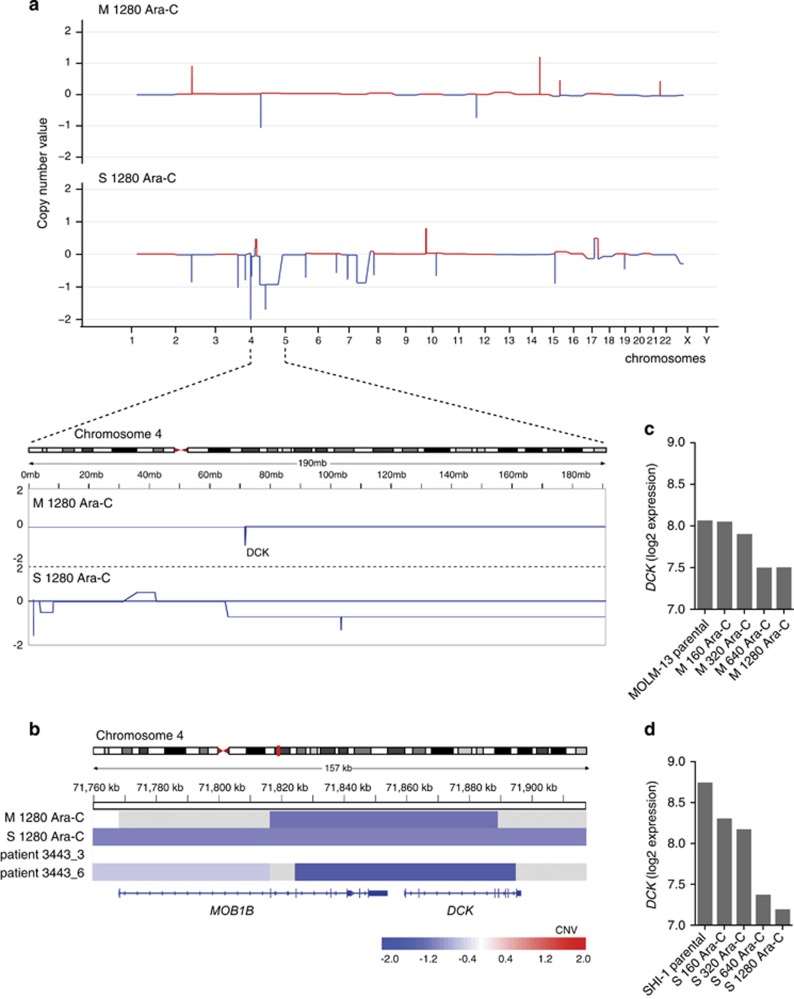
To identify copy number changes and mutations associated with cytarabine resistance, MOLM-13 and SHI-1 1280 Ara-C cells were subjected to whole-exome sequencing and compared with respective parental cells. Drug-resistant SHI-1 cells displayed copy number aberrations involving larger genomic regions as compared with the corresponding MOLM-13 cells (Supplementary Tables 3 and 4). However, we found shared copy number variations specific to cytarabine resistance in both model systems (Figure 2a), such as the copy number loss at 4q13.3 at the location of the DCK gene, previously reported to be responsible for cytarabine resistance.14
Figure 2.
Molecular profiling of cytarabine-resistant cells revealed DCK deletion. (a) Genome-wide copy number analysis by exome sequencing was performed with 1280 Ara-C-resistant MOLM-13 and SHI-1 variants using respective parental cells as controls. Individual chromosomes are presented on the x axis and copy number ratios on the y axis. (b) Copy number analysis of AML patient samples 3443_3 (obtained before cytarabine treatment) and 3443_6 (obtained after cytarabine treatment) showed acquisition of homozygous deletion of DCK gene acquired after the patient was treated with two cycles of cytarabine-based therapy. (c, d) Microarray-based expression of DCK in MOLM-13 and SHI-1 parental and cytarabine-resistant variants.
We also analyzed copy number changes and mutations from all available serial samples of AML patients. Serial samples taken from patient 3443 were designated as 3443_3 and 3443_6 (Table 1). 3443_6 sample, taken after two cycles of in vivo cytarabine-based treatment, showed homozygous deletion of DCK gene. Interestingly, the deletion breakpoints for the DCK gene in 3443_6 cells were at the same genetic location as in M 1280 Ara-C (Figure 2b). Moreover, gene expression data confirmed downregulation of DCK gene in all four cytarabine-resistant variants from MOLM-13 and SHI-1 compared with respective parental cells (Figures 2c and d; Supplementary Table 5).
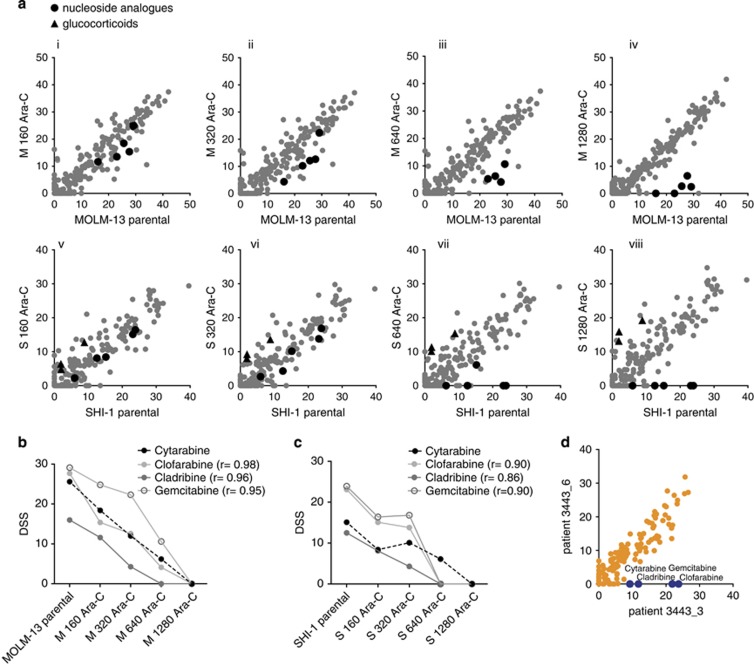
Loss of DCK function correlated with co-resistance to nucleoside analogs in cytarabine-resistant cells
MOLM-13 and SHI-1 parental and cytarabine-resistant variants were subjected to DSRT. The comparison of individual cytarabine-resistant variants with their respective parental cells showed consistent patterns of drug sensitivity and resistance (Figure 3a). The nucleoside analogs tested were clofarabine, cladribine and gemcitabine. Co-resistance patterns were confirmed by a significant correlation between cytarabine and individual nucleoside analogs in both MOLM-13 and SHI-1 variants (Figures 3b and c). Similarly, DSRT data of AML patient sample 3443_6 with the DCK deletion showed complete resistance to cytarabine along with gemcitabine, cladribine and clofarabine as compared with an earlier sample from the same patient (3443_3), which had no DCK deletion and showed moderate cytarabine sensitivity (Figure 3d).
Figure 3.
Nucleoside analogs showed stepwise co-resistance to cytarabine in AML cell lines and in an AML patient sample. (a) DSS for 250 drugs from individual MOLM-13 (Ai-Aiv) and SHI-1 (Av-Aviii) cytarabine-resistant variants were correlated with the corresponding parental cells to depict differential drug sensitivities and resistance patterns. DSS illustrates ex vivo sensitivity to the compound (high DSS meaning high sensitivity). Drugs showing co-resistance represented in black dots and vulnerabilities in black triangles in cytarabine-resistant variants compared with parental cells. (b, c) Cytarabine-resistant MOLM-13 and SHI-1 variants demonstrated consistent co-resistance pattern to nucleoside analogs by decreasing DSS values. Pearson correlation analysis was performed between cytarabine and individual nucleoside analogs; r indicated correlation values with P<0.05. (d) Comparison of DSS between AML patient samples 3443_3 and 3443_6 showed distinct pattern of co-resistance for nucleoside analogs, including cytarabine, gemcitabine, clofarabine and cladribine, shown in blue.
Systematic drug testing of the cytarabine-resistant cell line variants indicated acquisition of glucocorticoid sensitivity
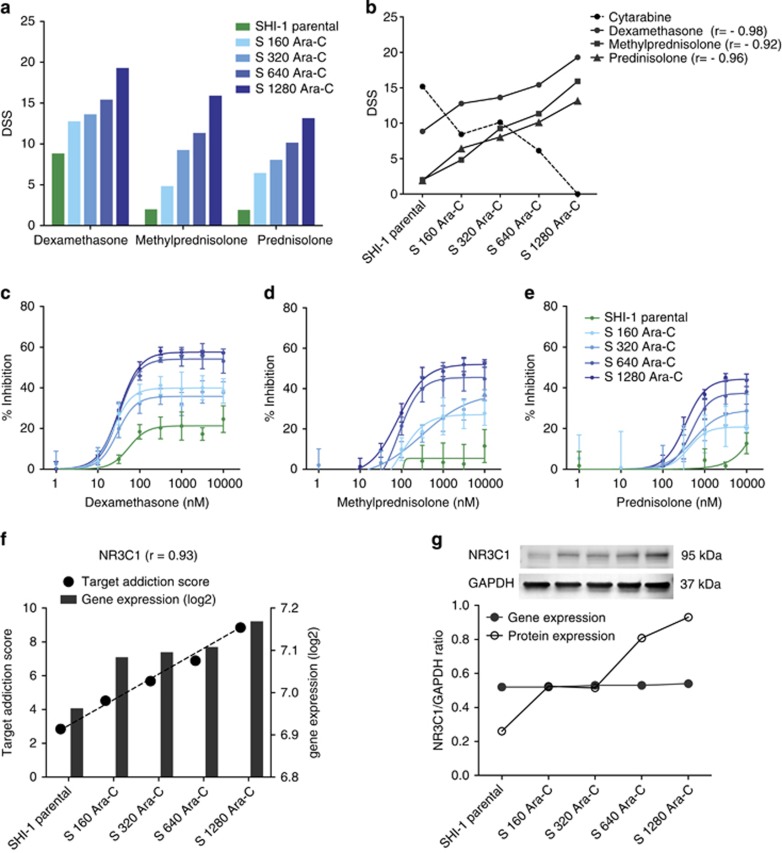
Interestingly, DSRT of all cytarabine-resistant SHI-1 cell line variants revealed systematic increase in sensitivity to glucocorticoids (Figure 4a). The glucocorticoids used in the drug testing included dexamethasone, methylprednisolone and prednisolone, which showed minimal efficacy in SHI-1 parental cells. The S 160 Ara-C, S 320 Ara-C, S 640 Ara-C and S 1280 Ara-C-resistant variants were increasingly sensitive to methylprednisolone, prednisolone and dexamethasone. The Pearson correlation between cytarabine and individual glucocorticoids (dexamethasone r= −0.98, methylprednisolone r= −0.92 and prednisolone r= −0.96) revealed a statistically significant (P<0.05) inverse relationship between acquired cytarabine resistance and glucocorticoid sensitivity (Figure 4b). The increasing glucocorticoid sensitivity was confirmed by repeating drug testing with a wide concentration range (nine doses between 1 and 10 000 nM) of glucocorticoids in cytarabine-resistant SHI-1 variants (Figures 4c–e). However, glucocorticoid sensitivity was not observed in MOLM-13 parental or cytarabine-resistant variants (Supplementary Figure 2A).
Figure 4.
Cytarabine-resistant SHI-1 cells exhibited enhanced sensitivity to glucocorticoids and upregulation of NR3C1. (a) SHI-1 parental and cytarabine-resistant variants showing sensitivity to three glucocorticoids—dexamethasone, methylprednisolone and prednisolone. (b) Correlation was calculated between DSS of cytarabine and individual glucocorticoids in SHI-1 cytarabine-resistant variants; r indicates Pearson correlation with P<0.05. (c, d, e) A targeted small-scale drug testing experiment was performed in SHI-1 parental and cytarabine-resistant variants to validate the high-throughput DSRT data on glucocorticoid sensitivity. Glucocorticoids were tested with nine doses in triplicates with same parameters as in the DSRT assay. The bars represent mean ±s.d. (f) TAS was calculated and correlated with gene expression profiles in SHI-1 parental and cytarabine-resistant variants; r indicates Spearman correlation, where P=0.02. (g) NR3C1 protein expression was analyzed using western blotting in SHI-1 parental and cytarabine-resistant cells. The integrated intensities of protein bands were quantified by Odyssey software and normalized against GAPDH.
NR3C1 (glucocorticoid receptor) upregulation is associated with glucocorticoid sensitivity in cytarabine-resistant SHI-1 variants
To define the potential mechanisms associated with glucocorticoid sensitivity, a network-based model was applied to reveal candidate molecular targets behind the observed drug response profiles in the cytarabine-resistant variants.29 Polypharmacological on- and off-target effects were modeled using TAS for each target protein on the basis of the DSRT data. Correlation between TAS and gene expression showed consistent increase in Nuclear Receptor Subfamily 3, Group C, Member 1 (NR3C1; Figure 4f). The integrated analysis identified NR3C1 as one of the top positively correlated genes (r=0.93, P=0.02) in SHI-1 cells (Supplementary Table 6). NR3C1 protein expression increased substantially compared gene expression with acquired cytarabine resistance (Figure 4g) and increased glucocorticoid sensitivity in SHI-1 cells (Supplementary Figure 3). However, NR3C1 gene or protein expression, along with glucocorticoid sensitivity, did not increase in MOLM-13 cytarabine-resistant variants compared with the parental cells (Supplementary Figure 2B and C).
DSRT of clinical samples suggested enhanced glucocorticoid efficacy in relapsed/refractory AML
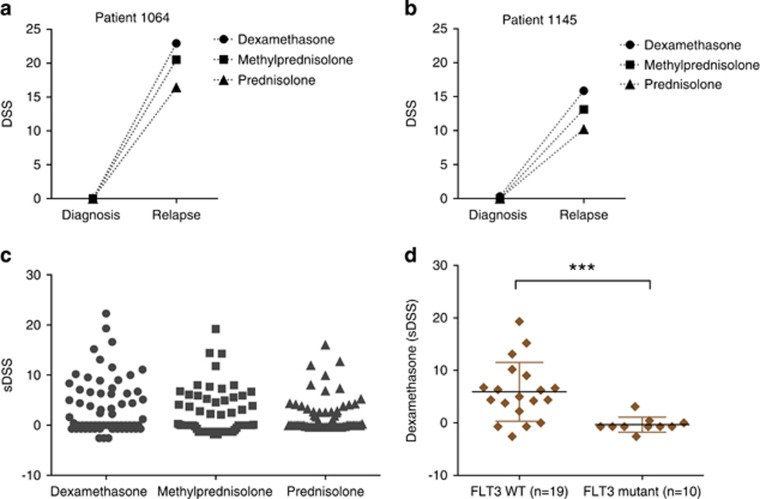
We assessed ex vivo drug sensitivity data available for paired diagnostic and relapsed samples from two patients 1064 and 1145. We compared drug responses for all three glucocorticoids—dexamethasone, methylprednisolone and prednisolone between these paired samples in both patients. We observed significant increase in efficacy of glucocorticoids in relapsed samples compared with their respective diagnostic samples in 1064 (P=0.009) and 1145 (P=0.013; Figures 5a and b). Cytarabine was included in the induction therapy for the patients, along with other chemotherapeutics (Table 1). However, it is challenging to study effect of individual drugs in AML patient samples as drugs are mostly given in combinations.
Figure 5.
Enhanced efficacy to glucocorticoids in chemorefractory AML patient samples revealed by ex vivo drug testing. (a, b) The DSS of dexamethasone, methylprednisolone and prednisolone were compared between diagnostic and relapsed samples in 1064 and 1145 AML patient cases using two-tailed student's t-test. (c) The drug testing was performed with 66 relapsed and refractory samples from 48 AML patients treated with cytarabine containing chemotherapy. The DSS of dexamethasone, methylprednisolone and prednisolone were normalized against 15 healthy bone marrow samples and quantified as selective DSS. Each data point represents drug response from individual patient sample. (d) Dexamethasone responses were compared between AML patient samples carrying FLT3 wild-type (n=19) and point and/or ITD mutation (n=10) using Mann–Whitney U-test.
Strong glucocorticoid sensitivity in a subset of AML patients with wild-type FLT3
We analyzed glucocorticoid sensitivity data in 66 chemorefractory samples from 48 individual AML patients. These samples include 18 samples from previously published study26 and 48 additional new samples. Eleven of the AML samples (16%) with selective DSS⩾10 were considered strong responders to glucocorticoid dexamethasone, whereas an additional 11 cases (32% together) showed moderate sensitivity to dexamethasone with selective DSS scores between 5 and 10 (Figure 5c). Sensitivity to glucocorticoids showed strong correlation with each other in AML patient samples, thus validating and confirming the observation (Supplementary Figure 4). We further explored molecular profiling data from 29 relapsed and refractory AML patient samples and observed that most of the strong dexamethasone responders carried wild-type Fms-Related Tyrosine Kinase 3 (FLT3). In contrast, samples that harbored either FLT3 point mutations or ITD were completely non-responsive to glucocorticoids (Figure 5d). Taken together, these findings suggest the therapeutic potential of glucocorticoids in a FLT3 wild-type subset of AML patients, resistant to cytarabine and standard chemotherapy.
Discussion
Here we tested 250 oncology drugs in cytarabine-resistant AML cell models as well as in chemorefractory AML patient samples ex vivo, and identified glucocorticoids as the drugs whose efficacy most consistently increased as resistance to cytarabine developed. This was only seen in patients with wild-type FLT3.
Synthetic glucocorticoids are structurally similar33 immunomodulatory agents mainly affecting cytokine production, cell cycle, oncogene expression and apoptosis regulation.34, 35 Although therapeutic mechanisms and functions of glucocorticoids are well known in ALL,36 it remain poorly understood in AML. Glucocorticoid mediated induction of apoptosis was reported in AML cell lines.37 Methylprednisolone was shown to induce differentiation in myeloid blasts in in vitro and in vivo AML cell models.38, 39 Clinical trials with chemotherapeutic agents and glucocorticoids resulted in significant response in a subset of AML patients suggesting supportive role of glucocorticoids.40, 41, 42 However, dexamethasone has shown to exert a cytoprotective effect when used in combination with standard chemotherapy drugs and contributes to chemotherapeutic resistance in ovarian cancer and in glioblastoma.43, 44 Therefore, glucocorticoids are known to exert both synergistic and antagonistic effects with other cytotoxic chemotherapeutic drugs across different cancer types.
Glucocorticoids induce apoptosis in leukemic cells through binding to glucocorticoid receptor and subsequently to two major transcription regulators NF-κB and AP-1.34, 35 In our unbiased approach using an integration of chemical biology and transcriptomic profiling, expression of the glucocorticoid receptor NR3C1 showed a modestly but significantly increased expression in the cell line variants with increasing resistance to cytarabine. At the protein level, this difference was more prominent. However, all clinical samples expressing the NR3C1 did not show statistically significant association with the glucocorticoid responsiveness using RNA-seq transcriptomics data. We did not find gain or amplification of 5q31 region including NR3C1 in cytarabine-resistant in vitro cells or clinical samples, which partially explains no substantial increase of NR3C1 gene expression.
Even though glucocorticoids have been used in AML clinical trials,40, 41 their therapeutic potential in cytarabine-resistant patients has not been previously described. Here we illustrate enhanced glucocorticoid sensitivity in 16% of cytarabine treated, relapsed or refractory patient samples. Although the patients received combinatorial therapy containing cytarabine, the AML cell line data suggest that cytarabine is alone sufficient to cause glucocorticoid sensitivity. Molecular profiling data further suggests that wild-type FLT3 is significantly associated with sensitivity to glucocorticoids in chemorefractory AML patients. In fact, none of the patients with FLT3-ITD or point mutations developed glucocorticoid sensitivity. Paired samples from two patients carrying wild-type FLT3 exhibited enhanced glucocorticoid sensitivity after acquired cytarabine resistance. This finding is consistent with enhanced sensitivity to glucocorticoids in SHI-1 cell line carrying wild-type FLT3, which was not observed in MOLM-13 harboring FLT3-ITD. A study reported direct interaction between FLT3 and dexamethasone to drive glucocorticoid signaling pathway.45 We hypothesize that mutant and constitutively active FLT3 changes glucocorticoid signaling-mediated apoptosis in cytarabine-resistant AML cells.
Acquisition of cytarabine resistance leading co-resistance to other nucleoside analogs corroborated with previous observations.7, 10 Although mutation and defective expression of the DCK gene is known in leukemia and lymphomas,7, 8, 9, 10, 14, 46 here, we report focal homozygous deletion of DCK for the first time in a leukemia patient along with the confirmation of nucleoside analog co-resistance. Loss of DCK function in MOLM-13 and SHI-1 cytarabine-resistant variants verifies their validity with previous studies.11, 12 We observed no genetic changes or aberrant expression in other genes previously reported to be involved in cytarabine resistance, such as SLC29A1, CDA, NT5C2 and NKG2D.12, 13, 14, 17
In conclusion, acquisition of cytarabine resistance is associated with an increase of glucocorticoid sensitivity in AML cell lines and AML patient cells. Importantly, glucocorticoids present safer treatment option compared with targeted drugs with toxic and short-lived clinical responses in AML. Our results support the concept that clinical studies are warranted to explore the effects of glucocorticoids in chemorefractory AML patients carrying wild-type FLT3.
Acknowledgments
The authors thank the patients for donating their samples to our research and staff of High Throughput Biomedicine and Sequencing Laboratory Units. The research was supported by Academy of Finland (Center of Excellence for Translational Cancer Biology, Grants 269862, 272437 and 295504 to TA, 277293 to KW), Cancer Society of Finland (OK, TA and KW), Sigrid Juselius Foundation, EU Systems Microscopy (FP7) and TEKES. The senior authors have received collaborative research grants for other projects as listed: OK received research funding from Pfizer, Roche, the IMI Predect consortium and is Board member and co-founder of bioinformatics company Medisapiens, Helsinki, Finland. KP received honoraria and research funding from Bristol-Myers Squibb, Celgene, Novartis and Pfizer. CH received honoraria from Celgene, Novartis and Roche and research funding from Celgene and Pfizer. MW received collaborative research funding from Pfizer and Bayer Pharma. KW received research funding from Pfizer.
Author contributions
DM, AM and OK designed the study; DM performed experiments and analyzed data; DM, AM and OK interpreted data and wrote the paper; DM and BY analyzed drug response and gene expression data; AK and SE analyzed sequencing data; TP, RK, SE, PO, MK and MMM obtained patient data; MK and KP obtained ethical permits, collected clinical samples and administered therapies; TA, KW, KP, CH, MK, MW, AM, OK provided critical review; and AM and OK supervised the study.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
The authors declare no conflict of interest.
Supplementary Material
References
- Rowe JM, Kim HT, Cassileth PA, Lazarus HM, Litzow MR, Wiernik PH et al. Adult patients with acute myeloid leukemia who achieve complete remission after 1 or 2 cycles of induction have a similar prognosis. Cancer 2010; 116: 5012–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cros E, Jordheim L, Dumontet C, Galmarini CM. Problems related to resistance to cytarabine in acute myeloid leukemia. Leuk Lymphoma 2004; 45: 1123–1132. [DOI] [PubMed] [Google Scholar]
- Tallman MS, Gilliland DG, Rowe JM. Drug therapy for acute myeloid leukemia. Blood 2005; 106: 1154–1163. [DOI] [PubMed] [Google Scholar]
- Rowe JM. Optimal induction and post-remission therapy for AML in first remission. ASH Education Program Book 2009; 2009: 396–405. [DOI] [PubMed] [Google Scholar]
- Wolach O, Itchaki G, Bar-Natan M, Yeshurun M, Ram R, Herscovici C et al. High-dose cytarabine as salvage therapy for relapsed or refractory acute myeloid leukemia-is more better or more of the same? Hematol Oncol 2015; 34: 28–35. [DOI] [PubMed] [Google Scholar]
- Matthews JP, Bishop JF, Young GAR, Juneja SK, Lowenthal RM, Garson OM et al. Patterns of failure with increasing intensification of induction chemotherapy for acute myeloid leukaemia. Br J Haematol 2001; 113: 727–736. [DOI] [PubMed] [Google Scholar]
- Cai J, Damaraju VL, Groulx N, Mowles D, Peng Y, Robins MJ et al. Two distinct molecular mechanisms underlying cytarabine resistance in human leukemic cells. Cancer Res 2008; 68: 2349–2357. [DOI] [PubMed] [Google Scholar]
- Klanova M, Lorkova L, Vit O, Maswabi B, Molinsky J, Pospisilova J et al. Downregulation of deoxycytidine kinase in cytarabine-resistant mantle cell lymphoma cells confers cross-resistance to nucleoside analogs gemcitabine, fludarabine and cladribine, but not to other classes of anti-lymphoma agents. Mol Cancer 2014; 13: 4598–13-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathe SK, Largaespada DA. Deoxycytidine kinase is downregulated in Ara-C-resistant acute myeloid leukemia murine cell lines. Leukemia 2010; 24: 1513–1515. [DOI] [PubMed] [Google Scholar]
- Nowak D, Liem NL, Mossner M, Klaumunzer M, Papa RA, Nowak V et al. Variegated clonality and rapid emergence of new molecular lesions in xenografts of acute lymphoblastic leukemia are associated with drug resistance. Exp Hematol 2015; 43: 32,43.e1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathe SK, Moriarity BS, Stoltenberg CB, Kurata M, Aumann NK, Rahrmann EP et al. Using RNA-seq and targeted nucleases to identify mechanisms of drug resistance in acute myeloid leukemia. Sci Rep 2014; 4: 6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmarini CM, Thomas X, Calvo F, Rousselot P, Jafaari AE, Cros E et al. Potential mechanisms of resistance to cytarabine in AML patients. Leuk Res 2002; 26: 621–629. [DOI] [PubMed] [Google Scholar]
- Bardenheuer W, Lehmberg K, Rattmann I, Brueckner A, Schneider A, Sorg UR et al. Resistance to cytarabine and gemcitabine and in vitro selection of transduced cells after retroviral expression of cytidine deaminase in human hematopoietic progenitor cells. Leukemia 2005; 19: 2281–2288. [DOI] [PubMed] [Google Scholar]
- Abraham A, Varatharajan S, Karathedath S, Philip C, Lakshmi KM, Jayavelu AK et al. RNA expression of genes involved in cytarabine metabolism and transport predicts cytarabine response in acute myeloid leukemia. Pharmacogenomics 2015; 16: 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith FJ, Bradbury DA, Zhu YM, Russell NH. Inhibition of bcl-2 with antisense oligonucleotides induces apoptosis and increases the sensitivity of AML blasts to Ara-C. Leukemia 1995; 9: 131–138. [PubMed] [Google Scholar]
- Saxena A, Viswanathan S, Moshynska O, Tandon P, Sankaran K, Sheridan DP. Mcl-1 and Bcl-2/Bax ratio are associated with treatment response but not with Rai stage in B-cell chronic lymphocytic leukemia. Am J Hematol 2004; 75: 22–33. [DOI] [PubMed] [Google Scholar]
- Ogbomo H, Michaelis M, Klassert D, Doerr HW, Cinatl J Jr. Resistance to cytarabine induces the up-regulation of nkg2d ligands and enhances natural killer cell lysis of leukemic cells. Neoplasia 2008; 10: 1402–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach D, Lengemann J, Voigt A, Hermann J, Zintl F, Sauerbrey A. Response to chemotherapy and expression of the genes encoding the multidrug resistance-associated proteins MRP2, MRP3, MRP4, MRP5, and SMRP in childhood acute myeloid leukemia. Clin Cancer Res 2003; 9: 1083–1086. [PubMed] [Google Scholar]
- Negoro E, Yamauchi T, Urasaki Y, Nishi R, Hori H, Ueda T. Characterization of cytarabine-resistant leukemic cell lines established from five different blood cell lineages using gene expression and proteomic analyses. Int J Oncol 2011; 38: 911–919. [DOI] [PubMed] [Google Scholar]
- Faderl S, Gandhi V, O'Brien S, Bonate P, Cortes J, Estey E et al. Results of a phase 1-2 study of clofarabine in combination with cytarabine (ara-C) in relapsed and refractory acute leukemias. Blood 2004; 105: 940–947. [DOI] [PubMed] [Google Scholar]
- Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, Gendron P, Romeo AA, Morris SJ et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature 2014; 511: 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobune M, Takimoto R, Murase K, Iyama S, Sato T, Kikuchi S et al. Drug resistance is dramatically restored by hedgehog inhibitors in CD34+ leukemic cells. Cancer Sci 2009; 100: 948–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Edwards H, Caldwell JT, Wang G, Taub JW, Ge Y. Obatoclax potentiates the cytotoxic effect of cytarabine on acute myeloid leukemia cells by enhancing DNA damage. Mol Oncol 2015; 9: 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi R, Yamauchi T, Negoro E, Takemura H, Ueda T. Combination of guanine arabinoside and Bcl-2 inhibitor YC137 overcomes the cytarabine resistance in HL-60 leukemia cell line. Cancer Sci 2013; 104: 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav B, Pemovska T, Szwajda A, Kulesskiy E, Kontro M, Karjalainen R et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci Rep 2014; 4: 5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemovska T, Kontro M, Yadav B, Edgren H, Eldfors S, Szwajda A et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov 2013; 3: 1416–1429. [DOI] [PubMed] [Google Scholar]
- Sulonen AM, Ellonen P, Almusa H, Lepisto M, Eldfors S, Hannula S et al. Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol 2011; 12: R94,2011–12-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes Fajardo KV, Adams D, NISC Comparative Sequencing ProgramMason CE, Sincan M, Tifft C et al. Detecting false-positive signals in exome sequencing. Hum Mutat 2012; 33: 609–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav B, Gopalacharyulu P, Pemovska T, Khan SA, Szwajda A, Tang J et al. From drug response profiling to target addiction scoring in cancer cell models. Dis Model Mech 2015; 8: 1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Szwajda A, Shakyawar S, Xu T, Hintsanen P, Wennerberg K et al. Making sense of large-scale kinase inhibitor bioactivity data sets: a comparative and integrative analysis. J Chem Inf Model 2014; 54: 735–743. [DOI] [PubMed] [Google Scholar]
- Stirewalt DL, Kopecky KJ, Meshinchi S, Appelbaum FR, Slovak ML, Willman CL et al. FLT3, RAS, and TP53 mutations in elderly patients with acute myeloid leukemia. Blood 2001; 97: 3589–3595. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013; 368: 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspers GJL, Veerman AJP, Popp-Snijders C, Lomecky M, Van Zantwijk CH, Swinkels LMJW et al. Comparison of the antileukemic activity in vitro of dexamethasone and prednisolone in childhood acute lymphoblastic leukemia. Med Pediatr Oncol 1996; 27: 114–121. [DOI] [PubMed] [Google Scholar]
- Greenstein S, Ghias K, Krett NL, Rosen ST. Mechanisms of glucocorticoid-mediated apoptosis in hematological malignancies. Clin Cancer Res 2002; 8: 1681–1694. [PubMed] [Google Scholar]
- Tissing WJE, Meijerink JPP, den Boer ML, Pieters R. Molecular determinants of glucocorticoid sensitivity and resistance in acute lymphoblastic leukemia. Leukemia 2003; 17: 17–25. [DOI] [PubMed] [Google Scholar]
- Gaynon PS, Lustig RH. The use of glucocorticoids in acute lymphoblastic leukemia of childhood. Molecular, cellular, and clinical considerations. J Pediatr Hematol Oncol 1995; 17: 1–12. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Ohki M, Nakagawa T, Honma Y. Glucocorticoids induce apoptosis in acute myeloid leukemia cell lines with a t(8;21) chromosome translocation. Leuk Res 1997; 21: 45–50. [DOI] [PubMed] [Google Scholar]
- Ozbek N, Erdemli E, Hicsonmez G, Okur H, Tekelioglu M. Effects of methylprednisolone on human myeloid leukemic cells in vitro. Am J Hematol 1999; 60: 255–259. [DOI] [PubMed] [Google Scholar]
- Hicsonmez G, Tuncer M, Toksoy HB, Yenicesu I, Cetin M. Differentiation of leukemic cells induced by short-course high-dose methylprednisolone in children with different subtypes of acute myeloblastic leukemia. Leuk Lymphoma 1999; 33: 573–580. [DOI] [PubMed] [Google Scholar]
- Morrison FS, Kopecky KJ, Head DR, Athens JW, Balcerzak SP, Gumbart C et al. Late intensification with POMP chemotherapy prolongs survival in acute myelogenous leukemia—results of a Southwest Oncology Group study of rubidazone versus adriamycin for remission induction, prophylactic intrathecal therapy, late intensification, and levamisole maintenance. Leukemia 1992; 6: 708–714. [PubMed] [Google Scholar]
- Nagura E, Kimura K, Yamada K, Ohta K, Maekawa T, Takaku F et al. Nationwide randomized comparative study of daunorubicin and aclarubicin in combination with behenoyl cytosine arabinoside, 6-mercaptopurine, and prednisolone for previously untreated acute myeloid leukemia. Cancer Chemother Pharmacol 1994; 34: 23–29. [DOI] [PubMed] [Google Scholar]
- Kolb EA, Steinherz PG. A new multidrug reinduction protocol with topotecan, vinorelbine, thiotepa, dexamethasone, and gemcitabine for relapsed or refractory acute leukemia. Leukemia 2003; 17: 1967–1972. [DOI] [PubMed] [Google Scholar]
- Mariotta M, Perewusnnyk G, Koechli OR, Little JB, von KD, Mirimanoff R et al. Dexamethasone-induced enhancement of resistance to ionizing radiation and chemotherapeutic agents in human tumor cells. Strahlentherapie und Onkologie 1999; 175: 392–396. [DOI] [PubMed] [Google Scholar]
- Naumann U, Durka S, Weller M. Dexamethasone-mediated protection from drug cytotoxicity: association with p21WAF1/CIP1 protein accumulation? Oncogene 1998; 17: 1567–1575. [DOI] [PubMed] [Google Scholar]
- Asadi A, Hedman E, Widen C, Zilliacus J, Gustafsson JA, Wikstrom AC. FMS-like tyrosine kinase 3 interacts with the glucocorticoid receptor complex and affects glucocorticoid dependent signaling. Biochem Biophys Res Commun 2008; 368: 569–574. [DOI] [PubMed] [Google Scholar]
- Song JH, Kim SH, Kweon SH, Lee TH, Kim HJ, Kim HJ et al. Defective expression of deoxycytidine kinase in cytarabine-resistant acute myeloid leukemia cells. Int J Oncol 2009; 34: 1165–1171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.