Abstract
Sterols are key cyclic triterpenoid lipid components of eukaryotic cellular membranes, which are synthesized through complex multi-enzyme pathways. Similar to most animals, Bathymodiolus mussels, which inhabit deep-sea chemosynthetic ecosystems and harbor methanotrophic and/or thiotrophic bacterial endosymbionts, possess cholesterol as their main sterol. Based on the stable carbon isotope analyses, it has been suggested that host Bathymodiolus mussels synthesize cholesterol using a sterol intermediate derived from the methanotrophic endosymbionts. To test this hypothesis, we sequenced the genome of the methanotrophic endosymbiont in Bathymodiolus platifrons. The genome sequence data demonstrated that the endosymbiont potentially generates up to 4,4-dimethyl-cholesta-8,14,24-trienol, a sterol intermediate in cholesterol biosynthesis, from methane. In addition, transcripts for a subset of the enzymes of the biosynthetic pathway to cholesterol downstream from a sterol intermediate derived from methanotroph endosymbionts were detected in our transcriptome data for B. platifrons. These findings suggest that this mussel can de novo synthesize cholesterol from methane in cooperation with the symbionts. By in situ hybridization analyses, we showed that genes associated with cholesterol biosynthesis from both host and endosymbionts were expressed exclusively in the gill epithelial bacteriocytes containing endosymbionts. Thus, cholesterol production is probably localized within these specialized cells of the gill. Considering that the host mussel cannot de novo synthesize cholesterol and depends largely on endosymbionts for nutrition, the capacity of endosymbionts to synthesize sterols may be important in establishing symbiont–host relationships in these chemosynthetic mussels.
Keywords: bacteriocyte, bivalve, chemosynthesis, methane seep, stable carbon isotopes, symbiosis
Introduction
Chemosynthetic ecosystems found at deep-sea hydrothermal vents and seeps depend on the biological conversion of carbon dioxide or methane into carbohydrates, such as sugars, using energy and the reducing power produced by the oxidation of reduced chemical compounds, including hydrogen sulfide and methane. Chemosynthetic bacteria, such as sulfur-oxidizing bacteria (thiotrophs) or methane-oxidizing bacteria (methanotrophs), are responsible for this type of carbohydrate production as primary producers, and endemic animals, such as bivalves (mussels and clams) and tubeworms, are highly dependent on these chemosynthetic bacteria for their survival in these ecosystems.
Mussels belonging to the genus Bathymodiolus are frequently found in chemosynthetic ecosystems as dominant animal species and harbor thiotrophs and/or methanotrophs as endosymbionts in specialized cells of gill tissues called bacteriocytes (Childress et al. 1986; Cavanaugh et al. 1987). Although Bathymodiolus species are considered to largely depend on these endosymbionts for nutrition, our knowledge regarding how the biochemical pathways of these host mussels and bacterial symbionts are interwoven is very limited (although Ponnudurai et al. 2017 recently analyzed the comprehensive metabolic interaction in the symbiosis of B. azoricus). For example, the identity of the metabolites that are supplied to the host mussels by their endosymbionts and how the hosts modify bacterially synthesized metabolite intermediates have yet to be conclusively elucidated. Nevertheless, Jahnke et al. (1995) reported that a methanotroph-bearing Bathymodiolus sp., which flourishes at hydrocarbon seep sites in the Gulf of Mexico, possesses abundant cholesterol, a major sterol end product in animals, containing highly 13C-depleted carbon atoms, which probably originate from thermogenic methane seeping up from the sediment. Furthermore, to generate cholesterol, it was suggested that the host mussel utilizes a sterol intermediate compound that is relatively rich in the gill tissues, which may be synthesized and supplied by the methanotrophic endosymbionts.
It is often considered that the presence of sterols is one of the major characteristics defining eukaryotes, because these cyclic triterpenoid lipids are essential for multiple eukaryote-specific biological processes, including endocytosis (Pichler and Riezman 2004). Indeed, steranes (the degraded and saturated derivatives of sterols) in sedimentary rocks are used as molecular fossil markers of eukaryotes. However, several free-living methanotrophic bacteria, together with a limited number of other distantly related bacterial species (such as the planctomycete Gemmata obscuriglobus), have been found to be exceptional in their ability to produce sterols (Bird et al. 1971; Schouten et al. 2000; Pearson et al. 2003). In the light of these findings, it is reasonable to expect that the endosymbionts in Bathymodiolus, which are closely related to these sterol-containing free-living methanotrophs (Fujiwara et al. 2000), also have the sterol biosynthetic pathway, supporting the aforementioned hypothesis proposed by Jahnke et al. (1995).
It is reasonable to assume that cholesterol biosynthesis in methanotroph-bearing Bathymodiolus could be more precisely understood by comprehensively identifying the genes associated with this metabolic pathway from both host and endosymbionts. Accordingly, in the present study, along with transcriptome sequencing of Bathymodiolus platifrons collected at methane seep sites in Sagami Bay, Japan, we sequenced the genome of its methanotrophic endosymbionts. Based on the obtained sequence data and determination of the stable carbon isotopic composition of cholesterol in this mussel, we decipher how cholesterol is synthesized within the mussel–symbiont pair, and further discuss the evolution of the chemosynthetic symbioses of Bathymodiolus mussels with special reference to sterol metabolism.
Materials and Methods
Sampling
Two Bathymodiolus species (B. platifrons and the closely related species B. japonicus) were collected using the remotely operated vehicle (ROV) Hyper Dolphin and the R/V Natsushima of the Japan Agency of Marine-Earth Science and Technology. The sampling sites were the Off Hatsushima seep sites in Sagami Bay at depths of 857 m (35°00.97′N, 139°13.32′E, Dive#1508) and 957 m (35°01.541′N, 139°22.383′E, Dive#1643), which were visited during cruises NT13-06 (March, 2013) and NT14-05 (April, 2014), respectively (supplementary table S1, Supplementary Material online). The samples of B. platifrons and B. japonicus obtained by Dive#1643 were used for RNA extraction, in situ hybridization, and/or isotope analyses, while the sample of B. platifrons obtained by Dive#1508 was used for DNA extraction. For DNA extraction, the gill tissues of B. platifrons were stored at −80 °C. For RNA extraction, some of the collected mussels were fixed in situ: the shells of mussels were partially crushed by the manipulator of the ROV, and the mussels were immediately soaked in a saturated ammonium sulfate solution on site. After recovery, they were dissected and transferred in RNAlater (Ambion, TX, USA) on board, incubated for 16 h at 4 °C, and stored at –80 °C.
Symbiont Genome Analyses
Genomic DNA of the methanotrophic endosymbionts was purified from the gill of a single individual of B. platifrons, as described previously (Kuwahara et al. 2007). Briefly, the gill was homogenized and tissue debris was removed by filtration through sterilized gauze and nylon filters with a pore sizes of 5 µm. The bacterial cells were collected by centrifugation (4000 × g for 15 min at 4 °C) and subjected to DNase digestion using DNase I (Takara, Japan) with a final concentration of 0.35 U/µl at 25 °C for 2 h to reduce host DNA. Symbiont DNA was then extracted from the pelleted bacterial cells using a DNeasy Blood and Tissue kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. DNA sample was fragmented into a mean length of 500 bp using Covaris acoustic shearing (Covaris, MA, USA). An Illumina library was constructed using the KAPA Hyper Prep Kit Illumina platform (Kapa Biosystems, MA, USA). An Illumina mate-pair library, of which insert size was 2–5 kb, was also constructed using Nextera Mate Pair Library Preparation Kit (Illumina, CA, USA). Both ends of inserts of these two libraries were sequenced using an Illumina MiSeqTM sequencer (Illumina), using a paired-end library with insert lengths of 400–600 bp. Adapters and low-quality sequences were removed from reads using Trimmomatic v0.33 (Bolger et al. 2014). De novo assembly was performed in CLC Genomics Workbench 9.0 assembler (QIAGEN) with 2 million of the trimmed paired-end reads (bubble and word size: 500 and 51). Assembly of the data resulted in 4,486 contigs with length >500 bp, totaling 7.31 Mb. Redundant contigs were removed using the reduction step in the Redundans pipeline (Pryszcz and Gabaldón 2016). For binning based on genomic signatures, marker genes, and contig coverage, the software MyCC (Lin and Liao 2016) was used with default parameters. As low coverage and short contigs are known to be error-prone, contigs with a length <1 kb and average coverage <3 were discarded from the binning. The two-dimensional scatter plots in MyCC binning showed that 1833 of the nonredundant contigs could be mainly separated into two groups (supplementary fig. S1a, Supplementary Material online). One of these (BIN1) included 1083 contigs (total length of 3.85 Mb) with high coverage (average sequence coverage is 68) and approximately 40% GC content. Taxonomic assignments of the coding genes in contigs were performed using the LCA assignment algorithm in MEGAN v6.10 (Huson et al. 2011) based on the top 100 hits for each gene in the nr database. In MEGAN analyses, many of these contigs were shown to encode genes corresponding to Gammaproteobacteria, or more specifically to the order Methylococcales (supplementary fig. S1b, Supplementary Material online). In the evaluation using CheckM (Parks et al. 2015), the completeness and contamination for the order Methylococcales was 94.75 and 1.3%, respectively. The second bin (BIN2) included 695 contigs (total length of 1.16 Mb) with approximately 29% GC content. Many contigs of BIN2 were shown to encode genes corresponding to Epsilonproteobacteria, or more specifically to the order Campylobacterales (supplementary fig. S1b, Supplementary Material online). The completeness and contamination were 45.3 and 7.51%, respectively. In addition to the methanotrophic endosymbionts, a species belonging to the Epsilonproteobacteria was recently reported to be associated with the gill epithelia of some Bathymodiolus species, including B. platifrons, as an epibiont (Assié et al. 2016). BIN2 was assumed to contain contigs derived from the epibiont. Finally, the contigs in BIN1 were scaffolded with mate-paired reads using the Redundans pipeline. We determined a composite genome of the methane-oxidizing endsymbionts of B. platifrons that consisted of 613 contigs (deposited into the DDBJ/EMBL/GenBank databases under accession nos. BDMN01000001–BDMN01000613) with a total length of 4,018,024 bp. The N50 was 12579 bp and the average coverage was 115. Evaluation with CheckM showed 94.75% completeness and 1.02% contamination. Thus, the composite genome was almost complete. Coding regions in the composite genome of endosymbionts were identified using a combination of MetaGeneMark (Zhu et al. 2010) and Glimmer-MG (Kelley et al. 2012). The deduced amino acid sequences were subjected to a BLASTP search against an NCBI nonredundant (nr) protein database. Functional assignments were manually conducted by homology searches against the Kyoto Encyclopedia of Genes and Genomes (KEGG) protein database.
Transcriptome Analyses
Total RNA was isolated from the gill tissues of B. platifrons (including endosymbionts) using the SV Total RNA Isolation System (Promega, WI, USA). Construction of cDNA libraries with a TruSeq RNA Library Preparation Kit v2 (Illumina) and paired-end sequencing with Illumina HiSeq 2000 (100 bp per read) were performed by Hokkaido System Science Co., Ltd. Six hundred and seventy-eight million raw sequencing read data were cleaned up using Trimmomatic to remove adapter sequences and low-quality bases. Filtered sequences were then assembled into 679,948 transcript contigs using Trinity v2.0.6 (Grabherr et al. 2011) with default settings. All of the transcript sequences were then taken into further process of redundancy removing using the tr2aacds pipeline, from the EvidentialGene package (Gilbert 2013). The pipeline selects the “best” set of de novo assembled transcripts, based on coding potential. The selected best set contained 95,631 transcript sequences. The length of the transcripts ranged from 224 to 26,866 bp, with an average length of 1,132 bp and N50 value of 1,844. Protein coding regions were predicted using the TransDecoder program (Haas et al. 2013). Functional annotations of the coding regions were performed by homology searches against nr and KEGG protein database with an e-value cut-off of 1e−4.
Identification and Stable Carbon Isotope Analysis of Sterols
Sterol extraction and stable carbon isotope analysis were performed according to the procedures of Chikaraishi (2006). Briefly, the frozen gill and mantle tissues of B. platifrons and B. japonicus were saponified with 0.5 M KOH in CH3OH/H2O (95/5, w/w) by refluxing for 4.5 h to hydrolyze ester/acetate bonds, followed by sonication with dichloromethane/methanol (2/1, v/v) to extract the hydrolyzed lipids. The sterol fraction was isolated by silica gel column chromatography, followed by acetylation with acetic anhydride/pyridine (1/1). Sterols were assigned by gas chromatography/mass spectrometry (GC/MS), using a 7890A gas chromatograph (GC) coupled to a 5975C mass selective detector (MSD) (Agilent Technologies, CA, USA). The isotopic composition of sterols was determined by gas chromatography/combustion/isotope ratio mass spectrometry (GC/C/IRMS) using a 7890N GC (Agilent Technologies) coupled to a DeltaplusXP isotope ratio mass spectrometer (IRMS) via a GC-Isolink interface (Thermo Fisher Scientific, MA, USA). The combustion was performed in a microvolume ceramic tube at 1,020 °C. The acetylated sterols were injected using a programmable temperature vaporizing injector (PTV, Gerstel, Mülheim an der Ruhr, Germany) into an HP-5ms capillary column (length, 30 m; I.D., 0.25 mm; film thickness, 0.1 μm; Agilent Technologies). The carrier gas (He) flow rate through the GC capillary column was controlled using a constant flow mode at 1.4 ml min−1. To assess the reproducibility of the isotope measurement and to obtain the isotopic composition of sterols, reference mixtures of 14 n-alkanes (from 18 to 36 carbon atoms) and 2 acetyl sterols (cholesterol and stigmasterol) having known δ13C values were analyzed prior to and following the sample runs. The isotopic composition was reported in per mil (‰) relative to Vienna-Peedee Belemnite (V-PDB), on scales normalized to known δ13C values of the reference n-alkanes and acetyl sterols. The contribution of carbon atoms incorporated during acetylation was corrected using an isotopic mass balance calculation. The analytical error in the isotope measurement was better than 0.3‰.
Phylogenetic Analyses
The deduced amino acid sequences of 11 proteins associated with the sterol biosynthesis from B. platifrons (retrieved from the gill transcriptome) or its endosymbionts (retrieved from the symbiont genome) were separately aligned with the corresponding sequences from phylogenetically diverse organisms using MAFFT v7.164b (Katoh and Standley 2013). The alignments were inspected by eye and ambiguously aligned sites were excluded prior to phylogenetic analyses with trimAl 1.2rev59 (Capella-Gutiérrez et al. 2009). The analyzed datasets had the following dimensions: farnesyl-diphosphate farnesyltransferase, 24 taxa, 249 sites; squalene monooxygenase, 26 taxa, 362 sites; oxidosqualene cyclase, 34 taxa, 557 sites; sterol 14-demethylase, 28 taxa, 402 sites; methylsterol monooxygenase, 32 taxa, 192 sites; 17beta-estradiol 17-dehydrogenase, 9 taxa, 256 sites; cholestenol Delta-isomerase, 16 taxa, 192 sites; Delta24-sterol reductase, 16 taxa, 403 sites; Delta7-sterol 5-desaturase, 23 taxa, 207 sites; 7-dehydrocholesterol reductase, 15 taxa, 352 sites; and sterol O-acyltransferase, 16 taxa, 315 sites. The alignment data have been uploaded on FigShare (https://figshare.com/s/2b6611daa5c8cd1fa784). For each single-gene dataset, the ML phylogenetic tree and corresponding bootstrap support values (1000 replicates) were calculated using RAxML v7.2.6 (Stamatakis 2006). The ML tree was selected from 20 heuristic tree searches initiated from randomized parsimony starting trees. In ML bootstrap analyses, a single tree search per replicate was performed. For these datasets, Bayesian analyses were also performed using MrBayes5D (Tanabe 2008). Six parallel Metropolis-coupled Markov chain Monte Carlo (MCMCMC) runs, each consisting of three heated and one cold chains with default chain temperatures, were run for 1,000,000 generations. Log-likelihood scores and trees with branch lengths were sampled every 1,000 generations. The first 250,000 generations were excluded as burn-in, and the remaining trees were summarized to obtain Bayesian posterior probabilities. Convergence of parallel MCMCMC runs was judged by the average standard deviation of split frequencies (ASDSF). For both ML and Bayesian analyses, the most appropriate models selected with Aminosan (Tanabe 2011) were applied (table 1).
Table 1.
Selected Models for Maximum-likelihood (RAxML) and Bayesian (MrBayes) Analyses of Sterol Biosynthesis Enzymes
| Genes | Models (RAxML) | Models (MrBayes) |
|---|---|---|
| Farnesyl-diphosphate farnesyltransferase | PROTGAMMALG | LG_Gamma |
| Squalene monooxygenase | PROTGAMMALG | LG_Gamma |
| Oxidosqualene cyclase | PROTGAMMALGF | LG+F_Gamma |
| Sterol 14-demethylase | PROTGAMMALG | LG_Gamma |
| Methylsterol monooxygenase | PROTGAMMAMTZOAF | mtZoa+F_Gamma |
| 17Beta-estradiol 17-dehydrogenase | PROTGAMMALG | LG_Gamma |
| Cholestenol Delta-isomerase | PROTGAMMALGF | LG+F_Gamma |
| Delta24-sterol reductase | PROTGAMMALG | LG_Gamma |
| Delta7-sterol 5-desaturase | PROTGAMMAMTZOA | mtZoa_Gamma |
| 7-Dehydrocholesterol reductase | PROTGAMMALGF | LG_Gamma |
| Sterol O-acyltransferase | PROTGAMMALGF | LG_Gamma |
In Situ Hybridization
The gills of mussels were cut out using a disposable scalpel and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 16 h at 4 °C, followed by stepwise dehydration in an ethanol series, and stored at −30 °C. Using 500 ng of total mRNA extracted from the gill tissues of B. platifrons (see above), cDNA was synthesized using a QuantiTect Reverse Transcription Kit (Qiagen). The PCR primer sets were designed to be specific to the genes for oxidosqualene cyclase, sterol 14-demethylase, and 16S rRNA from the methanotrophic endosymbionts of B. platifrons, as well as the gene for 7-dehydrocholesterol reductase from B. platifrons (table 2). The former three genes from the endosymbiont were PCR-amplified from genomic DNA extracted from the gill of B. platifrons (see above), whereas the host 7-dehydrocholesterol reductase gene was amplified from the gill cDNA, using Easy-A High-Fidelity PCR Cloning Enzyme (Agilent Technologies). The PCR fragments were cloned into the pTA2 vector (Toyobo, Osaka, Japan) and the clones were sequenced with an ABI PRIZM 3130xl Genetic Analyzer (Applied Biosystems, CA, USA) using a BigDye Terminator V3.1 Cycle Sequencing Kit (Applied Biosystems). Using 200 ng of the DNA fragments as templates, which were amplified from the plasmids containing each of the gene fragments, anti-sense and sense RNA probes were synthesized with T3 and T7 RNA polymerase, respectively, using 1× digoxigenin (DIG) or fluorescein RNA labeling mix (F. Hoffmann-La Roche Ltd., Basel, Switzerland) according to the manufacturer’s instructions. The sequences of probes are shown in supplementary figure S2, Supplementary Material online. In situ hybridization of gill transverse sections (6 μm) was carried out as described in Hongo et al. (2016), with the following modifications. Hybridization in the presence of both DIG-labeled (for the sterol biosynthesis genes) and fluorescein-labeled probes (for the 16S rRNA of the symbiont) and the following washing procedures were performed at 62 °C. After NBT/BCIP chromogenic staining to detect the probes for the sterol biosynthesis genes labeled with DIG, the sections were incubated with 1:2000 HRP-conjugated anti-fluorescein antibody (PerkinElmer, MA, USA) at 4 °C overnight, and signals for the 16S rRNA of the symbiont were detected with 1:50 fluorescein plus amplification reagent in amplification diluent buffer of the TSA plus cyanine 3/fluorescein kit (PerkinElmer). All sections were mounted in Vectashield with DAPI (Vector Laboratories, CA, USA) and covered with a coverslip. Images were obtained using an Olympus IX73 microscope (Olympus, Tokyo, Japan) equipped with an Olympus DP73 camera, and micrographs were processed with Adobe Photoshop CS5.1 (Adobe Systems, CA, USA).
Table 2.
PCR Primer Sets
| Genes | Forward (5'–3') | Reverse (5'–3') |
|---|---|---|
| Oxidosqualene cyclase | GCATATCGAAGGACAACCCA | CCTTGTTAATGCTGGCTGTTC |
| Sterol 14-demethylase | TGCACAGCAAACCTCGACTA | TTGCTTTCAACTTCTGTTTGCT |
| 16S rRNA | CTTGCTCCTGGCTGACGAGT | TAGATCGTCGCCTTGGTGAG |
| 7-Dehydrocholesterol reductase | GAAGCATGAATAAAGAAAACGTCTC | TTAAAATAAATACGGCACAATCCTG |
Results
Gene Identification
In the methanotrophic endosymbiont genome, all genes encoding enzymes involved in methane oxidation, the KDPG (2-keto-3-deoxy-6-phosphogluconate) aldolase/transaldolase variant of the ribulose monophosphate (RuMP) pathway (Anthony 1982), and the nonmevalonate (MEP/DOXP) pathway were identified (supplementary table S2, Supplementary Material online). Regarding the sterol biosynthetic pathway, homologs of farnesyl-diphosphate farnesyltransferase, squalene monooxygenase, oxidosqualene cyclase, and sterol 14-demethylase were detected in the endosymbiont genome, whereas those of methylsterol monooxygenase, 17beta-estradiol 17-dehydrogenase, cholestenol Delta-isomerase, Delta24-sterol reductase, Delta7-sterol 5-desaturase, 7-dehydrocholesterol reductase, and sterol O-acyltransferase (in addition to those of the four enzymes encoded in the symbiont genome mentioned above) were retrieved from the transcriptome (supplementary table S2, Supplementary Material online). Neither a homolog of Delta14-sterol reductase nor that of sterol-4alpha-carboxylate 3-dehydrogenase was found in either the gill transcriptome or the endosymbiont genome. It is possible that Delta14-sterol reductase and sterol-4alpha-carboxylate 3-dehydrogenase could not be detected based on homology searches due to their divergent sequences or considerably low expression (very low amount of transcripts). Thus, the mussel or symbiont may be responsible for the steps of cholesterol biosynthesis in question, in spite of our failure to detect these genes using blast searches.
Phylogenies of Genes Associated with Sterol Biosynthesis
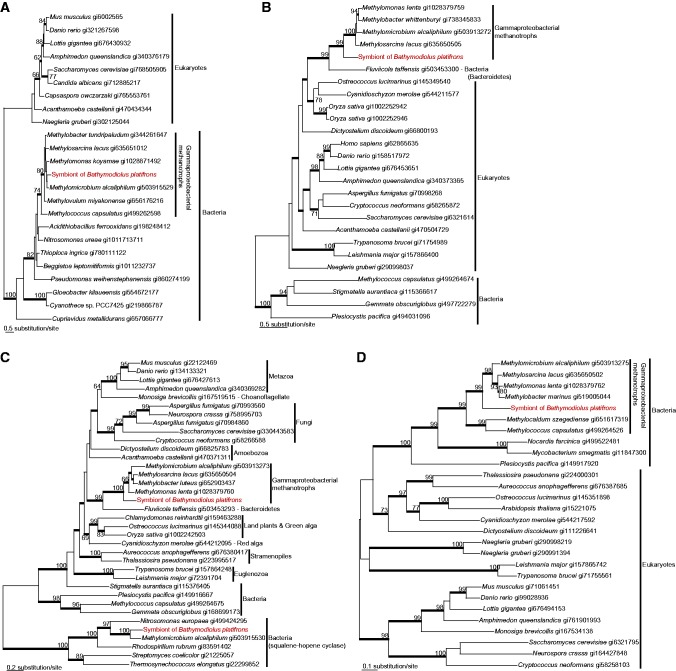
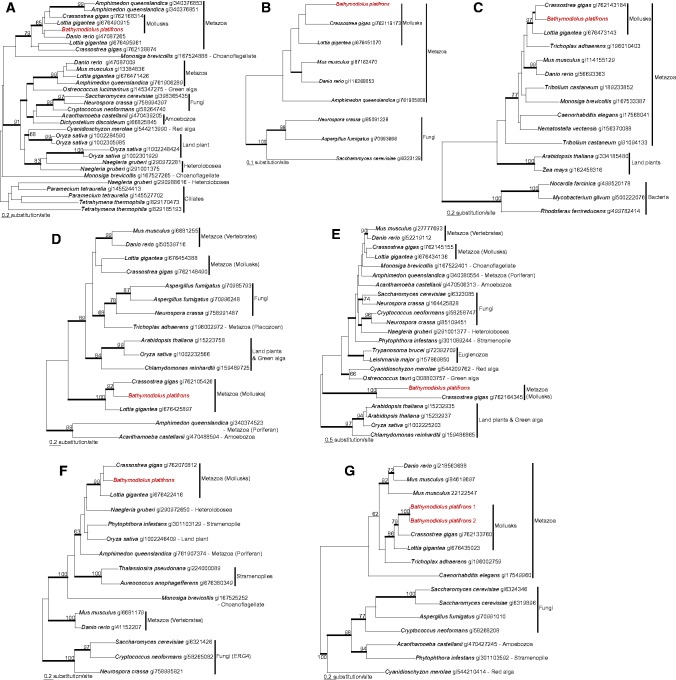
In the trees of farnesyl-diphosphate farnesyltransferase, squalene monooxygenase, oxidosqualene cyclase, and sterol 14-demethylase reconstructed in this study, the homologs of methanotrophic endosymbionts in B. platifrons were branched with those of the closely related Gammaproteobacterial methanotrophs with 80–100% RAxML bootstrap supports (BS) and 1.00 Bayesian posterior probability (PP) (fig. 1). In contrast, the phylogenies of methylsterol monooxygenase, 17beta-estradiol 17-dehydrogenase, cholestenol Delta-isomerase, Delta24-sterol reductase, Delta7-sterol 5-desaturase, 7-dehydrocholesterol reductase, and sterol O-acyltransferase showed that the respective homologs retrieved from the mussel transcriptome were nested with metazoans, or more specifically with mollusks (Crassostrea gigas and Lottia gigantea), with 79–100% RAxML BS and 1.00 Bayesian PP (fig. 2).
Fig. 1.
—Maximum-likelihood (ML) phylogenetic trees of farnesyl-diphosphate farnesyltransferase (a), squalene monooxygenase (b), oxidosqualene cyclase (c), and sterol 14-demethylase (d) inferred by RAxML. ML bootstrap supports are shown for bipartitions with support over 60%. Thick branches represent relationships with greater than 0.95 Bayesian posterior probabilities. The sequences of the methanotrophic endosymbiont of Bathymodiolus platifrons are shown in red typeface.
Fig. 2.
—Maximum-likelihood (ML) phylogenetic trees of methylsterol monooxygenase (a), 17beta-estradiol 17-dehydrogenase (b), cholestenol Delta-isomerase (c), Delta24-sterol reductase (d), Delta7-sterol 5-desaturase (e), 7-dehydrocholesterol reductase (f), and sterol O-acyltransferase (g) inferred by RAxML. ML bootstrap supports are shown for bipartitions with support over 60%. Thick branches represent relationships with greater than 0.95 Bayesian posterior probabilities. The sequences of Bathymodiolus platifrons are shown in red typeface.
Composition and Stable Carbon Isotope Signature of Sterols in Bathymodiolus
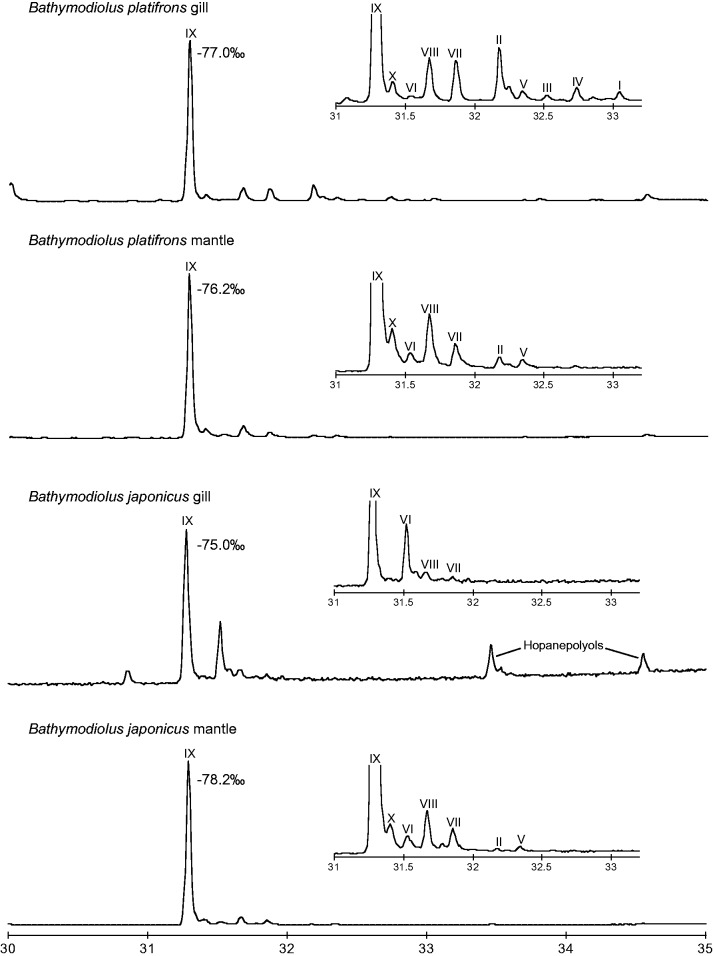
In total, nine sterols were identified in the gill and mantle tissues of B. platifrons and B. japonicas (fig. 3), with cholesterol being commonly abundant in all tissue samples. With the exception of cholesterol and cholestanol, all the detected sterols were found as intermediates in the cholesterol biosynthetic pathway. The δ13C values of cholesterol fell within a narrow range from −78.2‰ to −75.0‰ (−76.6‰ ± 1.3‰: mean ± 1σ SD). The δ13C values of sterols in the Bathymodiolus specimens showed substantial deviation from those in animals inhabiting surface ocean water (e.g., −20.3 ± 1.2‰; Chikaraishi 2006), but fell in the general δ13C range of biogenic methane (e.g., from −110‰ to −60‰; Whiticar 1999). No substantial difference in the δ13C value was found between gill and mantle tissues within a single mussel species or between the two mussel species.
Fig. 3.
—Total ion chromatograms of lipid extracts as acetates from gill and mantle tissues of two Bathymodiolus species (B. platifrons and B. japonicus) obtained by gas chromatography/mass spectrometry analyses. Detected sterols were I (lanosterol), II (4-methylzymosterol), III (4-methyl-cholesta-7,8(9),24-trienol), IV (4-methyl-cholesta-7,24-dienol), V (4-methyl-cholesta-7-enol), VI (zymosterol), VII (lathosterol), VIII (7-dehydrocholesterol), IX (cholesterol), and X (cholestanol). δ13C values of cholesterol in four samples are also shown.
Localization of Transcripts for Genes Associated with Sterol Biosynthesis
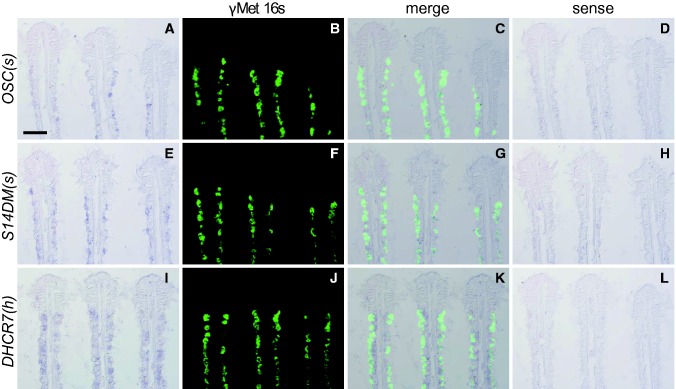
The frontal area of the Bathymodiolus gill filament consists of asymbiotic ciliated cells, whereas the inner area is composed of the symbiotic bacteriocytes and the asymbiotic intermediate cells (Fiala-Médioni et al. 1986). The signal of transcripts for the oxidosqualene cyclase gene from the endosymbionts was very weak but detectable in the apical area of the bacteriocytes (fig. 4a), where the signal of 16S rRNA from the endosymbionts was detected (fig. 4b). Similarly, the signal of transcripts for the sterol 14-demethylase gene from the endosymbionts was weak but detectable in the area (fig. 4e) where the endosymbionts were localized (fig. 4f). In contrast, the signal of transcripts of the 7-dehydrocholesterol reductase gene from the host was detected throughout the bacteriocytes, but not in the asymbiotic ciliated cells in the frontal area (fig. 4i). We were unable to determine whether this host gene is also expressed in the asymbiotic intermediate cells in the inner area due to the low resolution of our analysis. A sense probe was used as a control, and no positive signal was observed (fig. 4d, h, l).
Fig. 4.
—In situ hybridization for transcripts encoding oxidosqualene cyclase of the symbiont [OSC(s)], sterol 14-demethylase of the symbiont [S14DM(s)], and 7-dehydrocholesterol reductase of the host mussel [DHCR7(h)] using anti-sense probes (a, e, i) or control sense probes (d, h, l) with 16S rRNA of the symbiont (γMet 16s) (b, f, j) in the gill of Bathymodiolus platifrons. Differential interference contrast images with the anti-sense probes and fluorescence signals of 16S rRNA of the symbiont are merged in c, g, and k. The ciliated frontal surface of the gill filament is toward the top. Bar: 50 μm (applicable to all panels).
Discussion
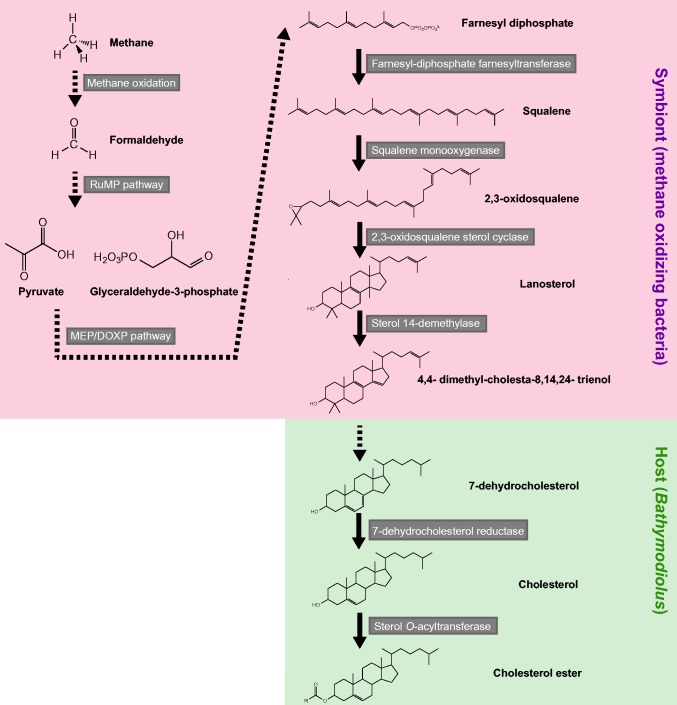
A large proportion of eukaryotes possess sterols that function to reinforce the cellular membrane. It has also been demonstrated that sterols are involved in the fluidity and permeability of eukaryotic cell membranes, and play key roles in various eukaryotic-specific cellular processes such as lipid raft-mediated signaling/membrane trafficking (Pichler and Riezman 2004). Major sterol end products, such as cholesterol in animals and β-sitosterol in terrestrial plants, are synthesized through complex multi-step pathways, including the epoxidation of squalene by squalene monooxygenase and the subsequent cyclization of epoxidized squalene to generate either lanosterol or cycloartenol by oxidosqualene cyclase (Bloch 1965; Summons et al. 2006). Among animals, some (e.g., mammals) can de novo synthesize cholesterol from low molecular weight precursors such as acetate, whereas others (e.g., insects) exclusively depend on dietary sterols due to the complete or partial absence of a cholesterol biosynthetic pathway. Several bivalve species, including mussels, have been suggested to be of the latter type based on radioisotope tracer experiments (Goad 1981; Knauer et al. 1998): it is considered that bivalves produce cholesterol by modification of exogenous sterols derived from dietary sources, such as microalgae, as shown by Giner et al. (2016). The genome sequence data of the Pacific oyster Crassostrea gigas reported by Zhang et al. (2012) also supports these findings: there are no genes encoding enzymes associated with upstream steps of the cholesterol biosynthesis pathway, including squalene monooxygenase and oxidosqualene cyclase, whereas genes for downstream steps of the pathway are present in this bivalve. Likewise, in the present study, only several genes for downstream steps leading to cholesterol were found in the transcriptome of B. platifrons, suggesting that this host mussel by itself cannot synthesize cholesterol de novo, but that it can convert exogenous sterols to cholesterol (fig. 5).
Fig. 5.
—A scheme for cholesterol biosynthesis in the Bathymodiolus–symbiont pair. The symbionts and host are responsible for the steps in the pink and green boxes, respectively. The range of dashed arrows indicates multiple steps of reactions.
The vast majority of bacteria are incapable of synthesizing sterols. Nevertheless, methanotrophs, such as Methylococcus capsulatus and Methylosphaera hansonii, have been shown to be exceptional in their ability to produce sterols (Bird et al. 1971; Schouten et al. 2000). In addition, homologs of the key enzyme for de novo sterol synthesis, oxidosqualene cyclase, have been detected in the genomes of more than 10 methanotrophic species (Wei et al. 2016). Based on the genome data for these methanotrophs, squalene, a precursor of sterols, could be built up with isopentenyl diphosphate (IPP), which is formed from pyruvate and glyceraldehyde-3-phosphate through the nonmevalonate (MEP/DOXP) pathway. Furthermore, pyruvate and glyceraldehyde-3-phosphate are probably generated through the RuMP pathway, in which formaldehyde produced by the oxidation of methane is used as a carbon source. In short, the above-mentioned methanotrophs potentially de novo synthesize sterols from environmental methane. In the genome sequence of the methanotrophic endosymbionts in B. platifrons obtained in the present study, we detected genes associated with the upstream steps of sterol biosynthetic pathway, as well as with the MEP/DOXP and RuMP pathways, suggesting that these endosymbionts can utilize methane for synthesis of a sterol intermediate compound used in the production of host cholesterol. This hypothesis is also supported by our stable carbon isotope data. Biogenic methane is generally depleted in 13C with δ13C value of −60‰ to −110‰ (Whiticar 1999). At the Hatsushima seep site, Tsunogai (2002) reported that the δ13C value of methane dissolved in the bottom seawater is −72‰. Thus, it is reasonable to assume that the highly 13C-depleted carbon atoms of cholesterol detected in B. platifrons and B. japonicas in the present study are derived from biogenic methane, which is assimilated by their methanotrophic endosymbionts.
Since the methanotrophic endosymbionts in B. platifrons probably possess a series of enzymes that generate up to 4,4-dimethyl-cholesta-8,14,24-trienol based on our genome data, this sterol compound could be transferred to the host mussel to produce cholesterol (fig. 5). Moreover, Jahnke et al. (1995) showed that based on the GC/MS analysis, 4-methyl-cholesta-8,14,24-trienol is relatively abundant in the gill tissues of Bathymodiolus sp. harboring methanotrophic endosymbionts when compared to other tissues (although how this mussel generates 4-methyl-cholesta-8,14,24-trienol from 4,4-dimethyl-cholesta-8,14,24-trienol or other sterol intermediates is unknown). Thus, this compound has been suggested to be the major end product of sterol biosynthesis in the endosymbionts, and this is subsequently converted to cholesterol by the host mussel. However, in our study, neither 4,4-dimethyl-cholesta-8,14,24-trienol nor 4-methyl-cholesta-8,14,24-trienol was detected in the gill tissues of either B. platifrons or B. japonicas, although minor sterol compounds in addition to cholesterol were identified in these Bathymodiolus species. It is unclear why our GC/MS results are inconsistent with those of Jahnke et al. (1995). One possibility is that the bacterially derived sterol(s) could not be detected because the mussel species investigated in this study rapidly metabolize the intermediate compound(s) into cholesterol. In the light of our in situ hybridization results, it is likely that cholesterol is synthesized exclusively in bacteriocytes in the gill tissues of B. platifrons. This means that the mussel host and methanotrophic endosymbionts collaboratively produce this end product within these specialized gill cells. Given such localized synthesis of cholesterol, this lipid synthesized in bacteriocytes may be esterified and carried by lipoproteins, making it soluble, for transport from bacteriocytes to other mussel cells or tissues. Jahnke et al. (1995) identified cholesteryl ester as well as other minor steryl esters in methanotroph-bearing Bathymodiolus sp., although we could not detect it due to the hydrolysis followed by acetylation in our protocol. The transcript for sterol O-acyltransferase, which forms cholesteryl esters from cholesterol, was retrieved from the gill transcriptome of B. platifrons, also supporting the presence of cholesteryl ester in this mussel.
In many cases, nutritional factors are closely associated with the establishment and maintenance of animal–microbe symbioses. These processes have been most intensively studied in insects (recently reviewed in Douglas 2016). For example, aphids, which are agriculturally important pests that feed exclusively on plant phloem sap, depend on the intracellular bacterium Buchnera aphidicola, which can synthesize essential amino acids that are absent or rare in phloem sap (Akman Gündüz and Douglas 2009). Furthermore, many sap-feeding psyllid species, including Ctenarytaina eucalypti and Heteropsylla cubana, harbor the “primary symbiont” Candidatus Carsonella ruddii along with a “secondary symbiont” (Buchner 1965). Genome data indicate that these two types of bacterial symbionts in C. eucalypti and H. cubana both independently complement biosynthesis pathways of amino acids essential for the host (Sloan and Moran 2012). Intriguingly, the Carsonella found in C. eucalypti and H. cubana contains the genes necessary to produce anthranilate, an intermediate of the tryptophan biosynthesis pathway, (trpEG), but lacks genes required to catalyze the synthesis of tryptophan from anthranilate (trpDCBA), whereas the secondary symbiont in these psyllids possesses the trpDCBA operon. These findings suggest that the pair of bacterial symbionts mutually compensate even multiple steps of one amino acid biosynthesis pathway, in addition to different amino acid pathways. Such partitioning in the tryptophan biosynthetic pathway has also been observed between pairs of symbionts in the mealybug Planococcus citri and the cedar aphid Cinara cedri (Gosalbes et al. 2008; Lamelas et al. 2011; McCutcheon and von Dohlen 2011). In addition, in host and symbiont pairs (e.g., aphids, psyllids, whiteflies, and mealybugs), the very last steps of essential amino acid biosynthesis are often carried out by host enzymes in the cytoplasm of bacteriocytes (Hansen and Moran 2011). However, to the best of our knowledge, there have been no reports regarding the mutual complementation of one biosynthetic pathway, in which upstream and downstream halves are conducted by symbiont and host, respectively, as found in the present study.
The chemosynthetic ability of bacterial symbionts is considered a major driving force in establishing and maintaining the symbioses among deep-sea animals inhabiting vents and seeps. However, our present findings may indicate that not all chemosynthetic bacteria are potential symbionts for these animals. A symbiont-free ancestor of Bathymodiolus (similar to Mytilus mussels) may have been a filter-feeding organism, which collected microalgae or detritus, and have utilized sterols derived from these foods. During the course of evolution, this ancestral mussel subsequently became nutritionally dependent on chemosynthetic endosymbionts rather than on sterol-containing prey organisms obtained from the surrounding environment. Therefore, sterol-producing bacteria should have been more suitable as partners for the host mussel than sterol-free bacteria. It is known that methanotrophs are mainly divided into type I belonging to Gammaproteobacteria (Methylococcaceae) and type II belonging to Alphaproteobacteria (Methylocystaceae). Type I methanotrophs potentially produce sterols, whereas type II members probably lack this metabolic capacity (Wei et al. 2016). Notably, Bathymodiolus species exclusively harbor type I rather than type II methanotrophs as endosymbionts (Fujiwara et al. 2000). Considering these points, the ability of de novo sterol synthesis as well as of chemosynthesis (using methane) of symbionts may have been important in the establishment and maintenance of the symbiosis in Bathymodiolus mussels. However, this hypothesis is not entirely straightforward. The vent mussel Bathymodiolus septemdierum and the seepage clam Calyptogena soyoae, both of which harbor thiotrophic endosymbionts and largely or exclusively depend on these for nutrition, contain sterols (Kawai et al. 2007). Nevertheless, in our preliminary analyses, no genes associated with de novo sterol synthesis have been detected in the genomes of these thiotrophic endosymbionts (Kuwahara et al. 2007; Ikuta et al. 2016), nor in the transcriptomes of the host bivalves (our unpublished data). How these thiotrophic endosymbiont-containing bivalves acquire sterols is still something of an enigma, and remains to be elucidated by biochemical and/or isotopic approaches in the future.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to the captain and crews of the R/V Natsushima and the operation team of the Hyper-Dolphin for their technical expertise. We also thank Mr Masaki Saito (JAMSTEC) for his technical support. This work was supported by a grant from the Japanese Society for the Promotion of Science awarded to K.T. (No. 15K07176).
Literature Cited
- Akman Gündüz E, Douglas AE.. 2009. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc Biol Sci. 276:987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C. 1982. The biochemistry of methylotrophs .New York: Academic Press. [Google Scholar]
- Assié A, et al. 2016. A specific and widespread association between deep-sea Bathymodiolus mussels and a novel family of Epsilon proteobacteria. Environ Microbiol Rep. 8:805–813. [DOI] [PubMed] [Google Scholar]
- Bird CW, et al. 1971. Steroids and squalene in Methylococcus capsulatus grown on methane. Nature 230:473–474. [DOI] [PubMed] [Google Scholar]
- Bloch K. 1965. The biological synthesis of cholesterol. Science 150:19–28. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. New York: Interscience. [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh CM, Levering PR, Maki JS, Mitchell R, Lidstrom ME.. 1987. Symbiosis of methylotrophic bacteria and deep-sea mussels. Nature 325:346–347. [Google Scholar]
- Chikaraishi Y. 2006. Carbon and hydrogen isotopic composition of sterols in natural marine brown and red macroalgae and associated shellfish. Org Geochem. 37:428–436. [Google Scholar]
- Childress JJ, et al. 1986. A methanotrophic marine molluscan (Bivalvia, Mytilidae) symbiosis: mussels fueled by gas. Science 233:1306–1308. [DOI] [PubMed] [Google Scholar]
- Douglas AE. 2016. How multi-partner endosymbioses function. Nat Rev Microbiol. 14:731–743. [DOI] [PubMed] [Google Scholar]
- Fiala-Médioni A, Metivier C, Herry A, Lepennec M.. 1986. Ultrastructure of the gill of the hydrothermal-vent mytilid Bathymodiolus sp. Mar Biol. 92:65–72. [Google Scholar]
- Fujiwara Y, et al. 2000. Phylogenetic characterization of endosymbionts in three hydrothermal vent mussels: influence on host distributions. Mar Ecol Prog Ser. 208:147–155. [Google Scholar]
- Gilbert D. 2013. Gene-omes built from mRNA seq not genome DNA. 7th annual arthropod genomics symposium. Notre Dame.
- Giner JL, Zhao H, Dixon MS, Wikfors GH.. 2016. Bioconversion of 13C-labeled microalgal phytosterols to cholesterol by the Northern Bay scallop, Argopecten irradians. Comp Biochem Physiol B Biochem Mol Biol. 192:1–8. [DOI] [PubMed] [Google Scholar]
- Goad LJ. 1981. Sterol biosynthesis and metabolism in marine invertebrates. Pure Appl Chem. 51:837–852. [Google Scholar]
- Gosalbes MJ, Lamelas A, Moya A, Latorre A.. 2008. The striking case of tryptophan provision in the cedar aphid Cinara cedri. J Bacteriol. 190:6026–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA.. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 108:2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongo Y, et al. 2016. Expression of genes involved in the uptake of inorganic carbon in the gill of a deep-sea vesicomyid clam harboring intracellular thioautotrophic bacteria. Gene 585:228–240. [DOI] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC.. 2011. Integrative analysis of environmental sequences using MEGAN 4. Genome Res. 21:1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta T, et al. 2016. Heterogeneous composition of key metabolic gene clusters in a vent mussel symbiont population. ISME J. 10:990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke LL, Summons RE, Dowling LM, Zahiralis KD.. 1995. Identification of methanotrophic lipid biomarkers in cold-seep mussel gills: chemical and isotopic analysis. Appl Environ Microbiol. 61:576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S, Takada Y, Tsuchida S, Kado R, Kimura J.. 2007. Sterols from bivalves Calyptogena soyoae and Bathymodiolus septemdierum living in deep sea. Fish Sci. 73:902–906. [Google Scholar]
- Kelley DR, Liu B, Delcher AL, Pop M, Salzberg SL.. 2012. Gene prediction with Glimmer for metagenomic sequences augmented by classification and clustering. Nucleic Acids Res. 40:e9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer J, Kerr RG, Lindley D, Southgate PC.. 1998. Sterol metabolism of Pacific oyster (Crassostrea gigas) spat. Comp Biochem Physiol B Biochem Mol Biol. 119:81–84. [Google Scholar]
- Kuwahara H, et al. 2007. Reduced genome of the thioautotrophic intracellular symbiont in a deep-sea clam, Calyptogena okutanii. Curr Biol. 17:881–886. [DOI] [PubMed] [Google Scholar]
- Lamelas A, et al. 2011. Serratia symbiotica from the aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet. 7:e1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Liao YC.. 2016. Accurate binning of metagenomic contigs via automated clustering sequences using information of genomic signatures and marker genes. Sci Rep. 6:24175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, von Dohlen CD.. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 21:1366–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, et al. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson A, Budin M, Brocks JJ.. 2003. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci USA. 100:15352–15357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler H, Riezman H.. 2004. Where sterols are required for endocytosis. Biochim Biophys Acta 1666:51–61. [DOI] [PubMed] [Google Scholar]
- Ponnudurai R, et al. 2017. Metabolic and physiological interdependencies in the Bathymodiolus azoricus symbiosis. ISME J. 11:463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryszcz LP, Gabaldón T.. 2016. Redundans: an assembly pipeline for highly heterozygous genomes. Nucleic Acids Res. 44:e113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten S, Bowman JP, Rijpstra WI, Sinninghe-Damsté JS.. 2000. Sterols in a psychrophilic methanotroph, Methylosphaera hansonii. FEMS Microbiol Lett. 186:193–195. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Moran NA.. 2012. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol. 29:3781–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Summons RE, Bradley AS, Jahnke LL, Waldbauer JR.. 2006. Steroids, triterpenoids and molecular oxygen. Philos Trans R Soc Lond B Biol Sci. 361:951–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe AS. 2008. MrBayes5D. [cited 2014 July 3]. Available from: http://www.fifthdimension.jp/products/mrbayes5d.
- Tanabe AS. 2011. Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Resour. 11:914–921. [DOI] [PubMed] [Google Scholar]
- Tsunogai U. 2002. Stable carbon isotopic studies of volatile components in seafloor venting hydrothermal and cold seep fluids, using CF-IRMS. Chikyukagaku 36:51–63. [in Japanese with English abstract]. [Google Scholar]
- Wei JH, Yin X, Welander PV.. 2016. Sterol synthesis in diverse bacteria. Front Microbiol. 7:990.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiticar MJ. 1999. Carbon and hydrogen isotope systematics of bacterial formation and oxidation of methane. Chem Geol. 161:291–314. [Google Scholar]
- Zhang G, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490:49–54. [DOI] [PubMed] [Google Scholar]
- Zhu W, Lomsadze A, Borodovsky M.. 2010. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38:e132.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.