Abstract
Chronic obstructive pulmonary disease (COPD) is a chronic inflammatory disease, and bacterial infection plays a role in its pathogenesis. Bacteria secrete nanometer-sized extracellular vesicles (EVs), which may induce more immune dysfunction and inflammation than the bacteria themselves. We hypothesized that the microbiome of lung EVs might have distinct characteristics depending on the presence of COPD and smoking status. We analyzed and compared the microbiomes of 13 nonsmokers with normal spirometry, 13 smokers with normal spirometry (healthy smokers) and 13 patients with COPD by using 16S ribosomal RNA gene sequencing of surgical lung tissue and lung EVs. Subjects were matched for age and sex in all groups and for smoking levels in the COPD and healthy smoker groups. Each group included 12 men and 1 woman with the same mean age of 65.5 years. In all groups, EVs consistently showed more operational taxonomic units (OTUs) than lung tissue. In the healthy smoker and COPD groups, EVs had a higher Shannon index and a lower Simpson index than lung tissue and this trend was more prominent in the COPD group. Principal component analysis (PCA) showed clusters based on sample type rather than participants' clinical characteristics. Stenotrophomonas, Propionibacterium and Alicyclobacillus were the most commonly found genera. Firmicutes were highly present in the EVs of the COPD group compared with other samples or groups. Our analysis of the lung microbiome revealed that the bacterial communities present in the EVs and in the COPD group possessed distinct characteristics with differences in the OTUs, diversity indexes and PCA clustering.
Introduction
Chronic obstructive pulmonary disease (COPD) is the third leading cause of death worldwide.1 Among several risk factors for COPD, cigarette smoking is the main factor. Smokers have a higher prevalence of lung function abnormalities and a greater COPD-related mortality rate than nonsmokers.2 However, smoking does not explain all of the aspects of this condition, because COPD can develop in nonsmokers, and more than half of smokers do not have COPD.3 Patients with COPD exhibit persistent inflammation despite smoking cessation.4 Similarly, accelerated loss of lung function may occur independently of smoking.5 Therefore, it is crucial to elucidate causative factors besides smoking to better understand and manage COPD.
COPD is characterized by chronic inflammation of the small airways. Repeated airway infection and hypersecretion of mucus have traditionally been thought to be the main features of COPD. In addition, respiratory tract infection is an important cause of acute exacerbation and progression of this disease.6 Exposure to infectious insults can trigger an immune reaction in the airway.7 However, it is difficult to characterize the bacterial community by using conventional culturing methods. Historically, the healthy lung has been presumed to be sterile; however, new observations have challenged this dogma.8 Novel techniques in culture-independent microbial identification have enabled the detection of complex microbial communities and have led to an increased understanding of their roles in various diseases.9, 10, 11 Pyrosequencing of 16S ribosomal RNA (rRNA) gene amplicons from bronchoalveolar lavage specimens, bronchial brushings and lung tissues from healthy individuals has revealed that various microorganisms exist in the healthy lower respiratory tract.12, 13, 14, 15
Bacteria secrete spherical shaped vesicles that are 20–200 nm in size and surrounded by a lipid bilayer into the extracellular milieu. These extracellular vesicles (EVs) can transmit virulence factors into host cells and modulate host defense and immune responses.16 A previous study has reported that EVs secreted by bacteria found in indoor dust contain LPS and can induce neutrophilic inflammation in the lungs of mice. That study has also suggested that patients with chronic respiratory diseases are highly sensitized to environmental EVs.17 To elucidate the relationship between bacterial inflammation and COPD, the contribution of the bacterial community present in the EVs should be considered.
We hypothesized that lung EVs might display specific microbiome characteristics in COPD. To test this possibility, we compared the microbiome data from three completely age- and sex-matched groups of nonsmokers, healthy smokers and COPD patients. For healthy smokers and COPD patients, the smoking amounts were also matched.
Materials and methods
Subjects and study design
Non-neoplastic lung tissues were obtained from 39 patients who underwent surgical resection for lung cancer. Shortly after lung resection, tissue blocks were placed into sterilized cryovials, and kept frozen in liquid nitrogen before analysis. This study was approved by the Institutional Review Board of Asan Medical Center (IRB 2013-0700), and all participants provided informed consent. The subjects were divided into three groups: nonsmokers (n=13), healthy smokers (n=13) and COPD patients (n=13). The nonsmoker group included subjects with no history of smoking in their lifetime and normal lung function. The healthy smoker group included individuals who had normal lung function and a history of smoking of more than 10 pack-years, and who had quit smoking <3 months prior to operation. COPD was defined as a ratio of <0.7 between forced expiratory volume in 1 s (FEV1) per forced vital capacity (FVC). The COPD group included individuals with the same smoking criteria as in the healthy smoker group. Nonsmokers, healthy smokers, and COPD patients were matched for age and sex. In addition, healthy smokers and COPD patients were matched for smoking intensity (pack-year). Sterile water served as the negative control. We analyzed the lung microbiome using 16S rRNA gene sequencing, from the lung tissue itself and from EVs derived from the lung tissue.
Extraction and characterization of EVs
EVs in the lung tissue were extracted by ultracentrifugation at 150 000 g for 3 h at 4 °C as previously described.18 Lung EVs were characterized by transmission electron microscopy, nonparticle tracking analysis and reverse transcriptase-polymerase chain reaction (RT-PCR).
DNA extraction and sequencing
DNA was extracted using a stool PowerWater DNA Isolation Kit (MO Bio, Carlsbad, CA, USA), and 20 ng aliquots of each DNA sample were then used in a 50 μl PCR reaction. The 16S universal primers 27F (5′-GAGTTTGATCMTGGCTCAG-3′), 518R (5′-WTTACCGCGGCTGCTGG-3′) were used for amplification of 16S rRNA genes with a FastStart High Fidelity PCR System (Roche, Mannheim, Germany). After amplification, sequencing was performed on a Genome Sequencer FLX plus (454 Life Sciences, Bradford, PA, USA) by Macrogen (Seoul, Korea).
Selection of 16S rRNA genes and OTU analysis
Through BLAST searches, all of the sequence reads were compared against the Silva rRNA database. Taxonomic assignment of these sequences was carried out using the rank of NCBI Taxonomy databases with an operational taxonomic unit (OTU) cutoff of 3%.
CD-HIT-OTU and Mothur software were used for clustering analysis19 and analyzing microbial communities.20
Biodiversity and community similarity analyses
Comparisons at the OTU level were performed between lung tissue and EVs from each individual and between lung tissue and EVs in nonsmokers vs healthy smokers vs COPD patients.21 The Shannon diversity index and the Simpson index were used for assessing species diversity.22 The beta diversity index was defined as the difference between the total number of species in two groups and the number of species common to both.23 Similarities between the microbial communities of the different groups were visualized using a dendrogram based on Yue and Clayton dissimilarity metrics. Principal component analysis (PCA) was used to identify which variables determined the difference among the six groups.
Statistical analysis
Statistical comparisons were performed using a nonparametric analysis of variance, and we corrected for multiple comparisons by using a step-down Tukey's correction. For the calculations, the R package (version 3.0.1) was used and P-values <0.05 were considered to indicate statistical significance. (Detailed methods are included in the Supplementary Information)
Results
Subjects
The clinical characteristics of the 39 patients enrolled in this study are presented in Table 1. These groups comprised age- and sex-matched nonsmokers, healthy smokers and COPD patients with moderate (n=11) and severe (n=2) disease. Seventy-eight samples were collected from 39 lung tissue samples and 39 lung EV samples of the study population. The mean age was 65.5 years (range, 53–76 years), and one woman was included in each of the three groups (Supplementary Table 1). The mean smoking level in the healthy smokers and COPD patients was 46.9±17.0 pack-years.
Table 1. Demographic and clinical characteristics of the enrolled subjects.
| Nonsmokers (n=13) | Healthy smokers (n=13) | COPD patients (n=13) | Total | |
|---|---|---|---|---|
| Age (years) | 65.5±7.8 | 65.5±7.8 | 65.5±7.8 | 65.5±7.6 |
| Sex (M:F) | 12:1 | 12:1 | 12:1 | 36:3 |
| Smoking (pack-year) | 0 | 50.0±16.8 | 43.8±17.3 | 46.9±17.0 |
| FEV1 (% of predicted) | 103.5±12.4 | 92.8±16.2 | 62.5±12.2a | 86.3±22.0 |
| FEV1 (l) | 3.0±0.6 | 2.7±0.4 | 1.9±0.1a | 2.5±0.7 |
| FVC (% of predicted) | 95.6±12.7b | 86.1±12.1 | 84.3±11.5b | 88.7±12.8 |
| FVC (l) | 3.8±0.8 | 3.5±0.6 | 3.5±0.7 | 3.6±0.7 |
| FEV1/FVC | 0.79±0.05 | 0.78±0.06 | 0.56±0.11a | 0.71±0.13 |
| DLCO (ml mmHg−1 min−1) | 21.3±5.2 | 18.4±5.3 | 12.8±3.1a | 17.3±5.7 |
| DLCO (% of predicted) | 105.5±22.5 | 91.6±22.1 | 61.5±9.5a | 84.9±26.1 |
| DLCO/VA (% of predicted) | 103.4±15.7 | 103.8±25.0 | 77.2±20.5a | 94.1±24.0 |
Abbreviations: COPD, chronic obstructive pulmonary disease; DLCO, diffusing capacity of the lung for carbon monoxide; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; VA, alveolar volume.
P<0.05 nonsmoker and healthy smoker vs COPD.
P<0.05 nonsmoker vs COPD.
Sample preparation and OTU analysis
Lung EVs were isolated from harvested lung tissues by using differential centrifugation. The presence of EVs was confirmed by electron microscopy (Supplementary Figure 1). After sequencing, 128 696 length trim reads, 316 694 chimeric reads and 66 632 ambiguous reads were removed.
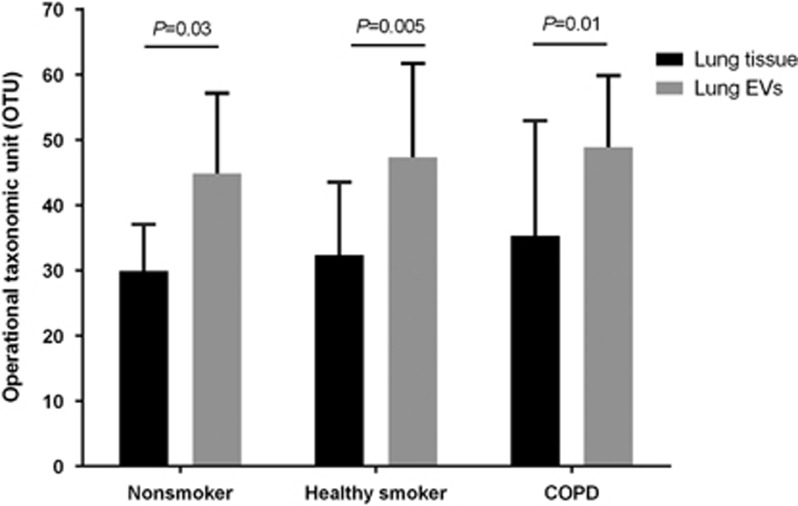
Out of the remaining sequences, 649 551 sequences were assigned to OTUs. Rarefaction curves at the genus level showed saturation, and further analysis for samples with unclear saturation did not provide additional OTUs (Supplementary Figure 2). A mean of 8328 sequences of each 16S rRNA gene were obtained from the 39 lung tissue and the 39 lung EV samples. When sequences were clustered at a 97% sequence identity, a total diversity indicated 468 species (Supplementary Table 2). OTU analysis showed significantly higher diversity in the lung EVs than in the lung tissue (46.7±13.6 vs 31.9±13.9, P<0.001). This trend was consistent across all groups (Figure 1).
Figure 1.
Quantitative comparison of OTUs between lung tissue and lung EVs. Lung EVs showed a significantly higher number of OTUs than lung tissue in all three groups. EV, extracellular vesicle; OTU, operational taxonomic unit.
Biodiversity
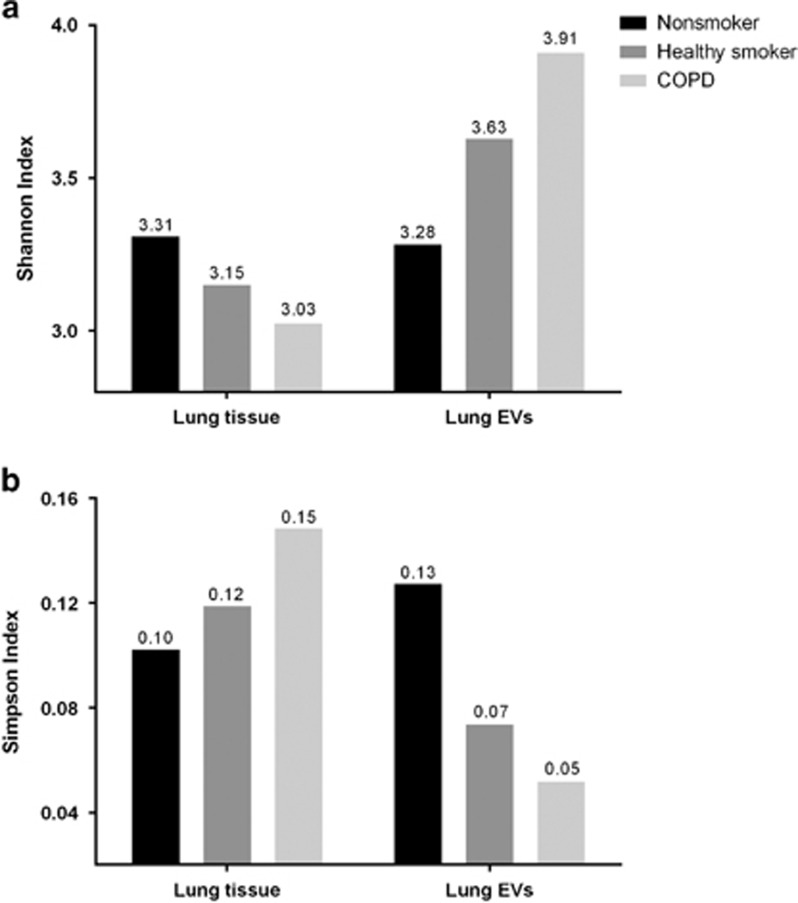
The Shannon diversity index for lung tissue demonstrated that the nonsmoker group (3.31) was the most diverse, whereas the COPD group (3.03) was the least diverse. In contrast, the Shannon diversity index for lung EVs showed that the nonsmoker group (3.28) was the least diverse, and the COPD group was the most diverse (3.91). The Simpson index indicated that the nonsmoker group had the highest evenness in the lung tissue, whereas the COPD group had the highest evenness in lung EVs. The difference in diversity indices between lung tissue and lung EVs was most prominent in the COPD group, followed by the healthy smoker and nonsmoker groups (Figure 2).
Figure 2.
Shannon and Simpson indices. (a) The Shannon index for lung tissue was highest in the nonsmoker group and lowest in the COPD group, thus indicating that the number of organisms was more heterogeneous in the nonsmoker group. The Shannon index for lung EVs showed opposite trends. (b) The Simpson index for lung tissue was highest in the COPD group and lowest in the nonsmoker group, thus indicating that there were some dominant organisms in the COPD group. The Simpson index for lung EVs showed opposite trends. COPD, chronic obstructive pulmonary disease; EV, extracellular vesicle.
The mean beta diversities for OTUs between lung tissue and lung EVs from each patient were 42.2±10.7 in the nonsmoker group, 45.9±12.8 in the healthy smoker group, and 47.4±12.4 in the COPD group. The COPD group showed the highest beta diversity between lung tissue and lung EVs (168 in nonsmokers, 161 in healthy smokers and 208 in the COPD group). The beta diversity indices were greater if both the groups and the sample types were different. The beta diversity percentages, which indicated a different proportion between two sample groups, were more than 50% except in one instance (Table 2).
Table 2. Beta diversities of the six study groups.
|
Nonsmokers |
Healthy smokers |
COPD patients |
||||
|---|---|---|---|---|---|---|
| Lung tissue | Lung EVs | Lung tissue | Lung EVs | Lung tissue | Lung EVs | |
| Lung tissue | ||||||
| Nonsmokers | 0 (0) | 168 (61) | 139 (57) | 173 (60) | 157 (61) | 190 (63) |
| Healthy smokers | 136 (57) | 150 (55) | 0 (0) | 161 (57) | 165 (63) | 174 (58) |
| COPD patients | 157 (61) | 197 (65) | 165 (63) | 212 (67) | 0 (0) | 207 (64) |
| Lung EVs | ||||||
| Nonsmokers | 168 (61) | 0 (0) | 150 (55) | 153 (51) | 197 (65) | 148 (48) |
| Healthy smokers | 173 (60) | 153 (51) | 161 (57) | 0 (0) | 212 (67) | 161 (50) |
| COPD patients | 190 (63) | 148 (48) | 174 (58) | 161 (50) | 207 (64) | 0 (0) |
Abbreviations: COPD, chronic obstructive pulmonary disease; EV, extracellular vesicle; OTU, operational taxonomic unit.
Presented as the number of unique OTUs (%).
Cluster analysis of the bacterial communities
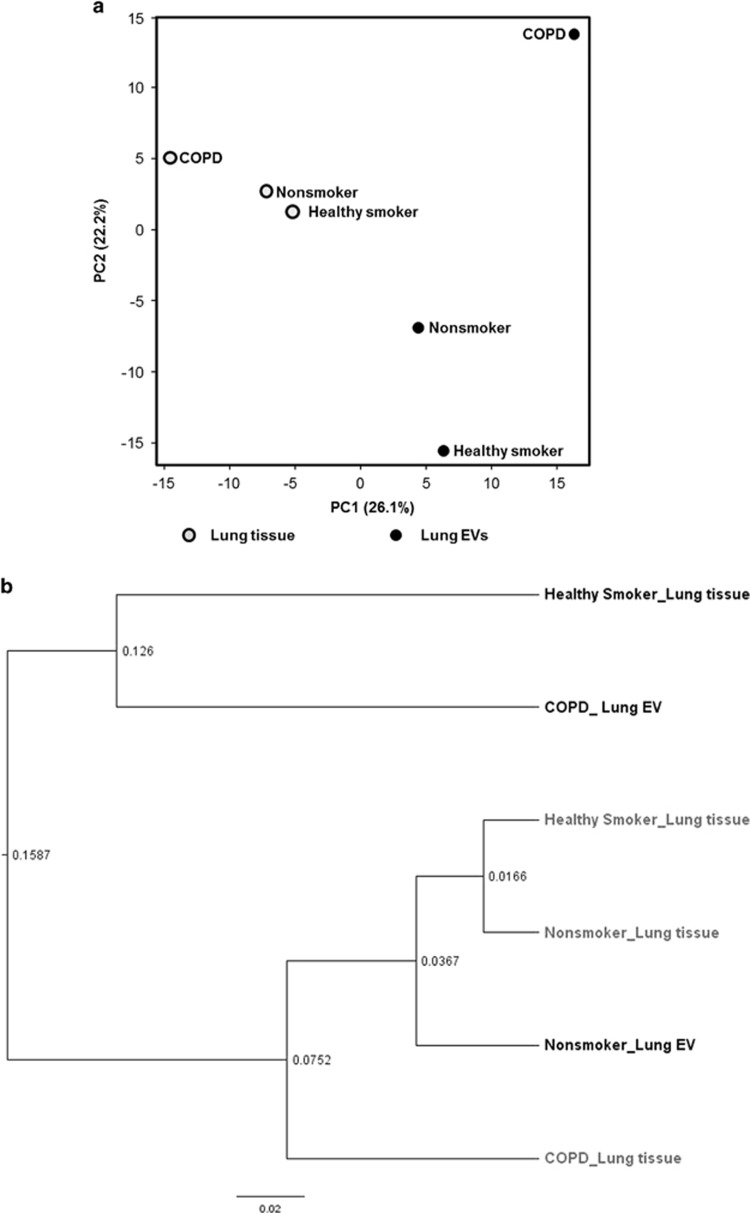
PCA was performed for all 16S rRNA gene reads clustered at a 97% similarity. The lung tissues of the three groups clustered, whereas the lung EVs were relatively more distant among groups and compared with the respective lung tissue. Among the EV samples, the nonsmoker samples showed greater similarity than the COPD samples. Cluster analysis indicated that the bacterial communities in the lung tissue and the lung EVs showed the greatest similarity in the nonsmoker group (Figure 3 and Supplementary Figure 3).
Figure 3.
Cluster analyses of the bacterial communities. (a) Principal component analysis with two components of PC1 26.1% and PC2 22.2% based on the composition and abundance of their bacterial communities. The lung EV groups were more distant not only from each other but also from the lung tissue from which they were derived. The groups were clustered relatively closely when lung tissue was analyzed. (b) Among the lung EV samples, the characteristics of nonsmokers were relatively closer to those of the lung tissues of the three groups. EV, extracellular vesicle; PC, principal component.
Characteristics of bacterial communities
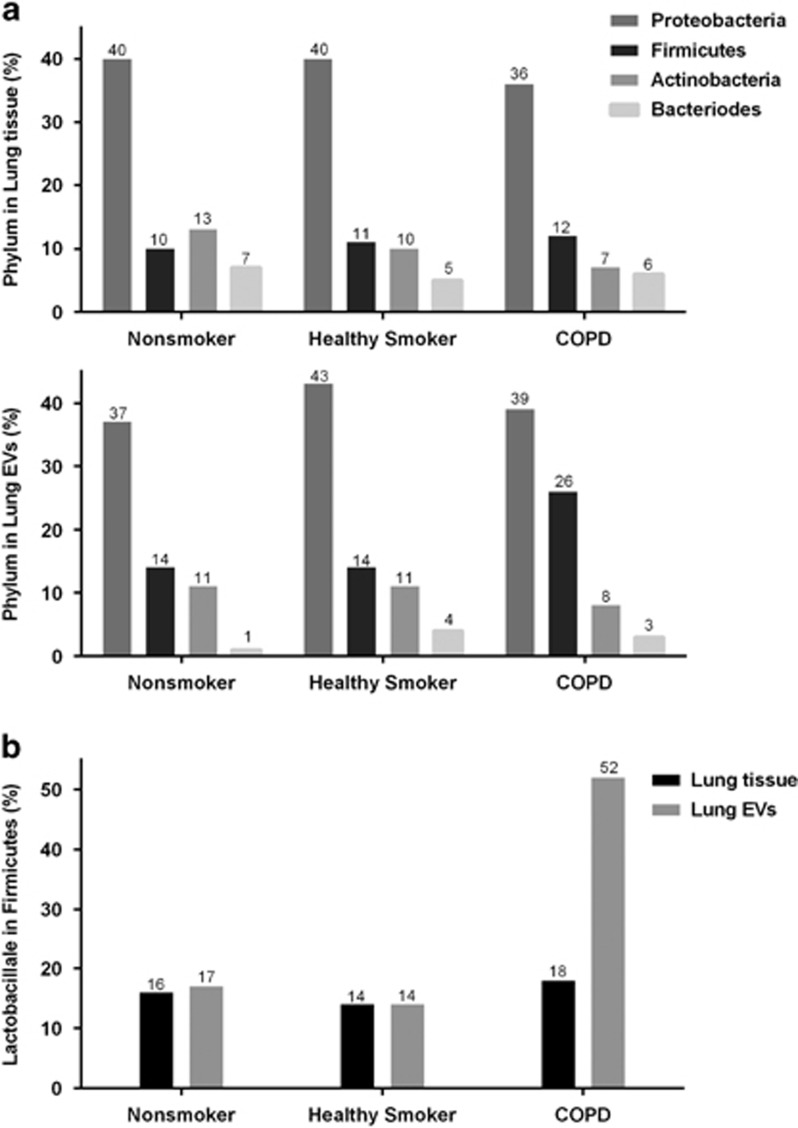
At the phylum level, the four most common phyla in all the samples were Proteobacteria, Firmicutes, Actinobacteria and Bacteroides, which together constituted 63–76% of the total number of organisms. Firmicutes showed a higher proportion in lung EVs of the COPD group. The proportion of Firmicutes in the COPD lung EVs was significantly higher than in the healthy smoker lung EVs (27.0% vs 13.4%, P=0.037, one-way analysis of variance and Tukey's post hoc analysis) or in the COPD lung tissue samples (27.0% vs 11.3%, P=0.026, paired t-test). The order Lactobacillales of phylum Firmicutes showed the greatest difference in prevalence between the COPD group and the other groups (Figure 4).
Figure 4.
The proportion of phyla and order Lactobacillales in Firmicutes. (a) The most common phyla in the three groups were Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes, which together constituted 95–99% of all organisms classified. Although the distributions were similar in all groups and samples, the prevalence of Firmicutes was higher in the COPD lung EVs. (b) The order Lactobacillales in the phylum Firmicutes showed a marked difference in prevalence between the COPD group and other groups and among sample types within the COPD group. COPD, chronic obstructive pulmonary disease; EV, extracellular vesicle.
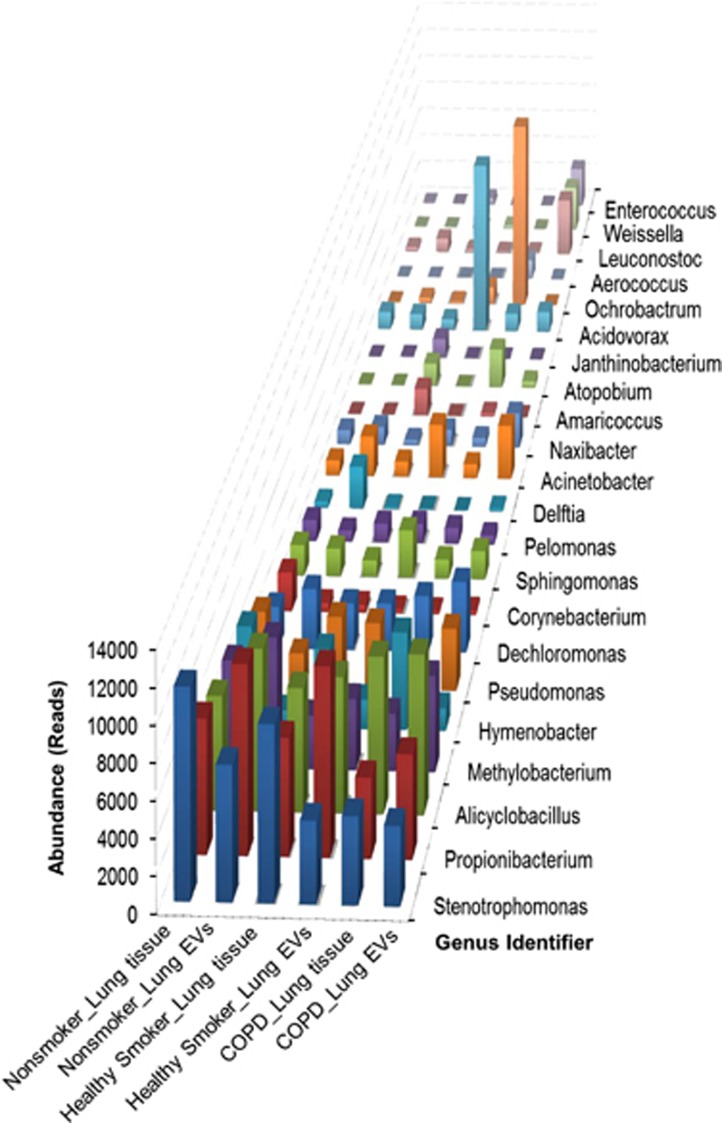
The same genera were found to be predominantly present in all the samples. Stenotrophomonas, Propionibacterium, Alicyclobacillus, Methylobacterium, Pseudomonas and Dechloromonas were found to be among the most common 10 genera in all the groups. Meanwhile, Ochrobactrum was found in the COPD lung tissue only (Figure 5). A Venn diagram for all the genera was created to illustrate the similarities between each group (Supplementary Figure 4), and a heat map showed the composition of the bacterial communities of each group (Supplementary Figure 5).
Figure 5.
Taxonomic assignment. Common genera were found in all groups; Stenotrophomonas, Propionibacterium, Alicyclobacillus, Methylobacterium, Pseudomonas and Dechloromonas were observed to be among the 10 most common genera.
Discussion
We present novel approaches for assessing the microbiome in the lung tissue of COPD patients. The bacterial communities derived from EVs as well as bacteria in the lung tissues were investigated and characterized. Lung EVs had more OTUs than lung tissue in all of our study groups. In terms of biodiversity indices, the microbiome data from the lung tissue and the lung EVs had different patterns; the Shannon index decreased from nonsmokers to COPD patients in lung tissue; in contrast, the Shannon index increased in lung EVs. The differences between lung tissue and lung EVs were greatest in the COPD group. This trend of an increase and a decrease in the Shannon index for lung EVs and lung tissue, respectively, was opposite in the Simpson index. The gap in the index between lung tissue and lung EVs was still greatest in the COPD group. The type of specimen (lung tissue or lung EVs) used for the microbiome analyses had a greater effect on PCA proximity than the patient group. Firmicutes were more prevalent in the lung EVs of COPD patients. In our present study cohort, Stenotrophomonas, Propionibacterium, Alicyclobacillus, Methylobacterium and Dechloromonas were the most common genera.
Several recent studies have reported on the relationship between COPD and the lung microbiome. Increased levels of the Firmicutes phylum have previously been found in the lung tissue of COPD patients, a result attributable to an increase in the Lactobacillus genus24 and consistent with our current findings. The outgrowth of Firmicutes in COPD patients may be explained by differences in pH, oxygenation levels and the temperature of the lung environment. In COPD, increased esophageal reflux25 may result in microbial immigration from the gut.26, 27 Firmicutes may be more likely than other species to adapt to the lung environment.28 In addition, our current findings showed that the differences in species were more prominent in lung EVs. A previous report has suggested that EVs from Lactobacillus may ameliorate helper type-1-dominated inflammatory responses,29 but their role in COPD is still not well established. Another study on human macrophages has indicated that Lactobacillus may play an anti-inflammatory role in smoke-associated diseases.30
A previous study has reported that the genus Novosphingobium is present in a relatively high abundance (>2% relative abundance) in patients with severe COPD.31 In our present study, Novosphingobium was present in all three groups. However, its relative abundance did not differ among groups and was below 1% in patients with COPD. Our current analyses differed from that in the previous study in terms of the severity of the COPD as well as in ethnicity, age and environment. According to the Shannon and Simpson index results in this study, lung tissue from our patients with COPD showed lower diversity and higher prevalence of certain organisms within the bacterial community. This trend was consistent with a previous observation that a few dominant genera emerge in COPD.12 However, the opposite trend was found in lung EVs. This difference may have resulted from the relative increase in Firmicutes, and accordingly the relative decrease in Proteobacteria.
This metagenomic analysis of EVs advances understanding of the association between the biodiversity of bacterial communities and COPD. One interesting finding was that lung EVs had more OTUs than the lung tissue itself, and some microorganisms were detected in EVs but not in the lung tissue. EVs can disseminate far from the direct site of bacterial colonization, into the urine, blood and several other organs.32 EVs have been shown to disrupt cell monolayers and to penetrate the epithelial cell layer.33 Therefore, the EVs that remain in vesicle form may originate from adjacent lobes or even from other organs. Another finding from our present study was that the sample type (lung tissue or EVs) had a greater effect on the bacterial community than the patient characteristics. This finding suggests that EVs have distinct characteristics from the lung tissue, some of which may originate from outside of the lung. EVs derived from bacteria have several functions: interbacterial communication, delivery of toxins, entry into cells, and activation of inflammation and innate immunity. Hence, we suggest that EVs are able to affect host cells and other bacteria even far from the colonized area. Recent reports have indicated that EVs as well as the microbiota play important roles in chronic inflammatory disease in the gut.34 COPD, a chronic airway inflammatory disease, may therefore be associated with dysbiosis of the bacterial community and EVs in the airway. Further study of lung-derived EVs may thus be a useful tool to explore the pathogenesis of COPD.
In the present study, sterile saline served as a negative control to exclude spurious sequences introduced from DNA kit contamination, which can be a problem in microbiome research.35 OTU analysis showed a much lower level of OTUs in the negative control compared with lung tissue, and lung EVs and PCA showed consistent clustering regardless of negative control. In addition, analyzing the data by excluding the taxa that were detected in the negative control sample showed the same results. Therefore, the taxa from lung tissue and lung EVs were not from contamination, and comparisons among the groups are accurate.
This study has several strengths and limitations of note. First, among our current study groups, the baseline demographic factors such as sex and age as well as smoking intensities in healthy smokers and the COPD patients were matched to minimize a confounding effect that might affect the bacterial community. Only one nonsmoker used inhaled corticosteroids for asthma, and the others had no medication history. Because the number of patients who had taken medications was very small, the effects of medication on the lung microbiome were insignificant in this study. Second, contamination from the upper airway tract is an important issue in lung microbiome studies.36 To avoid this issue, we harvested lung tissue from surgical resections by using an aseptic technique. To the best of our knowledge, this is the first investigation of the microbiome of lung tissues in chronic airway disease patients from an Asian population. Third, the bacterial growth conditions in the lower respiratory tract are not uniform; the type of lung segments used in a study can affect the composition of the microbiome.15 Lung tissue was harvested from the resected lobes for lung cancer, so we were unable to match the location of the tissue. Our specimens were located sufficiently far from the malignant lesions. Fourth, patients with very severe COPD were not able to be included in our series. Therefore, characterization according to COPD severity was not possible. Last, we used a cross-sectional design. As such, it remains unclear whether differences in the composition of the lung microbiome were a consequence of COPD or whether the changes in the lung microbiome composition resulted in the development of disease. Further animal studies or long-term clinical studies may help to elucidate this issue specifically. In addition, increasing the number of patients would make the results more convincing, but well-matched baseline characteristics and the use of surgical resection specimens still produced meaningful results.
In summary, bacterially derived EVs have distinctive characteristics in the lungs of nonsmokers, healthy smokers and patients with COPD. In addition, EV microbiomes are distinct from the whole lung tissue microbiomes in terms of OTUs, biodiversity, PCA clustering and dominant organisms. This information should contribute to knowledge of the involvement of lung microbiome in the development of COPD.
Acknowledgments
This study was supported by a grant (2015-613) from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea. The biospecimens and data used in this study were provided by the Asan Bio-Resource Center, Korea Biobank Network (2013-0816).
Author contributions
HJK performed the experiments and data analysis, and wrote the first draft; K-HK and Y-SK performed the experiments; J-PC prepared the figures; Y-KK helped design the vesicle model; SY provided bioinformatics support; LS and CSDC participated in critically revising the article for important intellectual content; JSL, Y-MO and S-DL made intellectual contributions and supervised the study; SWL conceived and designed the experiments, performed data analysis and wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
The authors declare no conflict of interest.
Supplementary Material
References
- Burney PGJ, Patel J, Newson R, Minelli C, Naghavi M. Global and regional trends in COPD mortality, 1990–2010. Eur Respir J 2015; 45: 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohansal R, Martinez-Camblor P, Agusti A, Buist AS, Mannino DM, Soriano JB. The natural history of chronic airflow obstruction revisited: an analysis of the Framingham offspring cohort. Am J Respir Crit Care Med 2009; 180: 3–10. [DOI] [PubMed] [Google Scholar]
- Salvi SS, Barnes PJ. Chronic obstructive pulmonary disease in non-smokers. Lancet 2009; 374: 733–743. [DOI] [PubMed] [Google Scholar]
- Rutgers SR, Postma DS, ten Hacken NH, Kauffman HF, van Der Mark TW, Koeter GH et al. Ongoing airway inflammation in patients with COPD who do not currently smoke. Thorax 2000; 55: 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli AE, Burrows B, Knudson RJ, Lyle SK, Lebowitz MD. Longitudinal changes in forced expiratory volume in one second in adults. Effects of smoking and smoking cessation. Am Rev Respir Dis 1987; 135: 794–799. [DOI] [PubMed] [Google Scholar]
- Sethi S. Bacterial infection and the pathogenesis of COPD. Chest 2000; 117: 286S–291S. [DOI] [PubMed] [Google Scholar]
- Cosio MG, Saetta M, Agusti A. Immunologic aspects of chronic obstructive pulmonary disease. N Engl J Med 2009; 360: 2445–2454. [DOI] [PubMed] [Google Scholar]
- Wilson R, Dowling RB, Jackson AD. The biology of bacterial colonization and invasion of the respiratory mucosa. Eur Respir J 1996; 9: 1523–1530. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al. A core gut microbiome in obese and lean twins. Nature 2009; 457: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006; 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- Ma C, Chen F, Zhang Y, Sun X, Tong P, Si Y et al. Comparison of oral microbial profiles between children with severe early childhood caries and caries-free children using the human oral microbe identification microarray. PLoS ONE 2015; 10: e0122075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA et al. Analysis of the lung microbiome in the "healthy" smoker and in COPD. PLoS ONE 2011; 6: e16384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG et al. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS ONE 2012; 7: e42786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med 2013; 187: 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 2011; 184: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol 2015; 15: 375–387. [DOI] [PubMed] [Google Scholar]
- Kim YS, Choi EJ, Lee WH, Choi SJ, Roh TY, Park J et al. Extracellular vesicles, especially derived from Gram-negative bacteria, in indoor dust induce neutrophilic pulmonary inflammation associated with both Th1 and Th17 cell responses. Clin Exp Allergy 2013; 43: 443–454. [DOI] [PubMed] [Google Scholar]
- Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S et al. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 2009; 9: 5425–5436. [DOI] [PubMed] [Google Scholar]
- Li W, Fu L, Niu B, Wu S, Wooley J. Ultrafast clustering algorithms for metagenomic sequence analysis. Brief Bioinform 2012; 13: 656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009; 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Statistics and partitioning of species diversity, and similarity among multiple communities. Oikos 1996; 76: 5–13. [Google Scholar]
- Magurran AE An index of diversity. In: Measuring Biological Diversity. Blackwell Science: Oxford, 2004, UK, pp 100–130.
- Veech JA, Summerville KS, Crist TO, Gering JC. The additive partitioning of species diversity: recent revival of an old idea. Oikos 2002; 99: 3–9. [Google Scholar]
- Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2012; 185: 1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee JH, Kim Y, Kim K, Oh YM, Yoo KH et al. Association between chronic obstructive pulmonary disease and gastroesophageal reflux disease: a national cross-sectional cohort study. BMC Pulm Med 2013; 13: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet 2014; 384: 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 2015; 6: e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsland BJ, Gollwitzer ES. Host-microorganism interactions in lung diseases. Nat Rev Immunol 2014; 14: 827–835. [DOI] [PubMed] [Google Scholar]
- Alpdundar E, Yildiz S, Bayyurt B, Ozcan M, Gucluler G, Bayik D et al Commensal bacteria-derived membrane vesicles suppress Th-1 dominated immune responses in vaccinated and tumor-bearing mice. Frontiers in Immunology Conference Abstract: 15th International Congress of Immunology; 22–27 August 2013; Milan, Italy, 2013.
- Mortaz E, Adcock IM, Ricciardolo FL, Varahram M, Jamaati H, Velayati AA et al. Anti-inflammatory effects of Lactobacillus Rahmnosus and Bifidobacterium Breve on cigarette smoke activated human macrophages. PLoS ONE 2015; 10: e0136455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutebemberwa A, Stevens MJ, Perez MJ, Smith LP, Sanders L, Cosgrove G et al. Novosphingobium and its potential role in chronic obstructive pulmonary diseases: insights from microbiome studies. PLoS ONE 2014; 9: e111150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorward DW, Schwan TG, Garon CF. Immune capture and detection of Borrelia burgdorferi antigens in urine, blood, or tissues from infected ticks, mice, dogs, and humans. J Clin Microbiol 1991; 29: 1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi B, Qi M, Kuramitsu HK. Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res Microbiol 2003; 154: 637–643. [DOI] [PubMed] [Google Scholar]
- Kang C-S, Ban M, Choi E-J, Moon H-G, Jeon J-S, Kim D-K et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS ONE 2013; 8: e76520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter S, Cox M, Turek E, Calus S, Cookson W, Moffatt M et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014; 12: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard AF, Staudinger BJ, Dowd SE, Joshi-Datar A, Wolcott RD, Aitken ML et al. Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota. Proc Natl Acad Sci USA 2012; 109: 13769–13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.