Abstract
Multicellularity in animals requires dynamic maintenance of cell-cell contacts. Intercellularly ligated cadherins recruit numerous proteins to form supramolecular complexes that connect with the actin cytoskeleton and support force transmission. However, the molecular organization within such structures remains unknown. Here we mapped protein organization in cadherin-based adhesions by superresolution microscopy, revealing a multi-compartment nanoscale architecture, with the plasma membrane-proximal cadherin-catenin compartment segregated from the actin cytoskeletal compartment, bridged by an interface zone containing vinculin. Vinculin position is determined by α-catenin, and upon activation, vinculin can extend ˜30 nm to bridge the cadherin-catenin and actin compartments, while modulating the nanoscale positions of the actin regulators, zyxin and VASP. Vinculin conformational activation requires tension and tyrosine phosphorylation, regulated by Abl kinase, and PTP1B phosphatase. Such modular architecture provides a structural framework for mechanical and biochemical signal integration by vinculin, which may differentially engage cadherin-catenin complexes with the actomyosin machinery to regulate cell adhesions.
Introduction
The question of how animal cells self-organize into complex and patterned structures at the tissue and organism levels are intrinsically multiscale, depending on an intricate interplay of local and long-range forces within tissues and cells, as well as exquisite coordination of sub-cellular programs ranging from genetic and signaling pathways to cell morphodynamic behaviors1,2. While recent advances in the understanding of these processes have prominently focused at the length scale of tissues and cells3,4, much has remained unexplored at the level of the very molecular machines that enable intercellular adhesions, cytomechanical adaptation, and mechanotransduction processes underlying these morphogenetic events. Cell-cell junctions mediated by the cadherin transmembrane receptors are among the most important molecular machinery that interlink and coordinate neighboring cells, participating in important cellular pathways including transcriptional control, cell polarization, cytoskeletal regulation, and cellular mechanotransduction5–10. Adhesions of cadherin recruit numerous proteins, collectively known as ‘cadhesome’11, to form supramolecular complexes closely associated with the actin cytoskeleton. However, the nanoscale dimension and the compositional complexity of the cadherin adhesions have long defied available structure-determination or imaging techniques, and thus the structural framework for understanding how such complex multiprotein assembly is physically organized to perform biological functions has not been available.
Previously, astigmatism-based 3-D superresolution microscopy12 has been applied to resolve nanocluster organization of cadherins in adherens junctions (AJs) where neighboring epithelial cells form contact sites13,14. However, the spatial resolution thus attained, >20-100 nm, poses a challenge for quantifying protein organization at the sub-20 nm molecular length scale. Likewise, it has been difficult to decipher molecular organization of cadhesome proteins from electron microscopy (EM) images15,16. Therefore, to provide a structural framework for understanding cadherin-based cell adhesions, we adopted a planarized biomimetic platform based on oriented cadherin-Fc arrayed on IgG-coated substrates17. This format confers a greater optical accessibility amenable to high-precision (sub-20 nm) superresolution fluorescence microscopy techniques18–22, allowing molecular scale interrogation with current fluorescent protein (FP) technologies.
In this study, we mapped protein organization within planar cadherin-based adhesions, observing a compartmentalized nanoscale architecture, whereby the plasma membrane-proximal cadherin-catenin compartment is physically segregated by ˜30 nm from the uppermost compartment containing actin and actin regulatory proteins, bridged by an interface compartment containing vinculin. We showed that the nanoscale positioning of vinculin is determined by α-catenin. Upon conformational activation, vinculin extends ˜30 nm to bridge the cadherin-catenin and actin compartments, while also modulating the nanoscale positions of the actin polymerization regulators zyxin and VASP. The extended conformation of vinculin requires both tension and tyrosine phosphorylation at residue Y822 by Abl kinase, while we also identified PTP1B as the tyrosine phosphatase that dephosphorylates vinculin. The observed multi-layer nanoscale architecture of cadherin-based adhesions appears to centrally position vinculin to act as an integrator of mechanical and biochemical signals, suggesting how the cadherin-based adhesions could selectively engage the actin cytoskeleton in response to regulatory input signals, effectively as a ‘molecular clutch’, to mediate intercellular interactions.
Results
Mapping protein position in planar cadherin-based adhesions by superresolution microscopy
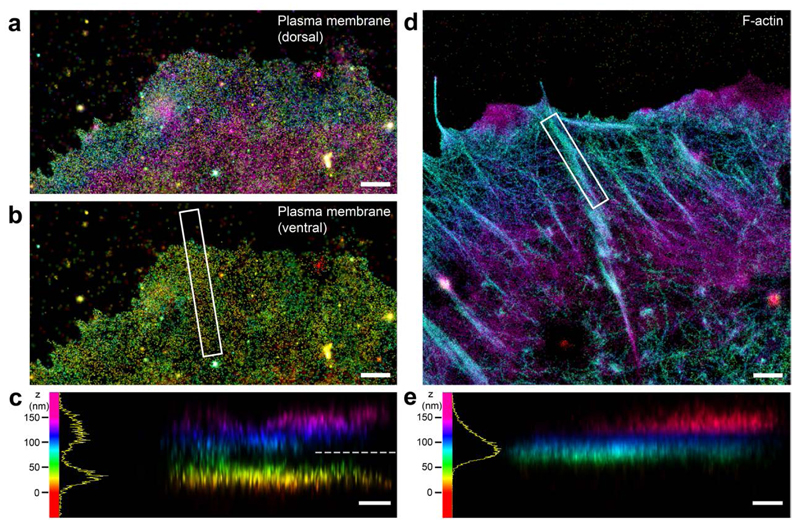
The multi-micron vertical (z) depth of the AJs in epithelial monolayer limited our ability to map molecular scale organization by astigmatism-based superresolution microscopy14 (Supplementary Fig. 1a). We noted that the planar cadherin-coated substrate format have been employed in earlier studies17,23–25 to obtain key molecular insights into interactions between cadherin and associated proteins. On such platform cells formed cadherin-based adhesions that recruited cadhesome proteins but not integrin-associated proteins (Supplementary Fig. 1b-c), suggesting that salient protein-protein interactions are likely recapitulated. To demarcate the plasma membrane position in this format, we first applied 3-D Interferometric PhotoActivated Localization Microscopy19 (Supplementary Fig. 2a-b) to image MDCK (Madin-Darby Canine Kidney) epithelial cells cultured on E-cadherin-coated substrate, using DiD membrane-targeting fluorophores26. This clearly resolved dorsal and ventral plasma membranes, with the z-position of the latter at ˜30-40 nm above the substrate (Fig. 1a-c). We then imaged filamentous (F)-actin using AlexaFluor 647-phalloidin, observing that F-actin bundles reside at a higher z-position, centering around ˜70-80 nm, (Fig. 1d-e). The spatial separation of ˜30 nm between the ventral plasma membrane and the actin cytoskeleton thereby minimizes direct cadherin-actin interaction. The F-actin angle of approach is nearly parallel to the adhesion plane (Supplementary Fig. 3a-d), geometrically comparable to the F-actin orientation around AJs4. Altogether these data are suggestive of the nanoscale similarity between planar cadherin adhesions and native cell-cell contacts.
Figure 1. iPALM imaging of plasma membrane marker and F-actin.
a-c, Plasma membrane of MDCK cell labelled by DiD fluorophore. a, Top view of dorsal plasma membrane; b, top view of ventral plasma membrane (z <80 nm); c, side view (white box in b). d-e, F-actin in MDCK cell labeled by Alexa Fluor 647-phalloidin. d, top view; e, side view of white box in d. Colours (hue scale in c, e, 0-150 nm) indicate the vertical (z) coordinate, relative to the substrate surface (z = 0 nm, red). Dashed line in c marks the z=80 nm threshold used for b. Histograms in c, e (bin size, 1 nm). Scale bars: 1 μm (a, b, d), 250 nm (c, e).
Nanoscale compartmentalization of E-cadherin-based adhesions
We next applied a surface-generated structured illumination technique20,21 (Supplementary Fig. 2c-g) to characterize nanoscale organization of FP-conjugated cadhesome proteins (Supplementary Fig. 4a). The fluorophore z-position relative to the substrate surface (z= 0 nm) was analyzed pixel-wise, with the median value, zcentre, for adhesion region-of-interests (ROI) used as the representative protein z-position27, while the z-position histograms denote the spatial distribution of proteins (Fig. 3, Supplementary Note 1.3, Supplementary Fig. 2h,i). We observed that E-cadherin (cytoplasmic domain GFP fusion) is positioned at zcentre = 46.6 nm, consistent with the dimensions of cadherin and other substrate components (Fig.2b, 3a).
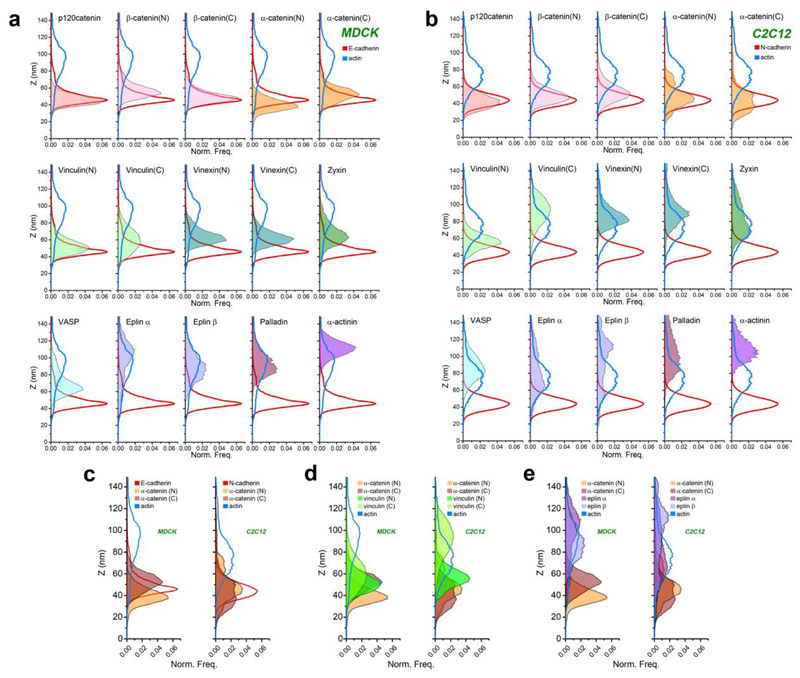
Figure 3. Z-dimension profiles of cadhesome proteins.
Normalized histogram of pixel z-position of indicated proteins (filled colors) in MDCK (a) and C2C12 (b) cells, relative to E- or N-cadherin (solid red) f-actin (solid blue). Integrated areas under each histogram are normalized to unity, such that each histogram reflects the probability distribution of locating a given protein as a function of z-position. c, d, e Comparison of protein z-profiles of c) α-catenin (N and C-terminal probes, shades of brown) and d) vinculin (N and C-terminal probes shades of green), or e) Eplins (α and β isoforms, shades of purple) between MDCK and C2C12 cells.
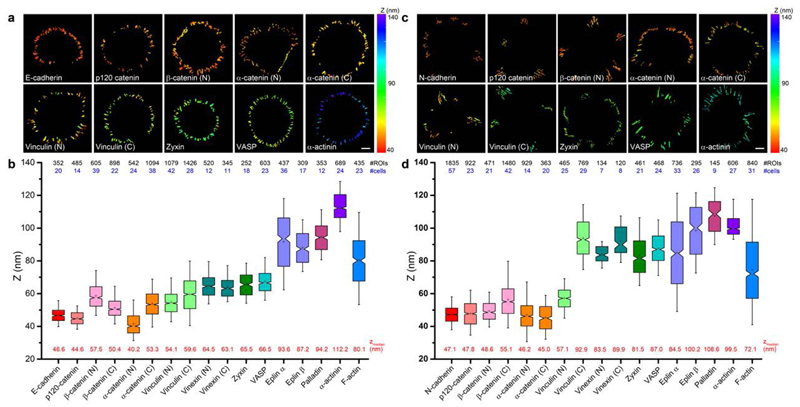
Figure 2. Protein stratifications in MDCK and C2C12 cadherin-based adhesions.
a and c, Topographic maps of protein z-positions (nanometers) in E-cadherin-based adhesions of MDCK cells (A) and N-cadherin-based adhesions of C2C12 cells. Colour bar indicates z-position relative to substrate surface. Scale bars: 10 μm. b and d, Notched box plots for z-position of indicated proteins: first and third quartiles, median and confidence intervals; whiskers, 5th and 95th percentiles. Median zcentre values are indicated below each box plot (red). n values are shown above each box plot and indicate the numbers of adhesions (#ROIs, black). Number of cells are indicated in blue. Statistics are described in Supplementary Tables 1-2.
Subsequently, we surveyed the nanoscale organization of key cadhesome proteins, observing a surprising degree of compartmentalization along the z-dimension that effectively spans between the plasma membrane and the actin cytoskeleton. Proteins observed in close proximity to cadherin include p120-catenin (44.6 nm), β-catenin (57.5 nm, N-terminus; 50.4 nm, C-terminus), and α-catenin (40.2 nm, N-terminus). Their z-positions are consistent with their close association with the E-cadherin cytodomain6, thus defining the cadherin-catenin compartment. In contrast, actin-binding proteins were observed at significantly elevated z-positions, largely coinciding with the actin cytoskeleton. Eplins were found at 93.6 nm (α-isoform) and 87.2 nm (β-isoform), palladin at 94.2 nm, and α-actinin at 112.2 nm. A number of proteins were observed at intermediate z-positions, including vinculin (54.1 nm, N-terminus), zyxin (65.5 nm), VASP (vasodilator-stimulated protein; 66.5 nm), and vinexin (64.5 nm, N-terminus; 63.1 nm, C-terminus) (Fig. 2a-b, 3a, Supplementary Figure 5, Supplementary Tables 1-2). Our measurements suggest that these centrally-positioned proteins likely play an important role as an interface compartment that mediates structural connection and mechanical coupling between the cadherin-catenin and the actomyosin compartments.
The conformation and nanoscale organization of α-catenin
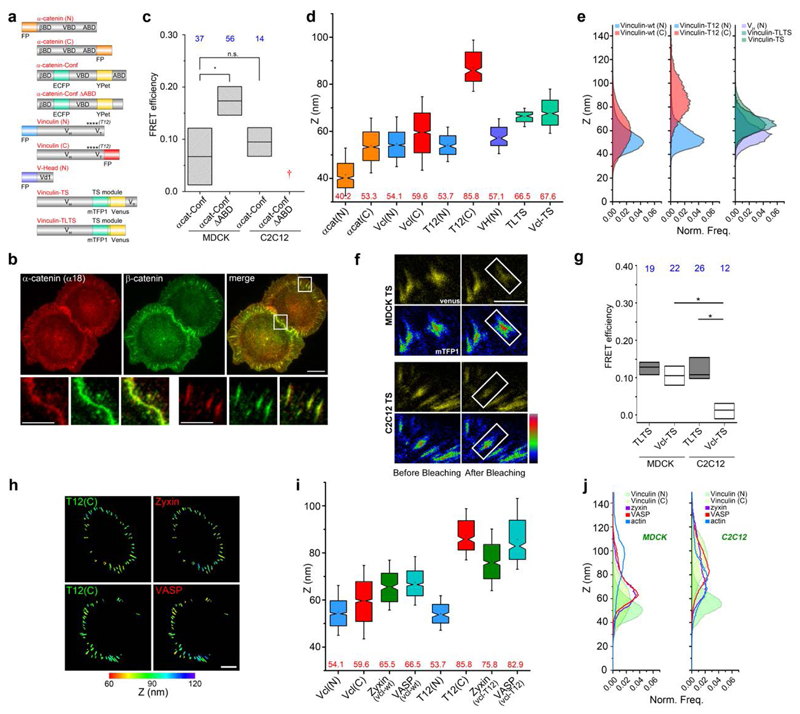
Since α-catenin and vinculin have been implicated as mechanotransducers28–31, we next investigated their configurations and spatial organization within the cadherin adhesions. Using a monoclonal antibody (α18) against the activated conformation of α-catenin32, we observed prominent staining (Fig. 4b, Supplementary Fig. 4d-e) consistent with measurements by fluorescence resonance energy transfer (FRET) conformation probe9 (Fig. 4c, Supplementary Fig. 6d). Furthermore, the high precision of our technique enables inference of protein orientation and/or conformation via the use of the N- and C-terminal FP fusion constructs (Supplementary Note 2, Supplementary Fig. 5e-h). We thus determined the C-terminal z-position of α-catenin, obtaining zcentre = 53.3 nm, compared to zcentre = 40.2 nm for the N-terminus, indicative of an oriented and activated configuration (Fig. 3c-e, Fig. 4d, Supplementary Note 3). We next probed the z-position of the α-catenin vinculin binding domain (VBD) by imaging vinculin head domain (Vd1, residue 1-258) N-terminal-tagged with GFP, observing the z-position of 57.1 nm closely overlapping with the α-catenin C-terminus, consistent with vinculin association to α-catenin (Fig. 4d). To further explore the role of α-catenin in vinculin positioning, we imaged vinculin-FP expressed in MDCK cells with stable α-catenin shRNA expression33 (Supplementary Fig. 4b). We found that with α-catenin depleted, vinculin localizes to a higher z-position within the actomyosin compartment, probably via the association with actin34 or actin-regulatory proteins such as VASP or α-actinin. On the other hand, upon re-expression of α-catenin-FP, the intermediate z-positioning of vinculin is restored (Supplementary Fig. 6a, Supplementary Note 3).
Figure 4. Nanoscale positions and conformations of vinculin and α-catenin in cadherin-based adhesions.
a, Diagrams for α-catenin and vinculin probes. FP: fluorescent protein (orange: α-catenin FP probes; blue, vinculin N-terminus; red, vinculin C-terminus); α-catenin-conf: α-catenin conformation sensor (teal: ECFP, yellow: YPet); α-catenin-conf ∆ABD: α-catenin conformation sensor with actin binding domain deletion; βBD, VBD, ABD: binding sites on α-catenin for β-catenin, vinculin, and actin, respectively; VH, VT, Vd1: vinculin head-domain, vinculin tail-domain, and the d1 sub-domain of VH, respectively; ****: approximated positions of point-mutations; vinculin-TS (Vcl-TS): vinculin tension-sensor (teal: mTFP1, yellow: Venus); vinculin-TLTS: Tail-less vinculin-TS. b, Immunofluorescence for activated α-catenin (α18) (red) and β-catenin (green) in MDCK cells on E-cadherin substrate. Bottom row, insets zoom-in: native cell-cell contacts and planar cadherin adhesions. Scale bars: 10 μm (insets: 5 μm). c, FRET efficiency of α-cat-conf and α-cat-conf ∆ABD. †: construct did not localise in C212. d, Box plots for zcentre of α-catenin and vinculin FP probes in MDCK. e, Z-profiles of vinculin probes in MDCK. N-terminal (blue), C-terminal (red), vinculin–TS and vinculin-TL-TS (shades of teal), vinculin head domain (VH, residue 1-258, purple) probes. f, FRET measurement of intra-vinculin tension. (Top) MDCK cells, (bottom) C2C12 cells expressing vinculin-TS, on E- or N-cadherin substrate, respectively. Rainbow look-up table (color bar) used for donor channel (mTFP1). Scale bar: 5 μm. g, FRET efficiency in MDCK and C2C12. h, Vinculin conformation modulates position of zyxin and VASP. Topographic map of z-positions: (left) vinculin-T12 C-terminal, (right) zyxin or VASP, co-expressed in MDCK cells. Color bar: z-position relative to substrate surface. Scale bar: 10 μm. i, Box plots for zcentre of vinculin, zyxin, and VASP (green and teal), with wild-type vinculin (Vcl-wt) or vinculin-T12. j Comparison of protein z-profiles for vinculin (N- and C- terminal probes, shades of green), and zyxin (purple) and VASP (red), overlay with f-actin z-profile (blue), between MDCK and C2C12 cells. c, g: Box plots for FRET efficiency (acceptor photobleaching): median, 1st and 3rd quartiles. *: p-value < 0.05. Number of cells (n values) in blue. d, i: Notched box plots: first and third quartiles, median and confidence intervals; whiskers, 5th and 95th percentiles, with n values (number of adhesions) in Supplementary Table 3.
Activated vinculin spans between the cadherin-catenin and actin cytoskeletal compartments
We next characterized how vinculin is organized within the cadherin adhesions. In MDCK, wild-type (wt) vinculin C-terminus was observed at zcentre = 59.6 nm, compared to 54.1 nm for the N-terminus (Fig. 2b). Since vinculin conformation is able to switch between the compact and the extended, uninhibited forms, the small N-C z-positional differences observed may correspond to either a relatively compact conformation, or an extended conformation that is oriented nearly parallel to the plasma membrane (<10°). To differentiate these, we first probed vinculin configuration by FRET. Using vinculin tension biosensor (vinculin-TS)29 or vinculin conformation biosensor35, we observed comparatively high FRET efficiencies, indicative of low vinculin tension and a relatively compact conformation, respectively (Fig. 4f-g, Supplementary Fig. 6b-c,e-h). Additionally, the z-position of the mTFP1 fluorophore within vinculin-TS (Fig . 4a) was mapped to report the z-position of the vinculin linker region, obtaining zcentre of 67.6 nm, further supporting a compact conformation of vinculin, with the linker region arching upward (Fig. 4d-e, Supplementary Fig. 5e-h, Supplementary Note 2). In contrast, when we probed the orientation of the constitutively active vinculin-T12 mutant36, we observed a drastic upshift of the C-terminal z-position (85.8 nm) relative to the N-terminus (53.7 nm) (Fig. 4d-e). Thus, activated vinculin effectively spans ˜30 nm or greater, a distance comparable to the fully extended length of vinculin37. Taken together, these results illustrate how α-catenin emplaces vinculin in the interface zone, a central position that enables activated vinculin to robustly couple the cadherin-catenin with the actomyosin compartments (Supplementary Note 3.4). We also note that a more subtle interplay between α-catenin, β-catenin, and vinculin configuration may also be present (Supplementary Fig. 6 a, Supplementary Note 3.2-3.3). Our approach may thus be of further use in unraveling how the cadherin:β-catenin:α-catenin module is structurally and mechanically integrated with the cortical actin cytoskeleton15,23,28, an important long-standing question, but that which is beyond the scope of the current study.
Vinculin conformation modulates nanoscale position of zyxin and VASP
We next investigated how vinculin conformational states may regulate the spatial organization of other cadhesome components. We performed 2-color nanoscale z-mapping experiments by imaging FP fusion of vinculin partners such as zyxin or VASP, together with vinculin-T12. As shown in Fig. 4h-j, zyxin and VASP, in the presence of endogenous vinculin (wt), were observed at zcentre = 65.5 nm and zcentre = 66.5 nm, respectively. In contrast, upon co-expression with vinculin-T12, we observed significant upshifts in their z-positions with zcentre = 75.8 nm for zyxin and zcentre = 82.9 nm for VASP. These results suggest that vinculin conformation may modulate the positioning of proteins such as zyxin and VASP that bind to its proline-rich linker region, which may in turn help promote actin polymerization in cadherin adhesions5 (Supplementary Note 3.4).
Vinculin conformational switch is regulated by Abl kinase and PTP1B phosphatase
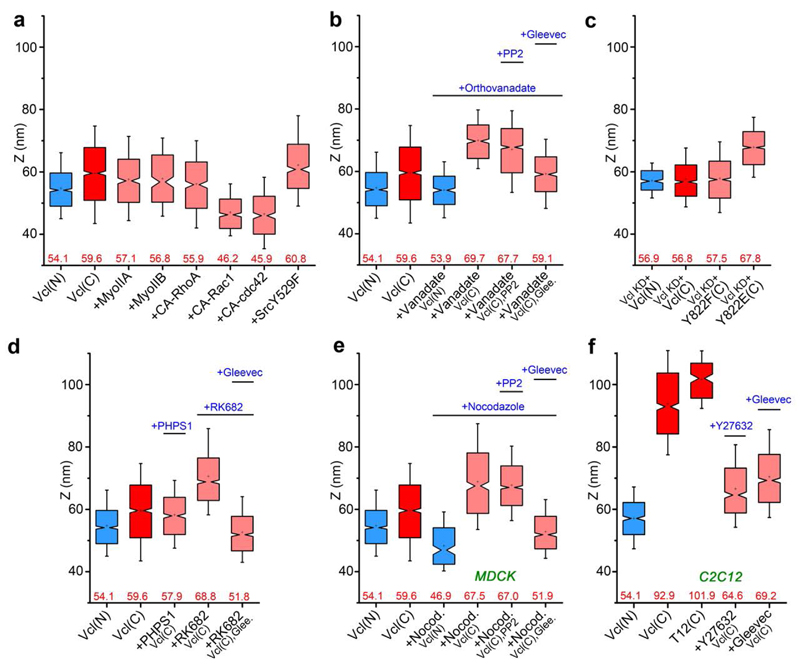
To probe how vinculin conformational transition is regulated, we mapped the C-terminal z-position of vinculin (wt) under various biochemical or pharmacological perturbations. We observed that vinculin remains in the compact state with the overexpression of contractility effectors such as myosin IIA, myosin IIB, activated Src, or constitutively-active RhoA. Interestingly, the overexpression of constitutively-active Rac1 or cdc42 led to even lower C-terminal z-positions of vinculin of ˜ 46 nm, coinciding with the membrane-proximal cadherin-catenin layer (Fig. 5a, Supplementary Table 3). This probably results from the vinculin-tail (VT) phospholipid interaction31, and will be investigated in a more thorough manner separately. Intriguingly, we found that the inhibition of tyrosine phosphatases by orthovanadate results in the upshift of the vinculin C-terminus to zcentre ˜70 nm, indicative of a more open conformation whereas the vinculin N-terminus remains at zcentre ˜54 nm (Fig. 5b). Since the phosphorylation of vinculin Y822 by Abl kinase has recently been shown to be specific to cell-cell contacts38, we mapped vinculin C-terminal z-position upon co-treatment of orthovanadate and Gleevec39, a specific Abl kinase inhibitor, observing the reversion of vinculin to the compact conformation (zcentre = 59.1 nm, Fig. 5b). In contrast, vinculin C-terminus remains at an elevated z-position upon co-treatment of orthovanadate and the Src kinase inhibitor PP2 (zcentre = 67.8 nm, Fig. 5b), thus arguing against the involvement of Src kinase. To assess the contribution of vinculin residue Y822, we expressed vinculin-FP constructs in MDCK cells with stable vinculin shRNA expression34 (vinculin-KD cells, Fig. 5c). For vinculin-wt, we observed a nearly similar z-positions of the N- and C-termini (zcentre = 56.9 nm and 56.8 nm, respectively), indicative of a compact conformation as expected. With the non-phosphorylatable mutation Y822F, the C-terminal z-position remained low (zcentre = 57.5 nm). In contrast, the phosphomimetic mutation Y822E resulted in the open conformation (zcentre = 67.8 nm). Thus, Abl kinase phosphorylation of Y822 appears to promote vinculin conformational activation.
Figure 5. Vinculin conformation is modulated by tyrosine phosphorylation and tension.
Notched box plots for vinculin z-positions. Blue and red: N- and C-terminal z-positions of vinculin wt coral: vinculin C-terminal z-position with perturbations. a, Overexpression of myosin IIA and IIB, constitutively active (CA) RhoGTPases (RhoA, Rac1, cdc42) and Src kinase. b, Treatment with orthovanadate, PP2, and Gleevec. c, Vinculin Y822 phosphorylation mutants (Y822F, and Y822E) in MDCK vinculin-KD cells. d, Treatment with PHPS1, RK682 and Gleevec. e, Vinculin in MDCK cells, treated with nocodazole and PP2 or Gleevec. f, Vinculin in C2C12 cells, treated with Y-27632 or Gleevec. Notched boxes in a-f indicate first and third quartiles, median and confidence intervals; whiskers, 5th and 95th percentiles. Numbers indicated in a-f: median zcentre (red). n values (number of adhesions) for each box plot, the number of cells, and statistical comparison are described in Supplementary Tables 3 and 4.
Our results also imply that in MDCK, vinculin Y822 may be maintained in the unphosphorylated state by an as-yet-unidentified tyrosine phosphatase. Previous studies have documented a few cadherin-associated tyrosine phosphatases such as SHP240 and PTP1B41. We therefore assayed for their involvement in vinculin conformational regulation by mapping vinculin C-terminal z-positions in the presence of their specific inhibitors. We observed that vinculin remains compact upon the treatment with PHPS1, a SHP2 inhibitor (zcentre = 57.9 nm), but opened up upon treatment with RK682, a PTP1B inhibitor, (zcentre = 68.8 nm, Fig. 5d). Furthermore, co-treatment of RK682 and Gleevec led to the compact conformation of vinculin. These results therefore implicate the Abl kinase/PTP1B phosphatase switch in regulating vinculin conformational opening in MDCK E-cadherin-based adhesions.
Nanoscale compartmentalization of N-cadherin-based adhesions
We next examined whether the nanoscale stratification of proteins observed in E-cadherin adhesions are present in other classical cadherin adhesions. We mapped cadhesome components in C2C12 mouse myoblast cells which form N-cadherin-based adhesions when cultured on oriented N-cadherin substrate17. Remarkably, we observed a comparable extent of nanoscale compartmentalization (Fig. 2c-d, 3b, Supplementary Fig. 5c-d). Most prominently, in contrast to the E-cadherin adhesions of MDCK cells, the wild-type vinculin is in a highly extended configuration in C2C12 cells, with the N- and C- termini at zcentre of 57.1 nm and 92.9 nm, respectively. Furthermore, zyxin, VASP, and vinexin were also observed at elevated z-positions of 81.5 nm, 87.0 nm, and 83.5/89.9 nm (vinexin N/C-termini), respectively, supporting the notion that their nanoscale positioning might be vinculin conformation-dependent.
Vinculin conformational switch integrates tension and phosphorylation signals to modulate the mechanical properties of cell-cell junctions
The comparatively high tension in C2C12 vinculin as measured by FRET (Fig. 4f-g, Supplementary Fig. 6b-c,d-h,j), together with the extended conformation (Fig. 2c-d, 3b), suggests that tension may also promote vinculin activation. Indeed, vinculin-T12 expressed in C2C12 adopts a similarly extended conformation with the C-terminal z-position of 102.0 nm, corroborating the activated state observed for wild-type vinculin (Fig. 5f). To test this further, we treated C2C12 cells with ROCK inhibitor (Y-27632) to reduce tension at the cadherin-based adhesions and imaged the vinculin C-terminal z-position, observing a significant downshift to 64.6 nm, suggestive of a more compact configuration. Interestingly, the treatment of C2C12 by Gleevec also results in a similar extent of z-downshift (69.2 nm). In light of these results, we treated MDCK cells with nocodazole, which upregulates contractility via microtubule depolymerization42, observing that vinculin C-terminal z-position upshifted to zcentre = 67.5 nm, indicative of a conformational opening (Fig. 5e). Likewise, upon co-treatment of nocodazole and Gleevec, vinculin reverted to the compact conformation (zcentre = 51.9 nm). Taken together, these results suggest that both tension and Y822 phosphorylation may be required for vinculin conformational activation.
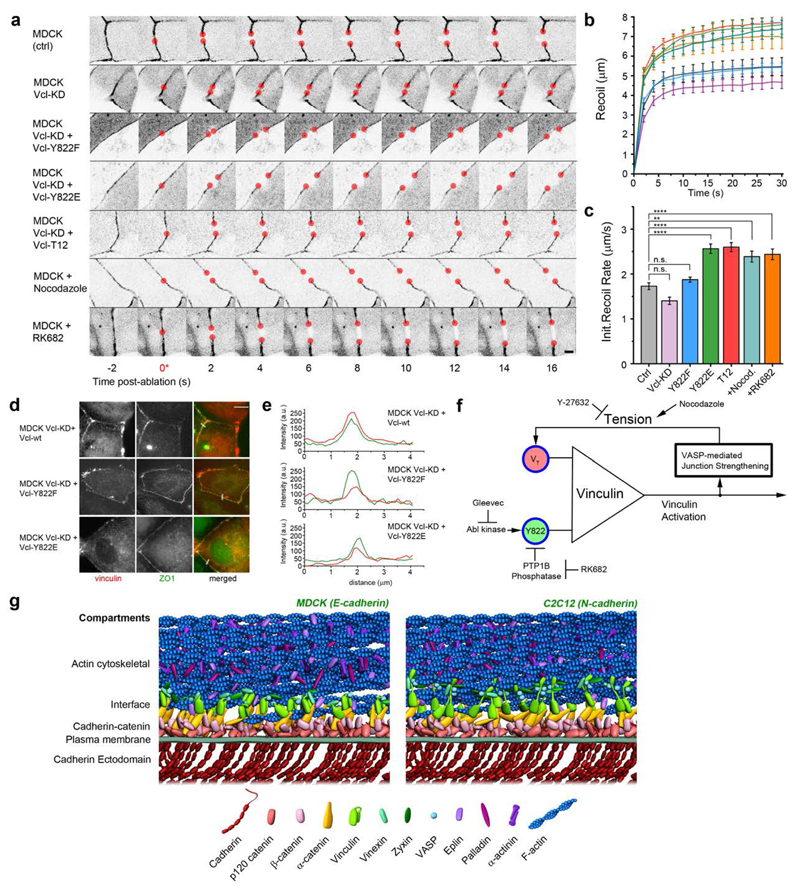
We next sought to ascertain whether the native AJs in epithelial monolayers are also regulated by the interplay of vinculin conformation, tension, and Y822 phosphorylation as delineated above. Using laser nanoscissor43 to probe tension in cell-cell contacts, we measured the dynamics of recoil upon junction scission in MDCK monolayer, observing that the treatment of cells with nocodazole to upregulate tension, or RK682 to inhibit Y822 dephosphorylation, resulted in higher initial recoil rates relative to control. Consistent with this, a similar effect is observed with greater recoil rates upon the expression of vinculin-T12 or Y822E, in comparison to the control, vinculin-KD, or Y822F (Fig. 6a-e, Supplementary Tables 6-7). Therefore a similar mechanism likely governs vinculin conformational regulation in both planar cadherin adhesions and native AJs of epithelial monolayers.
Figure 6. Integration of mechanical and biochemical signals by vinculin regulates mechanical properties of cell-cell contacts.
a, MDCK cells were cultured on fibronectin-coated substrate to form confluent monolayers, with cell-cell contacts labelled by ZO1 fused with either mEmerald or mCherry. Cells were co-transfected with vinculin constructs or treated with pharmacological inhibitors as indicated. Montage of consecutive frames (interval: 2 s) shown with junction excision at t= 0 s (*). Recoiling edges of the junctions (red circles) were used to quantify the recoil trajectory. Untreated MDCK (ctrl) were compared with MDCK stably expressing shRNA against vinculin (MDCK Vcl-KD), MDCK Vcl-KD rescued with Y822F, Y88E, or T12 vinculin mutants, MDCK cells treated with Nocodazole (to promote contractility, 10μM for 1 h), and RK682 (PTP1B inhibitor to inhibit Y822 dephosphorylation, 10 μg/ml for 1 h). Scale bar, 5 μm. Junction recoil (b) and initial recoil rate (c) upon laser ablation of native cell-cell junctions in MDCK epithelial monolayer, with vinculin mutants, or pharmacological treatment. Colors of plots in (b) correspond to bar graphs in (c). Ablation occurred at 0 s. Data in b and c represent mean +/- Error bars (s.e.m). n values: 19 (ctrl); 18 (Vcl-KD); 16 (Vcl-KD + Y822F); 17 (Vcl-KD+Y822E); 18 (Vcl-KD+Vcl-T12); 11 (+Nocodazole); 24 (+RK682). **: p < 0.005, ****: 5 x 10-5. Statistics in Supplementary Tables 6 and 7. d, Fluorescence micrographs of MDCK vinculin KD epithelial monolayer rescued with vinculin wt, vinculin Y822F (non-phosphorylatable), vinculin Y822E (phosphomimetic) fusion constructs to demonstrate their localization at native adherens junctions. Cells were co-transfected with ZO1 constructs. Scale bar: 10 μm. e, Fuorescence intensity line profiles along the lines ( white) in d, showing co-localization of vinculin (red) with ZO-1 (green) at the cell-cell junctions. f, Diagram of vinculin conformational regulation by tension and Y822 phosphorylation, and a putative positive feedback loop. g, Schematic models of protein organization in cadherin-based adhesions, with stratified compartments along the vertical axis at empirically determined z-positions, for MDCK E-cadherin-based adhesions (left, with compact vinculin) and C2C12 N-cadherin-based adhesions (right, with extended vinculin). Note that the model does not depict protein stoichiometry.
Discussion
Our study elucidated how proteins in cadherin-based adhesions are organized to form multi-layered supramolecular complexes that couple cadherin to the actin cytoskeleton. We propose that this stratified architecture (Fig. 6g) may provide the structural framework for regulatory complexity in response to diverse mechanical and signaling cues under different tissue contexts, developmental processes, or disease states1–4,6. For example, how vinculin appears to differentially engage the actin cytoskeleton can be conceptualized in terms of the molecular clutch, while how vinculin conformation is controlled by both mechanical (tension exerted via VT domain) and biochemical cues (Y822 phosphorylation/dephosphorylation by Abl kinase/PTP1B phosphatase) (Fig. 6d) is evocative of a rudimentary AND logic gate. Subsequent to both inputs, vinculin activation upshifts the position of proteins such as VASP, likely promoting further actin polymerization and a further increase in vinculin tension5, thereby resulting in a positive feedback loop, which may help impart robustness, bi-stability, and tunability44.
Interestingly, the nanoscale architecture of the cadherin-based cell adhesions is reminiscent of focal adhesions (FAs) characterized earlier18. Functionally, both these adhesion types serve to integrate the cortical actin cytoskeletal networks with membrane-anchored receptor complexes. As the integrins and cadherins are fundamentally dissimilar classes of receptors, we suggest that such differences necessitate a centrally positioned protein compartment to serve as a structural and mechanical interface, akin to a universal adaptor, so that the common actomyosin machinery can be coupled with different membrane-bound cell-adhesion apparatuses. Vinculin appears to be properly positioned for such a role, while also possessing an appropriate domain organization, with an actin binding tail domain (Vt) flexibly linked to the head domain (VH) which can alternately engage with α- or β-catenin45,46, in cadherin adhesions, or talin, in FAs47.
Nevertheless the molecular mechanisms governing vinculin spatial organization appears to diverge between the cadherin adhesions and FAs, despite the comparable stratified architecture. Unlike in FAs, where a large (270 kDa) and highly elongated protein, talin, plays a dominant structural role comprising the FA core27, directly tethering integrin to actin, in cadherin adhesions such singularly dominant scaffold is not present. Instead, cadherin-actin linkage appears to be mediated by a plurality of multi-protein complexes. For example, in addition to the cadherin:β-catenin:α-catenin complex, α-catenin:Eplin48 and β-catenin:vinculin46 have also been implicated. Furthermore, vinculin stoichiometry likely differs significantly between cadherin adhesions and FAs. In the former, the central placement of vinculin appears to be determined by a single binding site on α-catenin, while in the latter, at least 11 vinculin binding sites49 have been identified in talin. One may conjecture that this difference may reflect the contrasting mechanical context between cell-matrix and cell-cell adhesions. The rigidity of the extracellular matrix spans several orders of magnitude50, and thus variable and cooperative interaction between multiple vinculin binding sites on talin could play a role in fine-tuning FA mechanosensitivity over such a wide dynamic range. On the contrary, cell-cell junction forces are exerted between neighbouring cells, and thus much fewer number of vinculin binding sites may be required to respond to the narrower dynamic range of rigidity.
Our data, together with recent studies on vinculin in FAs and cell-cell contacts34,38,45, suggest that the conformational landscape of vinculin may be more complex than a simple 2-state on-off model. For example, studies in FAs using FRET biosensors for either conformation or tension revealed that while conformationally-open or high-tension vinculin (i.e. low FRET efficiency) were highly enriched at adhesion sites, high FRET efficiency vinculin biosensors also localize there29,35, suggesting that conformationally-compact or low-tension vinculin is capable of adhesion localization. In cell-cell contacts, our data shows that vinculin in a relatively compact, low-tension state is recruited to the MDCK E-cadherin adhesions via activated α-catenin. It is unclear yet how such pool of low-tension and compact vinculin differs structurally from the autoinhibited cytoplasmic pool. We surmise that vinculin could be partially unfurled such that α-catenin binding is enabled. This allows vinculin to be emplaced in the interface compartment, poised to engage the actin cytoskeleton, but remained in the unphosphorylated low-tension state due to the prevailing PTP1B activity. The relief of PTP1B enables Abl kinase phosphorylation of Y822 and an upward extension of the vinculin C-terminus by actomyosin contraction, although the extent of conformational opening in MDCK appears to be lower than fully active, probably due to the low tension generated by this cell type as documented previously28. In contrast, C2C12 vinculin may become fully activated due to both higher contractility and probably lower PTP1B activity (both PTP1B and Abl are expressed in these cell types, Supplementary Fig. 4c). At present, we do not know yet whether such differences reflect cell-type specificity or E-/N-cadherin specificity, and whether Y822 phosphorylation actuates vinculin conformational changes directly, or indirectly by recruiting vinculin binding partners to stabilize the open conformation. It is also unclear whether a similar phosphorylation-dependent conformation modulation is operational in FAs, since the Y822 phosphorylation is reportedly cell-cell contact specific38. For FAs, the presence of a large number of vinculin binding sites, such as on talin and paxillin31,47, would likely require the generation of proper molecular tools for a similar conformational analysis to be tractable.
Finally, we note that while the planar cadherin substrate has greatly facilitated nanoscale optical imaging, the adhesions thus formed probably differ from native cell-cell contacts in important aspects such as cadherin mobility, rigidity, and junctional topology, and thus not all aspects of in vivo cell-cell junction mechanics may be captured. As such, alternative explanations for the extended conformation of vinculin thus observed may include recruitment of as-yet-unexplored proteins, modulation of affinity to binding partners, indirect effects on cellular contractility, or other changes in protein stability that affect functions. Nevertheless, we suggest that the use of physically well-defined substrate can be highly valuable in revealing molecular-scale behaviors, particularly since native cell-cell contacts often undergo concerted and complicated changes in their mechanical, compositional, and morphological properties upon perturbations, which can greatly obfuscate molecular mechanistic dissection. Given the resolution limitation of current superresolution microscopy methods, an alternative approach to further probe native cell-cell contacts could be via correlative EM-superresolution microscopy techniques51.
Supplementary Material
Note: Supplementary Information is available in the online version of the paper
Acknowledgements
We thank Caroline Ajo-Franklin (Lawrence Berkeley National Laboratory), Gleb Shtengel, and Harald Hess (Howard Hughes Medical Institute, Janelia Research Campus) for generous help with superresolution microscopy instrumentation and analysis. We thank Hongying Chen (Protein Expression Facility, Mechanobiology Institute) for generation of mutant constructs, Hui Ting Ong and Aneesh Sathe (Mechanobiology Institute) for assistance with FRET analysis, Cindy Zhang (Mechanobiology Institute) for assistance with 3D graphical model, and Jie Yan (National University of Singapore) for critical reading of the manuscript. P.K., C.B., Y. Wang, A.R., T.S., and R.Z.B. are supported by Singapore National Research Foundation Fellowship awarded to P.K. (NRF-NRFF-2011-04), R.Z.B. (NRF-RF2009-RF001-074), and Competitive Research Programme (NRF2012NRF-CRP001-084) to P.K. and R.Z.B. Y.T. is supported by Mechanobiology Institute and National University of Singapore Startup Grants, and a Singapore Ministry of Education Tier 2 grant (MOE2015-T2-1-116). B.L. is supported by the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007–2013/ERC grant agreement no. 617233), the Mechanobiology Institute, and the Institut Universitaire de France, and USPC-NUS funding. B.L. and R.M.M. are also supported by CNRS, the Human Frontier Science Program (grant RGP0040/2012) and Agence Nationale de la Recherche (ANR 2010 Blan1515).
Footnotes
Author Contributions C.B. and Y.Wang performed the superresolution imaging experiments and conducted data analysis. C.B. and A.R. performed and analysed FRET experiments. C.B., A.R., Y.H., and Y.T designed and C.B. performed and analysed laser ablation experiments. Y.Wu and R.Z.B. performed imaging of Eph4 cell-cell junctions by astigmatism-based 3-D superresolution microscopy. C.B., T.S., M.B., M.W.D., B.L., and R.M.M. designed and generated fusion constructs, provided new reagents and analytical tools. C.B. and P.K. designed the study and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Heisenberg C-P, Bellaïche Y. Forces in tissue morphogenesis and patterning. Cell. 2013;153:948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Guillot C, Lecuit T. Mechanics of epithelial tissue homeostasis and morphogenesis. Science (New York, N.Y. 2013;340:1185–1189. doi: 10.1126/science.1235249. [DOI] [PubMed] [Google Scholar]
- 3.Lecuit T, Yap AS. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nature cell biology. 2015;17:533–539. doi: 10.1038/ncb3136. [DOI] [PubMed] [Google Scholar]
- 4.Takeichi M. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nature Reviews Molecular Cell Biology. 2014;15:397–410. doi: 10.1038/nrm3802. [DOI] [PubMed] [Google Scholar]
- 5.Leerberg JM, et al. Tension-sensitive actin assembly supports contractility at the epithelial zonula adherens. Curr Biol. 2014;24:1689–1699. doi: 10.1016/j.cub.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leckband D, de Rooij J. Cadherin adhesion and mechanotransduction. Annual review of cell and developmental biology. 2014;30:291–315. doi: 10.1146/annurev-cellbio-100913-013212. [DOI] [PubMed] [Google Scholar]
- 7.Niessen CM, Leckband D, Yap AS. Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol Rev. 2011;91:691–731. doi: 10.1152/physrev.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borghi N, et al. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci U S A. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TJ, et al. Dynamic visualization of alpha-catenin reveals rapid, reversible conformation switching between tension states. Curr Biol. 2015;25:218–224. doi: 10.1016/j.cub.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertocchi C, Vaman Rao M, Zaidel-Bar R. Regulation of adherens junction dynamics by phosphorylation switches. J Signal Transduct. 2012;2012:125295. doi: 10.1155/2012/125295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaidel-Bar R. Cadherin adhesome at a glance. Journal of cell science. 2013;126:373–378. doi: 10.1242/jcs.111559. [DOI] [PubMed] [Google Scholar]
- 12.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science (New York, N.Y. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Truong Quang BA, Mani M, Markova O, Lecuit T, Lenne PF. Principles of E-cadherin supramolecular organization in vivo. Curr Biol. 2013;23:2197–2207. doi: 10.1016/j.cub.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y, Kanchanawong P, Zaidel-Bar R. Actin-delimited adhesion-independent clustering of e-cadherin forms the nanoscale building blocks of adherens junctions. Developmental Cell. 2015;32:139–154. doi: 10.1016/j.devcel.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Buckley CD, et al. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science (New York, N.Y. 2014;346:1254211. doi: 10.1126/science.1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonemura S, Itoh M, Nagafuchi A, Tsukita S. Cell-to-cell adherens junction formation and actin filament organization: similarities and differences between non-polarized fibroblasts and polarized epithelial cells. Journal of cell science. 1995;108(Pt 1):127–142. doi: 10.1242/jcs.108.1.127. [DOI] [PubMed] [Google Scholar]
- 17.Gavard J, et al. Lamellipodium extension and cadherin adhesion: two cell responses to cadherin activation relying on distinct signalling pathways. Journal of cell science. 2004;117:257–270. doi: 10.1242/jcs.00857. [DOI] [PubMed] [Google Scholar]
- 18.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shtengel G, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ajo-Franklin CM, Ganesan PV, Boxer SG. Variable incidence angle fluorescence interference contrast microscopy for z-imaging single objects. Biophysical Journal. 2005;89:2759–2769. doi: 10.1529/biophysj.105.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paszek MJ, et al. Scanning angle interference microscopy reveals cell dynamics at the nanoscale. Nature Methods. 2012;9:825–827. doi: 10.1038/nmeth.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertocchi C, Goh WI, Zhang Z, Kanchanawong P. Nanoscale imaging by superresolution fluorescence microscopy and its emerging applications in biomedical research. Crit Rev Biomed Eng. 2013;41:281–308. doi: 10.1615/critrevbiomedeng.2014010448. [DOI] [PubMed] [Google Scholar]
- 23.Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kovacs EM, Goodwin M, Ali RG, Paterson AD, Yap AS. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr Biol. 2002;12:379–382. doi: 10.1016/s0960-9822(02)00661-9. [DOI] [PubMed] [Google Scholar]
- 25.Strale P-O, et al. The formation of ordered nanoclusters controls cadherin anchoring to actin and cell–cell contact fluidity. The Journal of cell biology. 2015;210:333–346. doi: 10.1083/jcb.201410111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shim SH, et al. Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc Natl Acad Sci U S A. 2012;109:13978–13983. doi: 10.1073/pnas.1201882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, et al. Talin determines the nanoscale architecture of focal adhesions. Proceedings of the National Academy of Sciences. 2015 doi: 10.1073/pnas.1512025112. 201512025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nature cell biology. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 29.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao M, et al. Force-dependent conformational switch of alpha-catenin controls vinculin binding. Nat Commun. 2014;5:4525. doi: 10.1038/ncomms5525. [DOI] [PubMed] [Google Scholar]
- 31.Peng X, Nelson ES, Maiers JL, DeMali KA. New insights into vinculin function and regulation. Int Rev Cell Mol Biol. 2011;287:191–231. doi: 10.1016/B978-0-12-386043-9.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagafuchi A, Tsukita S. The loss of the expression of α catenin, the 102 kD cadherin associated protein, in central nervous tissues during development. Dev Growth Differ. 1994;36:59–71. doi: 10.1111/j.1440-169X.1994.00059.x. [DOI] [PubMed] [Google Scholar]
- 33.Benjamin JM, et al. αE-catenin regulates actin dynamics independently of cadherin-mediated cell–cell adhesion. The Journal of cell biology. 2010;189:339–352. doi: 10.1083/jcb.200910041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sumida GM, Tomita TM, Shih W, Yamada S. Myosin II activity dependent and independent vinculin recruitment to the sites of E-cadherin-mediated cell-cell adhesion. BMC Cell Biol. 2011;12:48. doi: 10.1186/1471-2121-12-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Cohen DM, Choudhury DM, Kioka N, Craig SW. Spatial distribution and functional significance of activated vinculin in living cells. The Journal of cell biology. 2005;169:459–470. doi: 10.1083/jcb.200410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen DM, Chen H, Johnson RP, Choudhury B, Craig SW. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J Biol Chem. 2005;280:17109–17117. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- 37.Milam LM. Electron microscopy of rotary shadowed vinculin and vinculin complexes. J Mol Biol. 1985;184:543–545. doi: 10.1016/0022-2836(85)90301-8. [DOI] [PubMed] [Google Scholar]
- 38.Bays JL, et al. Vinculin phosphorylation differentially regulates mechanotransduction at cell–cell and cell–matrix adhesions. The Journal of cell biology. 2014;205:251–263. doi: 10.1083/jcb.201309092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagar B, et al. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer research. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 40.Ukropec JA, Hollinger MK, Salva SM, Woolkalis MJ. SHP2 association with VE-cadherin complexes in human endothelial cells is regulated by thrombin. Journal of Biological Chemistry. 2000;275:5983–5986. doi: 10.1074/jbc.275.8.5983. [DOI] [PubMed] [Google Scholar]
- 41.Xu G, Arregui C, Lilien J, Balsamo J. PTP1B modulates the association of β-catenin with N-cadherin through binding to an adjacent and partially overlapping target site. Journal of Biological Chemistry. 2002;277:49989–49997. doi: 10.1074/jbc.M206454200. [DOI] [PubMed] [Google Scholar]
- 42.Chang Y-C, Nalbant P, Birkenfeld J, Chang Z-F, Bokoch GM. GEF-H1 couples nocodazole-induced microtubule disassembly to cell contractility via RhoA. Molecular Biology of the Cell. 2008;19:2147–2153. doi: 10.1091/mbc.E07-12-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toyama Y, Peralta XG, Wells AR, Kiehart DP, Edwards GS. Apoptotic force and tissue dynamics during Drosophila embryogenesis. Science (New York N,Y. 2008;321:1683–1686. doi: 10.1126/science.1157052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai TY, et al. Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science (New York N,Y. 2008;321:126–129. doi: 10.1126/science.1156951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng X, Maiers JL, Choudhury D, Craig SW, DeMali KA. alpha-Catenin uses a novel mechanism to activate vinculin. J Biol Chem. 2012;287:7728–7737. doi: 10.1074/jbc.M111.297481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng X, Cuff LE, Lawton CD, DeMali KA. Vinculin regulates cell-surface E-cadherin expression by binding to beta-catenin. Journal of cell science. 2010;123:567–577. doi: 10.1242/jcs.056432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu Rev Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 48.Abe K, Takeichi M. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc Natl Acad Sci U S A. 2008;105:13–19. doi: 10.1073/pnas.0710504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gingras AR, et al. Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. The Journal of biological chemistry. 2005;280:37217–37224. doi: 10.1074/jbc.M508060200. [DOI] [PubMed] [Google Scholar]
- 50.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 51.Shtengel G, et al. Imaging cellular ultrastructure by PALM, iPALM, and correlative iPALM-EM. Methods Cell Biol. 2014;123:273–294. doi: 10.1016/B978-0-12-420138-5.00015-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.