Abstract
Introduction
The T to C transition at nucleotide 1565 of the human glycoprotein IIIa (ITGB3) gene represents a genetic polymorphism (PlA1/A2) that can influence both platelet activation and aggregation and that has been associated with many types of disease. Here, we present a newly designed multiplex tetra-primer amplification refractory mutation system – polymerase chain reaction (T-ARMS-PCR) for genotyping a single nucleotide polymorphism (SNP) (dbSNP ID: rs5918) in the human ITGB3 gene.
Material and methods
We set up T-ARMS-PCR for the rs5918 SNP in a single-step PCR and the results were validated by the PCR-RFLP method in 132 coronary artery disease (CAD) patients and 122 unrelated healthy individuals.
Results
Full accordance was found for genotype determination by the PCR-RFLP method. The multiple logistic regression analysis showed a significant association of the rs5918 polymorphism and CAD according to dominant and recessive models (dominant model OR: 2.40, 95% CI: 1.33–4.35; p = 0.003, recessive model OR: 4.71, 95% CI: 1.32–16.80; p = 0.0067).
Conclusions
Our T-ARMS-PCR in comparison with RFLP and allele-specific PCR is more advantageous because this PCR method allows the evaluation of both the wild type and the mutant allele in the same tube. Our results suggest that the rs5918 (PlA1/A2) polymorphism in the ITGB3 gene may contribute to the susceptibility of sporadic Iranian coronary artery disease (CAD) patients.
Keywords: glycoprotein IIIa PlA1/A2 polymorphism, rs5918, T-ARMS-PCR, coronary artery disease
Introduction
Coronary artery disease (CAD) is most commonly due to atherosclerotic occlusion of the coronary arteries and is the leading cause of death worldwide for both men and women [1]. Coronary artery disease occurs when plaque builds up inside the coronary arteries and can involve many blood vessels with a variety of presentations [2]. This can lead to coronary artery disease or heart failure and arrhythmias. The conventional risk factors for CAD such as elevated cholesterol, hypertension, obesity, and smoking were well associated with 30–40% increasing in mortality and morbidity [3]. Multiple epidemiological, family, and other factors have documented a genetic predisposition for CAD [4]. Most polygenic diseases such as CAD are due to several common genes and have their genetic predisposition transmitted by multiple genes [5]. In polygenic disorders many DNA markers and single nucleotide polymorphisms (SNPs) of unrelated individuals need to be analyzed by different methods. A greater frequency of a SNP in patients indicates the SNP is in close proximity to a genetic risk variant for the disease. These variants strongly show the importance of a genetic predisposition for CAD and also confirm that these SNPs are even more common than expected [6]. However, it is well recognized that certain genes will relate to plaque rupture and/or thrombosis, and the beginning of the process of atherosclerosis. Glycoprotein IIIa (GPIIIa) or the beta subunit of the platelet membrane adhesive protein receptor complex GP IIb/IIIa is coded by the ITGB3 gene, and is a surface protein found in various tissues, participating in cell-surface mediated signaling and cell adhesion [7]. The exons and introns of the entire ITGB3 gene have been demonstrated to contain many polymorphic regions, one of which was found to be associated with cardiovascular diseases. This common ITGB3 polymorphism at codon 33 of exon 2 (T1565C, dbSNP ID: rs5918) corresponds to the polymorphism of amino acid residues (leucine/proline) at position 33 (PlA1/A2) of the polypeptide chain. It has been reported that this SNP is a risk factor of many types of disease, such as myocardial infarction [8], coronary heart disease, type 2 diabetes, asthma [9, 10], many cancers including colon, non-Hodgkin lymphoma [11], breast [12–14] ovarian cancer [15] and kidney cancer [16]. It has also been documented that the platelets bearing the β3 subunit of integrin αIIbβ3 with a proline at position 33 are characterized by an increased risk for aggregation and immunogenic properties of platelets [3, 4]. Since the amount of platelet aggregates is increased in carriers of the PlA2 allele with CAD, it is of interest to determine whether this polymorphic variant is associated with this disease. The previous data on the association of the polymorphic marker PlA1/A2 of the ITGB3 gene with both arterial and venous thrombosis in various ethnic groups [5, 6] are contradictory [17, 18]. The presence of the PlA2 allele is associated with an increased binding affinity to fibrinogen as well as with platelet aggregability in response to epinephrine, adenosine diphosphate and collagen in vitro [19]. Several studies have suggested that the PlA2 allelic variant causes an altered sensitivity to aspirin and an increased sensitivity to platelet aggregation by various agonists [20, 21].
Single nucleotide polymorphism (SNP) genotyping has been done by PCR-RFLP, real time-PCR and direct sequencing methods [22–24]. These techniques have certain limitations and are relatively slow and very expensive, and fewer reactions are catalyzed per kit in comparison to the tetra primer-amplification refractory mutation system-PCR (T-ARMS-PCR). With this method, the genotyping could be done using only a thermocycler machine at the least time, and can be used to genotyping of essential SNPs in the entire genome.
In conventional ARMS PCR, the amplification of the normal and mutant allele is done in two separate reactions, but in T-ARMS-PCR, we can amplify both the normal and mutant allele with a control fragment in a single reaction. In the T-ARMS-PCR method two outer, non-allele-specific primers and two inner, allele-specific primers in opposite orientation to each other are used.
The outer primers amplify a large fragment of the target gene containing a variant nucleotide as a control fragment and smaller allele-specific amplicons with different sizes that can easily be discriminated in gel electrophoresis either as homozygous or heterozygous. A deliberate mismatch at position 2nd or 3rd nucleotide from the 3′ terminal end of the inner primers can improve allele specificity [25].
The results of a meta-analysis of research showed that the PlA2 variant was associated with an increased risk of coronary heart disease [26, 27]. Some reports suggest that PlA1/A2 heterozygotes are prone to thrombotic disease, whereas PlA1/A1 homozygotes may be prone to early atherosclerosis and more rapid progression of stable coronary artery disease [10]. Presently, we are developing a rapid single-step method using T-ARMS-PCR for detection of Leu33Pro (PlA1/A2), which is a possible SNP in cardiovascular diseases studies.
Material and methods
Patients and DNA extraction
This study was performed on 132 patients selected from 412 patients who referred to cardiac centers in Afshar Hospital (Yazd, Iran) due to symptoms of myocardial infarction between 2012 and 2015. Patients had major lesions (> 50% narrowing of luminal diameter) in one, two, or three vessels (LAD, LCX, and RCA) that were candidate vessels for coronary artery bypass graft surgery. CAD patients were identified according to the coronary angiography guidelines [28].
When up to 50% blockage was observed in the major epicardial coronaries and their branches associated with stenotic lesions, coronary arterial disease was considered to be present. We also chose 122 unrelated healthy individuals matched for age, sex, and ethnicity with normal or near-normal angiography reports (no lesion greater than 30%) as a control group (Table I). All of the patients and the control group were informed of the aims of the study and gave their informed consent for the genetic analysis. Genomic DNA was isolated from the peripheral blood samples using a DNA isolation kit (DNAfast Kit-Genfanavaran, Tehran, Iran).
Table I.
Clinical characteristics of coronary atherosclerosis patients
| Parameter | Patients (n = 132) | Controls (n = 122) | P-value |
|---|---|---|---|
| Male gender (%) | 75 | 71 | 0.670 |
| Age [years] | 52.6 ±6.8 | 52.6 ±6.9 | 0.56 |
| Smokers (%) | 28 | 23 | 0.517 |
| Body mass index [kg/m2] | 26.1 ±2.4 | 25.3 ±1.9 | 0.0012 |
| Cholesterol [mg/dl] | 214.9 ±52.8 | 181.1 ±36.8 | 0.001 |
| LDL-C [mg/dl] | 128.7 ±43.4 | 118.2 ±44.1 | 0.145 |
| HDL-C [mg/dl] | 45.4 ±8.5 | 46.9 ±11.2 | 0.456 |
| TGs [mg/dl] | 205.7 ±102.1 | 153.4 ±96.1 | 0.014 |
Primer design and bioinformatics analysis
A common missense rs5918 (PlA1/A2) polymorphism in the ITGB3 gene was selected (http://www.ncbi.nlm.nih.gov/books/SNP). The four primers used in this research were designed by the online website Primer1: http://primer1.soton.ac.uk/primer1.html. The specificity of the primers was checked by the ‘BLAST’ program at http://www.ncbi.nlm.nih.gov/blast (Table II). To enhance the specificity of inner primers, at the 3rd nucleotide from the 3’-terminus changes to destabilizing mismatch.
Table II.
PCR primers and conditions
| ITGB3 (SNP ID: rs5918) | Primer sequence | Temperature [ºC] | Amplicon size |
|---|---|---|---|
| Fo: 5’-GGATTATCCCAGGAAAGACCAC | 66 | T allele (284 bp) C allele (179 bp) Control band (424 bp) |
|
| Ro: 5’-GACTTCCTCCTCAGACCTCCAC-3’ | 70 | ||
| Fi: 5’-TGTCTTACAGGCCCTGCCGCT-3’ | 68 | ||
| Ri: 5’-GGTCACAGCGAGGTGAGCACG-3’ | 70 |
SWISS-MODEL (http://swissmodel.expasy.org) was used for protein modeling of mutant protein. SWISS-MODEL is a fully automated protein structure homology-modeling server. The PyMol software was used for visualizing the effect of the altered residue on the protein structure.
T-ARMS-PCR and PCR-RFLP analysis
Polymerase chain reaction was performed in a total volume of 12.5 µl containing 50 ng of template DNA, 250 nM of each primer, 250 µM dNTPs, 1.5 mM MgCl2, 1X buffer and 0.5 U of Taq polymerase (Yekta Tajhiz Azma, Tehran, Iran). PCR amplification (touchdown) was carried out at 95ºC for 2 min, followed by denaturation at 95ºC for 20 s, first annealing at 68ºC (10 cycles), further (25 cycles) annealing at 69ºC for 1 min and extension at 72ºC for 50 s, followed by a final extension for 5 min. Non-denaturation polyacrylamide gel electrophoresis (6%) and silver staining were used for detection of PCR products.
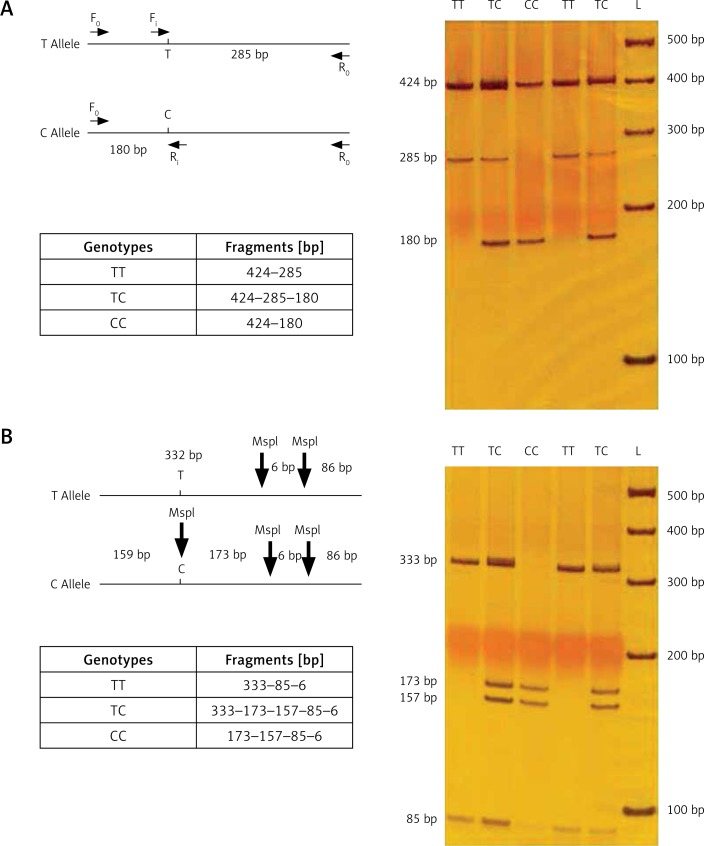
This procedure rendered 2 bands in homozygotes (PlA1/A1 resulting in 424 and 285 bp, and PlA1/A1 resulting in 424 and 180 bp) and 3 bands in heterozygotes (424, 285 and 180 bp). Figure 1 shows the products of tetra-primer ARMS-PCR for 5 different genomic DNA samples.
Figure 1.
A – Results of T-ARMS-PCR of ITGB3 polymorphism (rs5918); B – validation using PCR-RFLP by MspI restriction endonuclease
In this study, the concentration of PCR components such as primers, magnesium chloride, dNTP and Taq DNA polymerase were optimized. Beside PCR components, the PCR conditions were also optimized.
The forward (Fo) and reverse (Ro) primers were used to amplify the 424-bp fragment. The 424-bp product was digested with 5U MspI restriction enzyme for 16 h. Fragments were separated by electrophoresis using 6% polyacrylamide gel electrophoresis. The resulting fragments were 332, 86 and 6 bp for the PlA1 allele and 173, 159, 86 and 6 bp for the PlA2 allele (Figure 1).
Statistical analysis
The SPSS software (IBM SPSS 22, SPSS Inc., Chicago, IL., USA) was used for statistical analysis. The association between two groups was examined by the χ2 goodness-of-fit test. Multiple logistic regression models (co-dominant, dominant and recessive) were employed to analysis the genetic data. Values of p < 0.05 were regarded as statistically significant.
Results
Total DNA samples analyzed with T-ARMS-PCR were re-evaluated by PCR-RFLP analysis using the restriction enzyme MspI. We obtained 100% accordance between both methods for genotype determination. Mean age (mean ± SD) was 53.9 ±6.8 and 52.6 ±6.9 years for patients and controls, respectively. Coronary angiography revealed 132 patients (CAD+ group) with one vessel (LAD) (n = 31), two vessels (LCX) (n = 51), or three vessels (RCA) (n = 49) that were candidate vessels for coronary artery bypass grafting (CABG) and 122 patients (CAD– group) with no angiographically identified narrowing.
Genotype distributions and allelic frequencies of the ITGB3 polymorphism among CAD patients and controls are shown in Table III. The frequency of the ITGB3 C allele was significantly higher in the CAD patients than in the control group (p < 0.001).
Table III.
Genotype and allele variant frequencies in patients and controls
| ITGB3(rs5918) | Patients (n = 132) | Controls (n = 122) | OR (95% CI) | P-value |
|---|---|---|---|---|
| Codominant model: | ||||
| TT | 88 (66.67%) | 101 (85.2%) | 1 | |
| TC | 30 (22.7%) | 18 (14.8%) | 0.523 (0.273–1.002) | 0.051 |
| CC | 14 (10.6%) | 3 (0.0%) | 0.187 (0.052–0.671) | 0.010 |
| Dominant model: | ||||
| TT | 88 (66.7%) | 101 (82.8%) | ||
| TC + CC | 44 (33.3%) | 21 (17.2%) | 2.40 (1.33–4.35) | 0.003 |
| Recessive model: | ||||
| CC | 14 (10.6%) | 3 (2.5%) | ||
| TC + TT | 118 (89.4%) | 119 (97.5%) | 4.71 (1.32–1.608) | 0.0067 |
| Allele frequency: | ||||
| T | 206 (78.0%) | 220 (90.2%) | ||
| C | 58 (22.00%) | 24 (9.8%) | 0.387 (0.232–0.644) | < 0.001 |
OR – odds ratio, CI – confidence interval.
The multiple logistic regression analysis showed a significant association of the rs5918 polymorphism and CAD according to dominant and recessive models (dominant model OR: 2.40, 95% CI: 1.33–4.35; p = 0.003, recessive model OR: 4.71, 95% CI: 1.32–16.80; p = 0.0067) (Table III).
Discussion
So far, several variants of the ITGB3 gene have been described [27]. The correlation of the rs5918 polymorphism with the incidence of CAD was first reported by Marian et al. [29]. In the present study, the allele and genotype frequencies of the polymorphic marker PlA1/A2 of the ITGB3 gene were determined in a group of patients with CAD and control subjects. Weiss et al. demonstrated the association of the PlA2 allele with the risk of cardiovascular diseases [30]. PlA2 allele frequency was significantly higher in CAD patients than in control subjects. The frequency distributions of different genotypes and alleles using techniques for detection of the ITGB3 gene in other population studies are described in Table IV [22, 24, 31–33].
Table IV.
Distribution of ITGB3 polymorphism in current study in comparison with previously published data
| Study | Population | N | Genotype (%) | Allele (%) | Genotyping platform | |||
|---|---|---|---|---|---|---|---|---|
| (rs5918 SNP) | TT | TC | CC | T | C | |||
| Our study | Iranian | 254 | 74.4 | 18.9 | 6.7 | 83.9 | 16.1 | T-ARMS-PCR |
| Torabi et al. | Iranian | 200 | 70 | 14 | 16 | 77 | 23 | PCR-RFLP |
| Nikolajevic-Starcevic et al. | Slovenia | 342 | 47 | 36.8 | 16.2 | 65.5 | 34.5 | PCR-RFLP |
| Zhang et al. | Chinese | 622 | 96 | 4 | 0 | 98 | 2 | High-resolution melting analysis (HRM) |
| Yilmaz et al. | Turkish | 184 | 83.7 | 13 | 3.3 | 90.2 | 9.8 | PCR-RFLP |
| Bianconi et al. | Austrian | 109 | 79.8 | 17.4 | 2.8 | 88.5 | 11.5 | PCR-RFLP |
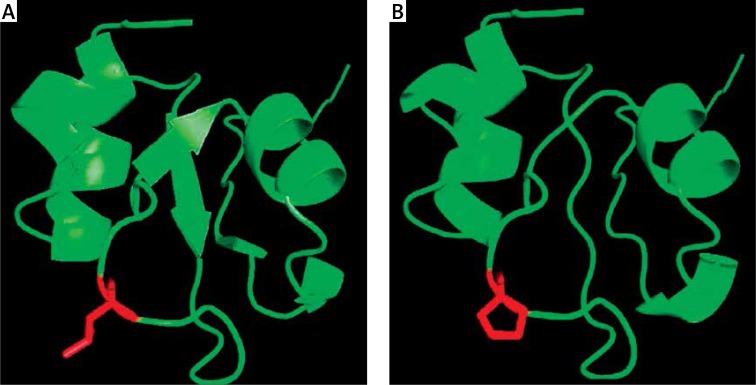
Protein modeling based on human ITGB3 protein structure (pdb id: 4g1m) shows that L33 is located on the beta strand of the extracellular domain. ITGB3 is a heterodimer composed of noncovalently associated a and b subunits. These subunits have a large extracellular domain, a single transmembrane region, and a short cytoplasmic domain [12, 33]. The L33P change lies in the N-terminal of ITGB3. An analysis of the ITGB3 wild-type and L33P mutant-derived protein structure using the SWISS-MODEL server indicates that three-dimensional structure of the protein is disrupted in the mutant (Figure 2).
Figure 2.
Molecular model of human ITGB3 protein base in PDB file 4g1m showing close-up of the L33 (A) and P 33 residues (B) using PyMol
In general, the allele and genotype frequency distributions in the Iranian population were similar to those in other populations. Thus, the results of our study suggest that the polymorphic variant PlA1/A2 is associated with CAD in the Iranian population.
The common ITGB3 studies have utilized MspI digestion to identify the rs5918 polymorphism. Because this polymorphism is a risk factor in many diseases including cardiovascular diseases and cancers, here we developed a rapid, sensitive and one-step tetra-primer PCR-ARMS method for detection of the rs5918 polymorphism.
The most critical step for successful development of the new multiplex tetra-primer amplification refractory mutation system-PCR method is the primer design. In this study, the primers for amplification of ITGB3 rs5918 were designed using the online website Primer1: http://primer1.soton.ac.uk/primer1.html. Both the outer primers were designed with 21 base pairs (bp) in length and the inner primers were designed with 22 bp with a guanine and adenine located at the 3rd nucleotide from the 3’-terminus for forward (wild type allele) and reverse (mutant allele) primers to increase their specificity for the template DNA.
Our T-ARMS-PCR in comparison with RFLP and allele-specific PCR is more advantageous because this PCR method allows the evaluation of both the wild type and mutant allele in the same tube. This assay is a reproducible and stable single-tube reaction.
In conclusion, our T-ARMS-PCR needs only a small amount of traditional PCR reagents, without special equipment. Our results suggest that the rs5918 (PlA1/A2) polymorphism in the ITGB3 gene may contribute to the susceptibility to sporadic CAD. Therefore, this procedure could be used in most clinical diagnostic laboratories.
Acknowledgments
This research was funded by Yazd University. We thank all the patients for providing blood samples for the scientific research, also the Especial Afshar Hospital (Yazd, Iran). The study was approved by Yazd University Human Research Ethics Committee.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Bolognese L. Changing patterns of ST elevation myocardial infarction epidemiology. Am Heart J. 2010;160:S1–3. doi: 10.1016/j.ahj.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Deakin SP, James RW. Genetic and environmental factors modulating serum concentrations and activities of the antioxidant enzyme paraoxonase-1. Clin Sci. 2004;107:435–47. doi: 10.1042/CS20040187. [DOI] [PubMed] [Google Scholar]
- 3.Franchini M, Peyvandi F, Mannucci PM. The genetic basis of coronary artery disease: from candidate genes to whole genome analysis. Trends Cardiovasc Med. 2008;18:157–62. doi: 10.1016/j.tcm.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Ozaki K, Tanaka T. Molecular genetics of coronary artery disease. J Hum Genet. 2016;61:71–7. doi: 10.1038/jhg.2015.70. [DOI] [PubMed] [Google Scholar]
- 5.Preuss M, Konig IR, Thompson JR, et al. Design of the Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Study: a genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ Cardiovasc Genet. 2010;3:475–483. doi: 10.1161/CIRCGENETICS.109.899443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musunuru K, Kathiresan S. HapMap and mapping genes for cardiovascular disease. Circ Cardiovasc Genet. 2008;1:66–71. doi: 10.1161/CIRCGENETICS.108.813675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett JS. Structure and function of the platelet integrin alphaIIbbeta3. J Clin Investig. 2005;115:3363–9. doi: 10.1172/JCI26989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Floyd CN, Mustafa A, Ferro A. The PlA1/A2 polymorphism of glycoprotein IIIa as a risk factor for myocardial infarction: a meta-analysis. PloS One. 2014;9:e101518. doi: 10.1371/journal.pone.0101518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimrin AB, Gidwitz S, Lord S, et al. The genomic organization of platelet glycoprotein IIIa. J Biol Chem. 1990;265:8590–5. [PubMed] [Google Scholar]
- 10.Mikkelsson J, Perola M, Laippala P, Penttila A, Karhunen PJ. Glycoprotein IIIa Pl(A1/A2) polymorphism and sudden cardiac death. J Am Coll Cardiol. 2000;36:1317–23. doi: 10.1016/s0735-1097(00)00871-8. [DOI] [PubMed] [Google Scholar]
- 11.Cerhan JR, Ansell SM, Fredericksen ZS, et al. Genetic variation in 1253 immune and inflammation genes and risk of non-Hodgkin lymphoma. Blood. 2007;110:4455–63. doi: 10.1182/blood-2007-05-088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bojesen SE, Kjaer SK, Hogdall EV, et al. Increased risk of ovarian cancer in integrin beta3 Leu33Pro homozygotes. Endocr Relat Cancer. 2005;12:945–52. doi: 10.1677/erc.1.01083. [DOI] [PubMed] [Google Scholar]
- 13.Langsenlehner U, Renner W, Yazdani-Biuki B, et al. Integrin alpha-2 and beta-3 gene polymorphisms and breast cancer risk. Breast Cancer Res Treat. 2006;97:67–72. doi: 10.1007/s10549-005-9089-4. [DOI] [PubMed] [Google Scholar]
- 14.Jin Q, Hemminki K, Grzybowska E, Klaes R, Soderberg M, Forsti A. Re: Integrin beta3 Leu33Pro homozygosity and risk of cancer. J Natl Cancer Instit. 2004;96:234–5. doi: 10.1093/jnci/djh032. [DOI] [PubMed] [Google Scholar]
- 15.Wang-Gohrke S, Chang-Claude J. Integrin beta3 Leu33Pro homozygosity and risk of cancer. J Natl Cancer Instit. 2005;97:778–9. doi: 10.1093/jnci/dji135. [DOI] [PubMed] [Google Scholar]
- 16.Kallio JP, Mikkelsson J, Tammela TL, Karhunen PJ, Kellokumpu-Lehtinen P. Genetic variation in platelet integrin alphabeta (GPIIb/IIIa) and the metastatic potential of renal cell carcinoma. BJU Intern. 2006;98:201–4. doi: 10.1111/j.1464-410X.2006.06196.x. [DOI] [PubMed] [Google Scholar]
- 17.Feng D, Lindpaintner K, Larson MG, et al. Increased platelet aggregability associated with platelet GPIIIa PlA2 polymorphism: the Framingham Offspring Study. Arterioscler Thromb Vasc Biol. 1999;19:1142–7. doi: 10.1161/01.atv.19.4.1142. [DOI] [PubMed] [Google Scholar]
- 18.Vijayan KV, Liu Y, Dong JF, Bray PF. Enhanced activation of mitogen-activated protein kinase and myosin light chain kinase by the Pro33 polymorphism of integrin beta 3. J Biol Chem. 2003;278:3860–7. doi: 10.1074/jbc.M208680200. [DOI] [PubMed] [Google Scholar]
- 19.Goodall AH, Curzen N, Panesar M, et al. Increased binding of fibrinogen to glycoprotein IIIa-proline33 (HPA-1b, PlA2, Zwb) positive platelets in patients with cardiovascular disease. Eur Heart J. 1999;20:742–7. doi: 10.1053/euhj.1998.1203. [DOI] [PubMed] [Google Scholar]
- 20.Papp E, Havasi V, Bene J, et al. Glycoprotein IIIA gene (PlA) polymorphism and aspirin resistance: is there any correlation? Ann Pharmacother. 2005;39:1013–8. doi: 10.1345/aph.1E227. [DOI] [PubMed] [Google Scholar]
- 21.Cooke GE, Liu-Stratton Y, Ferketich AK, et al. Effect of platelet antigen polymorphism on platelet inhibition by aspirin, clopidogrel, or their combination. J Am Coll Cardiol. 2006;47:541–6. doi: 10.1016/j.jacc.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 22.Nikolajevic-Starcevic J, Petrovic MG, Petrovic D. A1/A2 polymorphism of the glycoprotein IIIa gene and diabetic retinopathy in Caucasians with type 2 diabetes. Clin Exp Ophthalmol. 2011;39:665–72. doi: 10.1111/j.1442-9071.2011.02520.x. [DOI] [PubMed] [Google Scholar]
- 23.Jakubowska A, Rozkrut D, Antoniou A, Hamann U, Lubinski J. The Leu33Pro polymorphism in the ITGB3 gene does not modify BRCA1/2-associated breast or ovarian cancer risks: results from a multicenter study among 15,542 BRCA1 and BRCA2 mutation carriers. Breast Cancer Res Treat. 2010;121:639–49. doi: 10.1007/s10549-009-0595-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Han Y, Dong L, et al. Genetic variation of ITGB3 is associated with asthma in Chinese Han children. PloS One. 2013;8:e56914. doi: 10.1371/journal.pone.0056914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Etlik O, Koksal V, Arican-Baris ST, Baris I. Development and validation of a cost-effective in-house method, tetra-primer ARMS PCR assay, in genotyping of seven clinically important point mutations. Mol Cell Probes. 2011;25:177–81. doi: 10.1016/j.mcp.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Mikkelsson J, Perola M, Laippala P, et al. Glycoprotein IIIa Pl(A) polymorphism associates with progression of coronary artery disease and with myocardial infarction in an autopsy series of middle-aged men who died suddenly. Arterioscler Thromb Vasc Biol. 1999;19:2573–8. doi: 10.1161/01.atv.19.10.2573. [DOI] [PubMed] [Google Scholar]
- 27.Zhu MM, Weedon J, Clark LT. Meta-analysis of the association of platelet glycoprotein IIIa PlA1/A2 polymorphism with myocardial infarction. Am J Cardiol. 2000;86:1000–5. doi: 10.1016/s0002-9149(00)01136-x. [DOI] [PubMed] [Google Scholar]
- 28.Salonen R, Nyyssonen K, Porkkala E, et al. Kuopio Atherosclerosis Prevention Study (KAPS). A population-based primary preventive trial of the effect of LDL lowering on atherosclerotic progression in carotid and femoral arteries. Circulation. 1995;92:1758–64. doi: 10.1161/01.cir.92.7.1758. [DOI] [PubMed] [Google Scholar]
- 29.Marian AJ, Brugada R, Kleiman NS. Platelet glycoprotein IIIa PlA polymorphism and myocardial infarction. N Engl J Med. 1996;335:1071–2. doi: 10.1056/NEJM199610033351418. [DOI] [PubMed] [Google Scholar]
- 30.Weiss EJ, Bray PF, Tayback M, et al. A polymorphism of a platelet glycoprotein receptor as an inherited risk factor for coronary thrombosis. N Engl J Med. 1996;334:1090–4. doi: 10.1056/NEJM199604253341703. [DOI] [PubMed] [Google Scholar]
- 31.Torabi R, Zarei S, Zeraati H, et al. Combination of thrombophilic gene polymorphisms as a cause of increased the risk of recurrent pregnancy loss. J Reprod Infertil. 2012;13:89–94. [PMC free article] [PubMed] [Google Scholar]
- 32.Yilmaz U, Zeybek U, Kahraman OT, et al. Investigation of ICAM-1 and beta3 integrin gene variations in patients with brain tumors. Asian Pacif J Cancer Prevent. 2013;14:5929–34. doi: 10.7314/apjcp.2013.14.10.5929. [DOI] [PubMed] [Google Scholar]
- 33.Bianconi D, Schuler A, Pausz C, et al. Integrin beta-3 genetic variants and risk of venous thromboembolism in colorectal cancer patients. Thromb Res. 2015;136:865–9. doi: 10.1016/j.thromres.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]