Abstract
RNA-based vaccines have recently emerged as a promising alternative to the use of DNA-based and viral vector vaccines, in part because of the potential to simplify how vaccines are made and facilitate a rapid response to newly emerging infections. SAM vaccines are based on engineered self-amplifying mRNA (SAM) replicons encoding an Ag, and formulated with a synthetic delivery system, and they induce broad-based immune responses in preclinical animal models. In our study, in vivo imaging shows that after the immunization, SAM Ag expression has an initial gradual increase. Gene expression profiling in injection-site tissues from mice immunized with SAM-based vaccine revealed an early and robust induction of type I IFN and IFN-stimulated responses at the site of injection, concurrent with the preliminary reduced SAM Ag expression. This SAM vaccine-induced type I IFN response has the potential to provide an adjuvant effect on vaccine potency, or, conversely, it might establish a temporary state that limits the initial SAM-encoded Ag expression. To determine the role of the early type I IFN response, SAM vaccines were evaluated in IFN receptor knockout mice. Our data indicate that minimizing the early type I IFN responses may be a useful strategy to increase primary SAM expression and the resulting vaccine potency. RNA sequence modification, delivery optimization, or concurrent use of appropriate compounds might be some of the strategies to finalize this aim.
Introduction
Traditional vaccines are typically based on live-attenuated or inactivated pathogens, or subunit proteins derived from pathogens. Vaccines based on live-attenuated pathogens generally result in potent, long-lived immunity, but this approach is not always feasible due to issues of manufacturing or safety. Subunit vaccines based on polysaccharides or recombinant proteins can address the limitations of live-attenuated vaccines, but generally require the use of adjuvants to increase potency (1). Nucleic acid–based vaccines (viral vectors, plasmid DNA, and RNA vaccines) have the potential to provide the combined safety and effectiveness profiles of live-attenuated and subunit vaccines. Viral vectors and DNA vaccines have been in development for many years and broadly tested in human clinical trials, where they have been shown to be harmless and immunogenic (1).
Recent progress in nucleic acid vaccines has focused on RNA vaccines [for a review, see Ulmer and Geall (2)]. RNA vaccines obviate the potential safety risks associated with other nucleic acid–based vaccines (including genomic integration and cell transformation) (3) and avoid the limitation of antivector immunity that negatively impacts the potency of viral vectors (4). An additional potential benefit in the use of RNA vaccines compared with protein subunit vaccines is the ability to stimulate an innate immune response (5). Importantly, it has been established that pattern recognition receptors (PRRs), such as the endosomal TLR, TLR7, plays a significant role in activation of the innate immune response. TLR signaling pathways ultimately lead to dendritic cell (DC) maturation and Th cell activation, which is required for the T cell–dependent B cell activation, primarily through CD40–CD40L interaction and cytokine secretion. Second, TLRs expressed in B cells also have a direct role in B cell activation and Ab secretion (6). This function of TLRs may help to determine the microbial origin of Ags recognized by the BCR and help direct the response against infectious agents (6). RNA vaccines, particularly those derived from viral genomes, are a potent stimulus for PRRs and possibly eliminate the need for adjuvant codelivery required for subunit vaccines (7).
However, activation of the innate immune response by RNA vaccines is potentially a double-edged sword. Although systemic type I IFN activated by PRRs may facilitate the adaptive immune response, it may also inhibit the amplification of the RNA replicon and the expression of Ags encoded by self-amplifying vaccines, and thereby reducing efficacy. In this article, we report that a self-amplifying mRNA (SAM) vaccine elicits in a few hours an inflammatory response indicated by the upregulation of several IFN-stimulated genes (ISGs). Endosomal TLR7 in immune cells and cytoplasmic RIG-I–like receptors (RLRs) in nonimmune cells are SAM sensors, but the lack of one or the other is not relevant for the RNA in vivo expression. In contrast, we observed that SAM Ag expression and immunogenicity were both enhanced in the absence of IFN-α/β signaling, suggesting that reduction of early type I IFN responses could improve RNA vaccine potency. These results suggest that strategies to balance early innate immune activation to minimize interference by the IFN response, although maintaining the intrinsic adjuvant activity of the RNA molecule, could elicit a strong adaptive immune response.
Materials and Methods
Mice
Animals were housed in the Novartis Vaccines and Diagnostics Animal Facility, and experiments were approved and conducted according to the Novartis Animal Care and Use Committee in accordance with the requirements for the humane care and use of animals and all applicable local, state, and federal laws and regulations. Female mice 8–10 wk of age were used for all in vivo studies. BALB/c mice were purchased from The Jackson Laboratory (Bar Harbor, ME). TLR7rsq1 mice (C57BL/6 genetic background) were described previously (8, 9). Null type I IFN-α/β receptor (IFNAR) knockout (KO) mice (129/SvEv genetic background) were purchased from B&K Universal. MAVS and TLR3KO mice (mixed C57BL/6 and 129/SvEv genetic background; BL6) were purchased from The Jackson Laboratory. MyD88 KO mice (C57BL/6 genetic background) were generated by the Novartis Institutes for BioMedical Research (NIBR) Developmental and Molecular Pathways group (Cambridge, MA). All wild-type (WT) mice were purchased from The Jackson Laboratory.
Reagents and Abs
R848 was purchased from InvivoGen. The single-stranded oligoribonucleotide (ssRNA) R0006 (5′-UUGUUGUUGUUGUUGUUGUU-3′) containing a fully phosphothioate backbone and its fully 2′-O methylated form (TLR7 antagonist) were purchased from IDT (Coralville, IA) (9). R0006 and methylated R0006 were delivered with DOTAP (Roche, Switzerland) at 5 μg/ml. Poly(I:C) was purchased from InvivoGen (San Diego, CA) and used at 10 μg/ml. RNA/SAM and ΔRNA/SAM have been delivered at 5 μg/ml with DOTAP or with Lipofectamine 2000 (InvivoGen). Cytokines were quantified by ELISA (DuoSet or Quantikine kits; R&D Systems, Minneapolis, MN) or multiplexed cytokine analysis (Meso Scale Discovery, Rockville, MD) per the manufacturers’ protocols. The MAR1-5A3 Ab was purchased from BioXCell (BE0241; West Lebanon, NH), and 2 mg of the Ab was delivered i.p. per mouse 24 h before immunization with lipid nanoparticles (LNP)/RNA. The respiratory syncytial virus (RSV) F mAb (MAB7138P) for the immunoblot to detect RSV Fusion protein was purchased from Maine Biotechnology (Portland, ME).
RNA synthesis
DNA plasmids encoding the self-amplifying RNA (10) or a replication-deficient SAM (containing a 2-aa mutation in nonstructural protein 4 resulting in abolished polymerase activity (11) (ΔRNA) were linearized at the 3′ end immediately following the 3′ end of the self-amplifying RNA sequence by restriction digest. Phenol/chloroform-extracted, ethanol-precipitated, linearized DNA templates were transcribed into RNA using the MEGAscript T7 kit (Life Technologies) and purified by LiCl precipitation. RNA was then capped using the ScriptCap m7G Capping System (CellScript), purified by LiCl precipitation, and resuspended in 100 mM citrate buffer (pH 6). For the multicistronic vector, we fused the sequence FMVD 1D/2A, encoding for the earhkqkivapvkqtlnfdllklagdvesnpgp peptide (12), to the C terminus of luciferase, before the RSV F Ag.
LNP/RNA formulation
Encapsulation of RNA within LNPs was performed as previously described (10). In brief, the lipids 1,2-dilinoleyloxy-3-dimethylaminopropane (DLinDMA) (13) and 1,2-diastearoyl-sn-glycero-3-phosphocholine (DSPC; Genzyme), 1,2-dimyristoyl-sn-glycerol-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000] (ammonium salt) (PEG-DMG 2000; Avanti Polar Lipids), and cholesterol (Sigma-Aldrich) were combined at a molar ratio of 40:10:2:48 molar percent (DLinDMA:DSPC:PEG-DMG 2000:cholesterol) by an ethanol dilution process to produce the LNPs. Equal volumes of lipid (in ethanol) and RNA (in citrate buffer) were mixed, equilibrated at room temperature for 1 h, and further diluted with 1:1 v/v citrate buffer. The resulting LNPs were concentrated and dialyzed against 1× PBS using tangential flow filtration (Spectrum Labs). An 8:1 N:P (nitrogen on DLinDMA:phosphate on RNA) molar ratio was used.
Vaccine groups
For mouse vaccination experiments, five mice per group were immunized twice, with 1 μg per mouse RNA, at days 0 and 21. Vaccines were injected into both quadriceps (50 μl per side).
Viral replicon particles (VRPs) were generated in baby hamster kidney (BHK) cells as previously described (14). Ag-expressing (or reporter gene–expressing) replicons derived from the Venezuelan equine encephalitis virus genome containing the 3′ untranslated region and packaging signal of Sindbis virus (SV) were electroporated into BHK cells along with defective helper RNAs encoding the SV capsid and glycoprotein genes (10). VRPs in the culture media were harvested and titrated. For vaccination, 5 × 105 infectious VRPs were delivered bilaterally i.m. in mouse quadriceps.
The RSV F subunit vaccine (protein/aluminum hydroxide) was prepared as described by Swanson et al. (15). A plasmid encoding a codon-optimized, fusion peptide–deleted RSV F protein with a C terminus histidine tag was synthesized (GeneArt; Thermo Fisher Scientific, Waltham, MA), and recombinant F protein was expressed from a baculovirus expression vector in HiFive cells (Invitrogen). The expressed F protein was subsequently purified by nickel affinity and size-exclusion chromatography. For vaccination, RSV F trimers were adsorbed to aluminum hydroxide and at 3 μg injected bilaterally i.m. in mouse quadriceps. R848 was resuspended in PBS and at 100 μg injected bilaterally i.m. in mouse quadriceps.
In vivo imaging
For the luciferase reporter gene experiments, five mice per group were injected bilaterally i.m. on day 0. RNA was used at 1.5 μg per mouse. Before vaccination, mice were depilated. Mice were anesthetized [2% (v/v) isoflurane in oxygen] and then shaved. Ten minutes before imaging, mice were injected i.p. with 15 mg/kg d-luciferin solution (Perkin Elmer, Waltham, MA). Animals were then anesthetized [2% (v/v) isoflurane in oxygen] and transferred to the In Vivo Imaging System Spectrum imaging system (Caliper Life Sciences). Bioluminescence images were acquired with a cooled charge-coupled device camera from individual animals on days 1, 3, 7, 14, 21, 28, 35, and 49 after RNA vaccine administration.
Secreted embryonic alkaline phosphatase assay
For the secreted embryonic alkaline phosphatase (SEAP) reporter gene experiment, five mice per group were injected bilaterally i.m. Blood was collected by retro-orbital bleed or after mouse sacrifice on days 1, 3, and 7. A chemiluminescent assay (Phospha-Light system; Applied Biosystems, Foster City, CA) was used to analyze the serum for SEAP.
SEAP activity was measured using the Phospha-Light assay system (Applied Biosystems). In brief, serum was incubated with dilution buffer at 95°C for 30 min, cooled on ice, and then assay buffer added and samples incubated at room temperature for 5 min. Reaction buffer was added, and samples were incubated for 20 min in the dark. SEAP activity was measured using the EnVision 2104 Multilabel Reader (PerkinElmer).
RSV F immunogenicity and intracellular T cell analysis
WT and IFNAR KO mice were immunized (1 μg RNA; bilateral i.m. into quadriceps) on days 0 and 21: serum samples and spleens were collected 2 wk after the second immunization. Total IgG, IgG1, IgG2a titers, and T cell responses were determined as described previously (10). For T cell analysis, five spleens from identically vaccinated BALB/c mice were pooled, and single-cell suspensions were prepared. Two Ag-stimulated cultures and two unstimulated cultures were established for each splenocyte pool. Cultures contained 1 × 106 splenocytes, anti-CD28 mAb, and brefeldin A (BD Biosciences, San Jose CA). RSV F–specific T cells were stimulated with a pool of RSV F peptides representing aa sequences 85–93, 249–258, and 51–66. Unstimulated cultures did not contain peptides and were otherwise identical to the stimulated cultures. After culturing for 6 h at 37°C, cells were washed and then stained with Pacific Blue–labeled anti-CD4 and Alexa Fluor 700–labeled anti-CD8 mAbs (BD Biosciences). Cells were washed again and then fixed with Cytofix/Cytoperm (BD Biosciences) for 20 min. The fixed cells were then washed with Perm/Wash buffer (BD Biosciences) and stained with a mixture of PerCP/Cy5.5-labeled anti–IFN-γ (eBioscience), Alexa Fluor 488–labeled anti–TNF-α, allophycocyanin-labeled anti–IL-2, and PE-labeled anti–IL-5 (BD Biosciences). Cells were washed and then analyzed on an LSR II flow cytometer (BD Biosciences). FlowJo software was used to analyze the acquired data. The CD4+ and CD8+ T cell subsets were analyzed separately. For each subset in a given sample, the percent of cytokine-positive cells was determined. The net (%) Ag-specific T cells were calculated as the difference between the percent cytokine-positive cells in the Ag-stimulated cultures and the percent cytokine-positive cells in the unstimulated cultures. The 95% confidence limits for the percent Ag-specific cells were determined using standard statistical methods.
Statistical analysis
For the bioluminescence imaging, we used the Welch t test to compare the group immunized with SAM luciferase versus the group immunized with SAM luciferase/F.
For the IgG Ag-specific titer, we used the one-way ANOVA with Bonferroni multiple comparisons test. All statistical analyses were performed using GraphPad Prism 6 software.
Tissue extraction and cell differentiation
Splenocytes were isolated from TLR7rsq1 mutant mice and MyD88 KO (or appropriate WT mice) by mechanical dissociation (Miltenyi Biotec, Bergisch Gladbach, Germany). Cells were cultured in RPMI 1640, 2.5% FCS, and penicillin/streptomycin mix. Mouse embryonic fibroblasts (MEFs) from MAVS KO and WT were derived from 13.5-d-old embryos and used at very low passage. In brief, the head and vascular tissue were removed and embryos minced. The tissue was dissociated with 2× trypsin (Life Technologies) for 5 min in a 37°C incubator. The cells were then washed with culture media (DMEM containing 10% FBS, 1% penicillin/streptomycin, and 1% l-glutamine; all reagents were purchased from Life Technologies). Treatments were performed overnight in 96-well flat-bottom tissue culture plates at a cell density of 5 × 105 cells per well in 200 μl.
Cell lines
Human PBMCs were purified by Ficoll-Paque Plus separation (Amersham) from 50 ml of heparinized blood from healthy donors. RIG-I– and MDA5-deficient MEF cell lines were a kind gift from Dr. Akira’s laboratory (Laboratory of Host Defense, World Premier International Immunology Frontier Research Center, Osaka University, Osaka, Japan). MEF cell lines were generated from age-matched KO and WT mice as described previously (16). Human plasmacytoid DCs (pDCs) were purchased from All Cell (Chicago, IL). All the blood cells were maintained in RPMI 1640 containing 10% FBS, 1% penicillin/streptomycin, and 1% l-glutamine. MEFs were maintained in DMEM.
RNA isolation, labeling, and microarray analysis
BALB/c mice (three biological replicates per group) were injected bilaterally i.m. with 50 μl per quadriceps of PBS alone (control group), RNA alone (500 ng), LNP alone, or LNP-formulated RNA (LNP/RNA). A single quadriceps from each mouse was homogenized using QIAzol reagent and the FastPrep-24 instrument (MP Bio). Total RNA was purified using RNeasy columns according to the manufacturer’s instructions (Qiagen). RNA labeling, hybridization, and scanning were performed using methods, reagents, software, and hardware purchased from Agilent Technologies. In brief, 100 ng RNA was retrotranscribed, labeled using Cy3, and column purified (Qiagen). The efficiency of Cy3 dye incorporation was assessed on a NanoDrop 1000 instrument. Reactions with specific activities >6 pmol dye/μg cRNA were used for hybridization. cRNA (1.65 μg) was hybridized onto a 4 × 44 Whole Mouse Genome Microarray (G4122F). After scanning, images were analyzed using Feature Extraction 10.7.3.1 software.
Microarray data were analyzed using the R/Bioconductor Limma open-source package (17). In brief, each spot was background corrected and spot intensities between arrays normalized using the quantile method. The average normalized spot intensity from experimental replicates was determined, and differentially expressed genes (compared with the PBS control group) were calculated using a moderated t statistic (Empirical Bayes method) followed by Benjamini-Hochberg false discovery rate correction. Genes with a false discovery rate adjusted p ≤ 0.05 and a fold-change >2 or <2 (log2 = 1) were considered significantly changed. Functional clustering was performed using the DAVID online database (18, 19). Heat maps were generated using Spotfire 4.0.3 software.
Real-time PCR of cellular mRNAs
Total RNA (500 ng) from mouse quadriceps used for microarray analysis was reverse transcribed using the Quanta cDNA kit according to the manufacturer’s protocol. TaqMan primers for mouse IIGP1, IP10, Viperin, and 18S were purchased from Applied Biosystems. Real-time PCR (RT-PCR) was performed using an Applied Biosystems ViiA7 RT-PCR machine and the following thermocycling parameters: 95°C for 20 s for one cycle followed by 95°C for 1 s and 60°C for 20 s for 40 cycles. IIGP1, IP10, and Viperin mRNA levels were normalized to 18S mRNA levels, and the fold changes in mRNA levels were compared between vaccinated and unvaccinated groups. The fold change in mRNA levels was determined using the 2−ΔΔCt method (20) and plotted (mean ± SEM) using GraphPad Prism 6.02 software.
Western blotting
BHK cells were transfected with RNA (luciferase/F) delivered with Lipofectamine 2000 (Invitrogen). Twenty-four hours later, the cells were lysed in radioimmunoprecipitation assay buffer with freshly added protease inhibitors for 10 min on ice. Lysates were cleared by centrifugation at 13,000 rpm for 10 min, and supernatant was subjected to SDS-PAGE. After blotting, the membrane was probed with an anti-F mAb.
Results
Early in vivo Ag expression at site of injection
To determine the kinetics of in vivo vaccine Ag expression of a SAM vaccine vector, we used an In Vivo Imaging System. This strategy utilizes for bioluminescence imaging the enzymatic activity of the luciferase to catalyze light-producing oxidation of the substrate luciferin. Luciferin was administrated into BALB/c mice vaccinated with a luciferase-expressing SAM vaccine, and the light emitted was monitored and quantified in live mice. Previously, we have shown that this imaging system could detect bioluminesce after the i.m. administration of a SAM vector encoding luciferase, and bioluminescence persisted for almost 2 mo after vaccination (10).
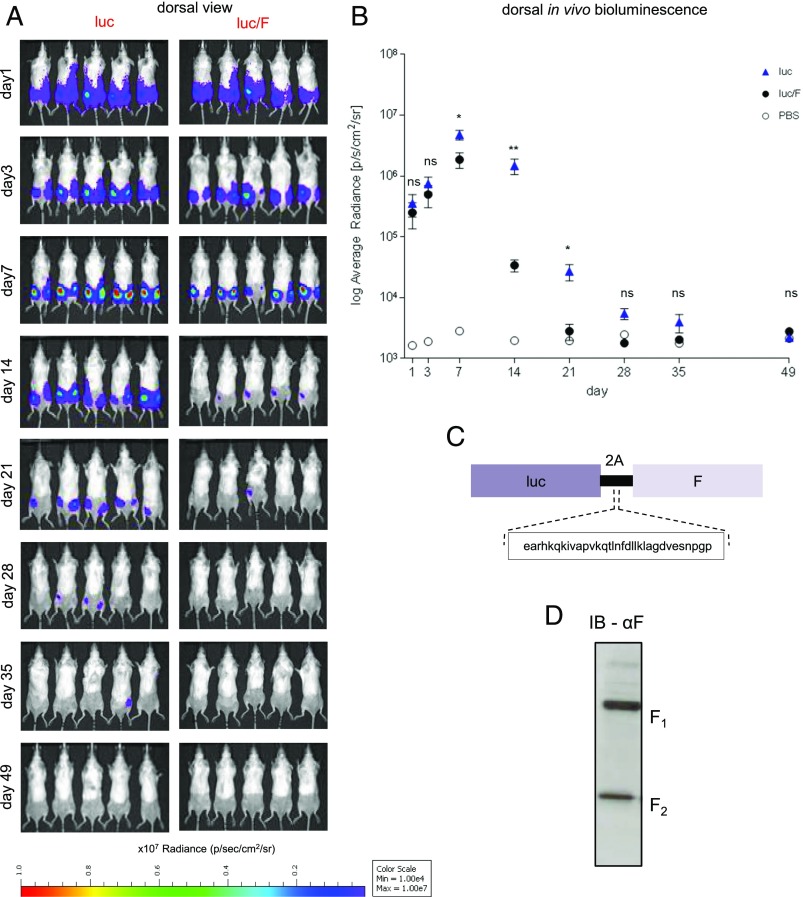
In this study, we monitored the early kinetics of SAM vaccine expression in mice. First, we examined the expression of luciferase, a cytoplasmic and poorly immunogenic protein. Luciferase was detected as early as day 1, increased at day 3, peaked at day 7, and returned to baseline after ∼1 mo (Fig. 1A, 1B, luciferase). To determine whether the nature of the Ag and immune responses directed against it affect the kinetics and magnitude of expression, we immunized a group of BALB/c mice with a SAM vaccine coexpressing luciferase and RSV F Ag (luciferase/F), a membrane-bound highly immunogenic viral protein (Fig. 1C). Immunoblot analysis of BHK cells transfected with SAM luciferase/F showed that RSV F was expressed as two characteristic fragments, F1 and F2, as the result of the cleavage of the precursor F0 for self-association and rearrangement into a stable postfusion trimer (Fig. 1D) (21). During the first week after the immunization, luciferase expression in mice immunized with SAM coexpressing luciferase and RSV F was similar to the levels in mice immunized with SAM expressing luciferase alone (Fig. 1A). This suggests that the concurrent expression of a highly immunogenic Ag such as RSV F does not affect the RNA persistence in cells, and that the early Ag expression is independent from the nature of the Ag. Intriguingly, at day 14, the signal dropped rapidly in the mice immunized with SAM luciferase/F (Fig. 1A, 1B), but not luciferase alone, indicating that the nature of the Ag can affect expression kinetics at later time points after administration (from days 7 to 14). This is reminiscent of previous findings obtained with DNA vaccines suggesting that an adaptive immune response directed toward the foreign protein expressed can lead to immune-mediated clearance of cells expressing the Ag (22).
FIGURE 1.
Mouse in vivo bioluminescence imaging after bilateral i.m. injection with SAM encoding firefly luciferase with or without RSV F. (A) Dorsal bioluminescence view of mice immunized with SAM expressing Luciferase (luc) or SAM coexpressing luciferase and RSV F (luc/F). (B) Plot of the average radiance calculated from the bioluminescence images. Each dot represents the mean of the dorsal photon count of 10 posterior legs per group of mice, expressed as photons per second per square centimeter per steradian (p/s/cm2/sr). The error bars represent the SEM. The bioluminescence intensity was evaluated at days 1, 3, 7, 14, 21, 28, 35, and 49 postimmunization. The limit of detection is 1 × 103 p/s/cm2/sr. *p < 0.05, **p < 0.01. (C) Schematic representation of the polycistronic luc/F RNA. (D) Immunoblot analysis of RSV F in BHK cell extracts transfected with the polycistronic RNA.
To evaluate whether this mechanism can be extended to SAM vaccine, on the day before the imaging we collected retro-orbital bleed from the mice immunized with luciferase/F (Fig. 1A) at days 6, 13, and 20, to avoid overstressing the animals. Animal whole blood was used to perform a pentamer analysis to evaluate F-specific CD8 response. Similar to the previous outcome with DNA vaccine (22), our pentamer analysis (Supplemental Fig. 1) indicated, over 1 wk after the immunization, that high-level expression of luciferase corresponded to imperceptible F-specific CD8+ T cell response. Over 2 wk following the immunization, the severe decline in luciferase activity corresponded at the increase of frequency of F-specific CD8-T lymphocytes in the peripheral blood. The chronological correlation between the hampering of vaccine Ag expression was consistent with a role for T cells in reducing vaccine Ag expression.
Whole-genome microarray analysis reveals a potent antiviral and inflammatory response at the site of injection
To evaluate the host response to the SAM vaccine at the site of injection, we vaccinated BALB/C mice i.m. bilaterally with PBS (negative control), RSV F–coding RNA alone (RNA), VRPs, LNP alone, or RSV F–coding RNA delivered with LNP (LNP/RNA). Quadriceps of vaccinated mice were harvested 24 h later, and total RNA was isolated and analyzed by whole mouse genome microarray.
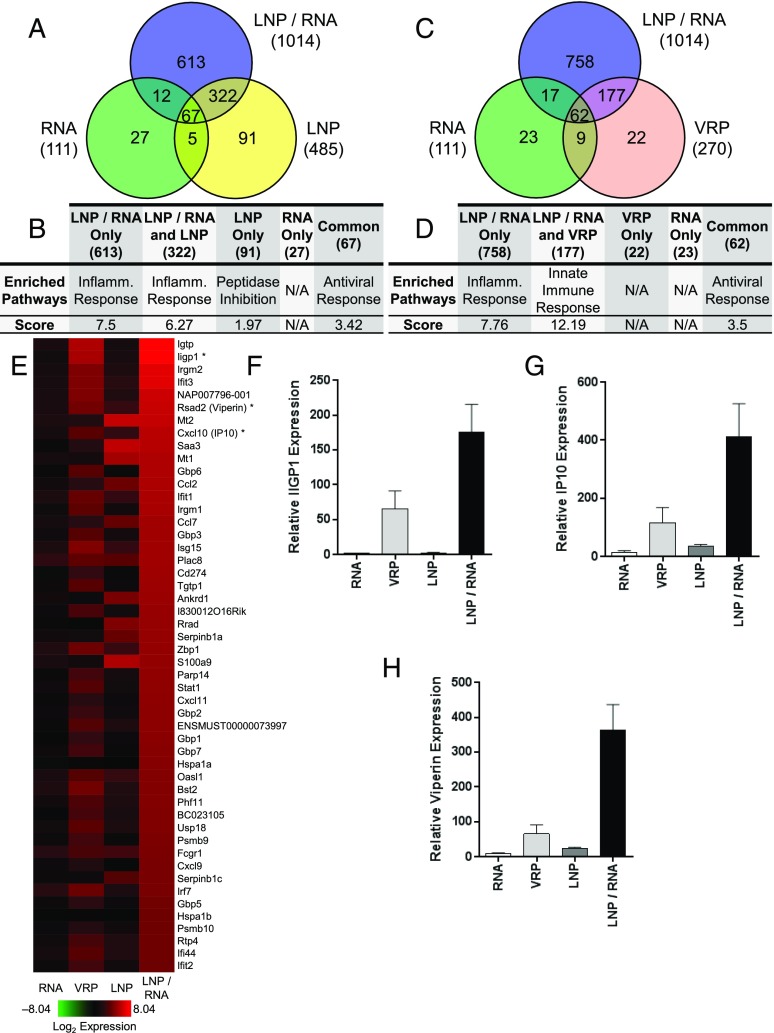
Changes of gene expression in vaccinated mice were considered significant if modulated with a p value ≤0.05 and an average intensity of log2 ratio ≥|2| compared with the PBS-injected muscle. The expression of a total of 1159 genes was significantly dysregulated by the various vaccines. Vaccination with RNA alone, LNP alone, or LNP/RNA resulted in partially overlapping patterns of host gene response (Fig. 2A). At 24 h postimmunization, a total of 1014, 485, and 111 genes were dysregulated, respectively, by LNP/RNA, LNP, and RNA. Genes regulated by LNP/RNA and RNA vaccinations appeared to be common in inflammatory and antiviral responses (Fig. 2B). Compared with RNA alone, VRP and LNP/RNA immunizations also revealed unique and overlapping signatures of gene expression (Fig. 2C). Functional clustering revealed that the inflammatory, innate immune, and antiviral responses were the most significantly enriched pathways (Fig. 2D).
FIGURE 2.
Transcription profiles induced by indicated groups in mouse quadriceps. Genes with an average log2 ratio ≥2 or ≤2 compared with PBS-immunized mice and an adjusted p ≤ 0.05 were considered significant. (A and C) Venn diagrams illustrating the number of genes modulated by RNA alone, LNP/RNA, or LNP (A) or by RNA alone, LNP/RNA, or VRP. The total number of significantly dysregulated genes by each group is listed in parentheses. (B and D) Functional clustering analysis of individual Venn diagram regions (total number of genes in parentheses) from (A) and (C) was performed using the DAVID online database. The prominent enriched pathways and associated enrichment scores are listed. (E) Heat map of the 45 most highly expressed genes. (F–H) RT-PCR of IIGP1, IP10, and Viperin confirming microarray results [denoted by an asterisk (*) in (E)]. Each column represents the mean with the SEM from three different animals.
Microarray analysis elucidated that replicon RNA delivered by VRP and LNP had the most potent impact on gene expression (Fig. 2E). Many of highly upregulated genes were ISGs that are also commonly upregulated after a viral infection. These genes included p47-GTPases (IGTP, IIGP1, TGTP1, IRGM1, and IRGM2), guanylate binding proteins (Gbp1, 3, 6, and 7), RSAD2 (Viperin), ISG15, OASL1, and the transcription factor STAT1. Another representative group of upregulated genes were members of the chemokine family (CCL2, CCL7, CXCL9, CXCL10, and CXCL11), indicating the presence of a local inflammatory response after SAM vaccine immunization (23). Immunization with replicon RNA alone had minimal impact on gene expression, but it did induce expression of several ISGs that are commonly upregulated after viral infection. To corroborate the microarray results, we verified expression of genes IIGP1, CXCL10 (IP10), and RSAD2 (Viperin) by real-time quantitative PCR (Fig. 2F, 2G). Similar to microarray results, mice immunized with LNP/RNA exhibited the largest induction of IIGP1, IP10, and Viperin expression followed by VRP, LNP alone, and RNA alone, respectively. In summary, gene expression analysis indicates an early induction of type 1 IFN at the site of injection of the SAM vaccine, similar to the host responses to a viral infection.
Gene expression time-course analysis reveals two phases of local immune responses
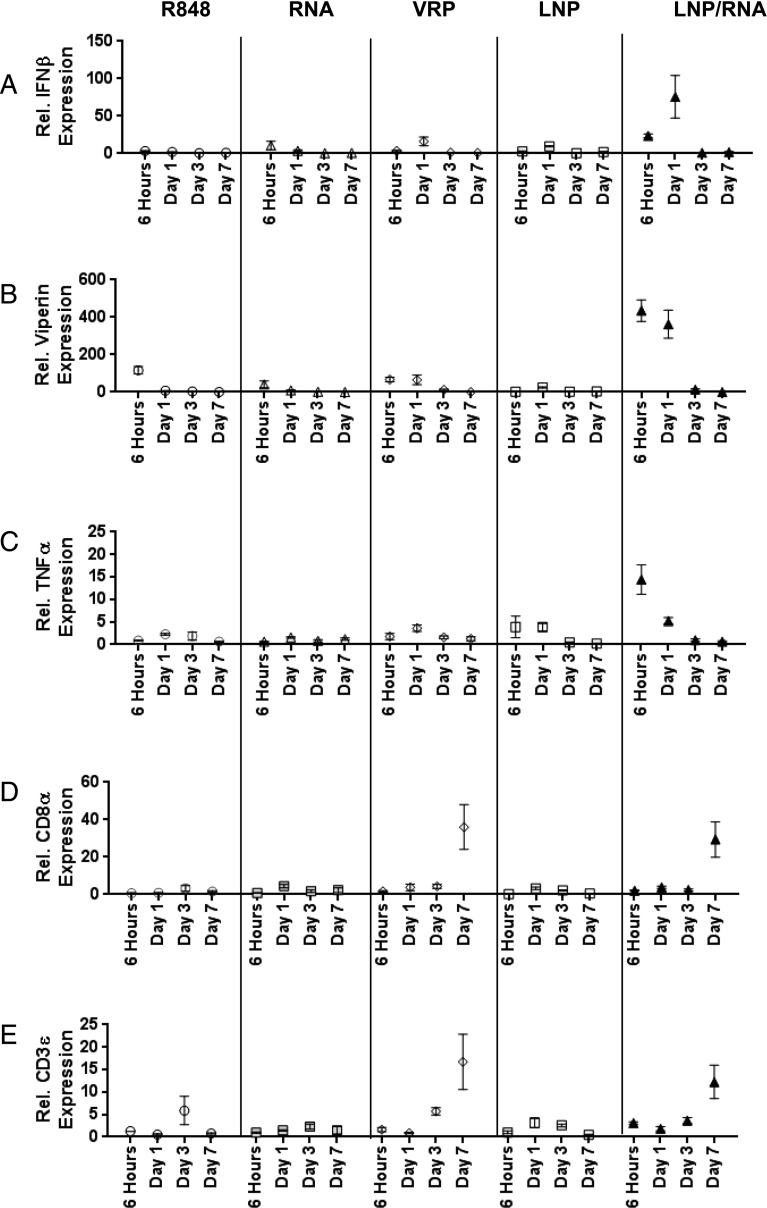
PRRs recognize exogenous pathogen-associated molecular patterns to facilitate the clearance of infectious agents. TLRs comprise a critical family of PRRs that activate early innate responses to invading microorganisms (24). Because TLR7 recognizes exogenous ssRNA, the involvement of this innate immune sensor in the host response to SAM vaccines was investigated. Mice were injected with PBS, RNA alone, LNP alone, RNA/LNP, VRP, or the TLR7 agonist R848 (25). Injected muscles were harvested at 6 h, day 1, day 3, or day 7, and expression of IFN-β, Viperin, TNF-α, CD8α, and CD3ϵ were analyzed by RT-PCR. IFN-β induction by LNP/RNA was apparent at 6 h and increased at day 1 (i.e., 75-fold, compared with the PBS control group) (Fig. 3A).
FIGURE 3.
RT-PCR time-course analysis of IFN- and T cell–associated genes. RNA harvested from mouse quadriceps after indicated vaccinations at 6 h, day 1, day 3, and day 7 was used for RT-PCR. Relative expression of IFN-β (A), Viperin (B), TNF-α (C), CD8α (D), and CD3ε (E) over PBS-injected mouse controls. Each dot represents the mean with SEM from three different animals.
At later time points (days 3 and 7), IFN-β mRNA levels returned to baseline for all groups. Similar induction patterns were also observed for Viperin and TNF-α expression (Fig. 3B, 3C). LNP/RNA immunization induced Viperin and TNF-α mRNA levels by 434- and 14-fold, respectively; all of the other vaccine groups did not show any significant change for these two genes. These findings confirm that the RNA vaccine induces a local, early antiviral response, and that this response is more potent than activation by the TLR7-specific agonist R848, suggesting that TLR7 may not be the major or the only PRR for induction of this response.
Because immunization by Ag-encoding RNA is able to induce functional, Ag-specific T cell responses, we evaluated the level of T cell markers, CD8α and CD3ε, by RT-PCR (10). The expression of both CD8α and CD3ε mRNA (Fig. 3D, 3E) were evident at day 7 after immunization with VRP-delivered replicon (36- and 17-fold, respectively) or LNP/RNA (29- and 12-fold, respectively). R848, RNA alone, and LNP alone were unable to induce any consistent change in the expression of these markers, indicating that T cell marker upregulation was elevated only in the groups where significant levels of Ag were expressed.
Mapping of endosomal RNA sensors highlights the role of TLR7 in immune cells
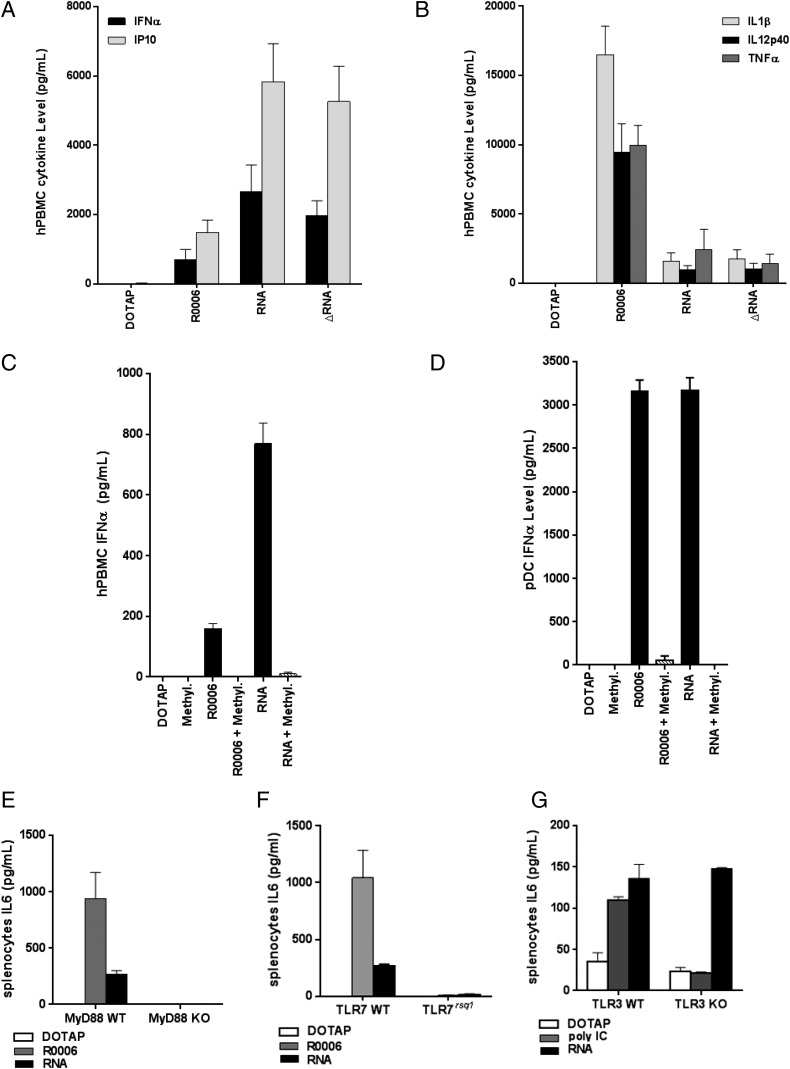
Because the genomic analysis of mouse muscle tissues at the site of vaccination revealed that SAM vaccine has a different signature from a small molecule (R848), TLR7 agonist (Fig. 3), the role of this receptor in RNA vaccine activity was further evaluated. We compared the cytokine pattern of human PBMCs (hPBMCs) stimulated with a TLR7 agonist, the single-stranded ribonucleotide R0006 (9), with the pattern induced with SAM (RNA) or an amplification-deficient version of SAM (ΔRNA). Both of the molecules were delivered with DOTAP, because this transfection reagent delivers RNA to the endosomes (26), where TLR7 is localized. Cytokines of hPBMCs from six donors were evaluated after 24 h of stimulation with the indicated molecules.
We observed that SAM was a strong inducer of IFN responses (IFN-α and IP10) (Fig. 4A) compared with the TLR7 agonist, and that the induction was independent of RNA replication. In contrast, both RNA and ΔRNA only slightly induced the expression of the proinflammatory cytokines IL-1β, IL-12p40, and TNF-α (Fig. 4B) compared with induction by R0006.
FIGURE 4.
Stimulation of hPBMCs with TLR agonists and RNA. IFN-related (A) and proinflammatory (B) cytokines induced after stimulation were measured using the mesoscale discovery platform. Each column represents the mean with SEM from six donors. IFN-α (C) in hPBMCs and IFN-α (D) in pDCs were evaluated as in (A) in the presence of methylated ssRNA. Each column represents the mean with SEM from two donors. Splenocytes from TLR7rsq1 (E), MyD88 KO (F), and TLR3 KO (G) were stimulated with the indicated TLR agonist or RNA, and IL-6 cytokines levels were measured 24 h later. Each column represents the mean with SEM from two different mice.
To further elucidate the role of TLR7 in SAM activity in hPBMCs, we took advantage of the fully methylated single-stranded oligonucleotide R0006 (Methyl.) (27). It has been reported that 2′-O methylation of any nucleotide in ssRNA can abrogate its immunostimulatory effect (28) and, as a result of higher receptor affinity, can compete with other immunostimulatory ssRNAs for TLR7 binding (29). As illustrated in Fig. 4C, 4D, and Supplemental Fig. 2, codelivery of methylated R0006 with SAM vaccines impaired IFN-α and proinflammatory cytokines induction in hPBMCs and IFN-α in pDCs, suggesting a role for engagement of the TLR7 signaling pathway. In addition, we used a mouse genetic mutant (TLR7rsq1) and KO models (MyD88 and TLR3) to elucidate the importance of the TLR7 pathway. TLR7rsq1 mice contain a point mutation within TLR7 that blocks the ability of the receptor to respond to ssRNA and small-molecule agonists (9). MyD88 is a key signaling adapter downstream of some TLRs including TLR7. We also assessed the possibility that TLR3 could respond to the SAM because TLR3 has been shown to recognize dsRNA after viral infection but is MYD88 independent (30).
Splenocytes from TLR7rsq1 mutant, MyD88 KO, or TLR3 KO mice were stimulated with RNA, R0006, or TLR3 agonist, and the induction of IL-6 was measured by ELISA 24 h later. Fig. 4E and 4F demonstrate that loss of TLR7 or MyD88 function abrogates the ability to respond to SAM because IL-6 levels were near background in splenocytes from TLR7 mutant and MyD88 KO mice. Conversely, IL-6 induction was not affected in splenocytes from TLR3 KO mice, suggesting that TLR3 does not play a role in sensing SAM vaccines (Fig. 4G).
Mapping of cytosolic RNA sensors highlights the role of RNA helicases in nonimmune cells
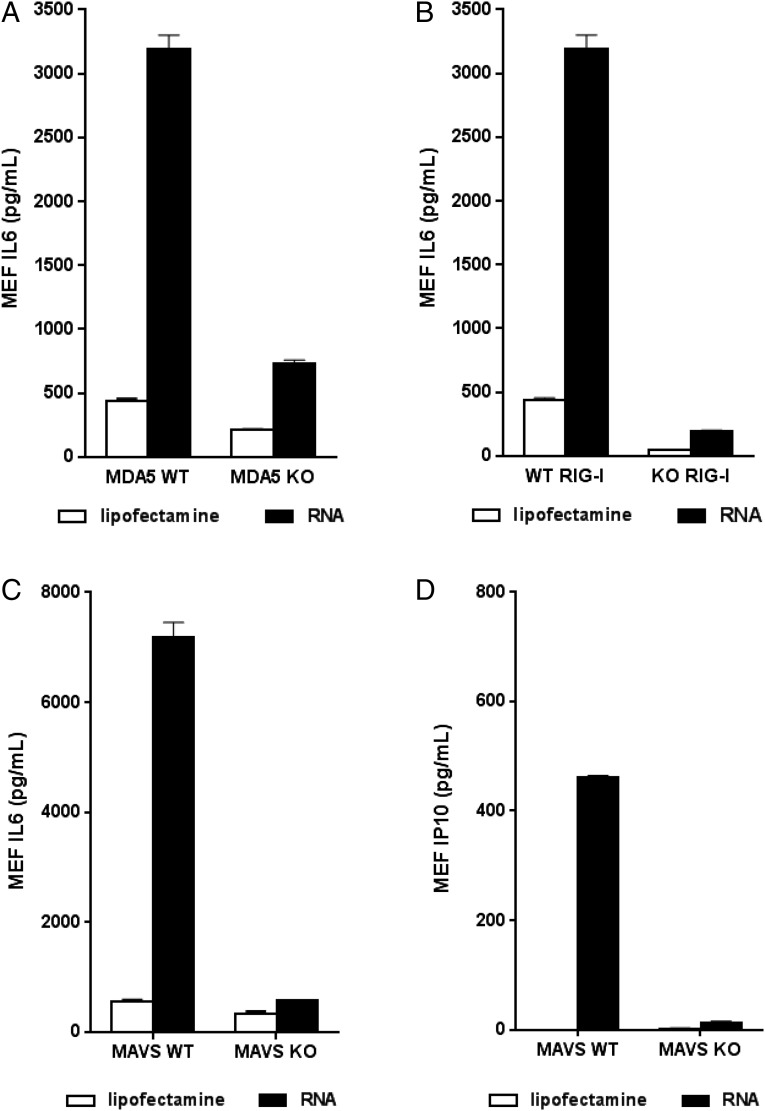
In addition to the role of TLRs as important mediators of antiviral responses, the DExD/H-box helicases RIG-I and MDA5 are a second key family of viral nucleic acid sensors that signal through the adapter protein MAVS leading to the activation of innate immune responses (31). To investigate the potential role of these cytoplasmic PRRs in the response to SAM vaccines, we transfected RIG-I and MDA5 WT or deficient MEFs (16) with lipofectamine, a carrier for cytosolic delivery (26), and measured IL-6 24 h later by ELISA. Fig. 5A and 5B illustrate that MEFs with functional RIG-I or MDA5, respectively, respond strongly, generating a large amount of IL-6 after RNA transfection. However, MEFs completely lacking functional RIG-I or MDA5 failed to respond to SAM.
FIGURE 5.
Evaluation of cytokine induction in PRR-deficient mice after replicon stimulation. Ex vivo MAVS KO MEFs were stimulated with replicon for 24 h, and IL-6 (A) and IP10 (B) were subsequently measured. RIG-I (C) and MDA5 (D) WT and deficient MEF cell lines were similarly stimulated with RNA, and IL-6 cytokine levels were measured. Each column represents the mean with SEM from two different mice.
We next asked whether MAVS-deficient cells respond to SAM. Embryos from MAVS KO mice were harvested, and MEFs were isolated and subsequently transfected with SAM as described earlier. Twenty four hours later, IL-6 levels in culture media were assayed by ELISA. The level of IL-6 generated by MAVS KO MEFs transfected with SAM RNA was markedly lower compared with MEFs from WT mice (Fig. 5C). Similarly, IP10 induction was much lower in KO mice (Fig. 5D). These data suggest that the RIG-I/MDA5 pathway plays an important role in the response of these cells to SAM vaccines.
SAM-driven Ag expression is enhanced in IFNAR KO and MAR1-5A3–treated mice
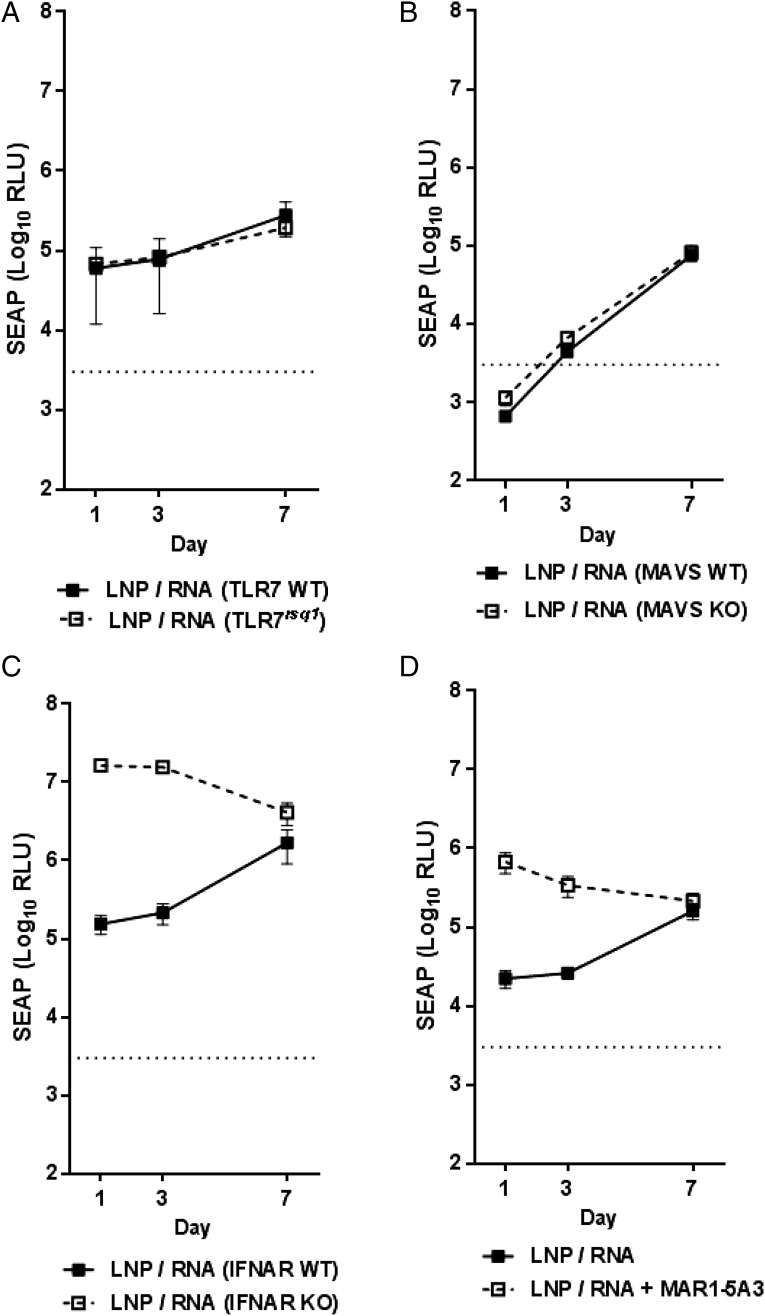
Because multiple PRRs sense SAM in vitro, we next determined whether Ag expression in KO mice deficient in these pathways was affected. SAM encoding SEAP delivered by LNP was used to immunize WT or KO mice. Sera were collected at days 1 and 3 by retro-orbital bleed, and mice were sacrificed at day 7 for the final bleed. Because splenocytes from TLR7rsq1 mice failed to respond to SAM (Fig. 4E), we evaluated SEAP expression in TLR7rsq1 mice. As illustrated in Fig. 6A, SEAP expression in both WT and KO mice were nearly identical at all times points after LNP/RNA immunization, suggesting that loss of TLR7 function had minimal impact on Ag expression from the SAM vaccine. To investigate SEAP expression after functionally limiting the ability of the RLR pathway (RIG-I and MDA5) to launch a response to SAM, we similarly immunized MAVS WT and KO mice and analyzed SEAP levels (Fig. 6B). Similar to TLR7rsq1 results, SEAP levels in MAVS WT and KO mice were similar at each time point after LNP delivery of SAM with only slighter higher SEAP levels at days 1 and 3.
FIGURE 6.
Evaluation of SEAP expression in KO mice models. TLR7rsq1 (A), MAVS KO (B), and IFNAR KO (C) or WT mice were immunized with VRP or LNP/RNA, and SEAP expression was measured at days 1, 3, and 7. SEAP expression was similarly evaluated after 24-h pretreatment of WT mice with MAR1-5A3 Ab (D). Each dot represents the mean with SEM from five animals. The dotted line represents the average alkaline phosphatase background from the PBS group.
TLR7 and the RLR pathway act to sense foreign nucleic acid within cells that have been infected by an invading pathogen and subsequently launch an innate immune response. Because microarray analyses indicated a strong local IFN-dependent response (Figs. 2, 3), we next evaluated whether inhibiting downstream antiviral responses could have an effect on Ag expression by using IFNAR KO mice (IFNAR null). Impaired type 1 IFN signaling and subsequent ISG activation had a striking effect on Ag levels after LNP delivery of SAM, resulting in a nearly 100-fold and 90-fold increase in Ag expression at days 1 and 3, respectively, over WT mice. By day 7, SEAP levels were similar in WT and KO mice (Fig. 6C).
It was reported previously that a mouse mAb specific for IFNAR (MAR1-5A3) can block type I IFN receptor signaling-induced antiviral responses (32). WT BALB/c mice were treated with the MAR1-5A3 Ab and subsequently vaccinated 24 h later, and sera were collected as described earlier. Early elevation of SEAP levels after SAM vaccine immunization in MAR1-5A3–treated mice was consistent with the findings obtained using IFNAR KO mice (Fig. 6D).
Increased Ag expression correlates with increased immunogenicity
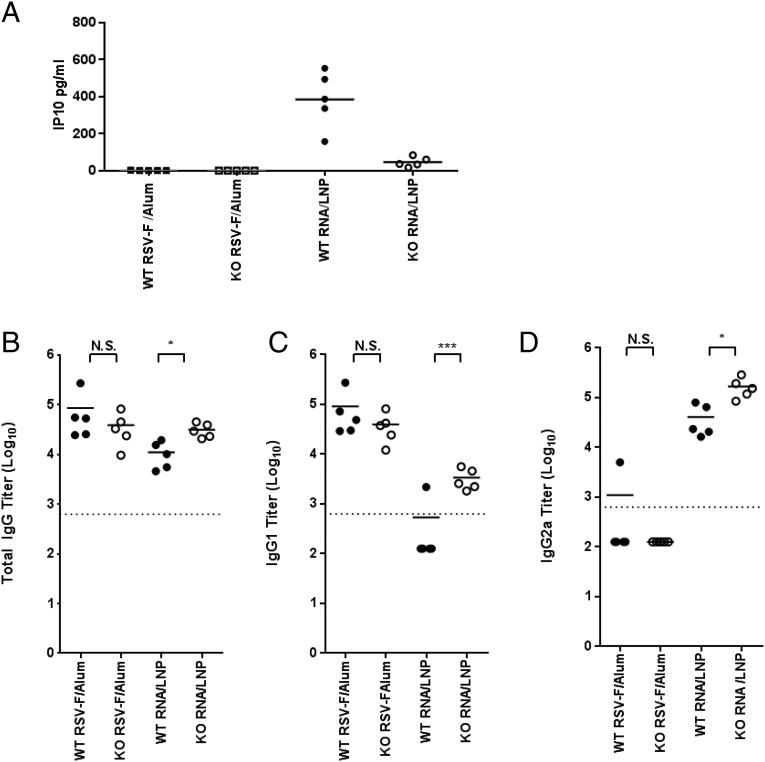
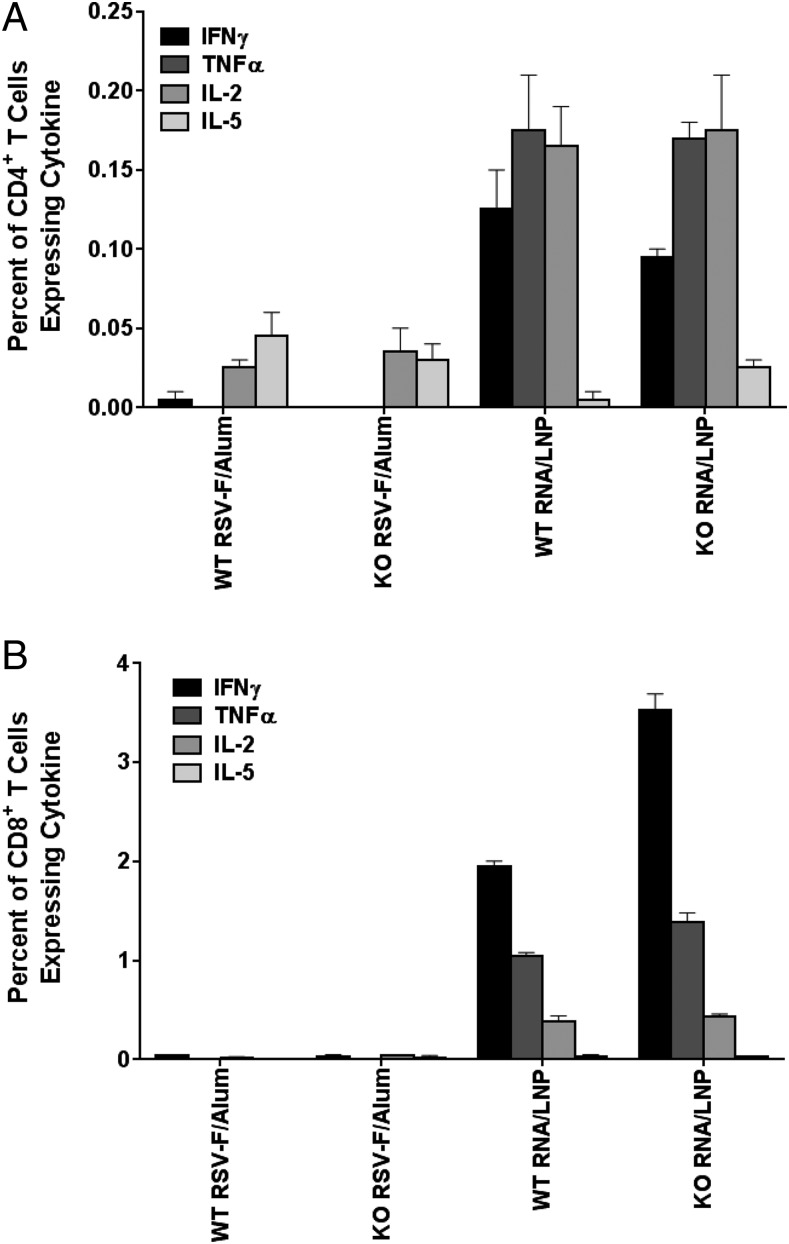
Because blocking IFN signaling (Fig. 6C) enhanced SEAP expression after SAM vaccination, it was possible this could lead to higher levels of Ag-specific immune responses. To address this possibility, we immunized IFNAR WT and KO mice with SAM encoding the respiratory syncytial virus (RSV) F protein, and total IgG, IgG1, and IgG2a titers were determined as previously described (10). In IFNR KO mice, early systemic IP10 release was blocked at 6 h after the immunization, confirming the absence of the IFN type I response (Fig. 7A). KO of IFNAR-mediated signaling resulted in a significant (p ˃ 0.1) increase in total IgG anti-RSV F Abs in mice immunized with the SAM vaccine (Fig. 7B). The levels of the IgG1 and IgG2a subclasses were also substantially increased (p < 0.05 and p < 0.001, respectively) in SAM vaccine-immunized IFNAR KO mice compared with WT controls (Fig. 7C, 7D). In contrast, an aluminum hydroxide adjuvanted recombinant protein-based RSV F vaccine induced equivalent Ab responses in WT and KO mice. Higher IgG2a titers compared with IgG1 observed for the LNP/RNA group are representative of a Th1 cell response. This is in agreement with previous findings for SAM vaccines (10). In addition, analysis of CD4+ (Fig. 8A) and CD8+ (Fig. 8B) T cell cytokine profiles revealed elevated IFN-γ levels in the LNP/RNA groups, which is consistent with a Th1 helper response; also, as expected, strong CD8+ T cell responses were evident in the SAM vaccine groups, but not in the subunit protein groups. The enhancing effect on T cell responses in IFNAR KO mice was less apparent than that on Abs, with a 1.8-fold greater increase exclusively for the subset of IFN-γ–positive CD8 T cells.
FIGURE 7.
IP10 response (6 h) in mice immunized with aluminum hydroxide/RSV F or RNA (A). Data are from individual mice (five per group, depicted as dots), and the geometric mean is represented by a solid line. Immunogenicity analysis of LNP/RNA candidate vaccine for RSV F. IFNAR WT or KO mice immunized with RNA/LNP or RSV F/aluminum hydroxide subunit vaccine and total IgG (B), IgG1 (C), and IgG2a (D) titers evaluated 2 wk after second immunization. Data are from individual mice (five per group, depicted as dots), and the geometric mean is represented by a solid line. Dotted lines indicate the limit of titer quantification (625 titer limit). To calculate geometric mean titers, we assigned titers <625 a value of 125. *p < 0.05, ***p < 0.001.
FIGURE 8.
Frequencies of F-specific, cytokine-producing T cells in mice spleens. CD4+ (A) and CD8+ (B) T cell cytokine profiling. Duplicate-cell suspensions from pooled spleens were cultured for 5 h in the presence of brefeldin A and anti-CD28 mAb and in the absence or presence of synthetic peptides representing immunodominant epitopes in the F protein. Cells were stained for cell surface CD4 and CD8 markers and for intracellular cytokines IL-2, IFN-γ, TNF-α, and IL-5, and were analyzed by flow cytometry. Each column represents a mean from pooled spleens of five mice. The error bar is a range from a duplicate culture.
Discussion
RNA-based therapeutics are demonstrating increased utility in a wide array of potential applications in modern biology, including gene therapy and vaccine indications. The synthetic methods to produce RNA, the broad effectiveness of RNA-based vaccines in animal models, and the potential to rapidly manufacture a large supply when needed (i.e., in response to pandemics) makes them an attractive platform technology. However, more efforts are needed to better understand the mechanism by which RNA vaccines work to rationally improve their efficacy (1, 2).
As with other types of nucleic acid vaccines, administration of SAM vaccines results in local Ag expression that increases in the week after the immunization (Fig. 1A, 1B). In addition, SAM vaccines induce an early antiviral-like response at the site of injection. Using whole-genome microarray analysis, we observed that many highly upregulated genes (Fig. 2E) are also commonly expressed at very high levels in host tissues after viral infections. Indeed, IRGs and GBPs belong to the same GTPase superfamily (33) and, with ISG15 (34) and Viperin (35), are IFN-inducible genes that are key mediators of the antiviral response. The transcription factor STAT1, an essential mediator of the host response to IFNs and a key player in pathogen resistance (36), was also found to be strongly upregulated. We observed that after SAM vaccination this “early” response reverted to the baseline by day 3 after SAM vaccination (Fig. 3A–C). Interestingly, the level of the T cell markers CD8α and CD3ε began to rise by day 7 (Fig. 3D, 3E), suggesting the presence of Ag-specific T cells infiltrating the site of injection. Further work is needed to better characterize the nature of the T cell infiltrate.
The SAM vaccine triggers an innate immune response, and several PRRs appear to play roles in generating a vaccine Ag-specific adaptive immune response. Using synthetic TLR7 agonists as benchmarks for receptor activation, we found that SAM was a highly potent activator of IFN responses (Fig. 4A). Moreover, IFN responses elicited in hPBMCs were solely the result of the presence of exogenous RNA and independent of RNA amplification. Furthermore, use of a methylated ssRNA TLR7 antagonist in hPBMCs (Fig. 4C, 4D) and murine splenocytes (Fig. 4E) from TLR7rsq1 mice revealed the central role of TLR7 as a sensor of SAM in immune cells. These findings are in agreement with previous reports for an mRNA-based tumor vaccine in which in vivo immune stimulation was TLR7 dependent (37). In addition, the cytoplasmic RLRs, RIG-I and MDA5, which are the key mediators of foreign nucleic acid sensing in nonimmune cells, were involved in SAM vaccine recognition, as demonstrated using RGI-I, MDA5, and MAVS KO mice.
Although the importance of the investigated PRRs in responding to SAM in vitro is clear, the lack of functional TLR7 or MAVS did not result in any apparent changes in reporter protein expression in situ after SAM vaccination (Fig. 6A). It is possible that, given the intrinsic complexity of an RNA molecule that can activate multiple PRRs, the loss of a single nucleic acid sensor mechanism by itself may not have a profound impact on Ag expression. However, the usual result of PRR-directed signaling activation is the establishment of an antiviral, type I IFN-mediated innate immune response (38). To investigate the role of IFN signaling, we took advantage of IFNAR KO mice, which are not able to respond to type I IFN. We found that reporter Ag expression was substantially higher in IFNAR KO mice after SAM vaccination (Fig. 6C), suggesting that blocking type I IFN signaling enhances Ag expression after RNA vaccine immunization. These results were confirmed using a mouse mAb specific for IFNAR (MAR1-5A3), which has been reported to potently block type I IFN receptor signaling-induced antiviral responses (32), to suppress IFN signaling in WT mice. The observed increase in Ag expression in mice with abrogated type I IFN signaling may be the result of increased translational efficiency of SAM-encoded Ag. Indeed, it has been reported that type I IFN impairs exogenous mRNA translation (39, 40). In addition, as reported by Cruz et al. (41), specific single mutations in nsP1 sequence of alphaviruses increase type I IFN levels in vitro as well as in vivo and determine the virus virulence attenuation. Based on this finding, we demonstrated (42) that specific single mutations in nsP1 of SAM, carried using viral delivery (VRP), increased IFN type I levels but reduced the RNA potency. In particular, the mutation A533I induced elevated type I IFN and decreased Ag expression of SAM in infected cells. Importantly, this mutation also reduced in vivo Ag expression and vaccine immunogenicity. We tested the impact of mutant A533I on SAM replicon using nonviral delivery. The result confirms our previous finding that, in transfected cells, the mutant A533I increases the levels of IFN type I but reduced the RNA potency (Supplemental Fig. 3). These data support our conclusion that elevated type I IFN responses impair SAM expression and potency.
This could also explain why the ratio of IgG2a to IgG1 (Fig. 7C, 7D) was not different between WT and IFNAR KO mice. Type I IFNs influence the replication and protein expression of RNA, and this could impact the magnitude of the immune response. This could explain its influence on different IgG subclasses levels, but not on their relative ratios.
Given the striking increase in reporter Ag expression after immunization of IFNAR KO mice, we investigated whether increased Ag expression could result in increased immunogenicity. Indeed, a SAM vaccine encoding RSV F protein was more potent for Ab and CD8+ T cell responses, exclusively induction of IFN-γ in IFNAR KO mice. These results suggest that the level of Ag expressed from the RNA vaccine plays an important role in vaccine efficacy, as has been reported for other nucleic acid vaccines. Kamrud et al. (43) demonstrated that increasing Ag expression from an alphavirus-based replicon vaccine resulted in markedly increased protection in immunized mice after challenge compared with an unmodified replicon. Such findings have been similarly demonstrated by multiple groups using DNA vaccines (43). IFNs regulate both innate and adaptive immune response, acting on NK cells, B cells, T cells, DCs, and phagocytic cells, but in addition to these effects, IFNs mediate many anticellular outcomes by modulating cell viability and function (44); hence transient impairment of the IFN type I response and the consequent innate resistance could improve vaccine effectiveness by multiple modes of action.
The prospects for SAM as a new vaccine platform technology are exciting, and the data are encouraging. Currently, there are two types of RNA vaccines: conventional, nonamplifying mRNA and RNA replicons derived from the genomes of positive stranded RNA viruses (2). Both approaches have their distinct advantages and limitations. The advantages of the conventional RNA vaccine approach include the simplicity of the construct and the small size of the RNA. However, the self-amplifying RNA vaccines can generate many more copies of RNA templates for Ag translation once launched. Therefore, the SAM vaccines can be significantly more potent than nonamplifying mRNA vaccines and can achieve a higher level of Ag expression with a lower amount of RNA molecules delivered. Insight into how RNA vaccines interact with host organisms and the subsequent entrainment of Ag-specific immunity will aid in the rational design of the next generation of vaccines. The data presented in this article demonstrate that modulating the early effects of the innate immune response is a key area for further investigation. Potential strategies to facilitate this in a vaccine include RNA sequence modification to generate IFN-insensitive RNA, novel formulations to optimize delivery of RNA vaccines in a way to minimize the consequences of interactions with innate immune sensors, and small-molecule modulators that target various points of the IFN signaling cascade.
Supplementary Material
Acknowledgments
We thank Joseph Loureiro (Developmental and Molecular Pathways, NIBR) for assistance with microarray data analysis, Vanessa Davis (Transgenic Breeding Service, NIBR) for assistance with transgenic and KO mice, Tina Scalzo for conducting the pentameric assay, and Ulrike Krause for editorial assistance. We are also indebted to Dr. Robert van den Berg for helpful discussions during the preparation of the manuscript. We thank the RNA Vaccine Platform Team and, in particular, Jacob Archer and Mithra Rothfeder for assistance producing RNA for this study, Luis Brito, Ayush Verma, and Nisha Chander for assistance providing formulation for this study, Christine Shaw for assisting with T cell assay analysis, and Swetha Sridhar for assisting in the primary cells isolation and cytokines ELISA.
This work was supported by the Defense Advanced Research Project Agency under agreement HR0011-12-3-001. This work was supported by Novartis Vaccines and Diagnostics, which was involved in all stages of the study conduct and analysis, now acquired by the GlaxoSmithKline group of companies.
The microarray data have been submitted to the Array Express database (https://www.ebi.ac.uk/fg/annotare/edit/4397/) under accession number E-MTAB-5603.
The online version of this article contains supplemental material.
- BHK
- baby hamster kidney
- DC
- dendritic cell
- DLinDMA
- 1,2-dilinoleyloxy-3-dimethylaminopropane
- DSPC
- 1,2-diastearoyl-sn-glycero-3-phosphocholine
- hPBMC
- human PBMC
- IFNAR
- IFN-α/β receptor
- ISG
- IFN-stimulated gene
- KO
- knockout
- LNP
- lipid nanoparticle
- MEF
- mouse embryonic fibroblast
- NIBR
- Novartis Institutes for BioMedical Research
- pDC
- plasmacytoid DC
- PRR
- pattern recognition receptor
- RLR
- RIG-I–like receptor
- RSV
- respiratory syncytial virus
- RT-PCR
- real-time PCR
- SAM
- self-amplifying mRNA
- SEAP
- secreted embryonic alkaline phosphatase
- VRP
- viral replicon particle
- WT
- wild-type.
Disclosures
T.P., A.-M.P., T.C., A.L.C., F.S.-S., K.R., J.C.D., G.M., G.R.O., A.J.G., D.Y., J.B.U., and C.I. were employees of Novartis at the time of this study. This study has been drafted after the acquisition of Novartis Vaccines by the GlaxoSmithKline group of companies in March 2015. T.P., D.Y., J.B.U., G.M., and C.I. are now employees of the GlaxoSmithKline group of companies. G.R.O. is listed as an inventor on a patent originally assigned to Novartis Vaccines and Diagnostics.
References
- 1.Geall A. J., Mandl C. W., Ulmer J. B. 2013. RNA: the new revolution in nucleic acid vaccines. Semin. Immunol. 25: 152–159. [DOI] [PubMed] [Google Scholar]
- 2.Ulmer J. B., Geall A. J. 2016. Recent innovations in mRNA vaccines. Curr. Opin. Immunol. 41: 18–22. [DOI] [PubMed] [Google Scholar]
- 3.Rayner J. O., Dryga S. A., Kamrud K. I. 2002. Alphavirus vectors and vaccination. Rev. Med. Virol. 12: 279–296. [DOI] [PubMed] [Google Scholar]
- 4.Fleeton M. N., Chen M., Berglund P., Rhodes G., Parker S. E., Murphy M., Atkins G. J., Liljeström P. 2001. Self-replicative RNA vaccines elicit protection against influenza A virus, respiratory syncytial virus, and a tickborne encephalitis virus. J. Infect. Dis. 183: 1395–1398. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki A., Medzhitov R. 2015. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16: 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasare C., Medzhitov R. 2005. Control of B-cell responses by Toll-like receptors. Nature 438: 364–368. [DOI] [PubMed] [Google Scholar]
- 7.Guy B. 2007. The perfect mix: recent progress in adjuvant research. Nat. Rev. Microbiol. 5: 505–517. [DOI] [PubMed] [Google Scholar]
- 8.Hoebe K., Beutler B. 2008. Forward genetic analysis of TLR-signaling pathways: an evaluation. Adv. Drug Deliv. Rev. 60: 824–829. [DOI] [PubMed] [Google Scholar]
- 9.Iavarone C., Ramsauer K., Kubarenko A. V., Debasitis J. C., Leykin I., Weber A. N., Siggs O. M., Beutler B., Zhang P., Otten G., et al. 2011. A point mutation in the amino terminus of TLR7 abolishes signaling without affecting ligand binding. J. Immunol. 186: 4213–4222. [DOI] [PubMed] [Google Scholar]
- 10.Geall A. J., Verma A., Otten G. R., Shaw C. A., Hekele A., Banerjee K., Cu Y., Beard C. W., Brito L. A., Krucker T., et al. 2012. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA 109: 14604–14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomar S., Hardy R. W., Smith J. L., Kuhn R. J. 2006. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J. Virol. 80: 9962–9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnelly M. L., Hughes L. E., Luke G., Mendoza H., ten Dam E., Gani D., Ryan M. D. 2001. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J. Gen. Virol. 82: 1027–1041. [DOI] [PubMed] [Google Scholar]
- 13.Heyes J., Palmer L., Bremner K., MacLachlan I. 2005. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release 107: 276–287. [DOI] [PubMed] [Google Scholar]
- 14.Perri S., Greer C. E., Thudium K., Doe B., Legg H., Liu H., Romero R. E., Tang Z., Bin Q., Dubensky T. W., Jr., et al. 2003. An alphavirus replicon particle chimera derived from venezuelan equine encephalitis and sindbis viruses is a potent gene-based vaccine delivery vector. J. Virol. 77: 10394–10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson K. A., Settembre E. C., Shaw C. A., Dey A. K., Rappuoli R., Mandl C. W., Dormitzer P. R., Carfi A. 2011. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc. Natl. Acad. Sci. USA 108: 9619–9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loo Y. M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M. A., García-Sastre A., Katze M. G., Gale M., Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smyth G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3. [DOI] [PubMed] [Google Scholar]
- 18.Huang da W., Sherman B. T., Lempicki R. A. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 19.Huang da W., Sherman B. T., Lempicki R. A. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 21.Szymczak-Workman A. L., Vignali K. M., Vignali D. A. 2012. Design and construction of 2A peptide-linked multicistronic vectors. Cold Spring Harb. Protoc. 2012: 199–204. [DOI] [PubMed] [Google Scholar]
- 22.Greenland J. R., Geiben R., Ghosh S., Pastor W. A., Letvin N. L. 2007. Plasmid DNA vaccine-elicited cellular immune responses limit in vivo vaccine antigen expression through Fas-mediated apoptosis. J. Immunol. 178: 5652–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borish L. C., Steinke J. W. 2003. 2. Cytokines and chemokines. J. Allergy Clin. Immunol. 111(Suppl.): S460–S475. [DOI] [PubMed] [Google Scholar]
- 24.Broz P., Monack D. M. 2013. Newly described pattern recognition receptors team up against intracellular pathogens. Nat. Rev. Immunol. 13: 551–565. [DOI] [PubMed] [Google Scholar]
- 25.Cervantes J. L., Weinerman B., Basole C., Salazar J. C. 2012. TLR8: the forgotten relative revindicated. Cell. Mol. Immunol. 9: 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsbach A., Müller C., Montino C., Kritzler A., Curdt R., Benahmed A., Jurk M., Vollmer J. 2012. Impact of delivery systems on siRNA immune activation and RNA interference. Immunol. Lett. 141: 169–180. [DOI] [PubMed] [Google Scholar]
- 27.Forsbach A., Nemorin J. G., Montino C., Müller C., Samulowitz U., Vicari A. P., Jurk M., Mutwiri G. K., Krieg A. M., Lipford G. B., Vollmer J. 2008. Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J. Immunol. 180: 3729–3738. [DOI] [PubMed] [Google Scholar]
- 28.Karikó K., Buckstein M., Ni H., Weissman D. 2005. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23: 165–175. [DOI] [PubMed] [Google Scholar]
- 29.Hamm S., Latz E., Hangel D., Müller T., Yu P., Golenbock D., Sparwasser T., Wagner H., Bauer S. 2010. Alternating 2′-O-ribose methylation is a universal approach for generating non-stimulatory siRNA by acting as TLR7 antagonist. Immunobiology 215: 559–569. [DOI] [PubMed] [Google Scholar]
- 30.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413: 732–738. [DOI] [PubMed] [Google Scholar]
- 31.Schlee M. 2013. Master sensors of pathogenic RNA - RIG-I like receptors. Immunobiology 218: 1322–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheehan K. C., Lai K. S., Dunn G. P., Bruce A. T., Diamond M. S., Heutel J. D., Dungo-Arthur C., Carrero J. A., White J. M., Hertzog P. J., Schreiber R. D. 2006. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 26: 804–819. [DOI] [PubMed] [Google Scholar]
- 33.Kim B. H., Shenoy A. R., Kumar P., Bradfield C. J., MacMicking J. D. 2012. IFN-inducible GTPases in host cell defense. Cell Host Microbe 12: 432–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao C., Collins M. N., Hsiang T. Y., Krug R. M. 2013. Interferon-induced ISG15 pathway: an ongoing virus-host battle. Trends Microbiol. 21: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattijssen S., Pruijn G. J. 2012. Viperin, a key player in the antiviral response. Microbes Infect. 14: 419–426. [DOI] [PubMed] [Google Scholar]
- 36.Najjar I., Fagard R. 2010. STAT1 and pathogens, not a friendly relationship. Biochimie 92: 425–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fotin-Mleczek M., Duchardt K. M., Lorenz C., Pfeiffer R., Ojkić-Zrna S., Probst J., Kallen K. J. 2011. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J. Immunother. 34: 1–15. [DOI] [PubMed] [Google Scholar]
- 38.Saito T., Gale M., Jr. 2007. Principles of intracellular viral recognition. Curr. Opin. Immunol. 19: 17–23. [DOI] [PubMed] [Google Scholar]
- 39.Pichlmair A., Reis e Sousa C. 2007. Innate recognition of viruses. Immunity 27: 370–383. [DOI] [PubMed] [Google Scholar]
- 40.Pollard C., Rejman J., De Haes W., Verrier B., Van Gulck E., Naessens T., De Smedt S., Bogaert P., Grooten J., Vanham G., De Koker S. 2013. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. 21: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cruz C. C., Suthar M. S., Montgomery S. A., Shabman R., Simmons J., Johnston R. E., Morrison T. E., Heise M. T. 2010. Modulation of type I IFN induction by a virulence determinant within the alphavirus nsP1 protein. Virology 399: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maruggi G., Shaw C. A., Otten G. R., Mason P. W., Beard C. W. 2013. Engineered alphavirus replicon vaccines based on known attenuated viral mutants show limited effects on immunogenicity. Virology 447: 254–264. [DOI] [PubMed] [Google Scholar]
- 43.Kamrud K. I., Custer M., Dudek J. M., Owens G., Alterson K. D., Lee J. S., Groebner J. L., Smith J. F. 2007. Alphavirus replicon approach to promoterless analysis of IRES elements. Virology 360: 376–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trinchieri G. 2010. Type I interferon: friend or foe? J. Exp. Med. 207: 2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.