Abstract
Antigen Receptor Galaxy (ARGalaxy) is a Web-based tool for analyses and visualization of TCR and BCR sequencing data of 13 species. ARGalaxy consists of four parts: the demultiplex tool, the international ImMunoGeneTics information system (IMGT) concatenate tool, the immune repertoire pipeline, and the somatic hypermutation (SHM) and class switch recombination (CSR) pipeline. Together they allow the analysis of all different aspects of the immune repertoire. All pipelines can be run independently or combined, depending on the available data and the question of interest. The demultiplex tool allows data trimming and demultiplexing, whereas with the concatenate tool multiple IMGT/HighV-QUEST output files can be merged into a single file. The immune repertoire pipeline is an extended version of our previously published ImmunoGlobulin Galaxy (IGGalaxy) virtual machine that was developed to visualize V(D)J gene usage. It allows analysis of both BCR and TCR rearrangements, visualizes CDR3 characteristics (length and amino acid usage) and junction characteristics, and calculates the diversity of the immune repertoire. Finally, ARGalaxy includes the newly developed SHM and CSR pipeline to analyze SHM and/or CSR in BCR rearrangements. It analyzes the frequency and patterns of SHM, Ag selection (including BASELINe), clonality (Change-O), and CSR. The functionality of the ARGalaxy tool is illustrated in several clinical examples of patients with primary immunodeficiencies. In conclusion, ARGalaxy is a novel tool for the analysis of the complete immune repertoire, which is applicable to many patient groups with disturbances in the immune repertoire such as autoimmune diseases, allergy, and leukemia, but it can also be used to address basic research questions in repertoire formation and selection.
Introduction
Every T and B cell expresses a unique Ag receptor, called TCR or BCR, respectively. Ag receptors are generated during B and T cell development by recombination of one V, one D, and one J gene on the Ag receptor loci. A large diversity of Ag receptors is created by making different combination of V, D, and J genes and by removal of nucleotides (deletions) and insertion of nontemplated (N) nucleotides at the junctional regions. This large diversity in Ag receptors is crucial for a proper immune defense against all different pathogens. After V(D)J recombination, T and B cells that express a functional Ag receptor on their membrane migrate to the periphery where they can encounter an Ag.
After Ag recognition, B cells migrate to the germinal center where they acquire somatic hypermutations (SHM) in the V region of the BCR, which change the affinity for their Ag. B cells with increased affinity are selected for survival, whereas cells with decreased affinity undergo apoptosis. Additionally, B cells can undergo class switch recombination (CSR) to replace the constant genes of their BCR, which changes the effector function of the Ab or Ig molecule, that is, the secreted form of the BCR.
Sequencing of Ag receptor genes has been done for many years to study the Ag receptor repertoire development in health and disease and to aid the diagnosis of, for example, leukemia and lymphomas. More recently, next-generation sequencing techniques have opened new doors for the analysis of the Ag receptor repertoire. Previous studies described 100–1000 reads at most, whereas now a single run can provide up to 1 million immune receptor sequences. This allows for more in-depth studies, but it also makes data analysis much more complex. Therefore, there is need for simple and robust pipelines that provide the analysis and visualization of all different aspects of the Ag receptor repertoire (1).
A variety of programs have been developed for high-throughput V(D)J gene assignment, as well as CDR3 identification and characterization, including international ImMunoGeneTics information system (IMGT)/HighV-QUEST, IgBLAST, JoinSolver, Ig analysis tool (IgAT), and Vidjil (2–6). Other tools can be used to define clones in the repertoire (Change-O) (4) or analyze Ag selection (IgAT and BASELINe) (5, 7) or CSR (8). We have previously published the ImmunoGlobulin Galaxy (IGGalaxy) tool that allows analysis of V, D, and J gene usage and CDR3 length (9). In this study, we describe the Antigen Receptor Galaxy (ARGalaxy), which is an open-source, user-friendly tool for analysis, visualization, and reporting of immune receptor repertoire data. The advantage of this tool is that it combines the analysis of V(D)J genes, CDR3, clonotypes, diversity, SHM, Ag selection, and CSR all in a single tool.
Materials and Methods
Patient samples
Peripheral blood was used from one ARTEMIS-deficient patient (ARTEMIS-13) (10), one UNG-deficient patient (UNG-2) (11), six XLF-deficient patients (XLF-1, XLF-5, XLF6-1, XLF6-2, and XLFP1, XLFP2) (12), four XRCC4-deficient patients (XRCC4-1, XRCC4-2, XRCC4-3, and XRCC4-4) (13), three LIG4-deficient patients (Ligase-IV-5, Ligase-IV-6, and Ligase-IV-7) (13), one NBS-deficient patient (NBS-4) (14), patients AT3 and AT4 (15) with ataxia telangiectasia (AT), and one patient suffering from chronic lymphocytic leukemia. Samples were obtained according to the guidelines of the Medical Ethics Committees of the Erasmus MC.
Repertoire sequencing using next-generation sequencing
For the analysis of the naive BCR repertoire, CD19+, CD27−, IgD+, CD3− naive B cells were FACS sorted. DNA was isolated using direct lysis (16), and IGH rearrangements were amplified and sequenced using Roche 454 sequencing as previously described (14, 15). In short, IGH rearrangements were amplified using a multiplex PCR using the forward VH1-6 FR1 and reverse JH consensus BIOMED-2 primers (17). These PCR products were purified and sequenced using Roche 454 sequencing as previously described (14, 15). In short, PCR products were purified by gel extraction (Qiagen, Valencia, CA) and Agencourt AMPure XP beads (Beckman Coulter, Fullerton, CA). Subsequently, the concentration of the PCR product was measured using the Quant-iT PicoGreen dsDNA assay (Invitrogen, Carlsbad, CA). The purified PCR products were sequenced on the 454 GS junior instrument using the Lib-A kit according the manufacturer’s recommendations. For the analysis of the Ag selected BCR repertoire, RNA was isolated from total PBMCs and RNA was extracted using the GenElute mammalian total RNA miniprep kit (Sigma-Aldrich, St. Louis, MO). Subsequently, cDNA was made using random primers and amplified using VH1-6 FR1 forward primers and either the CgCH (18) or the IGHA (19) reverse primer. The product was purified and sequenced as described above, using the Lib-A V2 kit for sequencing.
Data analysis
Sequences were demultiplexed based on their multiplex identifier sequence and 20–40 nt were trimmed from both sides to remove the primer sequence using the demultiplex tool. FASTA files were analyzed in IMGT/HighV-QUEST (selection of IMGT reference directory set: F+ORF+in-frame P with all alleles; search for insertions and deletions: yes; parameters for IMGT/Junction Analysis: default) (2). Subsequently, the IMGT/HighV-QUEST output files were analyzed in the immune repertoire pipeline and/or the SHM and CSR pipeline from ARGalaxy. We used the following data from healthy donors—NWK237 naive MID8 (Fig. 5A, 5C); NWK237 Ag selected (Fig. 5A–C); NWK276, NWK279, NWK180, NWK245, NWK301, NWK299, NWK3, NWK302, NWK162 (Fig. 5D, 5E); NWK31, NWK54, NWK65, NWK56 (Fig. 6A); NWK53, NWK43, NWK303, NWK383, NWK214 (Fig. 6B); Perio34, Perio37, NWK237, NWK397, NWK380, NWK299, NWK378, NWK377 (Fig. 6C)—from our previously published control data set (ENA PRJEB15348) (20). We used the immune repertoire pipeline to analyze the data from Fig. 5 using productive rearrangements and the clonotype definition (V, J, nucleotide CDR3) except for Fig. 5B, where we did not remove duplicates based on the clonotype definition. The SHM and CSR pipeline was used to analyze the data for Fig. 6 using the following settings: sequence start at FR1, productive, remove uniques, do not remove duplicates, >70% class, and >70% subclass.
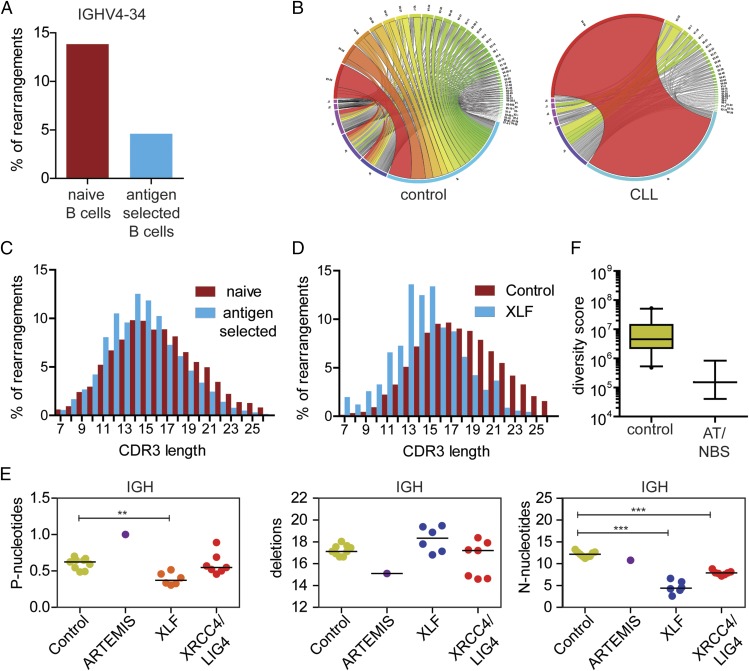
FIGURE 5.
Examples of patients with alterations in their Ag receptor repertoire as analyzed with the immune repertoire pipeline of ARGalaxy. (A) Example of a healthy control that has reduced use of IGHV4-34 in the Ag-selected B cell repertoire, as compared with the naive B cell repertoire. (B) Circos plots of VH-JH combinations of the Ag selected B cell repertoire of a chronic lymphoblastic leukemia (CLL) patient and a healthy control show clonal expansion in the CLL patient. (C) CDR3 length of IGH rearrangements in Ag-selected B cells are shorter compared with the CDR3 length of naive B cells of the same control. (D) CDR3 length distribution of IGH rearrangements in healthy controls and an XLF-deficient patient reveals reduced CDR3 length in the XLF patient. (E) Junction characteristics of naive B cell repertoire of healthy controls and patients with genetic defects in genes involved in V(D)J recombination. ARTEMIS deficiency leads to more palindromic (P) nucleotides, whereas XLF, XRCC4, and LIG 4 deficiency leads to fewer nontemplated (N) nucleotides. (F) Analysis of the diversity of the naive B cell repertoire shows reduced diversity in Nijmegen breakage syndrome (NBS) and AT patients.
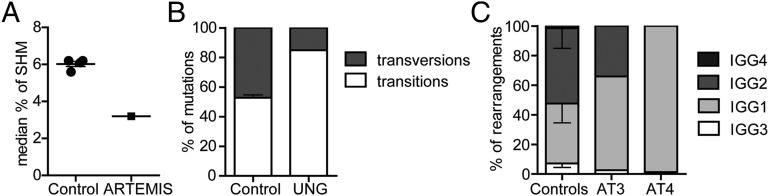
FIGURE 6.
Examples of Ag-experienced B cell repertoire in patients with altered SHM or CSR. (A) Analysis of SHM levels in an ARTEMIS-deficient patients and four age-matched healthy controls reveals reduced SHM in the ARTEMIS patient. (B) Analysis of SHM patterns shows that the UNG-deficient patient has reduced numbers of transversions at GC locations. (C) The distribution of transcripts between the different subclasses in two AT patients shows increased IGG1 transcripts in AT patients as compared with age-matched healthy controls.
Change-O
In the SHM and CSR pipeline Change-O (4) has been implemented for assigning clonal relationships. For calculations the nucleotide hamming distance substitution model was used with a complete distance of maximal three. For clonal assignment the first genes were used, and the distances were not normalized. In case of asymmetric distances, the minimal distance was used. With these settings clones are defined as sequences with the same V and J gene and junction length and a maximal of 3 nt difference between all CDR3 nucleotide sequences within a clone.
BASELINe
To determine the selection strength of the CDR and FR regions, the BASELINe tool was integrated into the SHM and CSR pipeline with the following settings: selection statistics: focused; SHM targeting model: human trinucleotide; custom boundaries: when the filter “sequence start at: leader” is chosen: 1:26:38:55:65:104:- or 27:27:38:55:65:104:- for the remaining options in the “sequence start at” filter. Please note that this means that when choosing the option CDR1 or FR2 in the sequence starts at filter, the CDR1 and FR2 are still included for the baseline calculations (7, 21).
Validation
The demultiplex tool, concatenate tool, the Immune Repertoire pipeline, and SHM and CSR pipeline are checked manually using Excel.
Results
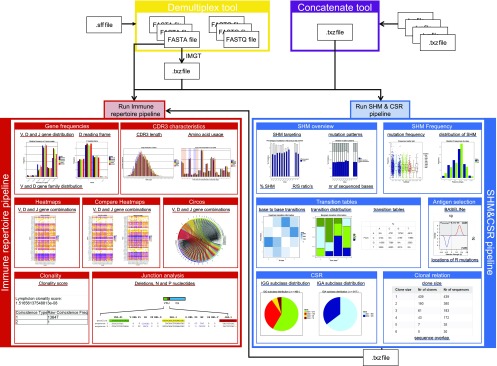
ARGalaxy provides a flexible, user-friendly tool that allows analysis of the complete immune repertoire. It is developed on the Galaxy platform and composed of a combination of Python and R components together, creating a straightforward data flow (22, 23). ARGalaxy consists of four parts: one tool for demultiplexing and sequence trimming of .sff files (the demultiplex tool); one tool that allows concatenating IMGT/High V-QUEST files (the IMGT concatenate tool); the immune repertoire pipeline, which allows for the study of V(D)J gene usage, CDR3 and junction characteristics, and the diversity of all immune receptor genes; and finally the SHM and CSR pipeline for the analysis of SHM and Ag selection in IGH, IGK, and IGL rearrangements, and CSR in the IGH rearrangements. All parts of the tool can be run independently or combined, depending on the available data and the question of interest. A complete overview of the workflow and output information of ARGalaxy can be found in Fig. 1. ARGalaxy can be installed on an existing Galaxy server through the Galaxy Tool Shed (https://toolshed.g2.bx.psu.edu/repository?repository_id=2e457d63170a4b1c&changeset_revision=28fbbdfd7a87) (24), and it is freely available via https://bioinf-galaxian.erasmusmc.nl/argalaxy/ (Fig. 2A). Tutorials and detailed descriptions for all filtering options and output data can be found on the front page of the Web site. Additionally, at the bottom of each page detailed descriptions of the filters and the results on that page can be found.
FIGURE 1.
Schematic overview of the ARGalaxy tool. ARGalaxy consists of four different parts (the demultiplex tool, the concatenate tool, the immune repertoire pipeline, and the SHM and CSR pipeline) that can be run independently of, or in combination with, each other.
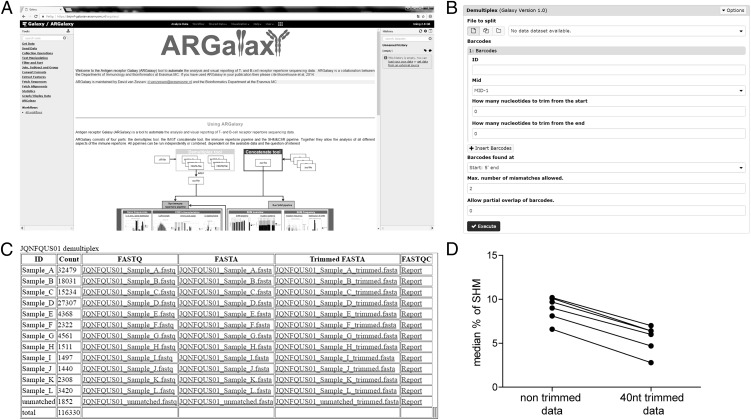
FIGURE 2.
Additional information on the demultiplex tool. (A) Main page of the ARGalaxy tool. (B) Main page of the demultiplex tool. (C) Results page of the demultiplex tool. (D) Data trimming is essential for the analysis of SHM frequency. SHM frequency in Ag-experienced B cells as analyzed using untrimmed and 40-nt trimmed data is shown.
The demultiplex tool
The demultiplex tool allows demultiplexing of .sff files based on the Roche multiplex identifier (MID) tags (Fig. 2B). This tool allows the user to select the appropriate MID tags, assign a sample name, and choose the amount of nucleotides to be trimmed from all sequences on either or both sides. It is possible to search for MID tags on either the 3′ or 5′ end and to adjust the number of allowed mismatches or the amount of partial overlap allowed between MID tags. This pipeline is based on freely available tools, including sff2fastq (https://github.com/indraniel/sff2fastq), FASTX-Toolkit software (http://hannonlab.cshl.edu/fastx_toolkit/commandline.html), and the FastQC tool to check the quality of your data (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Upon demultiplexing, data will be downloadable in FASTA and FASTQ formats (Fig. 2C). The FASTA files can be run in the immune repertoire pipeline, or they can be uploaded to IMGT/HighV-QUEST (2).
Data trimming using the demultiplex tool is an essential step in data preparation, as obtained reads often consist of template-specific sequences flanked by sequence tags or barcodes that are not template-specific but used to multiplex samples or are platform-specific. Additionally, Ag receptor rearrangements are frequently amplified with family or consensus primers. These primers are designed to limit the number of primers necessary to cover the high number of genes by allowing mismatches in primer binding. When including either the nontemplate-specific or the template-specific primer regions in downstream analysis, this can highly influence results. For example, the median percentage SHM in Ag-experienced B cells of healthy donors in trimmed data is significantly decreased compared with untrimmed data (Fig. 2D). Additionally, when the primer or tag regions contain stop codons, transcripts can be unfairly categorized as unproductive, also skewing analysis results.
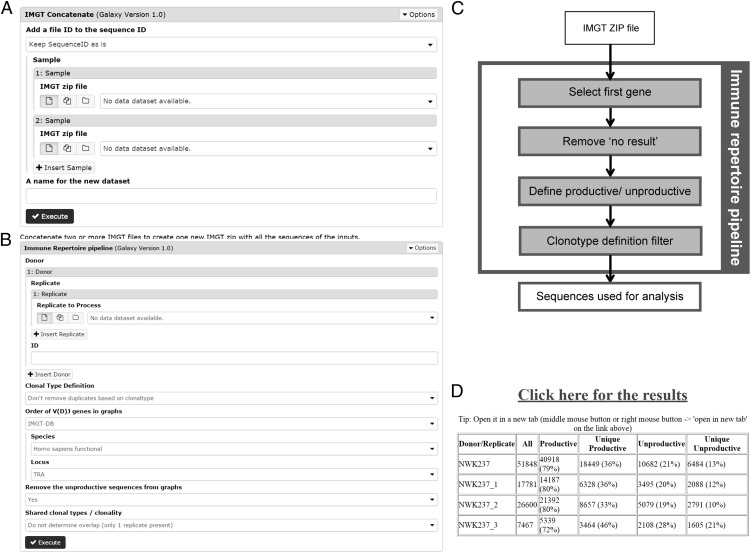
The concatenate tool
The concatenate tool allows combining of multiple .txz files (as downloaded from IMGT/High V-QUEST) into a single .txz file (Fig. 3A). This can for instance be used for combining sequencing data from a single patient that are obtained over multiple runs. The filter “add a file ID to the sequence ID” can be set to “add file ID to sequence ID to identify original file” to add a file specific ID to the sequence ID. This can be used to track from which original file a specific sequence originates.
FIGURE 3.
Additional information on the concatenate tool and the immune repertoire pipeline. (A) Main page of the concatenate tool. (B) Main page of the immune repertoire pipeline. (C) Flow chart with the filtering steps included in the immune repertoire pipeline. (D) Results page of the immune repertoire pipeline.
The immune repertoire pipeline
The immune repertoire pipeline is an extended version of the previously published ImmunoGlobulin Galaxy (IGGalaxy) pipeline and now allows the analysis of V(D)J gene usage, junction characteristics and the diversity of the repertoire (9). BCR (IGH, IGK, IGL) as well as TCR (TRA, TRB, TRD, TRG) data from up to 13 different species can now be analyzed.
Input files.
Three different input files can be used for the immune repertoire pipeline: FASTA files, .txz files from IMGT/HighV-QUEST (2), or output files from the SHM and CSR pipeline (see below). The FASTA sequences do not contain information about the CDR3 and V(D)J gene usage, and they will therefore be analyzed using IgBLAST and the corresponding BLAST database (ftp://ftp.ncbi.nih.gov/blast/executables/igblast/release/) that have been implemented in ARGalaxy (9). The .txz files from IMGT/HighV-QUEST and output files from the SHM and CSR pipeline already contain all the information provided by IMGT/HighV-QUEST, and the relevant information for the immune repertoire pipeline is extracted from the 11 IMGT/HighV-QUEST files (for details on columns used see Supplemental Table I). The immune repertoire pipeline allows simultaneous uploading of multiple replicates from the same donor (as needed for analysis of the diversity) and multiple donors can be analyzed in parallel (Fig. 3B).
Filtering options.
When analyzing IMGT/HighV-QUEST files, multiple filtering steps are performed in the immune repertoire pipeline to exclude sequencing reads where no rearrangement can be found and to prevent overrepresentation of rearrangements due to technical duplication and differences of RNA expression between cells (14). First, the pipeline only takes into account the best matched V, D, and J gene as assigned by IMGT/HighV-QUEST. Second, reads with “no results” as defined by IMGT/HighV-QUEST are removed from the data (Fig. 3C). Third, dependent on the research question, the “clonal type definition” filter can be used to filter out duplicate rearrangements. A duplicate can be defined based on the top V gene, the CDR3 nucleotide or amino acid sequence in combination with the top J gene, D gene, or both. The filter “remove the unproductive sequences from graphs” can be used to include or remove unproductive rearrangements. Unproductive rearrangements contain a stop codon or a frame shift and are derived from DNA from B or T cells that underwent a productive rearrangement on their second allele. These rearrangements have not been selected and therefore allow the user to study the direct result of V(D)J recombination without the influence of selection. The junction analysis tables (see description below) in the results section will always contain details on both unproductive and productive sequences. Details on the basic steps for sequence alignment and filtering when analyzing FASTA files using IgBLAST are previously described (9).
Furthermore, the immune repertoire pipeline also allows you to change the order of the V, D, and J genes used to visualize the data in the “gene frequency” results tab (see description below). This can be altered by changing the “order of V(D)J genes in graphs” option to “user defined” and entering the preferred gene order. This can be useful for visualizing the genes in order of their chromosomal location. In the default setting the order of the V, D, and J genes in the graphs will be based on the order of their gene name.
The immune repertoire pipeline also allows the identification of overlapping sequences between different replicates. The type of calculation performed is dependent on the setting of the “shared clonal types/clonality” filter. When only a single replicate is performed no overlap can be determined (“do not determine overlap”). When multiple replicates are performed the number of sequences that overlap between the replicates can be determined (“determine the number of sequences that share the same clonal type between the replicates”). When three or more replicates are analyzed the “determine clonality of the donor” option allows the calculation of the number of overlapping sequence and the clonality score as described by Boyd et al. (25).
Data visualization.
Upon analysis, the immune repertoire pipeline shows the total number of sequences for each sample and the number of productive and unproductive sequences that passed the chosen filter settings (Fig. 3D). After entering the results section, different tabs are shown with the various output data (Fig. 1). The “gene frequency” tab shows the distribution of top V, D, and J genes and gene families and gives information on the D-reading frame usage. The “CDR3 characteristics” tab gives the median and the distribution of CDR3 lengths and the amino acid usage in the CDR3. Information on the combinations of V, D, and J genes can be found in the “heatmaps” and the “circos” tabs (26). The “compare heatmaps” tab can be used to compare heatmaps if multiple donors are analyzed in parallel. Depending on the settings of the “shared clonal types/clonality” filter and the number of uploaded replicates, the “clonality” tab provides information on the diversity of the repertoire and the number of overlapping sequences. Results of the analysis of the junctions (number of nontemplated, palindromic, and deleted nucleotides) can be found in the “junction analysis” tab. This tab is not shown when using FASTA files as input files, because IgBLAST does not provide information on the junction characteristics. Under the “downloads” tab all data used for the above-described graphs can be found and downloaded for external analysis if needed. Details on all calculations used for the results and graphs of the immune repertoire pipeline can be found in Supplemental Table II.
The SHM and CSR pipeline
Input files.
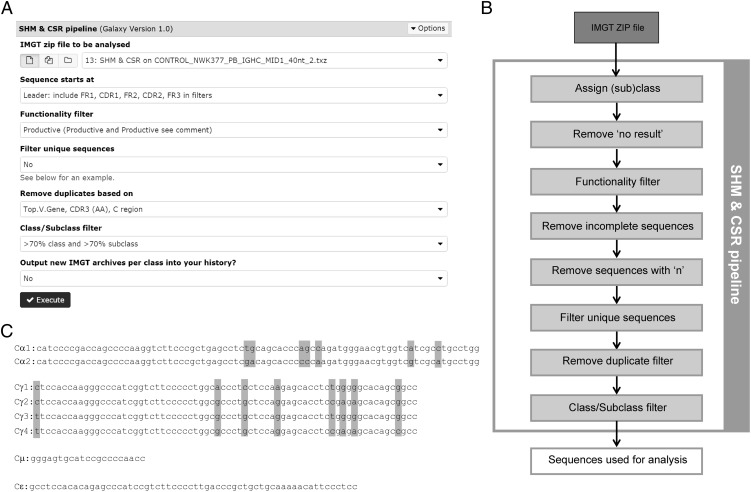
The SHM and CSR pipeline accepts IMGT/HighV-QUEST .zip and .txz as input files, as IMGT provides the most complete data set regarding SHM (2). Upon running the SHM and CSR pipeline, the relevant information on SHM is extracted from the 11 output files present in the IMGT/HighV-QUEST output files (Supplemental Table I), but the assignment of the subclasses and clonal relationship is performed separately and is based on the nucleotide sequences of the BCR rearrangements. When running the SHM and CSR pipeline several filter settings can be chosen for analysis (Fig. 4). All different filtering options are discussed below.
FIGURE 4.
Additional details on the SHM and CSR pipeline. (A) Main page of the SHM and CSR pipeline. (B) Flowchart showing the filtering steps performed in the SHM and CSR pipeline. (C) Alignment of the nucleotide sequences encoding the different (sub)classes in the SHM and CSR pipeline. The gray boxes indicate nucleotides that differ between subclasses.
Quality filtering.
The “sequence starts at” filter can be used to exclude FR or CDR regions from analysis when the sequence does not contain the complete VH gene, or when the VH primer is located in one of the FR or CDR regions. Furthermore, the “sequence starts at” filter also defines the region of the V gene that should be present in the analyzed region, and incomplete sequences (sequences missing a gene region [e.g., FR1/CDR1] that should be present) are removed from the analysis. Therefore setting this filter correctly is very important to prevent unfairly excluding sequences. Additionally, the pipeline automatically filters out sequences with ambiguous bases (uncalled “n” bases) in the analyzed region.
Filtering options.
The SHM and CSR pipeline allows several options for data filtering on functionality and unique rearrangements. The “functionality” filter allows filtering on productive rearrangements, unproductive rearrangements, or both.
The “filter unique sequences” option allows the inclusion of each unique sequence only once or the inclusion of only unique sequences present twice or more for further analysis. In this case, a unique sequence is defined by the nucleotide sequence of the above-defined analyzed region and the constant gene. When choosing the “remove uniques” option, first all sequences occurring only once (based on the nucleotide sequence of the above-analyzed regions) are removed. Afterward, all unique sequences are selected based on the analyzed region in combination with the subclass. Therefore, two sequences with the same nucleotide sequence but different subclasses will be both included in the analysis. This option can be used to differentiate between true SHM and sequencing errors as the likelihood of a sequencing error occurring twice in the exact same location in the same sequence is really small (27). When selecting the “keep unique” option, all duplicates are removed based on the combination of the above-selected region and the subclass. Selecting only unique sequences prevents overrepresentation of certain clones due to amplification biases. Additionally, or alternatively, the “remove duplicates bases on” filter can be used to only include a single rearrangement per clone. At this point, a clone can be identified based on the CDR3 sequence (amino acid or nucleotide) alone or in combination with the top V gene, the C region, or both. An overview of all filtering steps and their order are shown in Fig. 4B.
Subclass assignment.
During analysis the SHM amd CSR pipeline identifies human Cμ, Cα, Cγ, and Cε constant genes by a custom script specifically designed for human (sub)class assignment in repertoire data. In this script the reference sequences (Fig. 4C) for the subclasses (NG_001019) are divided into 8-nt chunks that overlap by 4 nt. These overlapping chunks are then individually aligned in the right order to each input sequence. The percentage of the chunks identified in each rearrangement is calculated in the “chunk hit percentage.” The “chunk hit percentage” gives a good separation between the Cμ, Cα, Cγ, and Cε genes, because they have little sequence similarity. In contrast, the Cα and Cγ subclasses are very homologous and only differ in a few nucleotides (Fig. 4C). To assign subclasses, the “nt hit percentage” is calculated. This percentage indicates how well the chunks covering the subclass-specific nucleotides match with the different subclasses. The “human class/subclass” filter can be used to define the stringency of the filter. The most stringent filter for the subclass is 70% “chunk hit percentage” and 70% “nt hit percentage,” which means that at least 70% of the 8 nt chunks have to align with the class and five out of seven subclass-specific nucleotides for Cα or six out of eight subclass-specific nucleotides of Cγ should match with the specific subclass. When using a constant primer that results in a shorter C sequences than the reference sequence (Fig. 4C), the “chunk hit percentage” will be lower because not all of the reference sequence can be aligned and the subclass may not be assigned.
We have compared our custom script with blastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) using a dataset of 245 unique IGHG and IGHA rearrangements in which the subclass was manually assigned. Our custom script correctly assigned all 245 IGH rearrangements, whereas blastn could not assign 3 of the 245 IGH rearrangements.
Filtered IMGT/HighV-QUEST output files.
If desired, additional output files (one for each class) can be obtained that contain information on the sequences that passed the selected filtering criteria. These files are in the same format as IMGT/HighV-QUEST output files and therefore can be analyzed in the immune repertoire pipeline or other analysis programs suitable for IMGT/HighV-QUEST output files, such as IgAT (5).
Data visualization.
Upon analysis, the SHM and CSR pipeline gives information on the number and percentage of sequences after each of the above described filtering steps. Subsequently, different tabs can be opened that contain the results (Fig. 1).
The “SHM overview” tab provides information on the number of sequences of the different (sub)classes. A table is included that provides for each (sub)class the number of mutations, the number of sequenced bases, and the average and median percentage of SHM. Additionally, the number of transitions and transversions at G/C and A/T locations and the amount of mutations located in AID (RGYW/WRCY) or pol η (WA/TW) motives are shown. Moreover, the number of replacement and silent mutations in both the FR and CDR region are shown, and the total number of sequenced A, T, C, and G nucleotides are given in the table. Bar graphs are included which visualize the percentage of mutations in either the AID (RGYW/WRCY) or pol η (WA/TW) motives or the relative and absolute number of mutations at A/T locations and transition and transversion mutations at G/C locations (Fig. 1).
In the “SHM frequency” tab graphs can be found on the percentage of SHM. The “transition tables” tab provides detailed tables and visualization of the nucleotide transitions. Information on the distribution of replacement mutations over the different amino acid locations and the results of analysis of selection strength using BASELINe (7) can be found in the “antigen selection” tab. The distribution of the sequences between the different subclasses is visualized in the “CSR” tab. Information about the number and size of clones in the analyzed sequences, as calculated using Change-O (4), can be found in the “clonal relation” tab. Furthermore, this tab also gives an overview table with the overlap of reads with the same CDR1–CDR3 sequence but different subclasses. All data used for making the different graphs and the results of clonal assignment as analyzed using Change-O can be found under the “downloads” tab. Details of all calculations performed in the SHM and CSR pipeline can be found in Supplemental Tables III and IV.
Applications of ARGalaxy
The Ag receptor repertoire can be studied for many different reasons. In our laboratory, we have mainly analyzed repertoire formation in healthy donors and patients with primary immunodeficiencies and/or autoimmunity. To illustrate the large number of applications of this tool, we included some examples of patients with alterations in their Ag receptor repertoire as analyzed using ARGalaxy (Figs. 5, 6).
V, D, and J gene usage.
One of the most frequently used parameters to study the immune repertoire is to analyze the frequency of the V, D, and J genes used. The frequency of these genes is not equal and can change upon Ag selection. For example, analysis of IGH gene frequency in naive and Ag-experienced B cells using ARGalaxy shows reduced IGHV4-34 gene frequency in the Ag-experienced repertoire (Fig. 5A).
Analysis of different V-D or V-J combinations can give information about restriction or clonality of the repertoire and can be visualized in the immune repertoire pipeline with heatmaps or circos plots (26). Fig. 5B shows a circos plot of VJ combinations in the Ag-experienced B cell repertoire of a patient with chronic lymphoblastic leukemia. In this case, a clear overrepresentation of one VJ combination is found, compared with the large variety of combinations found in the healthy donor.
CDR3 region.
The CDR3 region is the most variable part of Ag receptors and includes the junctional regions of the V, D, and J genes. The average CDR3 length of the BCR repertoire decreases after Ag exposure (Fig. 5C) (20, 28), and a long BCR CDR3 length is associated with autoreactivity (28). The CDR3 region is also often changed in patients with genetic defects in the V(D)J recombination process suffering from severe immunodeficiencies. These defects can effect both the CDR3 length and the composition of the junctional regions. Patients with XLF deficiency have a significantly shorter IGH CDR3 length compared with healthy controls (Fig. 5D) (20). Additionally, the junctional region can show alterations which are linked to their underlying genetic defect (Fig. 5E). ARTEMIS deficiency results in increased numbers of palindromic nucleotides, which are caused by defective opening of the hairpins formed on the coding ends of the junctions. XLF, XRCC4, and LIG4 form the ligation complex and genetic defects in these genes result in decreased number of nontemplated nucleotides (12, 13, 29, 30).
Diversity of the repertoire.
The immune repertoire tool also allows for the calculation of the diversity of the repertoire based on the presence of unique rearrangements in independent PCRs as was first described by Boyd et al. (25). Using ARGalaxy we have been able to show reduced diversity of the naive B cell repertoire in patients AT and Nijmegen breakage syndrome, which are both DNA repair deficiencies (Fig. 5F) (14).
SHM.
In the SHM and CSR pipeline different aspects of SHM can be studied, including the median percentage of SHM. Analyzing SHM in IGA transcripts of an ARTEMIS-deficient patient and four age-matched healthy controls reveals reduced SHM in the ARTEMIS-deficient patient as previously described (Fig. 6A) (10). Detailed analysis of the patterns of SHM can give insight into the repair pathways used to introduce SHM. UNG deficiency, for example, results in a reduction of transitions mutations at G/C base pair due to a block in base excision repair (Fig. 6B).
CSR.
Finally, CSR can also be affected by genetic defects in DNA repair or in patients that lack sufficient T cell help. For example, patients with AT have an increased frequency of IGG1 and IGG3 transcripts (Fig. 6C) (15).
Discussion
ARGalaxy is a user-friendly, open-source tool that allows fast and easy analysis of the complete immune repertoire. This tool is unique, as it provides both general information on V, D, and J gene usage, junction characteristics, and diversity in the immune repertoire pipeline, and detailed information about SHM, Ag selection, clonality, and CSR in the SHM and CSR pipeline. Open-source tools on Ag receptor data were implemented in ARGalaxy to make the analysis of Ag receptor data complete and fast. In the present study we described the set-up and workflow of ARGalaxy and included several clinical examples to illustrate its functionality, which focused on the field of primary immunodeficiencies. However, ARGalaxy is also applicable to many other patient groups such as autoimmune diseases, allergy, and leukemia. Additionally, it can be applied to more basic research questions such as the investigation of differences in repertoire between B cell subsets.
ARGalaxy extracts information from IMGT/HighV-QUEST output files. A similar tool that extracts information from these files is IgAT (5). This is a Microsoft Excel–based tool that can be used to analyze BCR L and H chain rearrangements, and it also visualizes V, D, and J gene usage, CDR3 and junction characteristics, and frequency of SHM. ARGalaxy complements IgAT because it provides analysis of both BCR and TCR rearrangement and importantly provides filtering of the immune repertoire data, which increases the reliability of the data. Additionally, the SHM and CSR pipeline provides additional information on the SHM patterns and the subclass distribution. Because IgAT also provides supplemental information to ARGalaxy, the filtered IMGT output files can be used to run IgAT on the filtered dataset to complete the detailed analysis of the immune repertoire.
When analyzing repertoire data using ARGalaxy, it is important to remove all adaptors, barcodes (such as multiplex identifier sequences), or primer sequences from all sequences before analysis in IMGT/HighV-QUEST. Depending on the primer location, IMGT/HighV-QUEST will include these regions in the alignment and therefore any stop codons in these regions lead to assignment of productive sequences as unproductive. Additionally, all mismatches in these regions will be assigned as SHM, which will significantly increase SHM levels in the analysis. Alternatively, the effect on the levels of SHM can be prevented by excluding the primers binding region and any regions upstream using the “sequence starts at” filter in the SHM and CSR pipeline. This, however, will not prevent the wrongful assignment of unproductive sequences.
The immune repertoire pipeline allows for basic data filtering to prevent overrepresentation of rearrangements due to amplification biases. In the SHM and CSR pipeline more advanced filtering options are included to allow better discrimination between sequencing errors and SHM. These filtering steps are not included in the immune repertoire pipeline, as single nucleotide differences are expected to have a very limited effect in assignment of V, D, or J genes. However, when combining the analysis of SHM or CSR with the analysis of V, D, and J segment usage it might be preferential to do both analyses on the exact same rearrangements. To allow for this, we have included the option in the SHM and CSR pipeline to create an output file with all rearrangements that passed the chosen filter settings. This output file can then be analyzed using the immune repertoire pipeline.
When analyzing SHM it is essential to be able to distinguish between sequencing errors and true SHM. To limit the effect of sequencing errors in the analysis of BCR repertoire data, we have included several additional filters to the SHM and CSR pipeline. This includes the removal of incomplete sequences or sequences containing an ambiguous base as automatically performed by the SHM and CSR pipeline. Additionally, the “only include sequences present twice or more” option in the “filter unique sequences” filter allows the inclusion of only unique sequences of which the exact same CDR1–CDR3 sequence occurs twice or more in the data. When using this setting it is important to realize that multiple sequences from a single clone are still included. In most cases this does not cause any problems, as data are often limited in their clonality. However, it is important to check for clonality of your data during analysis, as the presence of dominant clones can cause major skewing of the results. To prevent the presence of clonally related sequences in the filtered data the “remove duplicates bases on” filter can be applied to only include a single sequence per clone.
ARGalaxy was originally developed for the analysis of the human immune repertoire. However, as the format of the output files from IMGT/HighV-QUEST is the same for all species, it is also possible to analyze repertoire data from all other species implemented in IMGT/HighV-QUEST. The only limitation in the analysis of other species is the subclass assignment in the SHM and CSR pipeline, as our algorithm is specifically developed for assigning human (sub)classes. To prevent subclass assignment, we have added the “do not assign (sub)class” option in the SHM and CSR pipeline, which should be selected in the “human class/subclass” filter to prevent incorrect (sub)class assignment when analyzing nonhuman samples. Additionally, this option should be selected when analyzing BCR L chain data or BCR H chain data in which the (sub)class-specific sequence is absent.
In the SHM and CSR pipeline we have built in Change-O (4), a tool that allows the analysis of clonal relationship between sequences. In our pipeline this tool is applied to all sequences that have passed the chosen filter settings. Depending on the chosen filter settings, the results of Change-O will be only representative of the clonality within the filtered data and not the original sample. When the “filter unique sequences” or the “remove duplicates bases on” filter is applied, duplicate sequences are removed and therefore the clonality of the data does not represent the clonality of the original sample. Additionally, a high number of PCR cycles used to amplify the BCR rearrangements can cause nonlinear amplification of the input material, and therefore skewing of the clonality in the data as compared with the original material.
In conclusion, in this study we present ARGalaxy, a novel tool that allows the analysis of the complete immune repertoire of different species. This tool will be valuable for analysis of the immune repertoire in different research fields to gain more insight into the development of Ag-selected repertoire in health and disease.
Supplementary Material
Acknowledgments
We thank Steven Kleinstein, Jason Vander Heiden, Namita Gupta, Mohamed Uduman, and Susanna Marquez for allowing us to implement BASELINe and Change-O in ARGalaxy. The research for this study was (in part) performed within the framework of the Erasmus Postgraduate School of Molecular Medicine.
This work was supported by Dutch Organization for Scientific Research Vidi Grant 91712323 (to M.v.d.B.) and Veni Grant 91616058 (to P.A.v.S.).
See related articles in this issue: Langerak et al. (J. Immunol. 198, 3765; DOI: https://doi.org/10.4049/jimmunol.1602050) and Boyer et al. (J. Immunol. 198, 4148; DOI: https://doi.org/10.4049/jimmunol.1601924).
The online version of this article contains supplemental material.
- ARGalaxy
- Antigen Receptor Galaxy
- AT
- ataxia telangiectasia
- CSR
- class switch recombination
- IgAT
- Ig analysis tool
- IMGT
- international ImMunoGeneTics information system
- MID
- multiplex identifier
- SHM
- somatic hypermutation.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Langerak A. W., Brüggemann M., Davi F., Darzentas N., Gonzalez D., Cazzaniga G., Giudicelli V., Lefranc M.-P., Giraud M., Macintyre E. A., et al. 2017. High-throughput immunogenetics for clinical and research applications in immunohematology: potential and challenges. J. Immunol. 198: 3765–3774. [DOI] [PubMed] [Google Scholar]
- 2.Alamyar E., Duroux P., Lefranc M. P., Giudicelli V. 2012. IMGT® tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 882: 569–604. [DOI] [PubMed] [Google Scholar]
- 3.Ye J., Ma N., Madden T. L., Ostell J. M. 2013. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41: W34–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta N. T., Vander Heiden J. A., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S. H. 2015. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31: 3356–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogosch T., Kerzel S., Hoi K. H., Zhang Z., Maier R. F., Ippolito G. C., Zemlin M. 2012. Immunoglobulin analysis tool: a novel tool for the analysis of human and mouse heavy and light chain transcripts. Front. Immunol. 3: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giraud M., Salson M., Duez M., Villenet C., Quief S., Caillault A., Grardel N., Roumier C., Preudhomme C., Figeac M. 2014. Fast multiclonal clusterization of V(D)J recombinations from high-throughput sequencing. BMC Genomics 15: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yaari G., Uduman M., Kleinstein S. H. 2012. Quantifying selection in high-throughput immunoglobulin sequencing data sets. Nucleic Acids Res. 40: e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer F., Boutouil H., Dalloul I., Dalloul Z., Cook-Moreau J., Aldigier J.-C., Carrion C., Herve B., Scaon E., Cogné M., Péron S. 2017. CSReport: a new computational tool designed for automatic analysis of class switch recombination junctions sequenced by high-throughput sequencing. J. Immunol. 198: 4148–4155. [DOI] [PubMed] [Google Scholar]
- 9.Moorhouse M. J., van Zessen D., IJspeert H., Hiltemann S., Horsman S., van der Spek P. J., van der Burg M., Stubbs A. P. 2014. ImmunoGlobulin galaxy (IGGalaxy) for simple determination and quantitation of immunoglobulin heavy chain rearrangements from NGS. BMC Immunol. 15: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ijspeert H., Lankester A. C., van den Berg J. M., Wiegant W., van Zelm M. C., Weemaes C. M., Warris A., Pan-Hammarström Q., Pastink A., van Tol M. J., et al. 2011. Artemis splice defects cause atypical SCID and can be restored in vitro by an antisense oligonucleotide. Genes Immun. 12: 434–444. [DOI] [PubMed] [Google Scholar]
- 11.Cantaert T., Schickel J. N., Bannock J. M., Ng Y. S., Massad C., Delmotte F. R., Yamakawa N., Glauzy S., Chamberlain N., Kinnunen T., et al. 2016. Decreased somatic hypermutation induces an impaired peripheral B cell tolerance checkpoint. J. Clin. Invest. 126: 4289–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.IJspeert H., Rozmus J., Schwarz K., Warren R. L., van Zessen D., Holt R. A., Pico-Knijnenburg I., Simons E., Jerchel I., Wawer A., et al. 2016. XLF deficiency results in reduced N-nucleotide addition during V(D)J recombination. Blood 128: 650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray J. E., van der Burg M., IJspeert H., Carroll P., Wu Q., Ochi T., Leitch A., Miller E. S., Kysela B., Jawad A., et al. 2015. Mutations in the NHEJ component XRCC4 cause primordial dwarfism. Am. J. Hum. Genet. 96: 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.IJspeert H., Wentink M., van Zessen D., Driessen G. J., Dalm V. A., van Hagen M. P., Pico-Knijnenburg I., Simons E. J., van Dongen J. J., Stubbs A. P., van der Burg M. 2015. Strategies for B-cell receptor repertoire analysis in primary immunodeficiencies: from severe combined immunodeficiency to common variable immunodeficiency. Front. Immunol. 6: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driessen G. J., Ijspeert H., Weemaes C. M., Haraldsson A., Trip M., Warris A., van der Flier M., Wulffraat N., Verhagen M. M., Taylor M. A., et al. 2013. Antibody deficiency in patients with ataxia telangiectasia is caused by disturbed B- and T-cell homeostasis and reduced immune repertoire diversity. J. Allergy Clin. Immunol. 131: 1367–1375.e9. [DOI] [PubMed] [Google Scholar]
- 16.EU-supported EuroChimerism Consortium Project QLRT-2001-01485 2011. Standardization of DNA isolation from low cell numbers for chimerism analysis by PCR of short tandem repeats. Leukemia 25: 1467–1470. [DOI] [PubMed] [Google Scholar]
- 17.van Dongen J. J., Langerak A. W., Brüggemann M., Evans P. A., Hummel M., Lavender F. L., Delabesse E., Davi F., Schuuring E., García-Sanz R., et al. 2003. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 concerted action BMH4-CT98-3936. Leukemia 17: 2257–2317. [DOI] [PubMed] [Google Scholar]
- 18.Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M. C., Wardemann H. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berkowska M. A., Schickel J. N., Grosserichter-Wagener C., de Ridder D., Ng Y. S., van Dongen J. J., Meffre E., van Zelm M. C. 2015. Circulating human CD27-IgA+ memory B cells recognize bacteria with polyreactive Igs. J. Immunol. 195: 1417–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.IJspeert H., van Schouwenburg P. A., van Zessen D., Pico-Knijnenburg I., Driessen G. J., Stubbs A. P., van der Burg M. 2016. Evaluation of the antigen-experienced B-cell receptor repertoire in healthy children and adults. Front. Immunol. 7: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uduman M., Yaari G., Hershberg U., Stern J. A., Shlomchik M. J., Kleinstein S. H. 2011. Detecting selection in immunoglobulin sequences. Nucleic Acids Res. 39: W499–W504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blankenberg, D., G. Von Kuster, N. Coraor, G. Ananda, R. Lazarus, M. Mangan, A. Nekrutenko, and J. Taylor. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19: Unit 19.10.1–19.10.21. [DOI] [PMC free article] [PubMed]
- 23.Giardine B., Riemer C., Hardison R. C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., et al. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15: 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blankenberg D., Von Kuster G., Bouvier E., Baker D., Afgan E., Stoler N., Taylor J., Nekrutenko A., Galaxy Team 2014. Dissemination of scientific software with Galaxy ToolShed. Genome Biol. 15: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyd S. D., Marshall E. L., Merker J. D., Maniar J. M., Zhang L. N., Sahaf B., Jones C. D., Simen B. B., Hanczaruk B., Nguyen K. D., et al. 2009. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci. Transl. Med. 1: 12ra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S. J., Marra M. A. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yaari G., Vander Heiden J. A., Uduman M., Gadala-Maria D., Gupta N., Stern J. N., O’Connor K. C., Hafler D. A., Laserson U., Vigneault F., Kleinstein S. H. 2013. Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Front. Immunol. 4: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wardemann H., Yurasov S., Schaefer A., Young J. W., Meffre E., Nussenzweig M. C. 2003. Predominant autoantibody production by early human B cell precursors. Science 301: 1374–1377. [DOI] [PubMed] [Google Scholar]
- 29.van der Burg M., Verkaik N. S., den Dekker A. T., Barendregt B. H., Pico-Knijnenburg I., Tezcan I., vanDongen J. J., van Gent D. C. 2007. Defective Artemis nuclease is characterized by coding joints with microhomology in long palindromic-nucleotide stretches. Eur. J. Immunol. 37: 3522–3528. [DOI] [PubMed] [Google Scholar]
- 30.van der Burg M., van Veelen L. R., Verkaik N. S., Wiegant W. W., Hartwig N. G., Barendregt B. H., Brugmans L., Raams A., Jaspers N. G., Zdzienicka M. Z., et al. 2006. A new type of radiosensitive T−B−NK+ severe combined immunodeficiency caused by a LIG4 mutation. J. Clin. Invest. 116: 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.