Abstract
The perception of long non-coding RNAs as chunk RNA and transcriptional noise has been steadily replaced by their role as validated targets for a diverse set of physiological processes in the past few years. However, for the vast majority of lncRNAs their precise mode of action and physiological function remain to be uncovered. A large body of evidence has revealed their essential role in all stages of cancirogenesis and metastasis. In this review we focus on the role of lncRNAs in metastasis. We grouped selected lncRNAs into three categories based on in vitro and in vivo mode of action-related studies and clinical relevance for metastasis. Grouped according to their mode of action, in category I we discuss lncRNAs such as CCAT2, DREH, LET, NKILA, treRNA, HOTAIR, H19, FENDRR, lincROR, MALAT, GClnc1, BCAR4, SCHLAP1 and lncRNA ATP, all lncRNAs with in vitro and in vivo metastasis-related data and clinical significance. In category II we discuss lncRNAs CCAT1, PCAT1, PTENgp1, GPLINC, MEG3, ZEB2-AS, LCT13, ANRIL, NBAT1 and lncTCF7 all characterized by their mode of action in vitro and clinical significance, but pending or preliminary in vivo data. Finally, under category III, we discuss lncRNAs BANCR, FRLnc1, SPRY4-IT1 and LIMT with partially or poorly-resolved mode of action and varying degree of validation in clinical metastasis. Finally we discuss metastasis-related translational aspects of lncRNAs.
Keywords: Chromatin remodeling, decoy function, DNA-RNA interaction, epigenetic modification, guide function, nucleic acid-based therapy, protein-RNA interaction, RNA-RNA interaction, scaffold function, signaling networks, review
Metastasis is the major cause of death in patients with cancer. It is mediated by a multi-step process referred to as the metastatic cascade (1,2). Initial steps include local invasion and migration, angiogenesis, epithelial-mesenchymal transition (EMT) and intravasation. Tumor cells enter the circulation as single cells or circulating tumor cell clusters, are coated by platelets to escape an immune response and subsequently arrest in capillaries in distant organs as a prerequisite for extravasation. Colonization starts by homing of tumor cells in supporting niches of the organ parenchyma, followed by a latency phase which can last from several months to decades. A prerequiste for overt outgrowth of micrometastases is their adaptation to the local microenvironment and acquisition of colonisation-promoting traits (3-6). The pattern of colonized distant organs depends on the tumor type and can range from predominant spread to one organ and colonization of different types of organs sequentially or simultaneously (7). Several genes and their products have been identified to mediate crucial steps of the metastatic process such as metastasis initiation and progression, as well as organ-specific functions of metastasis (virulence) (8,9). These gene products include proteases, chemokines, cytokines and their receptors, angiogenic factors, intracellular and transmembrane kinases, adhesion molecules, components of the extracellular matrix (ECM), GPI-linked receptors and carbohydrate metabolism-related enzymes (8,9). More recently, an important impact of RNA-related molecules for metastasis has emerged. MicroRNAs (miRs) and other types of RNA modulate metastasis via regulatory networks (10,11). In this review we focus on the role of long non-coding RNAs (lncRNA) as promoters or inhibitors of metastasis in different tumor entities since there is an urgent need to define new targets for therapeutic intervention. With the exception of denosumab for treatment of bone metastases, all other agents evaluated in clinical studies for treatment of metastatic disease gave rise to mixed or disappointing results (6).
Long Non-coding RNA (lncrna) - General Features
Sequencing of the human genome resulted in identification of 20,000 protein-encoding genes which comprise less than 2% of the human genome. However, it was shown that 70% of the human genome is transcribed into RNA yielding many thousands of ncRNAs (12). They can be classified into several subtypes such as housekeeping RNAs, small non-coding RNA (sncRNA) and long non-coding RNAs (13,14). Examples for housekeeping RNAs are ribosomal RNA (rRNA), transfer RNAs (tRNAs), small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs). SncRNAs have a length of less than 200 nucleotides (nts), and include microRNAs (miRs), small interfering RNAs (siRNAs), circular RNAs, splice-site RNAs, promoter-associated short RNAs and 3’UTR-derived RNAs as examples. LncRNAs are arbitrarily defined by their size exceeding 200 nts and apparent lack of protein-coding capacity. 14,880 human lncRNAs have been identified in the context of the ENCORE project (15). LncRNAs are products of polymerse II (pol II), which are nearly always capped, polyadenylated and frequently spliced (16). They can be categorized into sense, antisense, bidirectional, intronic and intergenic lncRNAs (17). The identification of lncRNAs was accelerated by high-throughput RNA sequencing (RNA-Seq) and its combination with bioinformatic algorithms (18). Transcriptional profiling studies have underlined prevalence and function of lncRNAs in human cancer (19). Five thousand and thirty-seven tumor specimens across 13 cancer types from The Cancer Genome Atlas (TCGA) and 935 cell lines from the Cancer Cell Line Encyclopedia were sequenced. Significant deregulation in comparison to corresponding normal tissues was noted indicating tumor and lineage specific alterations, copy number changes, epigenetic modifications such as promoter methylation and cancer-associated single-nucleotide polymorphisms (SNPs).
LncRNAs exert their functions through interactions with other components such as proteins, RNAs and DNAs (20). RNA-protein, RNA-RNA and RNA-DNA interactions can be combined by a single lncRNA to build distinct functionality conferring complexes (20). As archetypic functions of lncRNAs, guide-, decoy- and scaffold functions have been identified (21). The guide function mediates recruitment of chromatin-modifiying enzymes to target genes in cis- or trans, chromatin remodeling and epigenetic regulation of target genes (22). The decoy function involves binding of miRs through lncRNAs (miR sponges), titration of transcription factors away from chromatin or recruitment of protein factors into nuclear subdomains (16). The scaffold function of lncRNAs can mediate stabilization of ribonucleoprotein complexes such as signaling modules and nuclear structures often accompanied by allosteric modification of protein activity (14).
Many of the functions of lncRNAs are transcription-related, such as forming chromatin-modifiying complexes, acting as transcriptional co-activators, mediating chromatin-looping by enhancer-derived lncRNAs or the functioning as natural antisense transcripts (NATs) (23,24). Modulation of transcription of oncogenes or tumor-suppressor genes is an oncology-related function of lncRNAs (25). Post-transcriptional functions of lncRNAs can trigger alternative splicing by pairing with other RNAs (16,26).
LncRNAs often associate with the polycomb repressive complexes (PRC1 and PRC2) and histone methyltransferase G9a to introduce post-translational modifications to specific amino acids in histone tails, resulting in negative regulation of RNA pol II. PRC1 is a multiprotein complex composed of proteins such as BMI1, RING1, RING2 and chromobox proteins (CBX) and is able to ubiquitinylate histone H2A at lysine 119 (26,27). PRC2 is assembled by EED, SUZ12 and histone methyltransferase EZH2 which trimethylates histone 3 lysine 27 (26,27). G9a introduces a repressive mark by trimethylation of histone 3 lysine 9 (28). In addition to mediating covalent modifications of histones, lncRNAs can be involved in modulation of nucleosome positioning to regulate expression of genes. An example is the SWI/SNF complex which mediates restructuring of chromatin based on hydrolysis of ATP (29).
The physiological function of lncRNAs was demonstrated by multiple knock-out models which indicated their requirement for life and brain development (30). Eighteen lncRNA knock-out strains were generated, focussing on intergenic lncRNAs whose deletion would not overlap known protein coding genes or other gene annotations. Peri- and postnatal phenotypes were observed in three mutant strains and growth defects in two additional strains. Knock-down of FENDRR, an lncRNA discussed later in this review, resulted in defects of the lung, gastrointestinal tract and heart in neonates (19) defining it as a functional moiety.
Metastasis-related lncRNAs - Category I
In this category we summarize metastasis-related lncRNA with data supporting mode of action (MOA), in vivo metastasis-related data and clinical data with respect to their role in metastasis. They are grouped according to their MOA. We discuss lncRNAs which function through RNA/protein interaction such as CCAT2, DREH, LET, NKILA and treRNA, or through epigenetic modification alone or in combination with other mechanisms such as HOTAIR, H19, FENDDR, lincROR, MALAT, GClnc1, BCAR4, SCHLAP1 and finally we discuss lncRNA ATP which functions by induction of miRs and mRNA stabilization.
Colon-cancer Associated Transcript 2 (CCAT2). CCAT2 was discovered in microsatellite-stable colorectal cancer (CRC) as a highly overexpressed lncRNA (31). CCAT2 is up-regulated in breast cancer and correlates with poor prognosis for a specific subgroup of patients (32,33). CCAT2 is localized mainly in the nucleus and its MOA is based on RNA-protein interaction as outlined in the following. A high-risk allele for CRC, SNP rs6983267 has been shown to produce more CCAT2 transcript than its corresponding allele (31,34). In vitro inhibition of CCAT2 decreases cell proliferation and invasion of both MCF7 and MBA-MB 231 breast cancer cells (32). In HCT116 colorectal cancer cells, transduced CCAT2 increases migration and in the highly metastatic colon cancer variant KM12SM, knock-down of CCAT2 reduces invasion (31). Mechanistic resolution points to activation of Wnt signaling by CCAT2 by physical interaction with transcription factor TCF7L2 (TCF4) (31). However, the mechanistic details of these findings have to be resolved. Wnt signaling leads to up-regulation of target genes such as c-MYC and CD44 as well as metastasis-promoting miR17-5p and miR20a. The involvement of Wnt signaling in metastasis is well documented (35,36) and aberrant Wnt-signaling in CRC has been observed previously (37). Interestingly, CCAT2 is also up-regulated by Wnt signaling, pointing to a feedback loop. Involvement of CCAT2 in metastasis has been shown with HCT116 cells. Injection of HCT116 cells overexpressing CCAT2 into the spleen of nude mice gave rise to higher incidence of liver metastasis and greater numbers of metastatic nodules (31).
LncRNA DREH (Down-regulated expression by HBx). DREH was discovered in a transgenic mouse model of hepatocellular carcinoma (HCC) induced by expression of Hepatitis B virus X protein (HBx) in the liver (39,39). DREH is down-regulated in murine tumor tissue compared to normal liver tissue as shown by comparison of the profile of lncRNAs in both types of tissues (38). The human ortholog of DREH is also down-regulated in human HBV-related HCC tissue and DREH expression levels correlate with relapse-free and overall survival (38). As outlined in the following, the MOA of DREH relies on RNA-protein interaction. The tumor-suppressive function of DREH is mediated by change of the structure of the cytoskeleton due to binding to vimentin and repression of vimentin expression (38). Vimentin is an intermediate filament protein which is known as a marker of mesenchymal cells and of cells undergoing epithelial mesenchymal transition (EMT) during metastatic progression (40-42). In addition to maintainance of cell morphology, vimentin is also involved in cell adhesion, migration, proliferation and signal transduction (43). Down-regulation of DREH in HCC cells by RNA interference promotes migration and invasion (38). DREH also has an impact on proliferation and metastasis in vivo. Hepa1-6 cells transfected with DREH exhibited drecreased tumor growth in nude mice (38). Tail vein injection of these cells gave rise to less pulmonary metastases in comparison to control cells (38). In an orthotopic liver cancer model in nude mice, Hepa1-6 cells transfected with DREH gave rise to reduced intrahepatic nodules as well as reduced metastasis in the abdominal cavity and wall, intestine and lymph nodes (38).
LncRNA LET (Low expression in tumor). LncRNA LET is down-regulated in hepatocellular carcinoma (HCC) and correlates with metastasis (44,45). Its MOA relies on binding to NF90 in a 1:1 stoichiometry resulting in ubiquitinylation and degradation of NF90 (2). NF90 is a double-stranded RNA binding protein implicated in stabilization, transport and translational control of target mRNAs including HIF-1α, cdc42 and dual specificity protein phosphatase DUSP (46) as well as mediating the degradation of these targets. HIF-1α (47,48), cdc42 (49) and DUSP (50) are validated mediators of metastasis. Down-regulation of lncRNA LET in HCC cell lines and patient’s tumors is mediated by histone deacetylase 3 (HDAC3) (45). Transwell experiments have indicated that hypoxia increases the invasive potential of HCC SMMC-7721 cells and up-regulation of lncRNA LET diminishes invasion in vitro and under hypoxic conditions (45). In vivo experiments with HCC cell lines SMMC-7721 and HCCLM3 as well as colon carcinoma SW480 cells with ectopic expression of lncRNA LET indicated that lncRNA LET inhibits lung colonization after tail vein injection (45). In an orthotopic liver xenograft model, lncRNA LET overexpression reduced hepatic invasion and abdominal metastasis (45). Furthermore, short hairpin ribonucleic acid (shRNA) directed against lncRNA LET increased lung metastasis after tail vein injection and increased hepatic and abdominal metastasis after orthotopic injection of HCC cell line Huh7 (45).
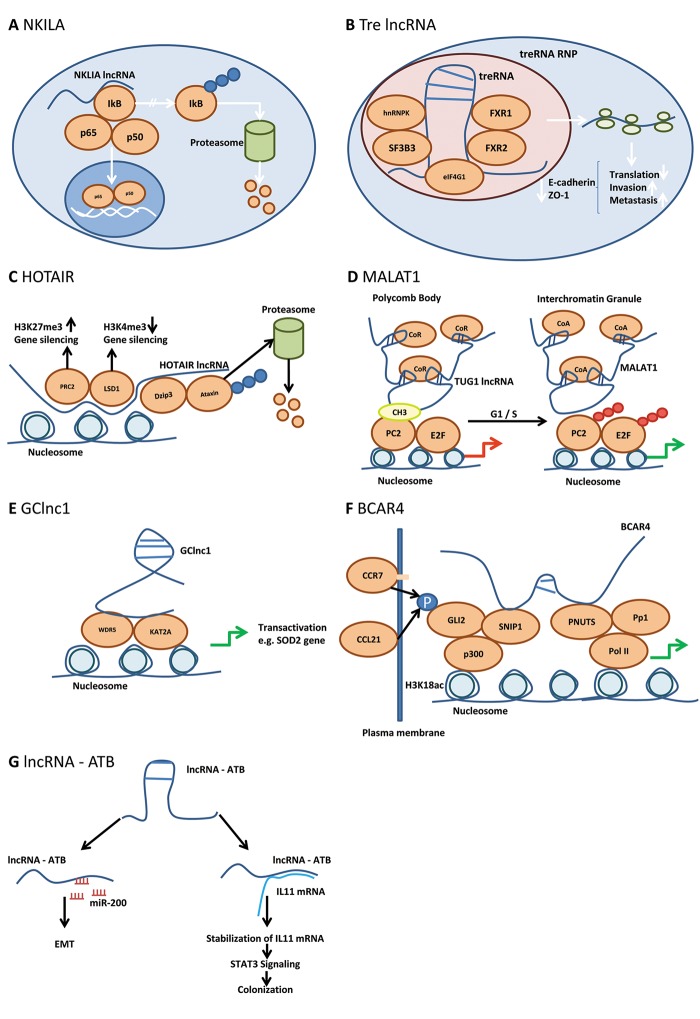
NKILA (NFĸB-activating lncRNA). As outlined in the following, NKILA is an inhibitor of metastasis in pre-clinical models in vitro and in vivo and its MOA relies on protein-RNA interaction. NKILA was found to be expressed at lower levels in breast carcinomas without distant or regional lymph node metastasis and further reduced in those with metastasis, predicting poor clinical outcome (51). NKILA was identified as a lncRNA induced by NF-ĸB-signaling activating inflammatory cytokines such as TNFα and IL-1β in MDA-MB 231 breast cancer cells by microchip-based technology (51-53). NF-ĸB is a mediator of invasiveness and NKILA acts as a negative regulator of NFĸB signaling (51,53). It stably associates with the NFĸB-IĸB complex and binds to two different sites of the homodimeric p65 subunit of NFĸB (54). Thus, IĸB kinase (IKK)-related phosphorylation sites on inhibitor of ĸB (IĸB) are masked and therefore NFĸB-signaling is inhibited due to interaction of NKILA with functional domains on signaling proteins (Figure 1A). It should be noted that abnormal NFĸB activation and poor patient outcome are based on degradation of NKILA by miRs 103/107 (55). In vitro, ectopic expression of NKILA in MDA-MB-231 cells reduces invasion, and vice versa the down-regulation in non-invasive MCF-7 cells increases invasion (51). These findings can be recapitulated in pre-clinical in vivo models. In MDA-MB-231 xenografts, ectopic expression of NKILA inhibits metastasis to the lungs, liver and lymph nodes and prolongs survival, whereas down-regulation of NKILA by shRNA in MCF-7 increases metastasis (51).
Figure 1. Mode of action of lncNKILA, Tre lncRNA, HOTAIR, MALAT1, GClncRNA, BCAR4 and lncRNA-ATP. (A) NKILA acts as a negative regulator of NFĸB signaling. IĸB, inhibitor of ĸB; NFĸB, nuclear factor ĸ; NKILA, NFĸB-activating lncRNA; p50 and p65, subunits of NFĸB. Blue circles indicate polyubiquitinylation. (B) Tre lncRNA inhibits translation of pro-metastatic mRNAs via RNP formation by a poorly resolved mode of action. eIF-4G1, translation initiation factor 4G1; FXR1, FXR3, fragile X mental retardation syndrome-related proteins 1, 3; hnRNPK, heterogeneous ribonucleoprotein complex K; SF3B3, splicing factor 3B subunit 3; treRNA, translational regulatory RNA; treRNA RNP, tre RNA ribonucleoprotein complex. (C) HOTAIR represses gene expression by recruitment of PRC2 and LSD1 by increasing the repression code H3K27me3 and decreasing the activation code H3K4me3. DZIP3, DAZ interacting protein 3 (E3 ubiquitin-protein ligase); HOTAIR, HOX antisense intergenic RNA; H3K4me3, histone 3 trimethylated lysine 4; H3K27me3, histone 3 trimethylated lysine 27; LSD1, lysine-specific demethylase 1; PRC2, polycomb repressive complex 2. Blue circles indicate polyubiquitinylation. (D) MALAT1 relieves gene repression in polycomb bodies by replacement of TUG1 lncRNA and recruitment of co-activators in Interchromatin granules thus promoting G1/S transition. CoA, co-activator; CoR, co-repressor; CH3, methyl group; E2F, transcription factor 2F; MALAT1, metastasisassociated lung adenocarcinoma transcript; Pc2, polycomb 2; Pc2-CH3, methylated Pc2; SUMO, sumoylated; lnc RNA TUG1, lnc RNA taurine-upregulated gene 1. Red/green arrow: inhibition/activation of transcription; red circles indicate sumoylation. (E) GClncRNA recruits histone modifiers such as WDR5 and KAT2A for transactivation of transcription of selected genes. GC lncRNA, Gastric cancer lncRNA; KAT2A, lysine acetyltransferase 2A; WDR5, WD repeat containing protein 5; SOD2, superoxide dismutase 2. Green arrow, indicates activation of transcription. (F) BCAR4 mediates activation of phospho-GLI-2 dependent target genes. BCAR4, Breast cancer anti-estrogen resistance; CCL21, chemokine (C-C motif) ligand 21; CCR7, CCchemokine receptor 7; GLI2, glioma-associated oncogene family zinc finger 2; p, phosphorylated; PNUTS, putative protein phosphatase 1 nuclear; Pol II, polymerase II; Pp1, phosphatase 1; p300, E1A binding protein 300; SNIP1, smad nuclear interacting protein 1. Black arrows indicate phosphorylation of GLI2 by CCR7/CCL21 interaction; green arrows indicate activation of transcription. (G) lncRNA – ATP mimics the prometastatic role of TGFβ biy inducing miR200 family members and promotes IL11 signaling. EMT, Epithelial mesenchymal transition; IL11, interleukin 11; lnc RNA – ATP, lncRNA activated by TGFβ; STAT3, signal transducer and activator of transcription 3; TGFβ, transforming growth factor β; blue circled P, phosphorylation.
Translational Regulatory RNA (treRNA). treRNA was originally identified as ncRNAa7 through genome wide computational analysis (56). It was shown to exert an enhancer-like function on expression of transcription factor snail in cis, however, the mechanism by which gene expression is enhanced is not yet resolved (56). Subsequently ncRNAa7 was renamed to treRNA because it regulates metastasis at the translational step based on RNA-protein interaction (57). treRNA was identified as an lncRNA overexpressed in breast cancer lymph node metastasis compared to matching primary breast tumors (57). Functional studies have delineated a metastasis-promoting function of treRNA in vitro and in vivo. Knock-down of treRNA in A549 lung cancer cells suppressed cell migration and invasion, but did not affect proliferation (57). Enforced expression of treRNA in non-invasive MCF-7 cells increased cell migration and invasion through Matrigel (57). Tail vein injection experiments of A549 cells and A549 siRNA treRNA expressing cells indicated abolishment of lung metastasis in mice injected with A549 siRNA treRNA cells (57). MCF-7 cells ectopically expressing treRNA gave rise to lung metastasis after implantation into the mammary fat pads, whereas MCF-7 cells did not colonize the lungs. A key mechanism for promotion of metastasis is based on treRNA-mediated suppression of translation of epithelial markers such as E-cadherin (57). treRNA is assembling a translation-modulating ribonucleoprotein complex involving several RNA binding and additional proteins (57). Knock-out experiments in MCF-7 cells expressing treRNA revealed that hnRNPK and FXR2 are required for the treRNA function on metastasis in vivo (58-60). Ribonucleoprotein complex RNPK (3,4) is involved in multiple steps of gene expression, including translation and FKR2 (60) can associate with ribosomes to form RNP complexes. In addition, translation initiation factor eIF-4G1 (61) was found to associate with the tre-RNA based ribonucleoprotein complex. The complex exerts its translation modulatory function by interaction with the 3’UTR of its target genes such as E-cadherin (Figure 1B). The precise mechanism of translation inhibition, however, remains to be resolved.
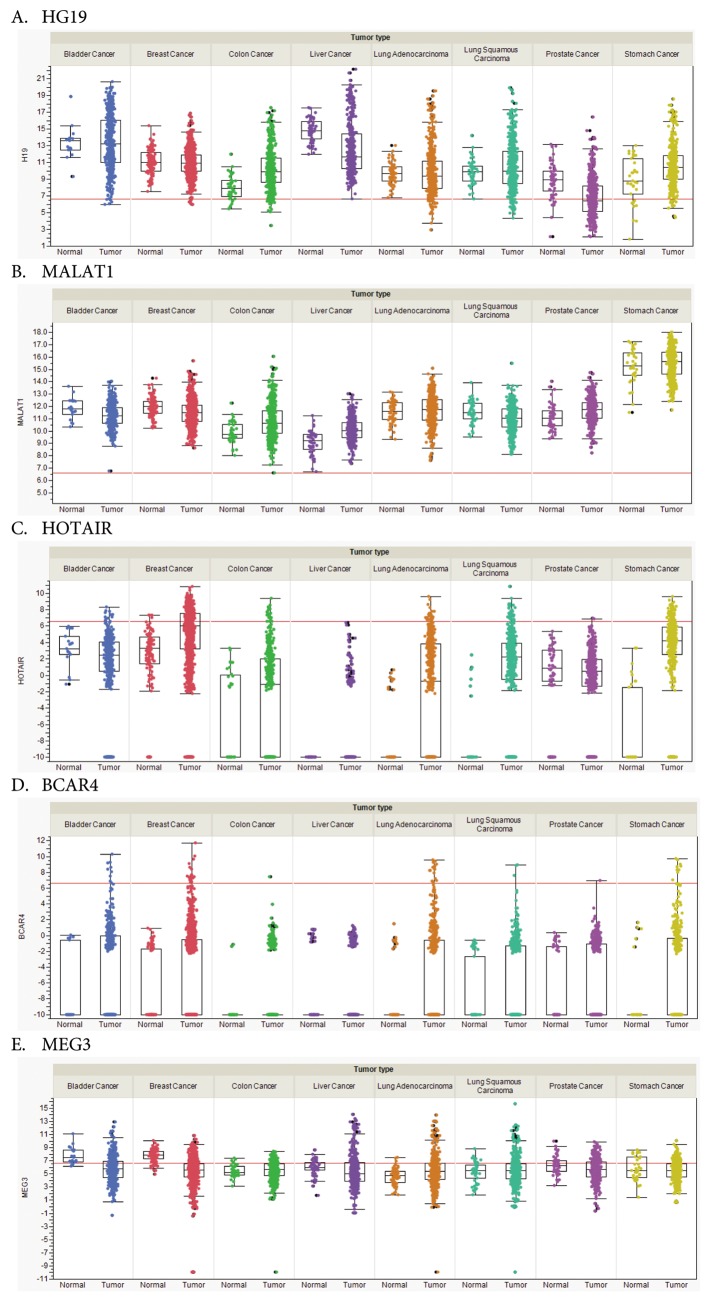
HOX Antisense Intergenic RNA (HOTAIR). Expression of HOTAIR correlates with metastasis and poor prognosis in breast, colon and lung cancer (62-64). HOTAIR is derived from HOX C, but functions in trans at the HOX D locus (65). HOTAIR recruits the PRC2 complex and the demethylase LSD1, promoting an increase in the repression code H3K27me3 and a decrease in the activation code H3K4me3 (66,67). In addition to these functions, in lung cancer, HOTAIR has been identified as a scaffold to facilitate the binding of E3 ubiquitinylases and corresponding substrates Ataxin-1 and Snurportin (Figure 1C) (68). Ataxin-1 is essential for lung alveolization and therefore, HOTAIR may promote dedifferentiation of lung epithelial cells (69). Forced expression of HOTAIR in non-transformed breast and lung cancer cell lines has been shown to promote growth in soft agar and invasion in matrigel (70). Another aspect of the MOA of HOTAIR was revealed by its binding to miR 331-3p, resulting in increased expression of HER2, promoting metastasis (71,72). There is also evidence that HOTAIR can interfere with DNA methylation, because depletion of HOTAIR in HepG2 HCC cells has been correlated with a decrease in phosphatase and tensin homolog (PTEN) promoter methylation (73). In vivo, the impact of HOTAIR on metastasis was demonstrated with human breast cancer cells after tail vein injection. Expression of HOTAIR in MDA-MB231 cells gave rise to an 8-10 fold increase of nodules in the lungs (70). Altogether, these results substantiate that HOTAIR-mediated reprogramming of the chromatin state can promote metastasis. TCGA-based transcript analysis confirms overexpression of HOTAIR in breast-, colon- and lung tumors and in addition HOTAIR is up-regulated in stomach cancer in comparison to matching normal tissues (Figure 2C).
Figure 2. RNA steady-state levels of selected RNAs in different tumor entities. (A) H19; (B) MALAT1; (C) HOTAIR; (D) BCAR4; (E) MEG3. Expression in tumor/matching normal samples is shown for selected cohorts derived from The Cancer Genome Atlas (TCGA) database. Bladder cancer (408/19), breast cancer (1,100/112), colon cancer (475/41), liver cancer (373/50), lung adenocarcinoma (517/59), lung squamous carcinoma (501/51), prostate cancer (498/52) and stomach cancer (415/35). Expression is measured as normalized read counts, the red lines indicate low versus higher expression (~100 counts). Expression data are shown as box blots where the line in the middle of the box represents the data median, the rectangles show the upper and lower quartile (therefore, 50% of all data points are included in the rectangle). All other data points, except for outliers lie within the upper and lower whiskers. A value of -10 indicates no expression in the corresponding sample. For some tumor types not a single sample expressing the corresponding lncRNA was found.
LncRNA H19. LncRNA H19 is an oncofetal antigen that is only marginally expressed in normal human tissues (74). It is up-regulated in several types of tumors such as colorectal cancer (CRC), HCC, breast- and bladder cancer and may be involved in metastasis of these types of tumors (75). Taken together, there is evidence that the functions of H19 are context specific, dependent on stage and type of tumors (75). In the following we focus on findings supporting the prometastatic function of H19. One has to keep in mind that processing of H19 gives rise to miR675, a 23 nt miR located at its 3’end and the residual sequences of H19, which leaves the functional contribution of both moieties difficult to differentiate in most of the reported experiments (75). Evidence for involvement of H19 in metastasis includes H19-mediated induction of genes mediating invasion, promotion of EMT and Wnt signaling. In T24 bladder carcinoma cells, H19 induces angiogenesis and metastasis promoting genes (74). In CRC cells, H19 activates EMT-related markers such as vimentin, ZEB1 and ZEB2, promoting cell migration by functioning as a sponge for miR-138 and miR-200a (76). Another invasion promoting ability of H19 is the activation of constitutive Wnt signaling (77). In bladder cancer T24 cells, H19 was shown to recruit histone lysine N-methyltransferase enhancer of zeste homolog 2 (EZH2) resulting in activation of Wnt/βcatenin signaling by inhibition of expression of Nkd1, an antagonist of Wnt signaling, through methylation of its promoter (77). In gastric cancer cells MKN45 and SGC7901, H19 was shown to be involved in proliferation, migration, invasion and metastasis (78). This was demonstrated through H19 knock-down in MKN45 cells and ectopic expression of H19 in SGC7-901 cells and their assessment in transwell and woundhealing assays (78). In this context, ISM1 was identified as a binding protein for H19 and CALN1 as a target gene for miR-675 (78). SGC7-901/H19 cells, subcutaneously injected into nude mice showed increased tumor growth and increased number of peritoneal nodules after injection into the peritoneal cavity (78). Reciprocal in vivo observations were made with MKN45 H19 knock-down cells (78). Assessment of these cell lines in additional metastasis-related models would be helpful for evaluation of the in vivo function of H19 in metastasis. TCGA-based transcript analysis indicates increased steady-state levels of H19 RNA in CRC and stomach cancer, but not in breast cancer as outlined above (Figure 2A).
FENDRR. FENDRR (79) is an lncRNA with decreased expression in gastric cancer correlating with poor prognosis (80). FENDRR mediates dsDNA-RNA triplex formation to epigenetically regulate expression of target genes by recruitment of PRC2 (80,81). Overexpression of FENDRR in gastric cancer cells suppresses migration in vitro by down-regulating fibronectin 1 (FN1) and MMP2/MMP9 expression (80). Down-regulation of FENDRR in gastric cancer cells was found to be due to histone acetylation by HDAC3. As previously outlined, FENDRR is essential for proper heart and body wall formation in the mouse (81). In vivo demonstration of FENDRR involvement in metastasis was performed by tail vein injection of MGC 803 gastric cancer cells stably transfected with a FENDRR expression vector. Ectopic expression of FENDRR significantly reduced the number of nodules in the lung compared to those in the control group (80).
Linc-ROR. Linc-ROR was originally identified as a lncRNA overexpressed in breast cancer tissues and TNBC versus corresponding normal tissues as well as in a panel of breast cancer cells in comparison to immortalized breast epithelial cells (82,83). Clinical studies correlating linc-ROR expression with survival and metastasis are still pending. Several preclinical studies are supportive regarding a possible role of linc-ROR in metastasis. For example, linc-ROR was shown to promote invasion and migration of breast cancer cells (82). Furthermore, linc-ROR can act as a promoter of EMT by functioning as a sponge for miR-205 and preventing degradation miR-205 target genes including EMT-inducer ZEB2 (82). An additional decoy-related functional contribution of linc-ROR was revealed in gastrointestinal cancer cells (84). Linc-ROR was shown to repel G9A methyltransferase promoting release of H3K9 methylation by binding to and activation of the tescalin promoter that mediates expression of tescalin, an EF-handed calcium-binding protein (85). In TNBC, linc-ROR was shown to be overexpressed and to act as a competitive RNA for miR-145, resulting in up-regulation of ADP-ribosylation factor 6 (ARF6), a small GTPase known as a regulator of breast cancer (83,86). In vivo experiments suggest a role of linc-ROR in metastasis, since tail vein injection of shROR expressing MDA-MB-231 cells exhibited fewer and smaller lung metastatic foci than controls (82). However, it is unclear if the effect was due to less metastasic or overall lower tumorigenic properties of these cells as the growth of primary tumors was also slower. Therefore, further experiments are needed to resolve the mode of action of linc-ROR in metastasis.
Metastasis-Associated Lung Adenocarcinoma Transcript (MALAT1). MALAT1 is also referred to as Nuclear Enriched Abundant Transcript 2 (NEAT2). MALAT1 expression is prognostic for survival of patients with stage I lung adenocarcinoma or squamous cell carcinoma and in bladder carcinoma its expression levels correlate with metastasis (87,88), furthermore MALAT1 is expressed in several types of cancer (89). The incidence of lymph-node metastasis in patients with high MALAT1 expression is higher than in patients with low MALAT1 expression across several types of tumors as shown in a meta-analysis of cancer patients in China (90). The involvement of MALAT1 in alternative splicing is still a controversial issue. This possible engagement was supported by the colocalization of MALAT1 to nuclear speckles, structures enriched in polyA mRNA and factors involved in mRNA processing, splicing and export. In HeLa cells, antisense oligonucleotide (ASO)-based depletion of MALAT1 has supported its involvement in alternative splicing by modulating serine/arginine (SR) splicing factor phosphorylation (91). However, in a panel of lung cancer cells, down-modulation of MALAT1 mRNA by introducing destabilizing elements using Zn-finger nucleases into its mRNA did not give rise to changes in the splicing pattern (92). The latter finding is compatible with MALAT1 knock-out mice which are viable and fertile and do not show any phenotype or splicing alterations, despite the strong evolutionary conservation of MALAT1 (93,94). However, involvement of MALAT1 in regulation of transcription is consistently supported by several investigations. An example is control of cell-cycle-related genes by their recruitment from transcriptionally inactive polycomb bodies (PcG) to transcriptionally active interchromatin granules (ICG) (95). This phenomenon is triggered by the methylation status of polycomb2 (Pc2), also referred to as CBX4, a member of the PRC1 complex (95). Methylated Pc2 localizes ncRNA TUG1 to the PcGs, whereas unmethylated Pc2 sequesters MALAT-1 into ICGs. Pc2 acts as a reader of the histone code through its chromodomain by binding to H2AK5ac and H2AK13ac, which are markers of gene activation and subsequent pol II activation (Figure 1D). Further evidence for involvement of MALAT1 in transcriptional regulation has been obtained in pancreatic cancer and bladder carcinoma cells. Ectopic expression of MALAT1 in pancreatic cancer cells is associated with loss of E-cadherin and gain of function such as N-cadherin, vimentin and TGFβ (96). MALAT1 knock-down in T24 bladder carcinoma cells leads to down-regulation of EMT promoting factors such as Slug, Zeb1 and Zeb2 as well as decreased nuclear localization of β-catenin (88). Metastasis-related experiments were performed with lung cancer cell lines A549 and EBC-1 in which MALAT1 is a driver of mobility (92). MALAT1 is required for effective lung nodule formation of A549 cells after tail vein injection. 80-90% reduction of the number of lung nodules was observed for A549 MALAT1 knock-out cells. In a model covering all steps of the metastatic cascade, EBC-1 cells were subcutaneously injected, the tumors were excised after five weeks of treatment with MALAT1 ASO and the lung metastases were assessed after 7 weeks without further ASO treatment. Fewer and smaller nodules were observed in the ASO group. It should be noted that MALAT1 does not effect cell proliferation of EBC-1 cells in vitro and only exerted a minor impact on growth of the primary tumor in vivo. TCGA-based transcript analysis does not support up-regulation of MALAT-1 in bladder und lung carcinoma as outlined above, however, increased expression in prostate and liver cancer in comparison to normal tissues was noticed (Figure 2B).
GClnc1. GClnc1 was identified by comparison of lncRNA expression profiling in gastric cancer and adjacent normal tissue and its up-regulation correlates with poor prognosis in patients with gastric cancer (97). In gastric cancer cell lines BGC 823, MKN 45 and GES-1, GClnc1 mediates proliferation and invasion as shown by a combination of ectopic expression and small interfering RNA (siRNA) experiments (97). Metastasis-promoting capability of GClnc1 was assessed in nude mice with GES-1 and MGC 803 cells after subcutaneous implantation and injection of adenovirus expressing GClnc1 or sh GClnc1. The experiments revealed a positive impact of GClnc1 on metastasis of these cell lines to the lungs (97). MOA studies revealed that GClnc1 acts as a scaffold for WDR5 (98) and KAT2A (99). WDR5 is a core subunit of the H3K4 methyltransferase complex and KAT2A functions as a histone acetyl transferase via H3K9 acetylation in the promoter region. One of the genes affected by GClnc1 is mitochondrial superoxide dismutase (SOD2). Binding of WDR5 and KAT2A to its promoter results in its transactivation due to increased H3K4 trimethylation and H3K9 acetylation (97) (Figure 1E). SOD2 has been shown to be elevated in gastric cancer and regulation of metastasis (100,101).
LncRNA BCAR4 (Breast Cancer Anti-estrogen Resistance). LncRNA BCAR4 was originally identified by its ability to confer resistance against anti-estrogens in breast cancer cells (102,103). BCAR4 also functions as a breast cancer oncogene (104) and its expression correlates with advanced breast cancer (105). The Oncomine database also showed a significant correlation between BRCA4 expression and metastatic colorectal, rectal, lung- and prostate cancer (105). Recently, a metastasis-promoting function of BCAR4 for breast cancer cells was uncovered based on cooperative regulation of downstream chemokine signals (105) as outlined in the following. Interaction between chemokine CCL21 and its receptor CCR7 (106) activates rho-interacting serine/threonine kinase 21, citron (CIT) (107), which phosphorylates transcription factor GLI-2 (108), which is subsequently translocated into the nucleus. BCAR4 is required for the transcriptional activation of phospho-GLI2-dependent target genes in breast cancer cells. In response to CCL21, BCAR4 binds to Smad Nuclear Interacting Protein 1 (SNIP1) (109) and serine/threonine phosphatase regulatory subunit 10 (PSP1R10) also known as PNUTS (110). Interaction of phospho-GLI2 with SNIP1 releases SNIP1-mediated inhibition of p300-dependent histone acetylation, which results in binding of PNUTS to H3K18ac, thereby releasing inhibition of pol II via activation of phosphatase PP1 (Figure 1F) (111). In vitro, BCAR4 acts as a promoter of invasion and migration of breast cancer cells (105). In vivo BCAR4 functions as a mediator of breast cancer metastasis in preclinical models. Lung metastasis of MDA-MB 231 LM2 cells harboring shRNA directed against BCAR4 was significantly reduced after their injection into mammary fat pads in comparison to control cells (105). Locked nucleic acids (LNAs) targeting BCAR4 reduced lung metastasis of MDA-MB 231 LM2 cells after i.v. administration twice a week for three weeks, underlining the importance of BCAR4 as a therapeutic target (105). TCGA-based transcript analysis confirms overexpression of BCAR4 in colon-, lung- and prostate cancer and in addition indicates overexpression in bladder-, breast- and stomach cancer in comparison to normal matching tissues (Figure 2D).
SCHLAP1 (Second Chromosome Locus Associated with Prostate-1). SCHLAP1 is expressed in 25% of prostate cancers and is a predictor of poor outcome, metastasis and mortality (112,113). SCHLAP1 interacts with chromatin complex SWI/SNF and binds to and disrupts function of SNF5, a core component of this complex (114). SCHLAP1 antagonizes the genome-wide localization, regulatory and tumor-suppressive functions of the SWI/SNF complex (115,116) due to its impaired ability to regulate gene expression properly. SCHLAP1 overexpressed in benign immortalized prostate cells increases invasion with no impact on proliferation (114). Likewise, knock-down of SCHLAP1 in 22Rv1 prostate cancer cells impairs cell invasion in vitro (115). Intra-cardiac injection of SCHLAP1 knock-down 22Rv1 cells abolished metastatic seeding at proximal (lungs) and distant sites (115). Altogether, these data support involvement of SCHLAP1 in metastasis of prostate cancer.
LncRNA-ATB (lncRNA activated by TGFβ). LncRNA-ATB is up-regulated by TGFβ and its high expression is a robust predictor of survival in patients with HCC (117). Its MOA is based on two different types of RNA-RNA interactions as outlined in the following. TGFβ is able to induce metastasis-suppressing as well as metastasis-promoting genes (118-120). LncRNA-ATB can mimic the pro-metastatic role of TGFβ by inducing miRs of the miR-200 family (121,122) and by promoting IL11 signaling (Figure 1G) (123). LncRNA-ATB up-regulates EMT- and invasion-promoting transcription factors ZEB1 and ZEB2 by competitively binding to miR200 family members, thus promoting an early step of the metastatic cascade (121,122). Additionally, lncRNA-ATB binds to IL11 mRNA resulting in its stabilization and increased expression and secretion, thus promoting IL11/STAT3 signaling which is essential for colonization of distant organs (Figure 1G) (123). In vitro, lncRNA-ATB incuces EMT and invasion after its overexpression in SMMC-7721 HCC cells (117). The previous findings are also reflected in preclinical in vivo models. Ectopic expression of lncRNA-ATB in HCC cells increased the number of CTCs and vice versa, its depletion resulted in decreased numbers of CTCs (117). In an orthotopic model with HCCLM6 cells, depletion of lncRNA-ATB resulted in inhibition of intrahepatic, mesenteric and pulmonary metastases (117). In a tail vein-injection metastasis experiment with SMMC-7721 cells overexpressing lncRNA-ATB, an increase in lung metastasis in comparison to control cells was noted (117).
Metastasis-related lncRNAs: Category II
In this category we summarize lncRNAs with MOA-supporting experiments and correlation with metastatic disease, but with pending experiments supporting metastasis-promoting role in preclinical in vivo models. We discuss CCAT1 and PCAT1 with a sponge-based mechanism, PTENpg1, GAPLINC and MEG3 with sponge function and additional MOA, ZEB2-AS, LCT13 and ANRIL with an antisense-based MOA as well as NBAT1 and lncTCF7 with a MOA due to epigenetic modifications.
Sponge-based mechanism. Colon cancer associated transcript 1 (CCAT1) is up-regulated in CRC, gastric cancer and HCC and correlates with bad prognosis in HCC (124-126). In CRC cells, CCAT1 mediates cell proliferation and enhanced transcription of c-MYC by regulating long-range chromatin interactions at the c-MYC locus (127). Prostate cancer associated transcript-1 (PCAT-1) was identified as a mediator of proliferation based on interaction with miR-3667-3p resulting in stabilization of c-MYC (128). However, a role of PCAT-1 in proliferation and migration has been described in NSCLC (129) and HCC (130).
Sponge-based and additional MOA. PTENgp1 is an lncRNA involved in modulating PI3K-AKT signaling, a pathway crucial for proliferation and metastasis, frequently activated by inactivation of tumor suppressor PTEN which results in PI3K phosphorylation and subsequent activation of AKT (131-133). PTENgp1 is encoded by PTENP1, a pseudogene of PTEN, exhibits tumor- and metastasis-suppressive activity via its 3’-UTR by acting as a sponge for PTEN degrading miR17 and miR21 (134). A further complexicity of this type of gene regulation is exerted by an antisense RNA to PTENgp1 that is expressed in two isoforms α and β (134). Isoform α functions in trans and localizes to the PTEN promoter and epigenetically modulates PTEN transcription by DNA methyltransferase 3a (DNMT3a) and EZH2 (134). Isoform β interacts with PTENgp1 through RNA-RNA pairing, facilitates PTENgp1 export to the cytoplasm and modulates its stability and sponge activity (134). Function of gastric adenocarcinoma predictive large intergenic non-coding RNA (GAPLINC) is based on a sponge-related and RNA-protein related MOA (135,136). GAPLINC is associated with shorter survival in a subset of patients with gastric cancer, which overexpress this lncRNA (135). GAPLINC mediates cell migration through regulation of CD44 acting as a sponge for miR 211-3p in gastric cancer cells (135). In CRC cells, GAPLINC promotes cell proliferation and invasion in vitro by binding to PTB-associated splicing factor (PSF) and non-POU-domain-containing octamer binding protein (NONO), factors which are involved in up-regulation of EMT-promoting factor SNAI-2 (136). For maternally expressed gene 3 (MEG3) lncRNA, sponge-related and RNA-DNA triplex formation based MOA has been described (137,138). MEG3 is decreased in gastric cancer patients and cell lines and down-regulation correlates with metastatic disease. Ectopic expression of MEG3 inhibits cell proliferation and migration and promotes apoptosis in several gastric cancer cell lines probably due to sequestration of oncogenic miR-181 (137). Another functional aspect of MEG3 was revealed in several types of cancer cell lines as its capacity to negatively regulate EMT-promoting TGFβ pathway genes (138). This function is based on guiding MEG3 to chromatin through RNA-DNA triplex formation at distal regulatory elements of these genes via GA-rich sequences and subsequent recruitment of PRC2 (138). TCGA-based transcript analysis could not confirm down-regulation of MEG3 in gastric cancer, however, we have noted down-regulation of MEG3 in bladder-, breast- and liver cancer in comparison to matching normal tissues (Figure 2E).
Antisense-based MOA. ZEB2-AS1, LCT13 and ANRIL are lncRNA with an antisense-based mechanism. ZEB2-AS1 is a natural antisense transcript that enables translation of ZEB2, an EMT-inducing transcription factor, which represses genes encoding functional proteins such as E-cadherin (139,140). Splicing of an intron in the 5’UTR of ZEB2 prevents translation of ZEB2 mRNA and ZEB2-AS1 abolishes splicing and enables translation of ZEB2 mRNA due to an internal ribosome binding site located in the intron (139,141). Possible clinical relevance of ZEB2-AS1 was demonstrated in urinary bladder cancer (UBC) (142). TGFβ1 secreted by fibroblasts was shown to induce ZEB2 expression, migration and EMT of bladder cancer cells dependent on ZEB2-AS1 (142). TGFβ1 expression and ZEB2-AS1 transcript levels were positively correlated in bladder carcinoma specimens, pointing to a role of the TGFβ1-ZEB2-AS1-ZEB2 axis in metastasis of this type of cancer (142). Another lncRNA in this category, LINE-1 chimeric transcript 13 (LCT13), inhibits expression of human tissue factor pathway inhibitor (TFPI-2) (143). It acts as a matrix-associated Kunitz inhibitor and its anti-metastatic effect is due to inhibiting the activation of zymogen matrix metalloproteinases involved in tumor progression, invasion and metastasis (144,145). LCT-13 induces silencing and deposition of repressive histone modifications in breast cancer and colon cancer cells (143). Clinical relevance of LCT-13 is suggested by the finding that in 56% (n=27) of colorectal tumors with reduced TFPI-2 expression, LCT13 transcripts were detected (143). Antisense non-coding RNA in the INK4 locus (ANRIL) is an lnc RNA whose expression level predicts poor prognosis in patients with HCC, NSCLC and ovarian cancer (146-148). ANRIL functions as a cis-regulator of the INK4a/ARF/INK4b locus of tumor suppressors by acting as an antisense RNA and by its capability to recruit PRC1 and PRC2 complexes, which mediate epigenetic regulation of this locus (149,150). In vitro experiments with HCC, NSCLC and ovarian cancer cell lines indicate that ANRIL is able to promote proliferation, migration and invasion (146-148,151). In thyroid carcinoma cells, these effects are based on repression of TGFβ1/Smad signaling and up-regulation of p15/INK4b. ANRIL silencing inhibited experimental metastasis of TPC-1 thyroid carcinoma cells and TGFβ1 siRNA was able to reverse the decline of visceral metastases seen by silencing ANRIL (151). Since the details of the MOA of ANRIL in thyroid carcinoma cells have not been worked-out yet and in vivo data have only been reported for thyroid carcinoma cells we have maintained ANRIL as a class II metastasis-related lncRNA.
MOA based on epigenetic modification. Neuroblastoma associated transcript-1 (NBAT-1) lncRNA and lncRNA T-cell factor 7 (lnc TCF-7) function by epigenetic modification of target genes. NBAT-1 exhibits a tumor-suppressive function in neuroblastoma and breast cancer and low expression of NBAT-1 correlates with poor survival in these types of tumors (152,153). Loss of NBAT-1 increases proliferation and migration of neuroblastoma cells due to expression of a set of genes which are repressed by NBAT-1 due to recruitment of EZH2, the catalytic component of PRC2 (152,153). NBAT-1 is critical for neuronal differentiation of neuroblastoma cells through its downstream effectors SOX9 and neuronal-specific transcription factor NRSF/REST. In addition, NBAT-1 inhibits invasion and proliferation of breast cancer cells due to activation of Wnt signaling through inactivation of dickkopf Wnt signaling inhibitor 1 (DKK1) via recruitment of PRC2 (153). The last lncRNA to be discussed is lncTCF-7, which has been studied in HCC and shown to be involved in renewal of human liver cancer stem cells (CSCs) and HCC aggressiveness through promotion of EMT (154,155). Self-renewal of liver CSCs is mediated through recruitment of the SWI/SNF chromatin remodeling complex resulting in activation of Wnt signaling through lncTCF-7 (154). Activation of EMT in HCC cells is initiated by inflammatory cytokine IL6 which activates lncTCF7 expression through binding of STAT3 to the lncTCF7 promoter (155). RNA interference based attenuation of lncTCF7 prevented IL6-induced EMT and invasion of SK-Hep1 HCC cells (155).
Metastasis-related lncRNAs: Category III
In this category we describe a melanoma, two gastric cancer-related and a breast-cancer related lncRNA with metastasis-modulating potential, which have only partially or poorly resolved MOA and varying degree of validation with respect to involvement in clinical metastasis. BANCR (BRAF-regulated lncRNA) was identified by RNA seq comparison of BRAF V600E transduced and control human melanocytes, and for insights in clinical revalance, BRAF-mutated human melanomas (156). Knock-down of BANCR reduces melanoma migration without affecting viability and proliferation. BANCR regulates a panel of genes involved in cell migration. In the less motile BANCR silenced melanoma cells, CXCL11 expression was down-regulated and impaired invasion could be rescued by exogeneous addition of CXCL11 (156). FRLnc1 (FOXM1-related lncRNA) was found to be induced by transcription factor FOXM1 in gastric cancer cells (157) and its steady-state mRNA levels are increased in 50% of gastric cancers. FRLnc1 increases expression of the EMT-mediators TGFβ1 and Twist and promotes migration of gastric cancer SGC 7901 cells ectopically transfected with FRLnc1, as shown by transwell and wound-healing assys (157). Tail vein injection experiments of these cells reveal promotion of lung metastases in comparison to control SGC 7901 cells (157). SPRY4-IT1 (SPRY4 intronic transcript 1) is derived from an intron of the SPRY gene and its decreased expression in gastric cancer correlates with overall survival (OS) and disease-free survival (DFS) (158,159). Its down-regulation in cancer tissue seems to be based on promoter methylation. Target validation experiments in SGC 7901, BGC 823 and MKN 45 gastric cancer cells point to its role in inhibition of proliferation, colony formation, cell migration and invasion and its capacity to up-regulate cyclin D1, MMP2, MMP9 and E-cadherin and to down-regulate vimentin (158,159). Ectopic expression of SPRY4-IT in BGC 823 cells reduced the number of metastatic nodules after teil vein injection (158).
As a prototype of a metastasis-suppressing lncRNA, lncRNA limiting metastasis (LIMT) was recently discovered (160). LIMT is down-regulated in aggressive basal-like breast cancer and is suppressed by EGF. The abundance of LIMT predicts clinical outcome of breast cancer patients (160). LIMT acts as an inhibitor of motility in vitro and inhibits lung metastasis after tail vein injection of MDA-MB-231 breast cancer cells (160). However, its MOA needs to be worked out in more detail.
Therapeutic Aspects
As outlined in the preceding chapters, metastasis-related lncRNAs can be either up- or down-regulated. Selection for therapeutic targets will be based on data including identification of tumor types with deregulated expression, abundance of expression and cellular localization. In addition, MOA studies such as identification of interacting partners such as DNA, RNA or protein, in vitro and in vivo metastasis-related assays and efficacy studies as well as correlation of expression with metastatic risk and survival in patients will all be important for target identification (161-165). In case of down-regulated metastasis-suppressing lncRNAs, substitution therapy with liposomes or nanoparticles delivering plasmid-based expression vectors or expression of the corresponding lncRNA with viral vectors are options for interference. However, optimisation of delivery is a pending issue. In case of overexpressed metastasis-promoting lncRNAs, silencing and MOA-dependent interference are possible options for therapeutic intervention. Tools for silencing are ASOs, liposome-delivered RNAi and vector-expressed shRNAs. ASO might be most suitable when the target functions in the nucleus, whereas duplex RNA may be the best choice when the target is located in the cytoplasm (166). These tools are the first option for inhibition of lncRNA/RNA or DNA interactions such as inhibition of lncRNA-DNA-DNA triple helix formation at genomic loci (167) or inhibition of natural antisense transcripts (168). An additional mode of intervention is to block the interaction of lncRNA with proteins mediated by domain-domain interactions involved in epi-genetic or translational control. Decoy RNAs derived by systemic evolution of ligands by experimental enrichment (SELEX) (169,170) or small molecules competing for domain-domain interaction by mimicry of the secondary structure of bindng partners are candidates for disrupting domain-domain interactions (165,171,172). Another option is inactivation of lncRNA structure due to blocking of correct folding (171). Proof-of-concept for inhibition of this type of interaction is the identification of aminoglycoside-based antibiotics that interact with ribosomal RNA (172). Availability of X-ray structures of interacting domains would be helpful to predict druggability with small molecules. Targeting lncRNAs by recruiting epigenetic modifiers may result in specificity issues, because lncRNA-epigenetic modifier interactions are involved in global reprograming of the genome. Targeting lncRNAs acting by a cis-mechanism would probably result in enhanced specificity compared to those acting by a trans-mechanism. Another critical issue is the poor conservation of the sequences of lncRNAs among different species since they are often derived from intronic or intergenic sequences. This issue has an impact on syngeneic metastasis models for evaluation of in vivo anti-metastatic efficacy and for toxicology studies of human lncRNAs in other species. As outlined, metastasis-suppressing or -promoting activity has been described for several lncRNAs. In order to design meaningful clinical studies, information which step(s) of the metastatic cascade are promoted or inhibited by individual lncRNAs would be very helpful. This issue holds true for all lncRNAs discussed in this review.
Nucleic acid-based approaches are under extensive clinical investigation. Two synthetic oligonucleotides have been approved by FDA and numerous ASO-based clinical trials are ongoing (173). At least 25 RNAi-based drug candidates are under clinical evaluation (174). Another lncRNA therapy-related aspect is plasmid-based therapy through expression of diphteria toxin-A in cancer cells under the control of the promoter of H19 lncRNA, which is an oncofetal antigen with strong specificity of expression in tumor cells. This approach is currently in Phase III clinical studies in patients with bladder cancer (175, www.biocancell.com).
The preclinical data should derive a picture, whether lnc-RNA-related therapeutic approaches will support therapy prior to dissemination, therapy after dissemination prior to formation of overt metastases, therapy after formation of overt metastases or for several of these scenarios (176). Expansion of our knowledge on context and tumor-stage dependent function of lncRNA is a further prerequisite for progress in translational exploitation of lncRNAs for treatment of metastatic disease.
References
- 1.Fidler IJ. The pathogenesis of cancer metastases: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 2.Langley RR, Fidler IJ. The seed and soil hypothesis revisited – the role of tumor-stroma interactions in metastasis to different organs. Int J Cancer. 2011;128:2527–2535. doi: 10.1002/ijc.26031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peinado H, Lavotshkin S, Lyden D. The secreted factors responsible for pre-metastatic niche formation: old sayings and new thoughts. Sem Cancer Biol. 2011;21:139–146. doi: 10.1016/j.semcancer.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat Rev Cancer. 2009;9:285–293. doi: 10.1038/nrc2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massague J, Obenauf AC. Metastatic colonization by circulating tumor cells. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16:201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obenauf AC, Massague J. Surviving at a distance: organ specific metastasis. Trends Cancer. 2015;1:76–91. doi: 10.1016/j.trecan.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang AC, Massague J. Molecular basis of metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanharanta S, Massague J. Control of metastatic progression by microRNA regulatory networks. Cancer Cell. 2013;24:410–421. [Google Scholar]
- 10.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouyssou JM, Manier S, Huynh D, Issa S, Roccaro AM, Ghodrial IM. Regulation of microRNAs in cancer metastasis. Biochim Biophys Acta. 2014;1846:255–265. doi: 10.1016/j.bbcan.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5–7. doi: 10.1038/nsmb.2942. [DOI] [PubMed] [Google Scholar]
- 13.Costa FF. Non-coding RNAs: Meet thy masters. Bioessays. 2010;32:599–608. doi: 10.1002/bies.200900112. [DOI] [PubMed] [Google Scholar]
- 14.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derrien T, Johnson R, Brussoti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1785–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ulitsky I, Bartel DP. LncRNAs: genomics, evolution and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauptman N, Glavac D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655–4669. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djebali S, Davies CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun j, Williams BA, Zaleski C, Rozowsky J, Röder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawerdena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shabab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarkis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, Fan L, Kandalaft LE, Tanyi JL, Li C, Yuan CX, Zhang D, Yuan H, Lu Y, Katsaros D, Huang Q, Montone K, Fan Y, Coukos G, Boyd J, Sood AK, Rebbeck T, Mills GB, Dang CV, Zhang L. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang KC, Chang HY. Molecular mechansisms of long noncoding RNAs. Moll Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Ann Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khorkova O, Hsiao J, Wahlstedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Del Rev. 2015;87:15–24. doi: 10.1016/j.addr.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meier K, Brehm A. Chromatin regulation: how complex does it get. Epigenetics. 2014;9:1485–1495. doi: 10.4161/15592294.2014.971580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondo Y, Shen L, Ahmed S, Boumber Y, Seikido Y, Haddad BR, Issa JP. Down-regulation of histone H3 lysine 9 methyltransferase induces centrosome disruption and chromosome instability in cancer cells. PLoS One. 2008;3:e2037. doi: 10.1371/journal.pone.0002037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sauvageau M, Golf LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li F, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D’Ecclesias MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, Nishida N, Gafa R, Song J, Guo Z, Ivan C, Barbarotto E, De Vries I, Zhang X, Ferracin M, Churchman M, van Galen JF, Beverloo BH, Shariati M, Haderk F, Estecio MR, Garcia-Manero G, Patijn GA, Gotley DC, Bhardwai V, Shureigi I, Sen S, Multani AS, Welsh J, Yamamoto K, Taniguchi I, Song MA, Gallinger S, Casey G, Thibodeau SN, LeMarchand L, Tiirikainen M, Mani SA, Zhang W, Davuluri RV, Mimori K, Mori M, Martens JW, Tomlinson I, Negrini M, Berindan-Neagoe I, Foekens JA, Hamilton SR, Lanza G, Kopetz S, Fodde R, Calin GA. CCAT2, a novel noncoding RNA mapping to 8q24 underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai Y, He J, Zhang D. Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the Wnt signaling pathway. Onco Targets Ther. 2015;8:2657–2664. doi: 10.2147/OTT.S90485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, Buiga R, Pop V, Irimie A, Fodde R, Bedrosian I, Martens JW, Foekens JA, Berindan-Neagoe I, Calin GA. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4:1748–1762. doi: 10.18632/oncotarget.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, Yao K, Kehoe SM, Lenz HJ, Haiman CA, Yan C, Henderson BE, Frenkel B, Barretina j, Bass A, Taberno J, Baselga J, Regan MM, Manak JR, Shivdasani R, Coetzee GA, Freedman ML. The 8q24 cancer risk variant rs69883267 shows long-range interaction with MYC in colon cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massague J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu M, Ting DT, Stott SL, Wittner BS, Ozsoak F, Paul S, Ciciliano JC, Smas ME, Winolur D, Gilman AJ, Ulman MJ, Xega K, Contino G, Alagesan B, Barannigan BW, Milos PM, Ryan DP, Sequist LV, Bardeesy N, Ramaswamy S, Toner M, Maheswaran S, Haber DA. RNA sequencing of pancreatic circulating tumor cells implicates WNT signaling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 38.Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN, Zhou WP, Sun SH. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression of HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. 2013;57:1882–1892. doi: 10.1002/hep.26195. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Cui F, Lv Y, Li C, Xu X, Deng C, Wang D, Sun Y, Hu G, Lang Z, Huang C, Yang X. HBsAg and HBx knocked into the p21 locus causes hepatocellular carcinoma in mice. Hepatology. 2004;39:318–324. doi: 10.1002/hep.20076. [DOI] [PubMed] [Google Scholar]
- 40.Chang L, Goldman RD. Intermediate filaments mediate cytoskeletal crosstalk. Nat Rev Mol Cell Biol. 2004;5:601–613. doi: 10.1038/nrm1438. [DOI] [PubMed] [Google Scholar]
- 41.Wu M, Bai X, Xu G, Wei J, Zhu J, Zhang Y, Li Q, Song A, Zhao L, Gang C, Han Z, Wang S, Zhou J, Lu Y, Ma D. Proteome analysis of human androgen-independent prostate cancer cell lines: variable metastatic potentials correlated with vimentin expression. Proteomics. 2007;7:1973–1983. doi: 10.1002/pmic.200600643. [DOI] [PubMed] [Google Scholar]
- 42.Mendez MG, Kojima S, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24:1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McInroy L, Määttä A. Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem Biophys Res Commun. 2007;360:109–114. doi: 10.1016/j.bbrc.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 44.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Shang YL, Gao CF, Zhang FR, Wang F, Sun SH. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 45.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1097. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 46.Kuwano Y, Pullmann R Jr, Marasa BS, Abdelmohsen K, Lee EK, Yang X, Martindale JL, Zhan M, Gorospe M. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 2010;38:225–238. doi: 10.1093/nar/gkp861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Q, Liu L, Zhao Y, Zhang Y, Wang D, Chen J, He Y, Wu J, Zhang Z, Liu Z. Hypoxia induces genomic DNA demethylation through the activation of HIF-1α and transcriptional up-regulation of MAT2A in hepatoma cells. Mol Cancer Ther. 2011;10:1113–1123. doi: 10.1158/1535-7163.MCT-10-1010. [DOI] [PubMed] [Google Scholar]
- 48.Jackson AL, Zhou B, Kim WY. HIF, hypoxia and the role of angiogenesis in non-small cell lung cancer. Exp Opin Ther Targets. 2010;14:1047–1057. doi: 10.1517/14728222.2010.511617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reymond N, Im JH, Garg R, Vega FM, Borda d’Agua B, Riou P, Cox S, Valderrama F, Muschel RJ, Ridley AJ. Cdc42 promotes transendothelial migration of cancer cells through β1 integrin. J Cell Biol. 2012;199:653–668. doi: 10.1083/jcb.201205169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stebbing J, Lit LC, Zhang H, Darrington RS, Melaiu O, Rudraraju B, Giamas G. The regulatory roles of phosphatases in cancer. Oncogene. 2014;33:939–953. doi: 10.1038/onc.2013.80. [DOI] [PubMed] [Google Scholar]
- 51.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. A cytoplasmic NF-ĸB interacting long noncoding RNA blocks IĸB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 52.Chaturvedi MM, Sung B, Yadav VR, Kannappan R, Aggarwal BB. NFĸB addiction and ist role in cancer: 'one size does not fit all'. Oncogene. 2011;30:1615–1630. doi: 10.1038/onc.2010.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huber MA, Azoitei N, Baumann P, Grünert S, Sommer A, Pehamberger H, Kraut N, Beug H, Wirth H. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J Clin Invest. 2004;114:569–581. doi: 10.1172/JCI21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen YQ, Ghosh S, Ghosh G. A novel DNA recognition mode by the NF-kappa B p65 homodimer. Nat Struct Biol. 1998;5:67–73. doi: 10.1038/nsb0198-67. [DOI] [PubMed] [Google Scholar]
- 55.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina m, Spruce T, Parenti AR, Daidone MG. Bicciato S, Piccolo S. A microRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 56.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gumireddy K, Li A, Yan J, Setoyama T, Johannes GJ, Orom UA, Tchou J, Liu Q, Zhang L, Speicher DW, Calin GA, Huang Q. Identification of a long non-coding RNA-associated RNP complex regulating metastasis after the translational step. EMBO J. 2013;32:2672–2684. doi: 10.1038/emboj.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudson CR, Wilce MC, Merrick WC, Howe PH. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell. 2011;41:419–431. doi: 10.1016/j.molcel.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomonaga T, Levens D. Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator. J Biol Chem. 1995;270:4875–4881. doi: 10.1074/jbc.270.9.4875. [DOI] [PubMed] [Google Scholar]
- 60.Hoogeveen AT, Willemsen R, Ostra BA. Fragile X syndrome related proteins and animal models. Microsc Res Tech. 2002;57:148–155. doi: 10.1002/jemt.10064. [DOI] [PubMed] [Google Scholar]
- 61.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chrisholm KM, Wan Y, Li R, Montgomery KD, Chang HY, West RB. Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One. 2012;7:e47998. doi: 10.1371/journal.pone.0047998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 64.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Function of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu L, Murat P, Matak-Vinkovic D, Murrell A, Balasubramanian S. Bibding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry. 2013;52:9519–9527. doi: 10.1021/bi401085h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale YL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, Wilson GM, Gorospe M. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. doi: 10.1038/ncomms3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee Y, Fryer JD, Kang H, Crespo-Barreto J, Bowman AB, Gao Y, Kahle JJ, Hong JS, Kheradmand F, Orr HT, Finegold MJ, Zoghbi HY. ATNX1 protein family and CIC regulate extracellular matrix remodeling and lung alveolarization. Dev Cell. 2011;21:746–757. doi: 10.1016/j.devcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, De W, Wang KM, Wang ZX. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yonemura Y, Ninomiya I, Yamaguchi A, Fushida S, Kimura H, Ohoyama S, Miyazaki I, Endou Y, Tanaka M, Sasaki T. Evaluation of immunoreactivity for erbB-2 proteins as a marker of poor short term prognosis in gastric cancer. Cancer Res. 1991;51:1034–1038. [PubMed] [Google Scholar]
- 73.Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, Liu M. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 74.Ayesh S, Matouk I, Schneider T, Ohana P, Laster M, Al-Sharef W, De-Groot N, Hochberg A. Possible physiological role of H19 RNA. Mol Carcinog. 2002;35:63–74. doi: 10.1002/mc.10075. [DOI] [PubMed] [Google Scholar]
- 75.Raveh E, Matouk U, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis – a proposed unifying theory. Mol Cancer. 2015;14:184. doi: 10.1186/s12943-015-0458-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 78.Li H, Yu B, Li J, Su L, Yan M, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khalil AM, Guttman M, Harte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Decreased expression of large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, Chen WM, Han L, Zhang EB, Kong R, De W, Shu YQ. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. doi: 10.1186/s13045-014-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grote P, Herrmann BG. The long non-coding RNA Fendrr links epigenetic control mechansisms to gene regulatory networks in mammalian embryogenesis. RNA Biol. 2013;10:1579–1585. doi: 10.4161/rna.26165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hou P, Zhao Y, Li Z, Ma M, Gao Y, Zhao L, Zhang Y, Huang B, Lu J. LincRNA ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA ROR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13:330–338. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fan J, Xing Y, Wen X, Jia R, Ni H, He J, Ding X, Pan H, Qian G, Ge S, Hoffman AR, Zhang H, Fan X. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 2015;16:139. doi: 10.1186/s13059-015-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Levay K, Slepak VZ. Tescalin is an essential factor in megakaryocytic differentiation associated with Ets family expression. J Clin Invest. 2007;117:2672–2683. doi: 10.1172/JCI27465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hashimoto S, Onodera Y, Hashimoto A, Tanaka M, Hamaguchi M, Yamada A, Sabe H. Requirement for Arf6 in breast cancer invasive activities. Proc Natl Acad Sci USA. 2004;101:6647–6652. doi: 10.1073/pnas.0401753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Müller-Tidow C. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 88.Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Up-regulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8:2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 89.Gutschner T, Hämmerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med. 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 90.Li Y, Yang Z, Wan X, Zhou J, Zhang Y, Ma H, Bai Y. Clinical prognostic value of metastasis-associated adenocarcinoma transcript 1 in various human cancers: an updated meta-analysis. Int J Biol Markers. 2016;31:e173–182. doi: 10.5301/jbm.5000185. [DOI] [PubMed] [Google Scholar]
- 91.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zörnig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhachacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yang L, Lin C, Liu W, Zhang Y, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Zhen G, Wang L. Elevated expression of long-noncoding RNA MALAT1 facilitates growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–2492. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]
- 97.Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, Sun D, Chen YX, Hong J, Chen H, Zou W, Fang JY. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a molecular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 98.Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol. 2006;13:704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tanner KG, Trievel RC, Kuo MH, Howard RM, Berger SL, Allis CD, Marmorstein R, Denu JM. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–18160. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 100.Janssen AM, Bosman CB, van Duijn W, Ostendorp-van de Ruit MM, Kubben FJ, Griffioen G, Lamers CB, van Krieken JH, van de Velde CJ, Verspaget HW. Superoxide dismutases in gastric and esophageal cancer and the prognostic impact in gastric cancer. Clin Cancer Res. 2000;6:3183–3192. [PubMed] [Google Scholar]
- 101.Chen PM, Wu TC, Wang YC, Cheng YW, Sheu GT, Chen CY, Lee H. Activation of NFĸB by SOD2 promotes the aggressiveness of lung adenocarcinoma by modulating NKK2-1 mediated IKKβ expression. Carcinogenesis. 2013;34:2655–2663. doi: 10.1093/carcin/bgt220. [DOI] [PubMed] [Google Scholar]
- 102.Meijer D, van Agthoven T, Bosma PT, Nooter K, Dorssers LC. Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Mol Cancer Res. 2006;4:379–386. doi: 10.1158/1541-7786.MCR-05-0156. [DOI] [PubMed] [Google Scholar]
- 103.Godinho MF, Sieuwerts AM, Look MP, Meijer D, Foekens JA, Dorssers LC, van Agthoven T. Relevance of BCAR4, a novel oncogene causing endocrine resistance in human breast cancer cells. Br J Cancer. 2010;103:1284–1291. doi: 10.1038/sj.bjc.6605884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Godinho M, Meijer D, Seytono-Han B, Dorssers LC, van Agthoven T. Characterization of BCAR4, a novel oncogene causing endocrine resistance in human breast cancer cells. J Cell Physiol. 2011;226:1741–1749. doi: 10.1002/jcp.22503. [DOI] [PubMed] [Google Scholar]
- 105.Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Quin L, Wie Y, Hawke DH, Hung MC, Lin C, Yang L. LncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li F, Zou Z, Suo N, Zhang Z, Wan F, Zhong G, Qu Y, Ntaka KS, Tian H. CCL21/CCR7 axis activating chemotaxis accompanied with epithelial-mesenchymal transition in human breast carcinoma. Med Oncol. 2014;31:180. doi: 10.1007/s12032-014-0180-8. [DOI] [PubMed] [Google Scholar]
- 107.Gai M, Camera P, Dema A, Bianchi F, Berto G, Scarpa E, Germena G, Di Cunto F. Citron kinase controls abscission through RhoA and anilin. Mol Biol Cell. 2011;22:3768–3778. doi: 10.1091/mbc.E10-12-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rubin LL, de Sauvage FJ. Targeting the hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- 109.Fuji M, Lyakh LA, Bracken CP, Fukuoka J, Hayakwa M, Tsukiyama T, Soll SJ, Harris M, Rocha S, Roche KC, Tominaga S, Jen J, Perkins ND, Lechleider RJ, Roberts AB. SNIP1 is a candidate modifier of the transcriptional activity of c-Myc on E box-dependent target genes. Mol Cell. 2006;24:771–783. doi: 10.1016/j.molcel.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 110.Ciurciu A, Duncalf L, Jonchere V, Lansdale N, Vasieva O, Glenday P, Rudenko A, Vissi E, Cobbe N, Alphey L, Bennett D. PNUTS/PP1 regulates RNAPII-mediated gene expression and is necessary for developmental growth. PLoS Genet. 2013;9:e1003885. doi: 10.1371/journal.pgen.1003885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Washington K, Ammosova T, Beullens M, Jerebtsova M, Kumar A, Bollen M, Nekhai S. Protein phosphatase-1 dephosphorylates the C-terminal domain of RNA polymerase II. J Biol Chem. 2002;277:40442–40448. doi: 10.1074/jbc.M205687200. [DOI] [PubMed] [Google Scholar]
- 112.Mehra R, Udager AM, Ahearn TU, Cao X, Feng FY, Loda M, Petimar JS, Kantoff P, Mucci LA, Chinnaiyan AM. Overexpression of the long non-coding RNA SCHLAP1 independently predicts lethal prostate cancer. Eur Urol. 2016;70:549–552. doi: 10.1016/j.eururo.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Prensner JR, Zhao S, Erho N, Schipper M, Iyer MK, Dhanasekaran SM, Magi-Galluzzi C, Mehra R, Sahu A, Siddiqui J, Davicioni E, Den RB, Dicker AP, Kames RJ, Wei JT, Klein EA, Jenkins RB, Chinnaiyan AM, Feng FY. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SCHLAP1. Lancet Oncol. 2014;15:1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, Jenkins RB, Triche TJ, Malik R, Bedenis R, McGregor N, Ma T, Chen W, Han S, Jing X, Cao X, Wang X, Chandler B, Yan W, Siddiqui J, Kunju LP, Dhanasekaran SM, Pienta KJ, Feng FY, Chinnaiyan AM. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 116.Hohmann AF, Vakoc CR. A rationale to target the SWI/SNF complex for cancer therapy. Trends Genet. 2014;30:356–363. doi: 10.1016/j.tig.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 118.Ikushima H, Miyazono K. TGFβ signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 119.Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pardali K, Moustakas A. Actions of TGF-beta as a tumor suppressor and pro-metastatic factor in human cancer. Biochim Biophys Acta. 2007;1775:21–62. doi: 10.1016/j.bbcan.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 121.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 122.Butz H, Racz K, Hunyady L, Patocs A. Crosstalk between TGF-β signaling and the microRNA machinery. Trends Pharmacol Sci. 2012;33:382–393. doi: 10.1016/j.tips.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 123.Putoczki TL, Thiem S, Loving A, Busuttil RA, Wilson NJ, Ziegler PK, Nguyen PM, Preaudet A, Farid R, Edwards KM, Boglev J, Luwor RB, Jarnicki A, Horst D, Boussioutas A, Heath JK, Sieber OM, Pleines I, Kile BT, Nash A, Greten FR, McKenzie BS, Ernst M. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell. 2013;24:257–271. doi: 10.1016/j.ccr.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 124.Alaiyan B, Ilyayev N, Stojadinovic A, Izadjoo M, Roistacher M, Pavlov V, Tzivin V, Halle D, Pan H, Trink B, Gure AO, Nissan A. Differential expression of colon cancer associated transcript (CCAT1) along the colonic adeno-carcinoma sequence. BMC Cancer. 2013;13:196. doi: 10.1186/1471-2407-13-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.He X, Tan X, Wang X, Jin H, Liu L, Ma L, Yu H, Fan Z. C-Myc activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumor Biol. 2014;35:12181–12188. doi: 10.1007/s13277-014-2526-4. [DOI] [PubMed] [Google Scholar]
- 126.Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. doi: 10.1186/s13046-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xiang J-F, Yin Q-F, Chen T, Yang Y, Zhang X-O, Wu Z, Zhang S, Weng H-B, Ge J, Lu X, Yang L, Chen L-L. Human colorectal specific CCAT1-L lncRNA regulates long-range chromatin interactions of the MYC locus. Cell Res. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V, Evans JR, Knudson KE, Paulsen MT, Ljungman M, Lawrence TS, Chinnaiyan AM, Feng FY. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–908. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhao B, Hou X, Zhan H. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in non-small cell lung carcinoma cells. Int J Clin Exp Med. 2015;8:18482–18487. [PMC free article] [PubMed] [Google Scholar]
- 130.Wen J, Xu J, Sun Q, Xing C, Yin W. Up-regulation of long noncoding RNA PCAT-1 contributes to cell proliferation, migration and apoptosis in hepatocellular carcinoma. Mol Med Rep. 2016;13:4481–4486. doi: 10.3892/mmr.2016.5075. [DOI] [PubMed] [Google Scholar]
- 131.Kanamori Y, Kigawa J, Itamoch H, Shimada M, Takahashi M, Kamazawa S, Sato S, Akeshima R, Terakawa N, Tang JM, He QY, Guo RX, Chang XJ. Correlation between loss of PTEN expression and Akt phosphorylation in endometrial carcinoma. Clin Cancer Res. 2001;7:892–895. [PubMed] [Google Scholar]
- 132.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumor suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 133.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumor biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Johnsson P, Ackley A, Vidarsdottir L, Liu WO, Corcoran M, Grander D, Morris KV. A pseudogene long-noncoding RNA network regulates PTEN transcription and translation in human cells. Nat Struct Mol Biol. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, Chen H, Hong J, Zou W, Chen Y, Xu J, Fang JY. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890–6892. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 136.Yang P, Chen T, Xu Z, Zhu H, Wang J, He Z. Long noncoding RNA GAPLINC promotes invasion in colrectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget. 2016;7:42183–42194. doi: 10.18632/oncotarget.9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, Yu J, Ma R. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, Moustakas A, Gyllensten U, Jones SJ, Gustafsson CM, Sims AH, Westerlund F, Gorab E, Kanduri C. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DANN triplex structures. Nat Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena T, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Vandewalle C, Comijn J, de Craene B, Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van Roy F, Berx G. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucl Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer. Hum Mol Genet. 2010;19:R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhuang Y, Lu Q, Shen B, Huang X, Shen L, Zheng X, Huang R, Yan J, Guo H. TGFβ1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci Rep. 2015;5:11924. doi: 10.1038/srep11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cruickshanks HA, Vadafar-Isfahari N, Dunican DS, Lee A, Sproul D, Lund JN, Meehan RR, Tufarelli C. Expression of large LINE-1-driven antisense RNA is linked to epigenetic silencing of metastasis suppressor gene TFPI-2 in cancer. Nucleic Acids Res. 2013;41:6857–6869. doi: 10.1093/nar/gkt438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chand HS, Du X, Ma D, Inzunza HD, Kamei S, Foster D, Brodie S, Kisiel W. The effect of human tissue factor pathway inhibitor-2 on the growth and metastasis of fibrosarcoma tumors in athymic mice. Blood. 2004;103:1069–1077. doi: 10.1182/blood-2003-06-1930. [DOI] [PubMed] [Google Scholar]
- 145.Sprecher CA, Kisiel W, Mathewes S, Foster DC. Molecular cloning, expression and partial characterization of a second human tissue-factor-pathway inhibitor. Proc Natl Acad Sci USA. 1994;91:3353–3357. doi: 10.1073/pnas.91.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hua L, Wang CY, Yao KH, Chen JT, Zhang JJ, Ma WL. High expression of long non-coding RNA ANRIL is associated with poor prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:3076–3082. [PMC free article] [PubMed] [Google Scholar]
- 147.Lin L, Gu ZT, Chen WH, Chao KJ. Increased expression of the long non-coding RNA ANRIL promotes lung cancer cell metastasis and correlates with poor prognosis. Diagn Pathol. 2015;10:14. doi: 10.1186/s13000-015-0247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Qiu JJ, Lin YY, Feng WW, Jin HY, Hua KQ. Long non-coding RNA ANRIL predicts poor prognosis and promotes invasion/metastasis in serous ovarian cancer. Int J Oncol. 2015;46:2497–2505. doi: 10.3892/ijo.2015.2943. [DOI] [PubMed] [Google Scholar]
- 149.Cabello AM, Raguz S, Zeng L, Muitaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Aguilo F, Zhou MM, Walsh MJ. Long noncoding RNA, polycomb and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011;71:5365–5369. doi: 10.1158/0008-5472.CAN-10-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhao JJ, Hao S, Wang LL, Hu CY, Zhang S, Guo LJ, Zhang G, Gao B, Jiang Y, Tian WG, Luo DL. Long non-coding RNA ANRIL promotes the invasion and metastasis of thyroid cancer cells through TGFβ/Smad signaling pathway. Oncotarget. 2016;7:57903–57918. doi: 10.18632/oncotarget.11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandru S, Ostensson M, Akyürek LM, Abrahamson J, Pfeifer S, Larsson E, Shi L, Peng Z, Fischer M, Martinsson T, Hedborg F, Kogner P, Kanduri C. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 153.Hu P, Chu J, Wu Y, Sun L, Lv X, Zhu Y, Li Y, Guo Q, Gong C, Liu B, Su S. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget. 2015;6:32410–32425. doi: 10.18632/oncotarget.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, Yan X, Ye B, Li C, Xia P, Zhang G, Tian Y, Chen R, Fan Z. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 155.Wu J, Zhang J, Shen B, Yin K, Xu J, Gao W, Zhang L. Long noncoding RNA lncTCF7, induced by IL6/STAT3 transactivation promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. J Exp Clin Cancer Res. 2015;34:116. doi: 10.1186/s13046-015-0229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Flockhart RJ, Webster DE, Qu K, Mascarenshas N, Kovalski J, Kretz M, Khavari PA. BRAFV600E remodels the melanocyte transcriptome and incuces BANCR to regulate melanoma cell migration. Genome Res. 2012;22:1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cai H, Chen J, He B, Li Q, Li Y, Gao Y. A FOXM1 related long non-coding RNA contributes to gastric cancer cell migration. Mol Cell Biochem. 2015;406:431–41. doi: 10.1007/s11010-015-2421-3. [DOI] [PubMed] [Google Scholar]
- 158.Xie M, Nie FQ, Sun M, Xia R, Liu YW, Zhou P, De W, Liu XH. Decreased long noncoding RNA SPRY4-IT1 contributing to cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J Transl Med. 2015;13:250. doi: 10.1186/s12967-015-0595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Peng W, Wu G, Fan H, Wu J, Feng J. Long noncoding RNA SPRY4-IT1 predicts poor patient prognosis and promotes tumorigenesis in gastric cancer. Tumor Biol. 2015;36:6751–6758. doi: 10.1007/s13277-015-3376-4. [DOI] [PubMed] [Google Scholar]
- 160.Sas-Chen A, Aure MR, Leibovich L, Carlvalho S, Enaka Y, Körner C, Polycarpu-Schwarz M, Lavi S, Nevo N, Kusnetzsov Y, Yuan J, Azuaje F, Oslo Breast Cancer Research Consortium (OSBREAC), Ulitsky I, Diederichs S, Wiemann S, Yakhini Z, Kristensen YN, Borrensen-Dale AS, Yarden Y. LIMT is a novel metastasis inhibiting lncRNA suppressed by EGF and down-regulated in aggressive breast cancer. EMBO Mol Med. 2016;8:1052–1064. doi: 10.15252/emmm.201606198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wahlestedt C. Targeting long non-coding RNA to therapeutically up-regulate gene expression. Nat Rev Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 162.Sanchez Y, Huarte M. Long non-coding RNAs: challenges for diagnosis and therapies. Nucl Acids Ther. 2013;23:15–20. doi: 10.1089/nat.2012.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fatemi RP, Velmeshev D, Fagihi MA. De-repressing lncRNA-targeted genes to up-regulate gene expression: Focus on small molecule therapeutics. Mol Ther Nucleic Acids. 2014;3:e196. doi: 10.1038/mtna.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fatima R, Akhade VS, Pal D, Rao SM. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther. 2015;3:5. doi: 10.1186/s40591-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci USA. 2010;107:22196–22202. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Modarressi F, Faghili MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional up-regulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Djordjevic M. SELEX experiments: new prospects, applications and data analysis in inferring regulatory pathways. Biomol Eng. 2007;24:179–189. doi: 10.1016/j.bioeng.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 170.Tsai MC, Spitake RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Colley SM, Iyer KR, Leedman PJ. The RNA coregulator SRA, its binding proteins and nuclear receptor signaling activity. IUBMB Life. 2008;60:159–164. doi: 10.1002/iub.22. [DOI] [PubMed] [Google Scholar]
- 172.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 173.McClorey G, Wood MJ. An overview of the clinical application of antisense oligonucleotides for RNA-targeting therapies. Curr Opin Pharmacol. 2015;24:52–58. doi: 10.1016/j.coph.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 174.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mat. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 175.Gofrit ON, Benjamin S, Halachmi S, Leibovitch I, Dotan Z, Lamm DL, Ehrlich N, Yutkin V, Ben-Am M, Hochberg A. DNA based therapy with diphteria toxin-A BC-819: a phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J Urol. 2014;191:1697–1702. doi: 10.1016/j.juro.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 176.Valastyan S, Weinberg R. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]