Abstract
Mitochondrial deficits or altered expressions of microRNAs are associated with the pathogenesis of various diseases, and microRNA-operated control of mitochondrial activity has been reported. Using a retrovirus-mediated short-hairpin RNA (shRNA) system, we observed that miR-24-mediated H2AX knockdown (H2AX-KD) impaired both mitochondria and the insulin signaling pathway. The overexpression of miR-24 decreased mitochondrial H2AX and disrupted mitochondrial function, as indicated by the ATP content, membrane potential and oxygen consumption. Similar mitochondrial damage was observed in shH2AX-mediated specific H2AX-KD cells. The H2AX-KD reduced the expression levels of mitochondrial transcription factor A (TFAM) and mitochondrial DNA-dependent transcripts. H2AX-KD mitochondria were swollen, and their cristae were destroyed. H2AX-KD also blocked the import of precursor proteins into mitochondria and the insulin-stimulated phosphorylation of IRS-1 (Y632) and Akt (S473 and T308). The rescue of H2AX, but not the nuclear form of ΔC24-H2AX, restored all features of miR-24- or shH2AX-mediated impairment of mitochondria. Hepatic miR-24 levels were significantly increased in db/db and ob/ob mice. A strong feedback loop may be present among miR-24, H2AX, mitochondria and the insulin signaling pathway. Our findings suggest that H2AX-targeting miR-24 may be a novel negative regulator of mitochondrial function and is implicated in the pathogenesis of insulin resistance.
Introduction
In the last decade, deficits in mitochondrial function have been linked to nearly all age-associated degenerative diseases, including insulin resistance,1, 2, 3 metabolic syndrome4, 5, 6 and vascular complications of diabetes.7, 8 Declines in oxidative phosphorylation (OXPHOS) or mitochondrial density have been observed in insulin-resistant individuals,3 the offspring of type II diabetic patients2, 9 and even normal senescent subjects.10, 11, 12, 13 Mitochondrial damage can be induced by genetic mutations14 and various exogenous chemicals.15, 16 However, the molecular mechanisms underlying the causes of mitochondrial dysfunction have not been clearly elucidated.
Mitochondria are dynamic organelles that are affected by mitochondrial DNA (mtDNA) and nuclear DNA (nDNA). Several nDNA-encoded factors control the expression of both mtDNA- and nDNA-encoded mitochondrial genes. Of these, the best-known genes are mitochondrial transcription factors A and B (TFAM and mtTFB), nuclear respiration factor-1 and -2 (NRF-1, NRF-2) and peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α). However, mitochondrial function is not always proportional to the expression level of these control factors. For example, mitochondrial density measured by electron microscopy is reduced in the skeletal muscles of the offspring of type II diabetic patients, but the expressions of PGC-1α, TFAM and NRF-1 are normal.17 This indicates that other factors may affect mitochondrial density or content. A wide range of proteomic, genomic and chemical approaches has been applied to identify novel mitochondrial modulators that are responsible for age- or disease-dependent changes, but so far, these entities remain unidentified.18, 19
We previously reported that histones were found in the mitochondrial proteome. H2A is an integral mitochondrial outer membrane protein with its N terminus protruding towards the cytoplasm.20 Here, we confirmed the mitochondrial localization of the H2A isoform H2AX. In the nucleus, H2AX has a critical role in DNA repair and genomic stability.21, 22, 23 Mice lacking H2AX show multiple phenotypic changes, including radiation sensitivity, growth retardation, immune deficiency and male infertility.24, 25 H2AX-deficient embryonic stem cells exhibit elevated levels of chromosomal aberrations.26 However, no function besides DNA repair and no location aside from the nucleus have previously been reported for H2AX.
H2AX has been reported as a target of microRNA-24.27 microRNAs (miRs) are potent post-transcriptional regulators of gene expression, but few targets or physiological implications of miRs have been analyzed in animals.28, 29 Most miRs are functionally associated with developmental processes, such as morphogenesis, neurogenesis and developmental timing.30 In addition, the altered expression of specific miRs (miR-143, -29, -126, -143, -124) is involved in the pathogenesis of several human diseases, including obesity31, 32, 33, diabetes34, 35, 36, cancer37, cardiac hypertrophy38 and neurodegeneration.39, 40, 41 In the present study, we found that miR-24 could negatively modulate mitochondrial function by targeting H2AX and consequently might be implicated in the development of diseases associated with mitochondrial dysfunction.
Materials and methods
Reagents
Dulbecco's Modified Eagle's Medium (DMEM) and fetal bovine serum (FBS) were purchased from Gibco-BRL (Grand Island, NY, USA). Dimethyl sulfoxide and all other chemicals were purchased from Sigma (St Louis, MO, USA). Double-stranded siRNAs targeting human H2AX (5′-caacaagaagacgcgaatc-3′), miR-24 (hsa-miR-24, hsa-miR-24-3p, 5′-tggctcagttcagcaggaacag-3′) and the scrambled control SCR (5′-aattctccgagcgtgtcacgt-3′) were synthesized by Samchulli, Seoul, Korea.
Cell culture and transient transfection
SK-Hep1 human hepatoma cells (ATCC, HTB-52) were cultured in DMEM supplemented with 10% FBS, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin at 37 °C and 5% CO2. For transient transfections, cells in 6-well plates (4 × 105 cells per well) were transfected with synthetic siRNA (100 pmole) using the GeneJammer transfection reagent (Agilent, Santa Clara, CA, USA). The cells were washed twice with phosphate-buffered saline (PBS) 4 h after transfection. The cells were then cultured in DMEM with 10% FBS for 48–72 h, harvested and assayed. For rescue experiments, stably expressing cells stably expressing shH2AX or miR-24 were transfected with pcDNA3.1-H2AX expression plasmids and selected using 800 μg ml−1 G418.
Retroviral transduction of shRNAs for H2AX and miR-24
Double-stranded DNA oligonucleotides for miR-24 containing the BamHI-miR-24-XhoI loop-antisense miR-24-T5-EcoRI sequences (5′-gatccg tggctcagttcagcaggaacag ttctcgaga ctgttcctgctgaactgagcca tttttggaag-3′) were cloned into the RNAi-Ready pSIREN-RetroQ vector (Clontech, Mountain View, CA, USA) to produce a retrovirus expressing miR-24 (Rv-miR-24) as short-hairpin RNA (shRNA) under the U6 promoter. Similarly, Rv-shRNA for H2AXi (Rv-shH2AX, 5′-gatccg caacaagaagacgcgaatc ttctcgaga gattcgcgtcttcttgttg tttttggaag-3′) and scrambled control shRNAs (Rv-shSCR, 5′-gatccg aattctccgagcgtgtcacgt ttctcgaga acgtgacacgctcggagaatt tttttggaag-3′) were produced. The BD RetroPack PT67 packaging cells (BD Biosciences, Billerica, MA, USA) were treated for 5 min with 25 μM chloroquine (Sigma) and transfected with 3 μg pSIREN-RetroQ-shRNA using the GeneJammer transfection reagent. At 48 h post-transfection, the virus population in the supernatant was harvested by filtration through a 0.45-μm syringe filter and centrifugation at 50 000 × g for 1.5 h. The pelleted virus was resuspended in one-tenth of the original volume of medium and incubated at 4 °C for several hours. To establish stably Rv-shRNA-infected cells expressing miR-24, shH2AX or shSCR, SK-Hep1 cells (0.8 × 106/60-mm plate) were infected with the recombinant retroviruses (1 × 105 pfu ml−1) using polybrene (8 μg ml−1 final concentration) and selected in media containing puromycin (1 μg ml−1) for 2 weeks.
Cell number
Cell numbers were determined using the ImageJ program. Briefly, cells were cultured on poly-L-lysine coated-cover slips in 6-well plates for various time periods. Then, the cells were stained with 2 μg ml−1 Hoechst 33342 for 10 min, fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. After washing with PBS, the coverslips were mounted with GEL/MOUNT (Biomeda, Foster city, CA, USA), and the fluorescence was visualized under a fluorescence microscope (Leica DMRBE, Ontario, NY, USA). The number of Hoechst-positive cells was counted automatically in 10 different areas (0.8 mm2 per area) using ImageJ.
Mitochondrial activity assays
The mitochondrial activity of the stably expressing cells was analyzed using a 96-well plate as described previously.42 The quantitative measurements of the lactate dehydrogenase (LDH) activity, lactate concentration, the pH of media, tetramethylrhodamine ethylester (TMRE, Molecular Probes, Eugene, OR, USA)-mediated mitochondrial membrane potential (ΔΨm), the 5,6-chloromethyl-2′,7′-dichlorofluorescin diacetate (CM-DCF-DA, Molecular Probes)- and MitoSox-mediated ROS content, Oxyblot and intracellular ATP content were probed. Briefly, cells (3 × 104 cells) were cultured for up to 72 h in 6-well plates with a medium change every 24 h. The LDH activity was measured using the TOX7 LDH assay kit (Sigma). The lactate concentration and culture media pH were measured using a lactate assay kit (BioVision, Mountain View, CA, USA) and a pH meter, respectively. Cells cultured in black 96-well culture plates were incubated with 200 nM TMRE or Hoechst 33342 (0.5 μM) at 37 °C for 30 min in phenol red-free SDM. The mitochondrial superoxide levels were measured after incubating cells with 5 μM MitoSox (Molecular Probes) for 10 min at 37 °C, and the samples were protected from light. The cells were washed with PBS and counterstained with 2 μg ml−1 Hoechst 33342 for 10 min, and the fluorescence intensities were determined at 510/580 nm. The MitoSox intensity was normalized to the Hoechst intensity. The extent of protein oxidation was assessed by measuring the carbonyl protein levels with an OxyBlot protein oxidation detection kit (Chemicon International, Temecula, CA, USA). The data are expressed as the means±s.e. of three independent experiments.
Oxygen consumption rate measurements
The oxygen consumption rate (OCR) of each OXPHOS complex was measured using Oxygraph-2 K (Oroboros, Innsbruck, Austria) as described previously.42 Cells were collected by trypsinization and resuspended in 1 ml of respiration media containing 100 μM p1, p5-di(adenosine)-5′-pentaphosphate and an adenosine kinase inhibitor. The cells (6 × 106) were permeabilized with 10 μg ml−1 digitonin in the Oxygraph-2 K chamber. The OCRs were determined as OXPHOS complex inhibitors and substrates were sequentially added as follows: 2 mM ADP, 8 mM malate and 20 mM glutamate for complex I; 1 μM rotenone, 10 μM succinate and 2.5 μM glycerol-3-phosphate for complexes II and III; 25 μM antimycin A, 80 μM ascorbate and 0.42 mM N,N,N,N-tetramethyl-p-phenylenediamine (TMPD) for complex IV; and 2.5 mM KCN for KCN-insensitive respiration.
The respiratory capacity and ATP turnover rate were determined using a Seahorse XF-24 analyzer (Seahorse Bioscience, Billerica, MA, USA). Cells (5 × 103 cells per well) were seeded in XF-24 microplates in 250 μl DMEM containing 10% FBS and incubated at 37 °C/5% CO2 for 24 h. The assays were initiated by replacing the medium in each well with 590 μl assay medium (DMEM without sodium bicarbonate) pre-warmed to 37 °C. After gentle mixing for 10 min in the XF-24 Analyzer, the basal OCR was measured for 3 min. Oligomycin (65 μl of 10 μg ml−1 stock), carbonylcyanide-p-trifluoromethoxyphenylhydrazone (FCCP, 73 μl of 3 μM stock) and rotenone (81 μl of 1 μM stock) were consecutively injected into each well to reach the desired final working concentration for quantifying the ATP turnover rate (basal OCR—oligomycin-OCR) and respiratory capacity (FCCP-OCR—rotenone-OCR). The OCR was calculated from 3-min measurement cycles. The respiration results were normalized to the cell number.43
Western blot analysis
Protein extracts were prepared from the cells using PRO-PREP lysis buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 2 mM MgCl2, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 5 μg ml−1 aprotinin, 5 μg ml−1 pepstatin A, 5 μg ml−1 leupeptin and 1% Triton X-100; iNtRON Biotech, Gyeonggi-do, Korea). A total of 25 μg of protein extract was separated on 12–15% sodium dodecyl sulfide (SDS) gels and analyzed using an enhanced chemiluminescence system (Amersham Bioscience, Piscataway, NJ, USA). Primary antibodies against mouse H2AX (R&D Systems, Minneapolis, MN, USA), H2A, PGC-1α (Santa Cruz Biotechnology, Santa Cruz, CA, USA), AKT1, pAKT, FOXO1, pFOXO1, AMPK, pAMPK (Cell Signaling Technology, Beverly, MA, USA), ND9, SDHA, UQCRC2, COXI, COXIV and ATPase α (Molecular Probes) were purchased from commercial sources as indicated. The rabbit polyclonal antibodies against human TFAM, NRF-1 and human H2AX were prepared in our laboratory.8 The H2AX-specific tail peptide (PKAPSGGKKATQASQE) was synthesized and conjugated with keyhole limpet hemocyanin to prepare the human H2AX antibody.44 An equivalent protein loading was verified using anti-β-actin antibodies (Sigma).
RNA preparation, semi-quantitative RT-PCR and real-time qPCR
The total RNA was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The total RNA (2 μg) was reverse transcribed using MMLV reverse transcriptase (Promega, Madison, WI, USA) and 250 ng of random primers (Invitrogen) according to the manufacturer's instructions. Real-time quantitative PCR was performed using the primers for H2AX (5′-cat gtc ggg ccg cgg caa-3′ and 5′-gtg gcg ctg gtc ttc ttg-3′ for human H2AX; 5′-ggc ctg tgg aca aga gtt cta t-3′ and 5′-gcc cat taa atc tcc cca ct-3′ for murine H2AX) or 18S rRNA (5′-gag cga aag cat ttg cca ag-3′ and 5′-ggc atc gtt tat ggt cgg aa-3′ for both human and murine 18S rRNA) on a Light Cycler 1.5 (Roche, Indianapolis, IN, USA) with SYBR Premix Ex Taq (TaKaRa, Shiga, Japan) at 95 °C for 10 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. The measurements were performed in duplicate for each sample. The H2AX mRNA quantity was corrected by the simultaneous measurement of nuclear DNA encoding 18S rRNA. The relative quantification of gene expression was determined using the 2-ΔΔCt method. The relative mRNA expression levels were presented as fold changes compared with that of the control condition.
We also carried out semi-quantitative RT-PCR to quantify the expression levels of mitochondria-related genes, including 13 mtDNA-encoded OXPHOS subunits, the nDNA-encoded OXPHOS subunits and the mitochondrial biogenesis controlling proteins. PCR products of these genes did not reach saturation levels within the cycle limits. The reaction products were examined using 1.2% agarose gel electrophoresis and normalized to the RT-PCR products for 18S rRNA. The presence of mtDNA was verified by PCR using a genomic DNA template and two different sets of primers, and the results were normalized to β-actin DNA. The primer information for RT-PCR and PCR is summarized in Supplementary Table 1.
The total RNA containing microRNAs was purified using the miRNeasy Mini Kit (Qiagen, Hilden, Germany). The reverse transcription of miRNA into cDNA was performed using the miScript Reverse Transcription Kit. The amount of miR-24 was quantified by real-time PCR performed on a Rotor-Gene Q cycler (Qiagen) using the miScript Primer Assay and miScript SYBR Green PCR Kits (Qiagen). The specific primer sets for quantification, MS00001827 for miR-24 and MS00033740 RNU6 control for normalization, were purchased from Qiagen.
Transmission electron microscopy
For morphological assessment of the mitochondria, the cell suspensions were fixed with 2% paraformaldehyde and 2% glutaraldehyde in 50 mM sodium cacodylate buffer (pH 7.2) for 4 h and washed with the same buffer. All specimens were exposed to 1% osmium tetroxide in 50 mM sodium cacodylate buffer (pH 7.2) for 2 h at 4 °C, rinsed, dehydrated with ethanol and flat-embedded in Spurr's embedding media. Ultrathin sections were stained with 2% uranyl acetate and Reynolds' lead citrate and then examined by transmission electron microscopy at 80 kV (JEM-1011, JEOL, Tokyo, Japan).45
Immunocytochemistry
Cells grown on glass coverslips were stained with MitoTracker Orange (Mito-T, Molecular Probes) at 300 nM for 20 min in complete medium containing 10% FBS. The cells were fixed for 5 min in 4% paraformaldehyde/PBS and permeabilized with ice-cold 0.1% Triton X-100 for 10 min. The cells were then covered with 5% BSA in Tris-buffered saline for 30 min at room temperature followed by incubation with the anti-H2AX antibodies (1:500, Cell Signaling Technology) for 1 h. After washing, the cells were probed with the appropriate secondary antibodies conjugated to Alexa Fluor 488 (1:1,000, Molecular Probes) for 1 h. The slides were then washed twice with PBS and mounted using DAKO fluorescent mounting medium (DAKO Corporation, Carpinteria, CA, USA). The specimens were viewed using a laser scanning confocal microscope (Carl Zeiss LSM510, Zena, Germany).
Luciferase assay for miR-24 target specificity
The short form (661 bp) of the human H2AX 3′-UTR46 was amplified by RT-PCR from the total RNA isolated from SK-Hep1 cells using the primers 5′-gacgaggagctcaacaa-3′ and 5′-ggttagctgcagaatt-3′. To produce the construct pCMV-luc-H2AX-3′UTR (Luc-H2AX-3′-UTR, full), the 664-bp PCR fragment of H2AX 3′-UTR was cloned into the XbaI site after the luciferase stop codon of pCMV-luc (a gift from Dr VN Kim, Seoul National University, Korea). Three different H2AX 3′-UTR fragments containing one of three putative miR-24 binding sites (MBS-A, B and C) were generated by PCR using Luc-H2AX-3′-UTR (full) and cloned into pCMV-luc to produce pCMV-luc-A, -B or -C (Luc-H2AX-3′-UTR: A, B, C). SK-Hep1 cells in 6-well plates were transiently co-transfected with the pSIREN-RetroQ plasmids (2 μg) containing either shSCR (pS-shSCR) or miR-24 (pS-miR-24) together with pCMV-luc (Luc, 100 ng) or Luc-H2AX-3′-UTR (100 ng) using the GeneJammer transfection reagent. The cells were harvested after 24 h of incubation in DMEM supplemented with 10% FBS and assayed for luciferase activity using a luciferase assay kit (Promega, Madison, WI, USA) and a luminometer (Berthold, Badwildbad, Germany). Because the LacZ vector (200 ng) was co-transfected with the reporter, the transfection efficiencies were normalized to β-galactosidase activity. All assays were performed in triplicate in three independent experiments.
Co-immunoprecipitation
Antibodies against TOM20 or H2AX were cross-linked to Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology) using dimethyl pimelimidate (Pierce, Rockford, IL, USA) according to the manufacturer's instructions. Cell lysates (500 μg) in IP buffer (50 mM Tris, pH 8, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF, 2 μg ml−1 leupeptin, 1 μg ml−1 pepstatin and 2 μg ml−1 aprotinin) were precleared with 1 μg of normal IgG for 1 h at 4 °C and incubated with TOM20- or H2AX-conjugated agarose beads under constant rotation overnight at 4 °C. The slurry was washed with IP buffer, boiled in 40 μl IP buffer and 10 μl SDS-PAGE loading buffer, and subjected to western blot analysis.
Mitochondrial protein import assay using isolated mitochondria
Mitochondria were isolated from cultured cells by differential centrifugation.20 Two different DsRed2 fusion constructs were cloned into a pcDNA3.1(+) vector under control of the T7 promoter (Invitrogen). MTS-DsRed2 protein (28.5 kDa) is DsRed2 conjugated to the 35-amino acid MTS of succinate dehydrogenase complex subunit C. H2AX-DsRed2 (41 kDa) is a fusion protein of H2AX at the N terminus of DsRed2. Both DsRed2 fusion proteins were synthesized using the TNT Coupled Reticulocyte Lysate System (Promega) according to the manufacturer's instructions. An in vitro import assay of hybrid proteins containing DsRed2 was performed as previously described.44 Briefly, we incubated newly in vitro-synthesized fusion proteins (3.75 μl in 50 μl TNT mixture) with 75 μg of mitochondria in 12.5 μl mitochondria isolation buffer (0.025 M Tris HCl, pH 7.4, 0.25 M sucrose and 1 mM EDTA) in 50 μl import assay buffer (50 mM HEPES, 0.47 M sucrose, 2.5 mM DTT, 2.5 mM MgCl2, 250 mM KCl, 1 mM ATP and 5 mM phosphoenolpyruvate) with 6.25 units of pyruvate kinase at 30 °C for 30 min. The assay mixture was divided into two aliquots and treated with or without proteinase K (0.2 mg ml−1 final concentration) for 30 min on ice. Mitochondria treated with FCCP (2 μM) were used as a negative control. The harvested mitochondria were subjected to 10% SDS-PAGE, and the imported fusion proteins were visualized by immunoblotting using anti-DsRed2 antibodies (Santa Cruz Biotechnology). Western blot analysis of H2AX and Hsp60 was performed to assess the presence of H2AX and equal mitochondrial loading, respectively.
Animals
Diabetic (db/db), obese (ob/ob) and control lean mice (C57BL/6, male, 15 weeks old, n=7 per group) were purchased from Orient-Bio, Korea and Shizuoka Laboratory Center, Hamamatsu, Japan. All animals were killed in the morning after a 1 week quarantine period, and the tissues were collected, frozen in liquid nitrogen and stored at −80 °C. The hepatic expression levels of H2AX and miR-24 were analyzed by western blot and real-time qPCR. All animal procedures were performed in accordance with the guidelines set forth by the Kyung Hee University Council Directive for the Proper Care and Use of Laboratory Animals.
Statistical analysis
All results are expressed as the mean±s.e. of the mean. Statistical differences between experimental groups were assessed by Student's t-tests using InStat (GraphPad Software, San Diego, CA, USA). Values of P<0.05 were considered statistically significant.
Results
Knock down of mitochondrial H2AX by miR-24 or shH2AX
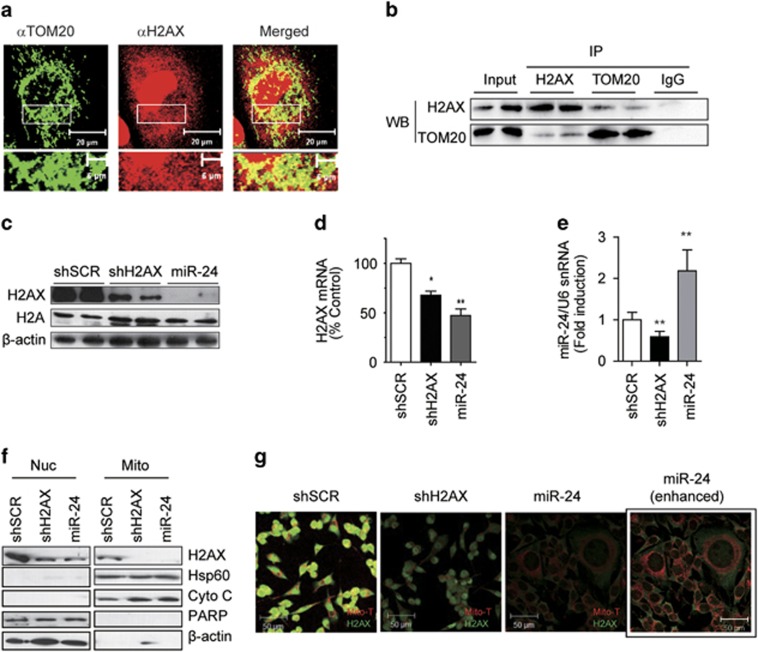
Immunostaining verified the mitochondrial localization of H2AX (Figure 1a). H2AX was co-immunoprecipitated with the mitochondrial transporter TOM20, showing their presence in a complex (Figure 1b). To investigate the role of H2AX in mitochondria, we applied a retroviral system expressing a short-hairpin RNA (Rv-shRNA) to deliver miR-24 or shH2AX inside cells for silencing H2AX. The miRNA miR-24 effectively reduced H2AX to a similar degree as shH2AX. Stably Rv-shRNA-infected cells had persistently decreased H2AX expression throughout the culture period without reducing the total H2A level (Figure 1c). In those cells, the H2AX mRNA levels were decreased by ~30–50%, suggesting that miR-24 mediated the degradation of H2AX mRNA in addition to halting translation (Figure 1d). The level of miR-24 experienced a 2-fold increase in miR-24 cells but a 40% decrease in shH2AX cells (Figure 1e). Because H2AX is present in both nuclei and mitochondria, we verified which H2AX was knocked-down by immunocytochemical staining and organelle fractionation. Both miR-24 and shH2AX suppressed the amount of H2AX protein in both organelles (Figure 1f). Importantly, we note here that MitoTracker (Mito-T) staining was significantly reduced in both H2AX knock down (KD) cells, and some miR-24-cells were much larger than shSCR- or shH2AX-cells (Figure 1g).
Figure 1.
Specific knockdown of human H2AX by miR-24. (a) Co-immunostaining of H2AX with the mitochondrial transporter TOM20 in SK-Hep1 cells. H2AX is present in the cytoplasm and is partially co-localized with TOM20 in the mitochondria. The area in the white rectangle is enlarged at the bottom of each picture. Scale bar=20 μm (top) or 5 μm (bottom). (b) Co-immunoprecipitation of H2AX and TOM20. SK-Hep1 cells were immunoprecipitated (IP) with antibodies against H2AX or TOM20 that were cross-linked to protein A/G beads. The eluted proteins were analyzed by western blot (WB) with antibodies against TOM20 or H2AX. (c) WB of whole-cell lysates from the stably Rv-shRNA-infected cells, using the anti-H2AX or pan-H2A antibody. shSCR, Rv-shRNA containing scrambled control siRNA; shH2AX, Rv-shRNA containing H2AX siRNA; miR-24, Rv-shRNA containing miR-24. (d) Real-time qRT-PCR of H2AX mRNA in stably Rv-shRNA-infected cells. The mRNA level was normalized by 18S rRNA. (e) Real-time qRT-PCR of miR-24 in stably Rv-shRNA-infected cells. The miR-24 level was normalized by U6 snRNA. (f) WB of organelle fractions from the stably Rv-shRNA-infected cells. Nuc, nuclei; Mito, mitochondria. Hsp60 and cytochrome c (Cyto C) are markers for the mitochondria and PARP is a nuclear marker. β-actin was used as a loading control. (g) Confocal images of the stably Rv-shRNA-infected cells. Cells were stained with MitoTracker (Mito-T, red), fixed and then stained with the anti-H2AX antibody (green). Box (miR-24 enhanced): the same miR-24 cells were exposed to 10-fold stronger laser excitation for better visualization. Scale bar=50 μm.
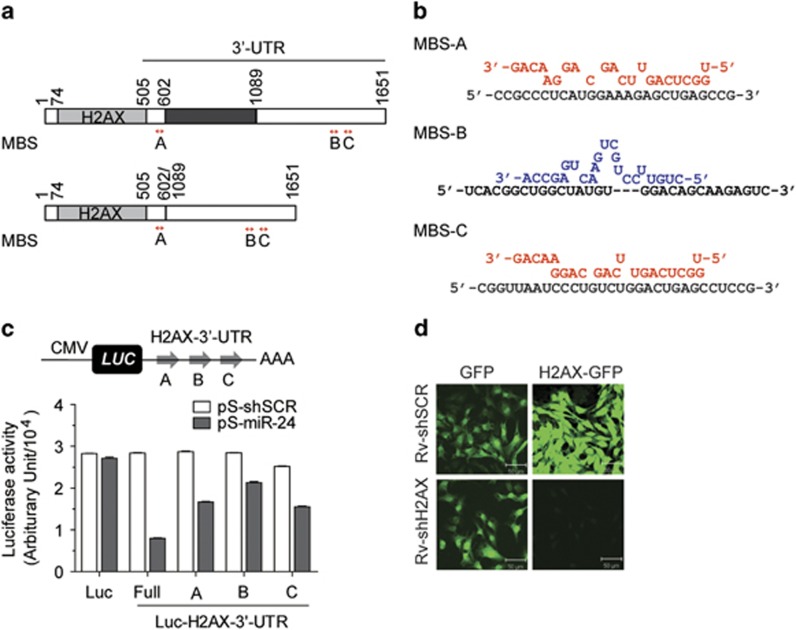
The target specificity of shH2AX and miR-24 was verified using a GFP-H2AX fusion protein and a luciferase reporter carrying the 3′-UTR of H2AX (Figure 2). It has been reported that there are two different lengths of H2AX 3'-UTR transcripts (Figure 2a).46 Importantly, both the long (1148 bp) and short (661 bp) 3′-UTRs contain all three predicted miR-24 binding sequences (MBS), which were designated A, B and C. Putative binding schemes of miR-24 to each of the MBS locations are presented in Figure 2b. Both the guide and passenger strands of pre-mature miRNAs could regulate the expression of their target genes.47 We predicted that the miR-24 guide strand targeted MBS-A and MBS-C of the H2AX 3′-UTR, whereas the miR-24 passenger strand targeted MBS-B. We constructed luciferase reporter plasmids containing the 661 bp H2AX 3′-UTR (Full) or each of the three MBS sites (A, B, C) at the 3′-end of the luciferase gene in a modified pGL3 vector (pCMV-luc, Luc). Then, co-transfection of the overexpression plasmids for either shSCR (pS-shSCR) or miR-24 (pS-miR-24), along with the luciferase reporters, was performed. The luciferase activity of all four Luc-H2AX-3′-UTR (Full, A, B, C) constructs was significantly decreased by pS-miR-24, demonstrating that miR-24 specifically targeted all three binding sites in the H2AX 3′-UTR (Figure 2c). The H2AX-GFP fluorescence intensity of the stably expressing cells that overexpress H2AX-GFP fusion protein was specifically decreased after infection with Rv-shH2AX (Figure 2d).
Figure 2.
Target specificity of miR-24 and shH2AX. (a) Diagram of the two different forms of H2AX 3′-UTR. Locations of three MBS (A, B, C) in the 3′-UTR of H2AX are depicted by arrows (red). Both the long (upper figure) and short 3′-UTR (lower figure) contain all three MBSs. (b) Putative binding schemes of miR-24. MBS-A and MBS-C are targets of miR-24 (red). MBS-B is a target of antisense miR-24 (blue). (c) SK-Hep1 cells were co-transfected with LacZ (β-gal) and pSIREN-RetroQ plasmids containing shSCR (pS-shSCR) or miR-24 (pS-miR-24) together with pCMV-luc (Luc) or four different pCMV-luc-H2AX-3′-UTRs (Luc-H2AX-3′-UTR) containing full-length (Full), MBS-A, MBS-B or MBS-C of the H2AX 3′-UTR. The luciferase activities were normalized to the β-gal activity. The data show the mean±s.e. of three independent duplicate experiments (n=6). (d) SK-Hep1 cells expressing GFP or H2AX-GFP chimeric protein were infected with Rv-shSCR or Rv-shH2AX and selected using 1 μg ml−1 puromycin. The confocal images of the stably expressing cells showed the specific knockdown of H2AX-GFP by shH2AX. Scale bar=50 μm.
Mitochondrial dysfunction in two groups of H2AX-KD cells
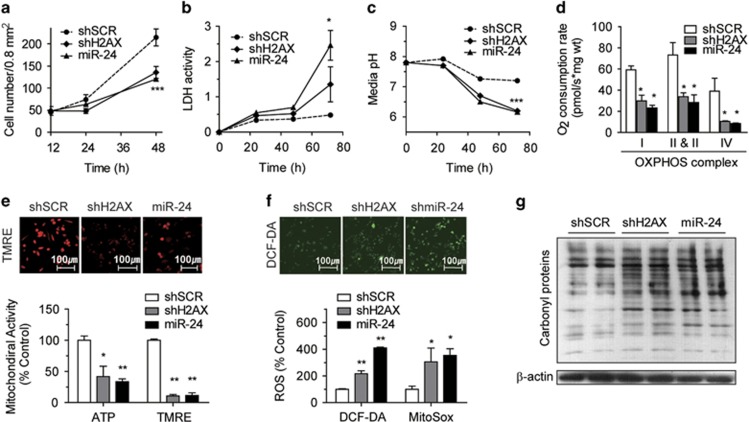
In addition to impairments in DNA repair, growth retardation, immune deficiency and male infertility have been reported in H2AX knock-out mice.25 Thus, we first assessed the growth rates of two populations of H2AX-KD cells. The proliferation rate of miR-24 and shH2AX cells were ~40–50% lower than that of shSCR control cells (Figure 3a). Both sets of cells showed an incubation time-dependent increase in LDH activity (Figure 3b) and acidification of the media pH (Figure 3c), suggesting that a blockage of mitochondrial function might induce anaerobic glycolysis. In fact, miR-24 and shH2AX was decreased >50% of the OCR by four OXPHOS complexes (Figure 3d), and they also reduced the TMRE-based mitochondrial membrane potential (ΔΨm) and intracellular adenosine-5′-triphosphate (ATP) content up to 60–70% (Figure 3e). When mitochondria become damaged, electrons leaking from the OXPHOS complex react with oxygen to produce reactive oxygen species (ROS). As expected, miR-24- or shH2AX-induced H2AX-KD enhanced productions of DCF-DA-ROS and MitoSox-mitochondrial superoxides (Figure 3f) and the levels of oxidized (carbonylated) proteins, as shown by Oxyblot (Figure 3g). We noted here that the characteristics of miR-24 cells were similar to those of shH2AX cells, a specific knockdown of H2AX, in most assayed aspects. This means that the major target of miR-24 should be H2AX.
Figure 3.
shH2AX- or miR-24-mediated H2AX knockdown leads to mitochondrial dysfunction. (a–c) Cell number (a), the LDH activity of cell lysates (b) and the pH of culture media (c) of stably Rv-shRNA-infected cells incubated for 0–72 h in complete media. (d) Oxygen consumption rate (OCR) of OXPHOS complexes of stably Rv-shRNA-infected cells that were incubated for 24 h in complete media. *P<0.05, **P<0.01 vs shSCR cells. (e) Mitochondrial activities of stably Rv-shRNA-infected cells. The cells were incubated for 24 h in complete media, and the intracellular ATP levels (ATP) and TMRE-mediated mitochondrial membrane potential (TMRE) were determined. Representative confocal images of TMRE-stained cells are shown. (f) Reactive oxygen species (ROS) generation of stably Rv-shRNA-infected cells. The cells were incubated for 24 h in complete media, and the levels of DCF-DA-mediated ROS (DCF-DA) and MitoSox-mediated mitochondrial superoxide (MitoSox) were measured. Representative confocal images of the DCF-DA-stained cells are shown. (g) OxyBlot determination of the carbonyl protein levels, reflecting the degree of total protein oxidation. Equal protein loading was confirmed using antibodies against β-actin. All data are presented as the mean±s.e. (n=5–6).
Alterations in mitochondrial gene expressions and morphology of H2AX-KD cells
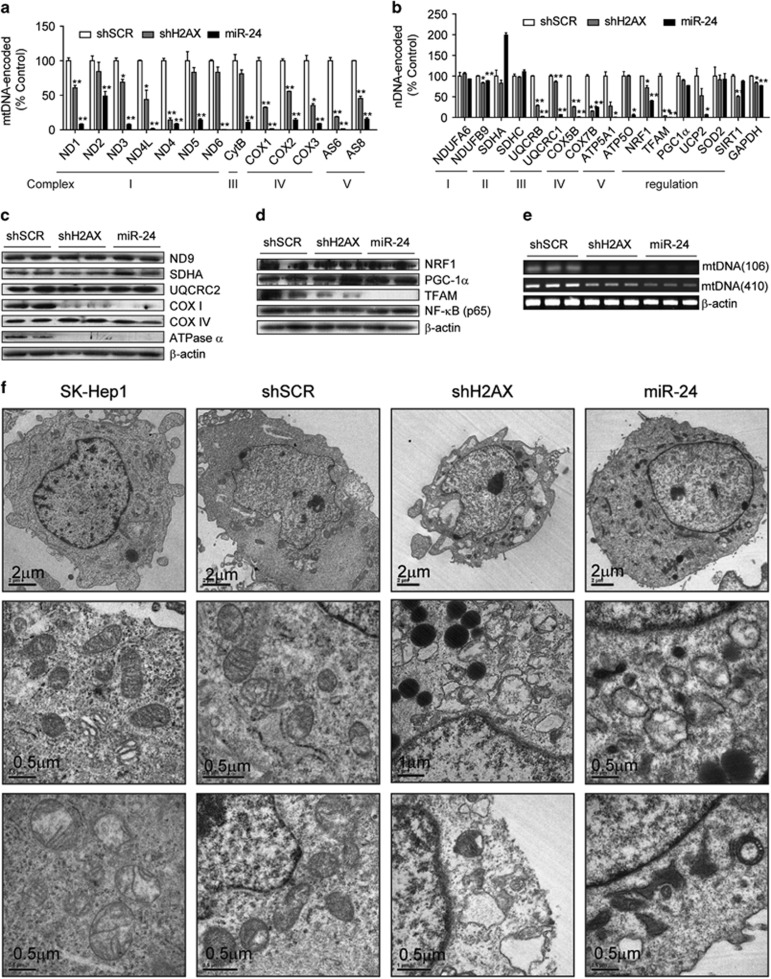
Next, we performed semi-quantitative reverse transcription-polymerase chain reaction (semi-qRT-PCR) or western blot analyses for 13 mtDNA-encoded OXPHOS genes (mtOXPHOS), 10 nDNA-encoded OXPHOS genes (nuOXPHOS) and 6 mitochondrial control genes. The miRNA miR-24 almost abolished the transcripts of all mtOXPHOS genes, and shH2AX decreased the levels of most of them (Figure 4a). However, the effects of miR-24 and shH2AX on nDNA-encoded genes were varied. They either decreased the transcripts of nuOXPHOS (UQCRB, COX5B, COX7B, ATP5A1, ATP5O) or mitochondrial control genes (NRF-1, TFAM, UCP2) or did not affect other genes (NDUFA6, NDUFB9, SDHC, PGC-1α, SOD2) (Figure 4b). Western blotting showed that miR-24 and shH2AX resulted in a specific decrease in the expression of COX I and ATPase α, but not in the expression of ND9, SDHA and COX IV (Figure 4c). Among the tested mitochondrial control proteins, only TFAM showed a severe decrease in both mRNA and protein levels (Figures 4b and d). As TFAM decreased, the mtDNA copy numbers were also reduced (Figure 4e). When the morphology and density of the mitochondria were examined using transmission electron microscopy, the mitochondria of shH2AX and miR-24 cells appeared round, swollen and less electron dense compared with control cells (Figure 4f), which is similar to other mtDNA-depleted cells.48
Figure 4.
H2AX knockdown disrupted mitochondrial mRNA/protein expression and the mitochondrial ultrastructure. (a, b) Semi-quantitative RT-PCR of mRNA in the stably Rv-shRNA-infected cells cultured for 24 h in complete media. (a) 13 mtDNA-encoded OXPHOS subunits, (b) 10 nuclear DNA-encoded OXPHOS subunits and 6 mitochondrial biogenesis control genes. All data are presented as the mean±s.e. (n=4). (c, d) Western blot analysis of the OXPHOS subunits (c) and mitochondrial biogenesis control proteins (d). (e) Quantification of mtDNA. The mtDNA regions encoding COXI (106 bp) or the D-loop region (410 bp) were PCR-amplified from genomic DNA. (f) Electron micrographs of the mitochondria. The designated stably Rv-shRNA-infected cells were fixed and examined by electron microscopy at magnifications of × 6000 (Scale bar=2 μm) and × 50,000 (Scale bar=0.5 μm).
H2AX, but not ΔC24-H2AX, rescued the mitochondrial defects
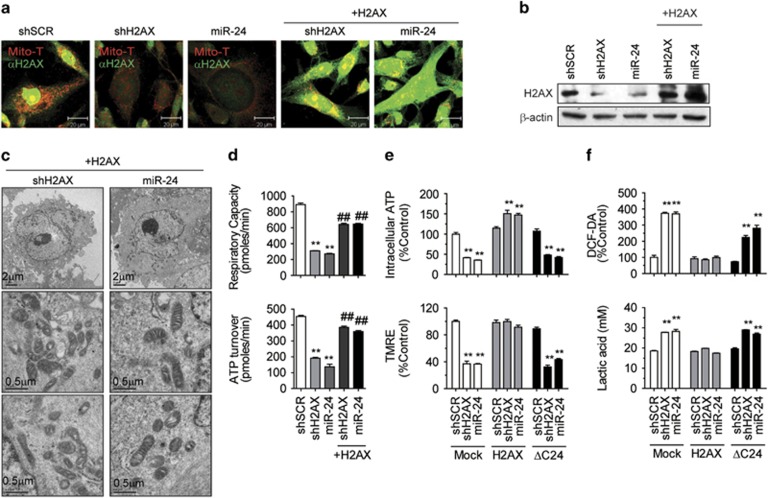
We next tested whether H2AX deficiency is solely responsible for the miR-24-induced mitochondrial damage by performing rescue experiments. We stably transfected pcDNA3.1-H2AX into shH2AX- or miR-24-cells and confirmed the successful rescue of H2AX expression (Figures 5a and b; Supplementary Figure 1a). The H2AX-rescued cells showed normal mitochondrial morphology in EM images (Figure 5c). They also had restored FCCP-induced respiratory capacity and an oligomycin-mediated ATP turnover rate at normal levels (Figure 5d).
Figure 5.
Ectopic expression of mitochondrial H2AX-rescued mitochondrial function. (a) Immunocytochemical analysis of H2AX with MitoTracker staining. The stably Rv-shRNA-infected cells expressing shSCR or shH2AX, miR-24 were transfected with pcDNA3.1-H2AX (+H2AX) and selected for using G418 (800 μg ml−1). The cells were stained with MitoTracker (Mito-T, red) and the anti-H2AX antibody (green). The two confocal images were merged. Scale bar=20 μm. (b) Western blot of H2AX-rescued cells. (c) Recovery of the mitochondrial ultrastructure in H2AX-rescued cells shown at magnifications of × 6,000 (Scale bar=2 μm) and × 50,000 (Scale bar=0.5 μm). (d) H2AX rescued the oxygen consumption rate (OCR). The total respiratory capacity (FCCP-induced OCR) and ATP turnover rate (basal OCR—oligomycin-inhibited OCR) of the mitochondria were determined using a Seahorse XF-24 analyzer. (e) Recovery of the mitochondrial activities by H2AX, but not by ΔC24-H2AX. The intracellular ATP and TMRE-mitochondrial membrane potential of stably expressing cells were compared. (f) Normalization of ROS and lactic acid by H2AX, but not by ΔC24-H2AX. The DCF-DA-mediated ROS and lactic acid content of stably expressing cells were compared. All data are presented as the mean±s.e. (n=3–6). *P<0.05, **P<0.01 vs shSCR. #P<0.05, ##P<0.01 vs stable parental H2AX-KD cells.
As H2AX does not contain a notable MTS and the C-terminus is responsible for mitochondrial transport,20 we constructed the nuclear form of H2AX instead of the mitochondrial form of H2AX. A C-terminally truncated form of H2AX (deletion of 24 amino acids from 120-143, ΔC24) was localized only in the nucleus and not in the mitochondria.20 ΔC24 was used as nuclear H2AX. The overexpression of ΔC24 failed to restore intracellular ATP content, ΔΨm (TMRE), DCF-DA-mediated ROS and lactic acids to normal levels (Figures 5e and f; Supplementary Figure 1b). These results indicated that only mitochondrial H2AX, but not nuclear H2AX, was responsible for modulating mitochondrial activities.
Deficient protein import of mitochondria isolated from H2AX-KD cells
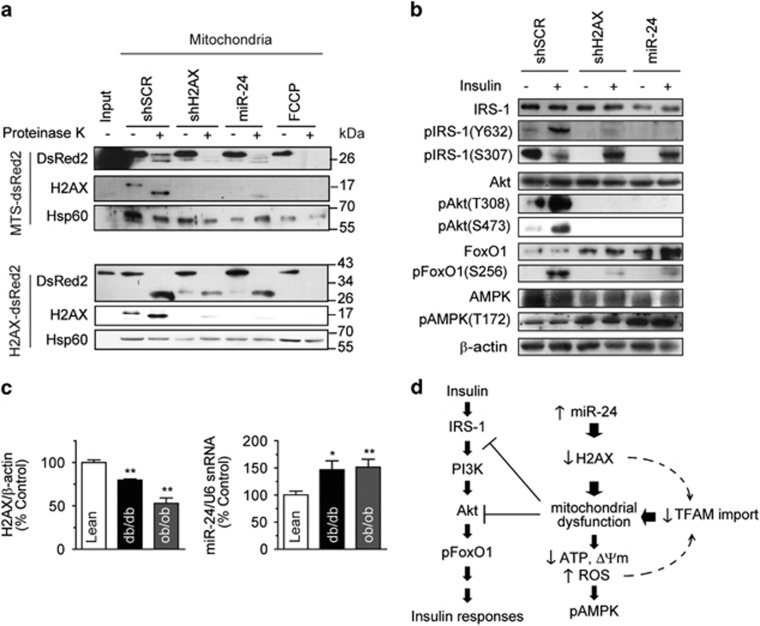
Two important issues that needed to be resolved were the mechanism by which H2AX is transported into the mitochondria, as H2AX does not contain a notable mitochondrial-targeting sequence (MTS), and how H2AX controls mitochondrial activities. Because mitochondrial H2AX (mtH2AX) forms a complex with TOM20 on the mitochondrial surface, we hypothesized that mtH2AX might be involved in the mitochondrial import of cytoplasmic precursor proteins. The conventional mitochondrial import assay was not possible because the H2AX protein does not contain the amino acids for proper labeling. Instead, we constructed two different artificial mitochondrial precursor proteins fused to the DsRed2 fluorescent protein, MTS-DsRed2 (28.5 kDa) and H2AX-DsRed2 (41 kDa). Mitochondria isolated from shSCR-, shH2AX- or miR-24-cells were incubated with the newly synthesized fusion proteins. The DsRed2 proteins that were transported into mitochondria were analyzed by immunoblotting of the re-isolated mitochondria (Figure 6a). Both MTS-DsRed2 and H2AX-DsRed2 were transported into control shSCR-mitochondria, processed and protected from proteinase K digestion. MTS-DsRed2 import into either shH2AX- or miR-24-mitochondria was completely abolished, and H2AX-DsRed2 import was reduced up to 80%. The molecular weight of proteinase K-resistant DsRed2 in H2AX-DsRed2 import was approximately 26 kDa, indicating that H2AX was removed from the fusion protein in the mitochondria. The FCCP-treated mitochondria completely lost its mitochondrial import capability. We concluded that shH2AX- and miR-24-mitochondria should be defective in the import of nuclear-encoded proteins, such as TFAM and H2AX. In addition, H2AX itself is able to translocate the DsRed2 cargo protein into the mitochondria.
Figure 6.
H2AX knockdown impaired insulin signaling and mitochondrial protein import. (a) Mitochondrial import assay. Mitochondria isolated from the stably Rv-shRNA-infected cells expressing shSCR or shH2AX, miR-24 were incubated for 30 min at 30 °C with newly synthesized MTS-DsRed2 (28.5 kDa) or H2AX-DsRed2 (41 kDa). FCCP-treated mitochondria (FCCP) of the stably Rv-shSCR-infected cells were used as the negative control. After incubation with or without proteinase K, which digests any proteins outside the mitochondria, the mitochondrial lysates were subjected to western blotting using DsRed2 antibody. ‘Input' indicates the proteins used for import produced by in vitro transcription/translation. H2AX and Hsp60 were used as mitochondrial controls. (b) Western blotting of insulin signaling molecules in the stably Rv-shRNA-infected cells with or without 100 nM insulin stimulation for 15 min. (c) Hepatic expression of H2AX and miR-24 in 15-week-old C57BL/6 lean, diabetic (db/db) or obese (ob/ob) mice. Left panel: western blot quantification of H2AX in liver lysates normalized to β-actin. Right panel: real-time qPCR of endogenous hepatic miR-24 levels normalized using U6 snRNA. Mean±s.e. (n=7). *P<0.05, **P<0.01 vs WT or lean mice. (d) Schematic model of the relationship among H2AX knockdown, the mitochondrion and the insulin signaling pathway. Mitochondrial deficiency blocks Akt and IRS-1, which may be aggravated by cellular aging caused by reduced TFAM import.
Insulin signaling pathway defects in H2AX-KD cells
To investigate whether H2AX-KD-mediated mitochondrial dysfunction altered the insulin signaling pathway, we determined the phosphorylation state of insulin signaling molecules in these cells with or without insulin stimulation. The overexpression of miR-24 or shH2AX repressed insulin-stimulated pIRS-1(Y632), pAkt(T308), pAkt(S473) and pFoxO1(S256) to similar degrees (Figure 6b). In these stably expressing cells, pAMPK(T172) was enhanced independently of insulin stimulation, implying that H2AX-KD-mediated mitochondrial dysfunction might constitutively activate AMPK. However, the AMPK activation was not enough to recover the mitochondrial damage. The results suggested that the miR-24- or shH2AX-induced mitochondrial damage may block the phosphorylation of Akt and IRS-1 in the insulin signaling pathway, similar to insulin resistance. This agrees with our previous report showing the cross-talk between IRS-1/Akt and the mitochondria.8, 42 The toxin-induced mitochondrial deficits suppressed the phosphorylation of Akt and IRS-1. The present results confirmed that IRS-1 and Akt might be the cross-talk points between insulin signaling and mitochondrial dysfunction.
Hepatic H2AX was reduced in diabetic and obese mice
To validate the roles of H2AX and miR-24 in disease models, we determined the levels of H2AX and miR-24 expression in diabetic (db/db) and obese (ob/ob) mouse livers. Although these mice are leptin receptor- or leptin-deficient genetic models, they show hepatic insulin resistance and mitochondrial dysfunction.49, 50, 51, 52 Western blots of H2AX revealed that the H2AX protein levels were decreased, and the real-time qPCR of miR-24 showed the miR-24 levels were increased in these mice (Figure 6c).
Considering our data, we thought that the enhanced miR-24 expression may suppress H2AX, leading to mitochondrial import deficiency. Blocking the transport of mitochondrial proteins into the mitochondria, including TFAM, may aggravate mitochondrial damage, resulting in the inactivation of insulin signaling at Akt and IRS-1. The possible connections among the proteins are summarized in Figure 6d.
Discussion
The present study demonstrated novel functions of mitochondrial H2AX and its post-transcriptional regulator miR-24 in the regulation of mitochondrial activity. Specifically, mitochondrial H2AX is involved in the import of precursor proteins into the mitochondria. The increased expression of miR-24 eliminated mitochondrial H2AX, leading to defects in mitochondrial protein import. Insufficient TFAM transport subsequently induced mitochondrial damage in a feedback loop, as depicted in Figure 6d.
The term ‘mitochondrial H2AX (mtH2AX)' may be confusing because mitochondria have been previously thought to be histone-free organelles. As mtH2AX does not bind to mtDNA in the mitochondrial matrix,53 the biological role of mtH2AX was considered to be distinct from the DNA repair activity of H2AX in the nucleus. Instead, mtH2AX binds to TOM20, a component of the mitochondrial import machinery, and has a critical role in the post-translational transport of mitochondrial precursor proteins into the mitochondria (Figure 6a). Our results demonstrated that both shH2AX and miR-24 primarily suppressed mtH2AX and inhibited the mitochondrial transport of the cargo protein DsRed2 ligated to MTS or H2AX in vitro (Figure 6a). Among the nDNA-encoded mitochondrial proteins tested, the most affected were TFAM and ATPase α (Figure 4). It is likely that the downregulation of TFAM in mtH2AX KD cells might contribute to the reduced mtDNA contents and the levels of 13 mtDNA-encoded OXPHOS mRNAs. The expression level of PGC-1α, a mitochondrial biogenesis activator, was not significantly affected because it controls mitochondrial proteins in the nucleus. We suspect that miR-24-induced mitochondrial damage might be initially mild but becomes exacerbated with time. This might be why miR-24 and shH2AX cells shared many characteristics, such as a rapid acidification of culture media, growth retardation, decreased expression of mtDNA-encoded genes, repressed mitochondrial activities and swollen mitochondria, with mtDNA-depleted ρ0 cells.18 We believe that the primary target of miR-24 is mtH2AX and that the chronic action of miR-24 leads to progressively severe mitochondrial abnormalities that are associated with disease pathogenesis.
It has been reported that miR-24 upregulation in terminally differentiated hematopoietic cell lines decreases H2AX mRNA and protein levels, leading to the suppression of DNA repair induced by γ-irradiation or genotoxic drugs.54 The overexpression of a miR-24 mimic results in the downregulation of 248 genes involved in DNA repair and G1 arrest in cell cycle control.55 Our study also demonstrated that miR-24 and shH2AX retarded the proliferation rates of the cells (Figure 3a). The mitochondrial dysfunction of H2AX-KD cells might be another contributing factor for growth retardation. It would be interesting to test if miR-24 cells might be defective in proliferation and DNA repair if they encounter genotoxic stimuli compared with control cells.
Several publications have suggested that stress-induced miR-24 may provoke diseases such as cancer56 and cardiac hypertrophy.38, 57, 58 The results of the current study add insulin resistance to the list of miR-24-related diseases. It has been reported that hepatic mitochondrial dysfunction precedes insulin resistance and hepatic steatosis.59 To better understand the interactions among H2AX, miR-24 and mitochondrial pathogenesis in disease models, the levels of miR-24 and H2AX were determined in genetically modified mice (ob/ob and db/db). We demonstrated a 1.5-fold increase in miR-24 and a 20–50% decrease in H2AX in the liver of ob/ob or db/db mice (Figure 6c). Jordan et al. reported that the expression of miR-143 was upregulated 2-fold in the liver of db/db, ob/ob, or high-fat diet-induced obese mice compared with control mice.60 They found that miR-143 downregulated oxysterol-binding protein related protein 8 (ORP8) and inhibited insulin-stimulated Akt activation and glucose metabolism. We previously reported that Akt inhibition caused the inactivation of mitochondrial function and vice versa.8 Therefore, if obesity-induced miR-143 overexpression initiates a blockage in the insulin signaling pathway through Akt, subsequent mitochondrial damage may induce the overproduction of miR-24. This assumption is derived from the observation that miR-24 is upregulated by exogenous stimuli, such as phorbol ester, ROS (H2O2) or hemin.54, 55 Various sources of mitochondrial damage ranging from DNA mutations to environmental toxins block electron transfer through OXPHOS complexes and induce profound ROS generation. It is reasonable to suggest that mitochondrial damage-induced ROS leads to miR-24 upregulation. A balance may be present among miR-24, H2AX, mitochondria, insulin signaling and obesity that is interconnected. Thus, if one of these factors is disturbed, it will cause an imbalance in the whole cycle, which will become progressively aggravated in the absence of intervention.
Multiple studies have shown that deficiencies of mtDNA or TFAM in various tissues can cause a wide range of diseases, depending on tissue type. However, it is not understood why mitochondrial activities are decreased in patients with metabolic diseases and in normal senescent subjects. Our data imply that miR-24 and mtH2AX may be the missing link between mitochondrial dysfunction and insulin resistance. The reduction of mitochondrial function through the upregulation of miR-24 and a decrease in H2AX might result in the development of metabolic disorders in senescent subjects. Collectively, we hypothesize that miR-24 and H2AX might be novel therapeutic targets for age-related diseases associated with mitochondrial dysfunction.
Acknowledgments
We thank Tong-Kon Yu (University of Ulsan, Korea) for excellent technical assistance. This study was supported by a grant (HI14C2700) from the Korean Health Technology R&D Project, Ministry of Health & Welfare and partly by a grant (10051960) from the Technology Innovation Program from the Ministry of Trade, Industry & Energy of Korea.
Author contributions
JHJ, YCK, YP and SK planned and performed the experiments, and analyzed the data. YKP supervised the project, designed the experiments and wrote the manuscript with comments from the coauthors. All authors collaborated on the work.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
The authors declare no conflict of interest.
Supplementary Material
References
- Park KS, Nam KJ, Kim JW, Lee YB, Han CY, Jeong JK et al. Depletion of mitochondrial DNA alters glucose metabolism in SK-Hep1 cells. Am J Physiol Endocrinol Metab 2001; 280: E1007–E1014. [DOI] [PubMed] [Google Scholar]
- Song J, Oh JY, Sung YA, Pak YK, Park KS, Lee HK. Peripheral blood mitochondrial DNA content is related to insulin sensitivity in offspring of type 2 diabetic patients. Diabetes Care 2001; 24: 865–869. [DOI] [PubMed] [Google Scholar]
- Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 2003; 300: 1140–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Park KS, Cho YM, Lee YY, Pak YK. Mitochondria-based model for fetal origin of adult disease and insulin resistance. Ann N Y Acad Sci 2005; 1042: 1–18. [DOI] [PubMed] [Google Scholar]
- Lee YY, Park KS, Pak YK, Lee HK. The role of mitochondrial DNA in the development of type 2 diabetes caused by fetal malnutrition. J Nutr Biochem 2005; 16: 195–204. [DOI] [PubMed] [Google Scholar]
- Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science 2005; 307: 384–387. [DOI] [PubMed] [Google Scholar]
- Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ Res 2008; 102: 401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SY, Choi YS, Koo HJ, Jeong JH, Park WH, Kim M et al. Mitochondrial dysfunction enhances the migration of vascular smooth muscles cells via suppression of Akt phosphorylation. Biochim Biophys Acta 2010; 1800: 275–281. [DOI] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med 2004; 350: 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging and development. Cell 2006; 125: 1241–1252. [DOI] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE et al. Mitochondrial DNA mutations, oxidative stress and apoptosis in mammalian aging. Science 2005; 309: 481–484. [DOI] [PubMed] [Google Scholar]
- Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science 1999; 286: 774–779. [DOI] [PubMed] [Google Scholar]
- Cardellach F, Galofre J, Cusso R, Urbano-Marquez A. Decline in skeletal muscle mitochondrial respiration chain function with ageing. Lancet 1989; 2: 44–45. [DOI] [PubMed] [Google Scholar]
- Koo HJ, Piao Y, Pak YK. Endoplasmic reticulum stress impairs insulin signaling through mitochondrial damage in SH-SY5Y cells. Neurosignals 2012; 20: 265–280. [DOI] [PubMed] [Google Scholar]
- Wagner BK, Kitami T, Gilbert TJ, Peck D, Ramanathan A, Schreiber SL et al. Large-scale chemical dissection of mitochondrial function. Nat Biotechnol 2008; 26: 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Ahn SY, Song IC, Chung MH, Jang HC, Park KS et al. Chronic exposure to the herbicide, atrazine, causes mitochondrial dysfunction and insulin resistance. PLoS ONE 2009; 4: e5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino K, Petersen KF, Dufour S, Befroy D, Frattini J, Shatzkes N et al. Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J Clin Invest 2005; 115: 3587–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J, Jeong JH, Baek JH, Koo HJ, Park WH, Yang JS et al. Network clustering revealed the systemic alterations of mitochondrial protein expression. PLoS Comput Biol 2011; 7: e1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S et al. Characterization of the human heart mitochondrial proteome. Nat Biotechnol 2003; 21: 281–286. [DOI] [PubMed] [Google Scholar]
- Choi YS, Jeong JH, Min HK, Jung HJ, Hwang D, Lee SW et al. Shot-gun proteomic analysis of mitochondrial D-loop DNA binding proteins: identification of mitochondrial histones. Mol Biosyst 2011; 7: 1523–1536. [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 2005; 123: 1213–1226. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273: 5858–5868. [DOI] [PubMed] [Google Scholar]
- Fillingham J, Keogh MC, Krogan NJ. GammaH2AX and its role in DNA double-strand break repair. Biochem Cell Biol 2006; 84: 568–577. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci USA 2002; 99: 8173–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA et al. Genomic instability in mice lacking histone H2AX. Science 2002; 296: 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijima Y, Sakasai R, Okita N, Asahina K, Mizutani S, Teraoka H. Phosphorylation of histone H2AX at M phase in human cells without DNA damage response. Biochem Biophys Res Commun 2005; 336: 807–812. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 2003; 115: 787–798. [DOI] [PubMed] [Google Scholar]
- Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev 2005; 15: 410–415. [DOI] [PubMed] [Google Scholar]
- Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev 2005; 15: 563–568. [DOI] [PubMed] [Google Scholar]
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development 2005; 132: 4653–4662. [DOI] [PubMed] [Google Scholar]
- Caruso P, Dempsie Y, Stevens HC, McDonald RA, Long L, Lu R et al. A role for miR-145 in pulmonary arterial hypertension: evidence from mouse models and patient samples. Circ Res 2012; 111: 290–300. [DOI] [PubMed] [Google Scholar]
- Aurora AB, Mahmoud AI, Luo X, Johnson BA, van Rooij E, Matsuzaki S et al. MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca(2)(+) overload and cell death. J Clin Invest 2012; 122: 1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM et al. Inhibition of miR-15 protects against cardiac ischemic injury. Circ Res 2012; 110: 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X et al. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell 2012; 149: 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation 2011; 124: 1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010; 107: 810–817. [DOI] [PubMed] [Google Scholar]
- Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest 2010; 120: 3912–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA 2006; 103: 18255–18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 2004; 279: 52361–52365. [DOI] [PubMed] [Google Scholar]
- Gauthier BR, Wollheim CB. MicroRNAs: ‘ribo-regulators' of glucose homeostasis. Nat Med 2006; 12: 36–38. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 2007; 27: 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao Y, Kim HG, Oh MS, Pak YK. Overexpression of TFAM, NRF-1 and myr-AKT protects the MPP(+)-induced mitochondrial dysfunctions in neuronal cells. Biochim Biophys Acta 2012; 1820: 577–585. [DOI] [PubMed] [Google Scholar]
- Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol 2009; 21: 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak YK, Weiner H. Import of chemically synthesized signal peptides into rat liver mitochondria. J Biol Chem 1990; 265: 14298–14307. [PubMed] [Google Scholar]
- Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E et al. Modulation of K-Ras-dependent lung tumorigenesis by microRNA-21. Cancer Cell 2010; 18: 282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res 2008; 103: 919–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Liu N, Olson EN. MicroRNAs flex their muscles. Trends Genet 2008; 24: 159–166. [DOI] [PubMed] [Google Scholar]
- Challagundla KB, Sun XX, Zhang X, DeVine T, Zhang Q, Sears RC et al. Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol Cell Biol 2011; 31: 4007–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RC, Castellani LW, Hosseini M, Ben-Zeev O, Mao HZ, Weinstein MM et al. Early hepatic insulin resistance precedes the onset of diabetes in obese C57BLKS-db/db mice. Diabetes 2010; 59: 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentin R, Benhamed F, Hainault I, Fauveau V, Foufelle F, Dyck JR et al. Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes 2006; 55: 2159–2170. [DOI] [PubMed] [Google Scholar]
- Garcia-Ruiz I, Rodriguez-Juan C, Diaz-Sanjuan T, Martinez MA, Munoz-Yague T, Solis-Herruzo JA. Effects of rosiglitazone on the liver histology and mitochondrial function in ob/ob mice. Hepatology 2007; 46: 414–423. [DOI] [PubMed] [Google Scholar]
- Shao J, Yamashita H, Qiao L, Friedman JE. Decreased Akt kinase activity and insulin resistance in C57BL/KsJ-Leprdb/db mice. J Endocrinol 2000; 167: 107–115. [DOI] [PubMed] [Google Scholar]
- Choi YS, Hoon Jeong J, Min HK, Jung HJ, Hwang D, Lee SW et al. Shot-gun proteomic analysis of mitochondrial D-loop DNA binding proteins: identification of mitochondrial histones. Mol Biosyst 2011; 7: 1523–1536. [DOI] [PubMed] [Google Scholar]
- Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol 2009; 16: 492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to ‘seedless' 3' UTR microRNA recognition elements. Mol Cell 2009; 35: 610–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang T, Wakita T, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology 2010; 398: 57–67. [DOI] [PubMed] [Google Scholar]
- Divakaran V, Mann DL. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ Res 2008; 103: 1072–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res 2007; 100: 416–424. [DOI] [PubMed] [Google Scholar]
- Rector RS, Thyfault JP, Uptergrove GM, Morris EM, Naples SP, Borengasser SJ et al. Mitochondrial dysfunction precedes insulin resistance and hepatic steatosis and contributes to the natural history of non-alcoholic fatty liver disease in an obese rodent model. J Hepatol 2010; 52: 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan SD, Kruger M, Willmes DM, Redemann N, Wunderlich FT, Bronneke HS et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol 2011; 13: 434–446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.