Abstract
Background
The objectives of this study were to determine if C. difficile is present in the food of hospitalized patients, and to estimate the risk of subsequent colonization associated with the presence of C. difficile in food.
Methods
This was a prospective cohort study of inpatients at a university-affiliated tertiary care center. Enrolled patients submitted a portion of food from each meal. Patient stool specimens and/or rectal swabs were collected at enrollment, every three days thereafter, and at discharge, and were cultured for C. difficile. Clinical data were reviewed for evidence of infection due to C. difficile. A stochastic, discrete event model was developed to predict exposure to C. difficile from food, and the estimated number of new colonization events from food exposures per 1,000 admissions was determined.
Results
One-hundred forty-nine (149) patients were enrolled and 910 food specimens were obtained. Two food specimens from two patients were positive for C. difficile (0.2% of food samples; 1.3% of patients). Neither of the two patients was colonized at baseline with C. difficile. Discharge colonization status was available for one of the two patients and was negative. Neither was diagnosed with CDI while hospitalized or during the year before or after study enrollment. Stochastic modeling indicated contaminated hospital food would be responsible for <1 newly colonized patient per 1,000 hospital admissions.
Conclusions
The recovery of C. difficile from the food of hospitalized patients was rare. Modeling suggests hospital food is unlikely to be a source of C. difficile acquisition.
Keywords: Clostridium difficile, food, colonization
INTRODUCTION
Clostridium difficile infection is the most common healthcare associated infection in the US, and is associated with significant patient morbidity, mortality, and high attributable acute care hospital costs.1–3 Given the continued high incidence and severity of clinical outcomes associated with CDI, measures to prevent CDI are an area of ongoing interest. Current strategies for CDI prevention are focused on interrupting the cycle of transmission from individuals with CDI; however, it is important to evaluate other potential modes for C. difficile acquisition.
While CDI is primarily associated with healthcare facilities, the precise source of C. difficile exposure is unknown. Recent studies have found that only 15–25% of CDI cases could be attributed to ward-based or patient to patient transmission, indicating that there may be other sources of C. difficile acquisition in the hospital.4;5 A potential reservoir and source for C. difficile acquisition is the food of hospitalized patients. C. difficile has been isolated from retail foods world-wide, including ground meats, poultry, and vegetables.6–17 The spores of C. difficile are heat-resistant, and thus may have the potential to survive cooking temperatures.10 Given the presence of C. difficile in food and its heat-resistant qualities, it is theoretically possible that hospitalized patients could be exposed to C. difficile from their food. We conducted a prospective cohort study with the objectives of determining whether C. difficile was present in the food of hospitalized patients, and to estimate the risk of colonization associated with the presence of C. difficile in the food of hospitalized patients.
METHODS
Setting
This prospective cohort study was conducted at Barnes-Jewish Hospital (BJH), a 1,250 bed tertiary care center in St. Louis, MO, from May 2011 to July 2012, in conjunction with a study of C. difficile colonization in hospitalized patients.18;19 The study was approved by the Washington University Human Research Protection Office.
Subjects
Subjects ≥18 years old admitted to the medical and surgical wards with a projected length of stay (LOS) ≥3 days and no diarrhea were invited to participate; all provided written, informed consent.
Data Sources and Statistical Analyses
Data collected included demographics, comorbidities, and CDI diagnoses from one year prior to enrollment to one year after enrollment. Data sources included patient interviews, chart review, and Medical Informatics queries. Data analyses were descriptive. SPSS version 21 (IBM, Armonk, NY) was used. The model was implemented in NetLogo (version 5.1). R (R Foundation for Statistical Computing, Vienna, Austria) was used for model parameterization and output analysis.
Specimen Collection
Stool or rectal swab specimens were collected from patients upon study enrollment, every three days, and at discharge. Rectal swabs (ESwab, Becton, Dickinson, & Company, Franklin Lakes, NJ) were obtained from patients unable to provide a stool specimen within 48 hours of admission or 24 hours of a post-admission specimen collection time.20–22
Patients were provided with a cooler and four sterile specimen cups labeled breakfast, lunch, supper, or snack. Patients were instructed to place a piece of food from everything they ate into the corresponding container. If they did not eat a particular meal, no food was collected. As patients placed all components of their meal into the same container, there were multiple types of food per container.
Food specimens were transported to the laboratory and frozen at −30°C. Prior to culture, food specimens were thawed and the food types were documented. The food specimen was combined with 10 mL of sterile water and homogenized for approximately 1 minute.
Microbiological Analysis
1 mL of food homogenate was added to cycloserine-cefoxitin mannitol broth with taurocholate, lysozyme, and cysteine (CCMB-TAL, Anaerobe Systems, Morgan Hill, CA), and the broth was sub-cultured to pre-reduced blood agar (Becton, Dickinson, & Company, Franklin Lakes, NJ) as previously described.20 C. difficile in food was quantified by weighing initial specimens, processing via heat shock, plating onto a pre-reduced blood agar plate then streaking for isolation. Colonies per gram of food were calculated. Additionally, food was diluted in in CCMB-TAL broth in a series of 5, 10-fold dilutions to approximate the burden of C. difficile. C. difficile negative and positive controls were included with every set of cultures to monitor for contamination. Ribotyping was performed on all C. difficile isolates as previously described.23
Stool and rectal swabs were cultured for C. difficile using CCMB-TAL using methods previously published.20 Isolates were evaluated for the presence of tcdA, tcdB, and binary toxin genes (cdtA/cdtB) by multiplex PCR as previously described.18 Isolates were also characterized by PCR ribotyping for strain comparison.23
Model Overview
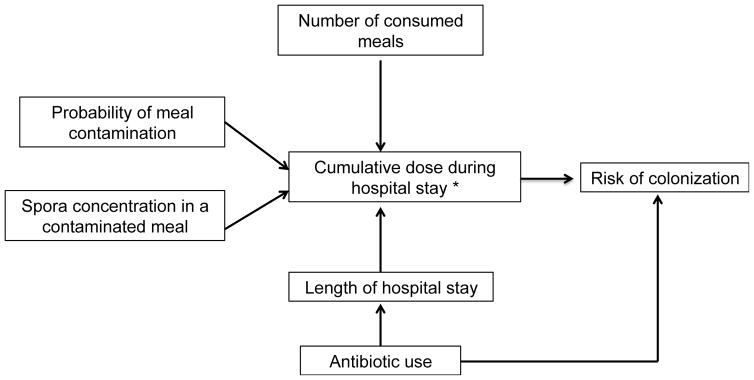
To estimate the risk of C. difficile acquisition associated with exposure to C. difficile-contaminated food during a hospital stay, we developed a stochastic, individual-based model that simulated the flow of patients admitted to BJH, antimicrobial exposures, number of meals eaten per day, and concentration of C. difficile in food (Figure 1). A formal description of the model, code and parameters are available at http://www.lanzaslab.org/research/cdifficile#food. The hospital model simulated the 171 BJH general medicine hospital ward beds. Each patient was followed from admission to discharge. On admission, each patient was assigned a LOS. LOS depended on whether the patient received antibiotic treatment based on the distribution of LOS from 11,046 admissions from these ward to several distributions using maximum likelihood methods.24 Based on these data, antibiotic use was used as a marker for longer LOS. Antibiotic use was also a marker for susceptibility to C. difficile colonization. The log-normal distribution, which provided the best fit, was used to parameterize LOS (Table 1).
Figure 1. Exposure Assessment.
*Cumulative dose calculated by total number of spores present in all contaminated meals eaten in a hospital stay.
Table 1.
Summary of functions and probability distributions for the exposure model
| Description | Functions and Probability Distributions | |
|---|---|---|
| Antibiotic prescription probability | Uniform (0.3, 0.7) | |
| Distribution of length of stay for patients who do not receive antibiotic | Log-normal (0.84, 0.77) | |
| Distribution of length of stay for patients who receive antibiotic | Log-normal (1.54, 0.87) | |
| N of meals (X) received by a patient daily | P(X=0)=0.441, P(X=1)=0.175, P(X=2)=0.264, P(X=3)=0.112, P(X=4)=0.008 | |
| Probability that a meal is contaminated with C. difficile spores | Uniform (0.0005, 0.0035) | |
| Distribution of the number of spores found on a contaminated meal | Poisson-lognormal (2, 0.8) | |
| Relationship between cumulative dose in a hospital stay and probability of colonization |
|
The parameters needed to characterize the probability distributions are indicated between brackets. For the uniform distribution (a,b); a is the minimum value and b is the maximum value. For the log-normal and Poisson-lognormal distributions (μ, σ); μ is the log mean and σ is the log standard deviation.
The number of meals consumed by a patient daily and the probability that a meal was contaminated were based on this study's results (Table 1). A Poisson log-normal distribution was used to simulate the number of spores per contaminated meal. This distribution is often used to describe microbial counts.25 The parameters of the distribution were chosen to generate a mean number of spores of approximately 10 CFU/gm, as this was the limit of detection of the culture methods. Data from a clinical trial in which healthy adults received escalating doses of nontoxigenic C. difficile spores were used to estimate the probability of C. difficile colonization upon dose exposure.26 We used logistic regression to model the data from study subjects in cohort 4 who received 5 days of pretreatment with oral vancomycin prior to receive a daily dose of 104, 106 or 108 spores for 14 days. Because of the repeated measurements on the same subjects, the binary correlated data were analyzed by means of the generalized estimating equation27 as implemented in package geepack in R.28
The model was iterated 5,000 times to assure output convergence. Each iteration simulated the patients’ admissions to the wards for one year. In each iteration, model inputs described as probability distributions were sampled and fed to the model. The model outcomes were the number of patients exposed to C. difficile through food and the number of colonization events due to food exposure per 1,000 admissions.
RESULTS
Enrollment and Demographics
One hundred forty-nine (149) patients were enrolled, and food specimens from 910 meals were obtained and cultured for C. difficile. Patient characteristics are in Table 2. The vast majority of patients had healthcare exposures within the previous 90 days (n=136, 91%), but only two patients had a history of CDI within the previous year (none within 60 days prior to enrollment).
Table 2.
Patient Characteristics (n=149)
| Variable | N (%) or median (range) |
|---|---|
|
| |
| Age (years) | 55 (23–90) |
|
| |
| Length of stay (days) | 4.0 (0.4–292) |
|
| |
| Female | 80 (54) |
|
| |
| Non-white race | 43 (29) |
|
| |
| Healthcare worker | 20 (13) |
|
| |
| Lives with a healthcare worker | 19 (13) |
|
| |
| Spends ≥2 hours/week visiting a healthcare facility | 16 (11) |
|
| |
| Admitted to medicine service | 145 (97) |
|
| |
| Admitted from | |
| Home | 99 (66) |
| Healthcare facility | 50 (34) |
|
| |
| Reason for admission | |
| Infection | 45 (30) |
| Exacerbation of chronic condition | 54 (36) |
| Elective surgery | 2 (1) |
| New medical or surgical problem | 48 (32) |
|
| |
| Any healthcare exposures in previous 90 days | 136 (91) |
|
| |
| Diabetes mellitus | 58 (39) |
|
| |
| Congestive heart failure | 27 (18) |
|
| |
| Liver disease | 28 (19) |
|
| |
| Chronic renal insufficiency | 17 (11) |
|
| |
| Chronic lung disease | 25 (17) |
|
| |
| HIV | 3 (2) |
|
| |
| Solid organ transplant | 8 (5) |
|
| |
| Stem cell transplant | 0 |
|
| |
| Solid malignancy | 24 (16) |
|
| |
| Hematologic malignancy | 2 (1) |
|
| |
| Other immunocompromised | 20 (13) |
|
| |
| Inflammatory bowel disease | 5 (3) |
|
| |
| Surgery in previous 90 days | 15 (10) |
|
| |
| Upper endoscopy performed during hospitalization | 9 (6) |
|
| |
| Lower endoscopy performed during hospitalization | 8 (5) |
|
| |
| History of CDI in the year pre-enrollment | 2 (1) |
|
| |
| CDI diagnosis within 1 year post-enrollment | 0 (0) |
|
| |
| Colonized with C. difficile on admission | 34 (23) |
|
| |
| Colonized with C. difficile on discharge* | 35 (24) |
Discharge colonization status was unknown for 8 (5%) patients.
Food Cultures
Toxigenic C. difficile was recovered from two food specimens from two separate patients, representing 0.2% of food cultures and 1.3% of patients (Table 3). The food items that tested positive were a gelatin dessert (ribotype 001) and a sample consisting of vegetables/bread/grains (ribotype 027). The concentration of C. difficile spores recovered from the positive food samples was ≤10 CFU/mL. C. difficile was successfully recovered from all positive controls, and there was no growth in any of the negative controls.
Table 3.
Types of Food Positive for Clostridium difficile, by food typea
| Food Item | Total (n=910) | C. difficile (n [%]) |
|---|---|---|
| Meat | 308 | 0 |
| Poultry | 142 | 0 |
| Fruit | 179 | 0 |
| Vegetables | 455 | 1(<1)b |
| Nuts | 1 | 0 |
| Dairy/eggs | 210 | 0 |
| Bread/grains | 376 | 1 (<1)b |
| Otherc | 200 | 1 (1)d |
Percentages are percent of positive samples / all food items of that type
The positive specimens for vegetables and bread/grains were combined in a single specimen cup
e.g., veggie burger, sauce/gravy, pudding, jelly, fish, cake
Gelatin dessert
C. difficile Colonization and CDI
Neither of the two patients exposed to C. difficile in food was colonized at baseline with C. difficile. A discharge stool specimen was available for one of the two patients and was negative. Neither was diagnosed with CDI during their hospitalization or during the year before or after study enrollment. No patients in the study developed CDI within a year of discharge.
Exposure Modeling
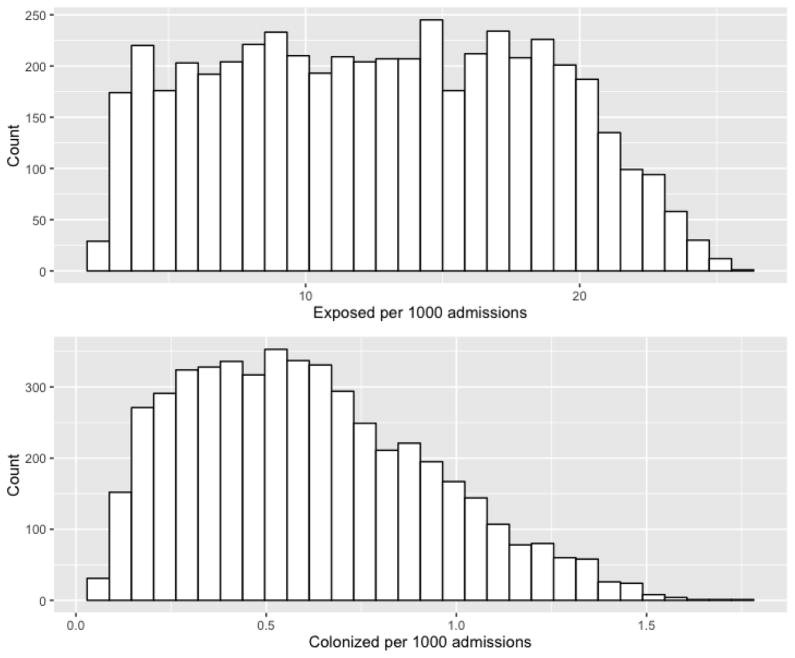
A summary of the functions and probability distributions for the exposure model are detailed in Table 1. On 44.1% of days, no meals were eaten; on 17.5% of days 1 meal was eaten; on 24.4% of days 2 meals were eaten; on 11.2% of days 3 meals were eaten; and on 0.8% of days 4 meals were eaten. Reasons for missing meals were variable, but included instructions to take nothing by mouth in preparation for an upcoming procedure(s) or lack of appetite. The mean number of patients who were exposed to C. difficile through food was 12.70 per 1,000 admissions (95% confidence interval 12.542–12.858). The minimum and maximum simulated values were 2.34 and 25.85 exposed patients per 1,000 admissions, respectively (Figure 2). The mean number of predicted colonization events was 0.609 per 1,000 admissions (95% confidence interval 0.600–0.618), and the median number was 0.57. The minimum and maximum simulated colonization events were 0.04 and 1.73 per 1,000 admissions, respectively (Figure 2).
Figure 2. Histograms for the simulated number of patients exposed to C. difficile in food and colonization events due to exposure to C. difficile spores in food*.
* Counts are the number of exposed and colonized patients in each histogram bar.
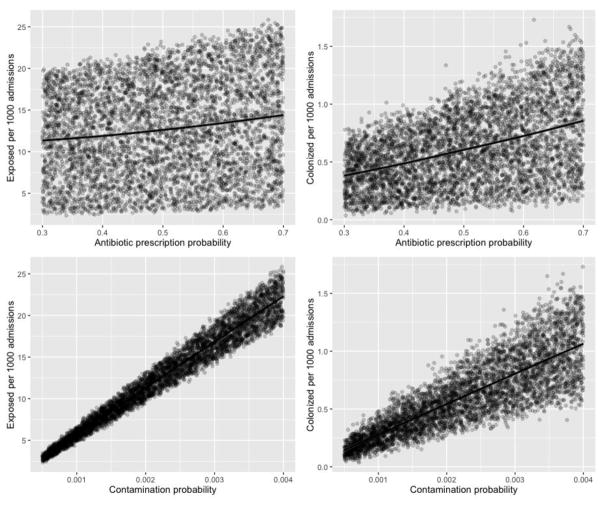
Both the predicted number of exposed and colonized patients was highly influenced by the probability of meal contamination (Figure 3). A 0.1% increase in the probability of meal contamination resulted in an increase of 5.5 exposures and 0.26 colonization events per 1,000 admissions (Figure 3). Overall, the simulated number of spores in contaminated food was low, reflecting the low counts recovered from hospital food. As a result, on average fewer than 5% of the patients exposed to C. difficile became colonized. Antibiotic prescription probability had a marginal effect on the number of predicted exposed and colonized patients compared to probability of meal contamination (Figure 3).
Figure 3. Scatterplotsa between the hospital-level variablesb and model outcomesc.
aThe points indicate individual simulations (total number of simulations = 5000) and the line indicates the linear trend between variables
bAntibiotic prescription and meal contamination probability
cNumber of exposed and colonized patients per 1000 admissions
DISCUSSION
In this study of C. difficile in the food of hospitalized patients, recovery of toxigenic C. difficile was rare, with only 0.2% of food specimens from testing positive for C. difficile with a low estimated concentration (≤10 CFU/mL). Stated differently, 1.5% of patients ingested food from which C. difficile was recovered. Based on this, theoretically hundreds of hospitalized patients could be exposed to C. difficile from food and develop CDI every year at BJH, which in 2014 alone had >53,300 inpatient admissions. Thus, our objective was to model the likelihood of C. difficile acquisition from food in the hospital setting. Using a similar modeling framework based on BJH data, we previously predicted that on average there were approximately 100 new colonization events per 1,000 admissions.29 In this study, we found at <1 new colonization events per 1,000 admissions, C. difficile acquisition linked to contaminated food was likely uncommon. The results of the modeling indicate that acquisition of C. difficile from food is likely a rare event at BJH, and that the food of hospitalized patients was not a significant source of new C. difficile colonization.
Most previously published studies of C. difficile in food have focused on retail meat products, with prevalence rates reported in Europe and Canada ranging from 2.7% in chicken, 4.3%–20% in beef/pork, and US studies with rates as high as >40% (ground meats).8;10–12 The presence of C. difficile in foods of non-animal origin (e.g., fruits, vegetables, grains) has not been fully explored, with a Scottish study indicating that 7.5% of salads and a French study reporting 2.9% of raw vegetables were contaminated with C. difficile.6–8 These previous studies were based on singular food types, rather than a mixture of foods that constitute a meal that a patient might eat. In our study, we included all food that the patient would be consuming during that meal, which would better represent a hospitalized patient’s actual C. difficile exposure.
Although we were able to recover C. difficile from the food of hospitalized patients, this does not equate directly to C. difficile being a foodborne pathogen in the healthcare setting. The source of contamination is not known; contamination may have occurred at the food source (farm, factory), food handler, food transporter, and/or from the patient handling the food. The results of our study are consistent with those of Rodriguez et al, who found C. difficile in <1% of food samples collected from the kitchens of a Belgian nursing home.13 C. difficile was isolated from only one meal sample composed of pork sausage, mustard sauce, and carrot salad.13 Together, our study and the Rodriquez study suggest that C. difficile is present in hospital foods, but at lower rates compared to retail foods. The reason for this discrepancy is unclear. Although C. difficile spores can survive cooking temperatures, it is possible that soaking, washing, and/or cooking food reduces the C. difficile burden and may have accounted for this difference.10
This study had limitations: it was a single center study, and results may not be generalizable to all institutions. Regarding specimen collection, patients placed food into containers for culture, potentially introducing variability. However, this provided a practical method of obtaining food samples that were actually consumed by the patient. There are no data available to indicate whether or not C. difficile is evenly distributed in food. Thus the amount of food included in each food cultures may have impacted our findings, especially in the setting of a low contamination burden. In previously published studies the amount of food cultured varies widely, from 1 g samples to complete pieces of meat.14;17 Previous studies were limited to specific food types, however this study focused on the variety of foods that a patient ingests per meal, providing a more realistic estimate of a patient’s entire meal.
A strength of our study was the collection of food that was actually consumed, rather than a single food type. In the studies of retail food products, the food would have likely been washed and/or cooked prior to consumption; therefore the prevalence of C. difficile detected may not represent what individuals would have consumed. Additional strengths include the collection of clinical data to examine CDI in the year post-enrollment and culture of food specimens along with the culture of stool specimens throughout the patients’ hospitalizations. This allowed us to link C. difficile food contamination with acquisition. Our laboratory standards were rigorous, and we included both positive and negative controls to ensure against laboratory contamination.
C. difficile is an ubiquitous organism, and only a minority of new C. difficile acquisitions in the hospital have been linked to another patient with CDI.4;5;30;31 Therefore, understanding all potential sources of C. difficile exposure in the hospital is necessary to inform prevention measures for CDI. Our findings indicate that food is unlikely to be a significant source of C. difficile acquisition in hospitalized patients. Towards the goal of CDI prevention, future studies aimed at understanding modes of C. difficile transmission and acquisition are necessary.
Acknowledgments
Financial Support: This study was supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK): P30DK52574. JHK was supported by the Washington University Institute of Clinical and Translational Sciences grant UL1TR000448, sub-award KL2TR000450, from the National Center for Advancing Translational Sciences of the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH. CL was supported by R01GM113239 from the joint NSF/NIGMS Mathematical Biology Program.
Footnotes
Conflict of Interest: CAB has received research support from bioMerieux, Cepheid, and Accelerate Diagnostics. ERD has been a consultant to Sanofi Pasteur, Merck, Summit, Alere, Valenva and Rebiotix
Reference List
- 1.Antibiotic Resistance Threats in the United States. Centers for Disease Control and Prevention; 2013. [Accessed 1-9-2016]. http://www.cdc.gov/drugresistance/threat-report-2013/. Published 2014. [Google Scholar]
- 2.Lessa FC, Mu Y, Bamberg WM, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kwon JH, Olsen MA, Dubberke ER. The Morbidity, Mortality, and Costs Associated with Clostridium difficile Infection. Infect Dis Clin North Am. 2015;29:123–134. doi: 10.1016/j.idc.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Walker AS, Eyre DW, Wyllie DH, et al. Characterisation of Clostridium difficile Hospital Ward-Based Transmission Using Extensive Epidemiological Data and Molecular Typing. PLoS Med. 2012;9:e1001172. doi: 10.1371/journal.pmed.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grundmann H, Barwolff S, Tami A, et al. How many infections are caused by patient-to-patient transmission in intensive care units? Crit Care Med. 2005;33:946–951. doi: 10.1097/01.ccm.0000163223.26234.56. [DOI] [PubMed] [Google Scholar]
- 6.Bakri MM, Brown DJ, Butcher JP, Sutherland AD. Clostridium difficile in ready-to-eat salads, Scotland. Emerg Infect Dis. 2009;15:817–818. doi: 10.3201/eid1505.081186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckert C, Burghoffer B, Barbut F. Contamination of ready-to-eat raw vegetables with Clostridium difficile in France. J Med Microbiol. 2013;62:1435–1438. doi: 10.1099/jmm.0.056358-0. [DOI] [PubMed] [Google Scholar]
- 8.Gould LH, Limbago B. Clostridium difficile in food and domestic animals: a new foodborne pathogen? Clin Infect Dis. 2010;51:577–582. doi: 10.1086/655692. [DOI] [PubMed] [Google Scholar]
- 9.Jobstl M, Heuberger S, Indra A, Nepf R, Kofer J, Wagner M. Clostridium difficile in raw products of animal origin. Int J Food Microbiol. 2010;138:172–175. doi: 10.1016/j.ijfoodmicro.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 10.Lund BM, Peck MW. A possible route for foodborne transmission of Clostridium difficile? Foodborne Pathog Dis. 2015;12:177–182. doi: 10.1089/fpd.2014.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Palacios A, Staempfli HR, Duffield T, Weese JS. Clostridium difficile in retail ground meat, Canada. Emerg Infect Dis. 2007;13:485–487. doi: 10.3201/eid1303.060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Palacios A, Reid-Smith RJ, Staempfli HR, et al. Possible seasonality of Clostridium difficile in retail meat, Canada. Emerg Infect Dis. 2009;15:802–805. doi: 10.3201/eid1505.081084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez C, Korsak N, Taminiau B, et al. Clostridium difficile from food and surface samples in a Belgian nursing home: an unlikely source of contamination. Anaerobe. 2015;32:87–89. doi: 10.1016/j.anaerobe.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Songer JG, Trinh HT, Killgore GE, Thompson AD, McDonald LC, Limbago BM. Clostridium difficile in retail meat products, USA, 2007. Emerg Infect Dis. 2009;15:819–821. doi: 10.3201/eid1505.081071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Von Abercron SM, Karlsson F, Wigh GT, Wierup M, Krovacek K. Low occurrence of Clostridium difficile in retail ground meat in Sweden. J Food Prot. 2009;72:1732–1734. doi: 10.4315/0362-028x-72.8.1732. [DOI] [PubMed] [Google Scholar]
- 16.Weese JS, Avery BP, Rousseau J, Reid-Smith RJ. Detection and enumeration of Clostridium difficile spores in retail beef and pork. Appl Environ Microbiol. 2009;75:5009–5011. doi: 10.1128/AEM.00480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weese JS, Reid-Smith RJ, Avery BP, Rousseau J. Detection and characterization of Clostridium difficile in retail chicken. Lett Appl Microbiol. 2010;50:362–365. doi: 10.1111/j.1472-765X.2010.02802.x. [DOI] [PubMed] [Google Scholar]
- 18.Alasmari F, Seiler SM, Hink T, Burnham CA, Dubberke ER. Prevalence and risk factors for asymptomatic Clostridium difficile carriage. Clin Infect Dis. 2014;59:216–222. doi: 10.1093/cid/ciu258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubberke ER, Reske KA, Seiler S, Hink T, Kwon JH, Burnham CA. Risk Factors for Acquisition and Loss of C difficile Colonization in Hospitalized Patients. Antimicrob Agents Chemother. 2015;59:4533–4543. doi: 10.1128/AAC.00642-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hink T, Burnham CA, Dubberke ER. A systematic evaluation of methods to optimize culture-based recovery of Clostridium difficile from stool specimens. Anaerobe. 2013;19:39–43. doi: 10.1016/j.anaerobe.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McFarland LV, Coyle MB, Kremer WH, Stamm WE. Rectal swab cultures for Clostridium difficile surveillance studies. J Clin Microbiol. 1987;25:2241–2242. doi: 10.1128/jcm.25.11.2241-2242.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kundrapu S, Sunkesula VC, Jury LA, Sethi AK, Donskey CJ. Utility of perirectal swab specimens for diagnosis of Clostridium difficile infection. Clin Infect Dis. 2012;55:1527–1530. doi: 10.1093/cid/cis707. [DOI] [PubMed] [Google Scholar]
- 23.Westblade LF, Chamberland RR, Maccannell D, et al. Development and evaluation of a novel, semiautomated Clostridium difficile typing platform. J Clin Microbiol. 2013;51:621–624. doi: 10.1128/JCM.02627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delignette-Muller ML, Dutang C. fitdistrplus: An R Package for Fitting Distributions. Journal of Statistical Software. 2015:64. [Google Scholar]
- 25.Gonzales-Barron U, Butler F. A comparison between the discrete Poisson-gamma and Poisson-lognormal distributions to characterise microbial counts in foods. Food Control. 2011;22:1279–1286. [Google Scholar]
- 26.Villano SA, Seiberling M, Tatarowicz W, Monnot-Chase E, Gerding DN. Evaluation of an oral suspension of VP20621, spores of nontoxigenic Clostridium difficile strain M3, in healthy subjects. Antimicrob Agents Chemother. 2012;56:5224–5229. doi: 10.1128/AAC.00913-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosmer DW, Lemeshow S, Sturdivant RX. Applied Logistic Regression. 3. Hoboken, New Jersey: John Wiley & Sons, Inc; 2013. [Google Scholar]
- 28.Halekoh U, Hojsgaard S, Yan J. The R Package geepack for Generalized Estimating Equations. Journal of Statistical Software. 2016:15. [Google Scholar]
- 29.Lanzas C, Dubberke ER. Effectiveness of screening hospital admissions to detect asymptomatic carriers of Clostridium difficile: a modeling evaluation. Infect Control Hosp Epidemiol. 2014;35:1043–1050. doi: 10.1086/677162. [DOI] [PubMed] [Google Scholar]
- 30.Fekety R, Kim KH, Brown D, Batts DH, Cudmore M, Silva J., Jr Epidemiology of antibiotic-associated colitis; isolation of Clostridium difficile from the hospital environment. Am J Med. 1981;70:906–908. doi: 10.1016/0002-9343(81)90553-2. [DOI] [PubMed] [Google Scholar]
- 31.McFarland LV, Mulligan ME, Kwok RY, Stamm WE. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med. 1989;320:204–210. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]