Abstract
The genetic changes underlying the initial steps of animal domestication are still poorly understood. We generated a high-quality reference genome for rabbit and compared it to resequencing data from populations of wild and domestic rabbits. We identified over 100 selective sweeps specific to domestic rabbits, but only a relatively small number of fixed (or nearly fixed) SNPs for derived alleles. SNPs with marked allele frequency differences between wild and domestic rabbits were enriched for conserved non-coding sites. Enrichment analyses suggest that genes affecting brain and neuronal development have often been targeted during domestication. We propose that due to a truly complex genetic background, tame behavior in rabbits and other domestic animals evolved by shifts in allele frequencies at many loci, rather than by critical changes at only a few ‘domestication loci’.
Introduction
Domestication of animals - the evolution of wild species into tame forms – has resulted in striking changes in behavior, morphology, physiology and reproduction (1). However, the genetic underpinnings of the initial steps of animal domestication are poorly understood but likely involved changes in behavior that allowed the animals to survive and reproduce under conditions that might be too stressful for wild animals. Given the differences in behavior between wild and domesticated animals, we investigated to what extent this process involved fixation of new mutations with large phenotypic effects as opposed to selection on standing variation. Such studies are hampered in most domestic animals due to ancient domestication events, extinct wild ancestors, or geographically widespread wild ancestors.
Rabbit domestication was initiated in monasteries in Southern France as recent as ~1,400 years ago (2). At this time, wild rabbits were mostly restricted to the Iberian Peninsula, where two subspecies occurred (Oryctolagus cuniculus cuniculus and O. c. algirus), and to France, colonized by O. c. cuniculus (Fig. 1). Additionally, the area of origin of domestic rabbit is still populated with wild rabbits related to the ancestors of the domestic rabbit (3). This recent and well-defined origin provides a major advantage for inferring genetic changes underlying domestication.
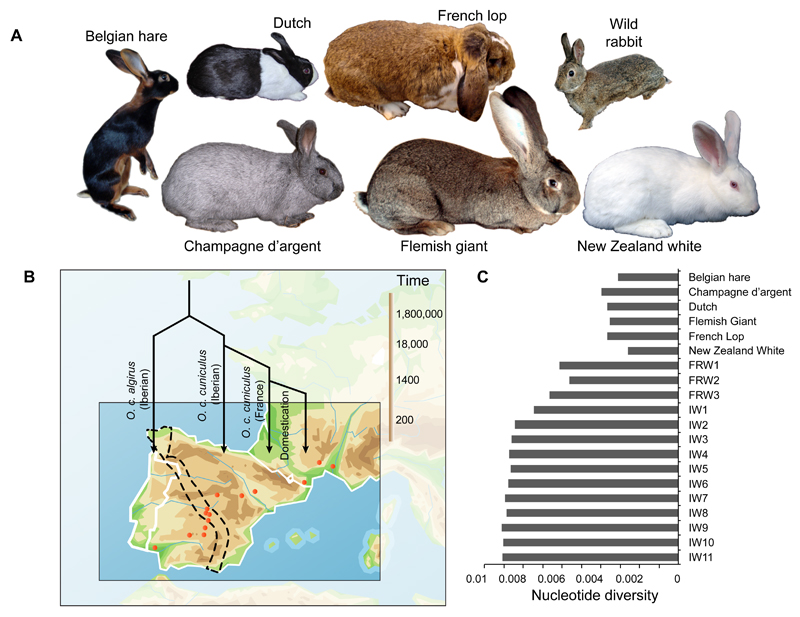
Fig. 1. Experimental design and population data.
(A) Images of the six rabbit breeds, sized to reflect differences in body weight, included in the study and of a wild rabbit. (B) Map of the Iberian Peninsula and Southern France with sample locations marked (orange dots). Demographic history of this species is indicated and a logarithmic time scale is indicated to the left. The hybrid zone between the two subspecies is marked with dashes. (C) Nucleotide diversities in domestic and wild populations. The French (FRW1-3) and Iberian (IW1-11) wild rabbit populations are ordered according to a northeast to southwest transection. Sample locations are provided in Table S4.
A female rabbit genome was Sanger sequenced and assembled (4). The draft OryCun2.0 assembly size is 2.66 Gb with a contig N50 size of 64.7 kb and a scaffold N50 size of 35.9 Mb (Tables S1-S2). The genome assembly was annotated using the Ensembl gene annotation pipeline (Ensembl release 73, Sept. 2013) and with both rabbit RNA-seq data and the annotation of human orthologs (4) (Table S3). Our analysis of rabbit domestication used Ensembl annotations as well as a custom pipeline for annotation of UTRs (168,286 unique features), non-coding RNA (n=9,666) and non-coding conserved elements (2,518,476 unique features).
To identify genomic regions under selection during domestication we performed whole genome resequencing (10X coverage) of pooled samples (Table S4) of six different breeds of domestic rabbits (Fig. 1A), three pools of wild rabbits from Southern France, and 11 pools of wild rabbits from the Iberian Peninsula, representing both subspecies (Fig. 1B). We also sequenced a close relative, snowshoe hare (Lepus americanus), to deduce the ancestral state at polymorphic sites. Short sequence reads were aligned to our assembly; SNP calling resulted in the identification of 50 million high quality SNPs and 5.6 million insertion/deletion polymorphisms after filtering (Table S5). The numbers of SNPs at non-coding conserved sites and in coding sequences were 719,911and 154,489, respectively. The per site nucleotide diversity (π) within populations of wild rabbits was in the range of 0.6-0.9% (Fig. 1C). Thus, rabbit is one of the most polymorphic mammals sequenced so far, presumably due to a larger long-term effective population size relative to other sequenced mammals (5). Identity scores confirm that the domestic rabbit is most closely related to wild rabbits from Southern France (Fig. S1A), and we inferred a strong correlation (r = 0.94) in allele frequencies at most loci between these groups (Fig. S1B). The average nucleotide diversity of each sequenced population is consistent with a bottleneck and reduction in genetic diversity when rabbits from the Iberian Peninsula colonized Southern France and a second bottleneck during domestication (3)(Figs. 1B,C).
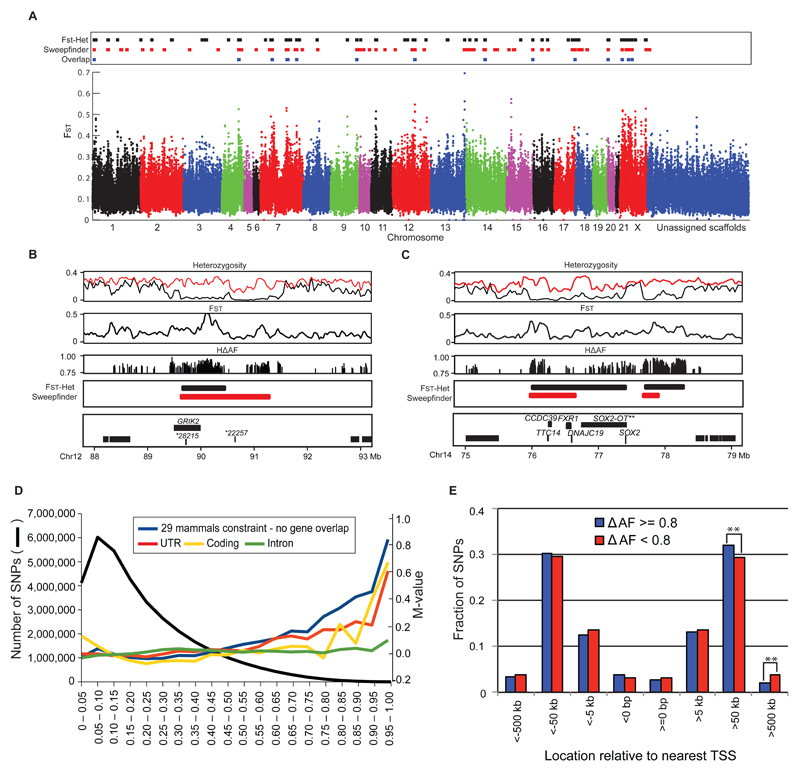
Selective sweeps occur when beneficial genetic variants increase in frequency due to positive selection together with linked neutral sequence variants (6). This results in genomic islands of reduced heterozygosity, and increased differentiation between populations around the selected site. We compared genetic diversity between domestic rabbits as one group to wild rabbits representing 14 different locations in France and the Iberian Peninsula. We calculated FST values between wild and domestic rabbits, and average pooled heterozygosity (H) in domestic rabbits, in 50 kb sliding windows across the genome (hereafter referred to as FST-H outlier approach). We identified 78 outliers with FST>0.35 and H<0.05 (Figs. 2A, S2, Database S1). We also used SweepFinder (7), which calculates maximum composite likelihoods for the presence of a selective sweep, taking into account the background pattern of genetic variation in the data, and with a significance threshold set by coalescent simulations incorporating the recent demographic history of domestic and wild rabbits (Figs. S3, S4, Databases S1, S2) (4). This analysis resulted in the identification of 78 significant sweeps (Fig. 2A, Database S1). Thirty-one (40%) of these were also detected with the FST-H approach (Fig. 2A). This incomplete overlap is likely explained by the fact that SweepFinder primarily assesses the distribution of genetic diversity within the selected population, while the FST-H analysis identifies the most differentiated regions of the genome between wild and domestic rabbits. We carried out an additional screen using targeted sequence capture on an independent sample of individual French wild and domestic rabbits. We targeted over 6 Mb of DNA sequence split into 5,000 1.2 kb intronic fragments that were distributed across the genome and selected independently of the genome resequencing results above. Coalescent simulations, using the targeted resequencing dataset and incorporating the recent demographic history of domestic rabbit as a null model (Figs. S3, S4, Databases S1, S2) (4), confirmed the majority of sweeps detected in our whole genome resequencing approach (76.0% with SweepFinder and 73.7% with FST-H outlier regions, excluding regions without targeted fragments), a highly significant overlap (Fisher’s exact test, P<1x10-5 for both tests). Furthermore, 26 of the 31 sweep regions detected with both SweepFinder and the FST-H approach were targeted in the capture experiment, 23 of these (88.5%) were confirmed.
Fig. 2. Selective sweep and delta allele frequency analyses.
(A) Plot of FST values between wild and domestic rabbits. Sweeps detected with the FST-H outlier approach, SweepFinder and their overlaps are marked on top. Unassigned scaffolds were not included in the analysis. (B) and (C) Selective sweeps at GRIK2 (B) and SOX2 (C). Heterozygosity plots for wild (red) and domestic (black) rabbits together with plots of FST values and SNPs with ΔAF>0.75 (HΔAF). The bottom panel shows putative sweep regions, detected with the FST-H outlier approach and SweepFinder, marked with horizontal bars. Gene annotations in sweep regions are indicated; *represents ENSOCUT000000. **SOX2-OT represents the manually annotated SOX2 overlapping transcript (4). (D) Number of SNPs in non-overlapping ΔAF bins (black lines, left y-axis). M-values (log2-fold changes) of the relative frequencies of SNPs at non-coding evolutionary conserved sites, in untranslated regions (UTR), exons and introns according to ΔAF bins (colored lines, right y-axis); M-values were calculated with the average frequency of the corresponding annotation category as reference. (E) Location of SNPs at conserved non-coding sites with ΔAF≥0.8 SNPs (n=1,635) and with ΔAF<0.8 SNPs (n=502,343) in relation to the transcription start site (TSS) of the most closely linked gene; **, P<0.01).
An example of a selective sweep overlapping the 3’-part of GRIK2 (glutamate receptor, ionotropic, kainate 2) is shown in Fig. 2B. Parts of this region have low heterozygosity in domestic rabbits, and at position chr12:90,153,383 bp domestic rabbits carry a nearly fixed derived allele at a site with 100% sequence conservation among 29 mammals except for the allele present in domestic rabbits (8) suggesting functional significance. GRIK2 encodes a subunit of a glutamate receptor highly expressed in brain and has been associated with recessive mental retardation in humans (9). Both SweepFinder and the FST–H outlier analysis identified two sweeps near SOX2 (SRY-BOX 2) separated by a region of high heterozygosity (Fig. 2C). SOX2 encodes a transcription factor that is required for stem-cell maintenance (10).
Given the comprehensive sampling in our study and the correlation in allele frequencies between domestic and French wild rabbits (Fig. S1B), highly differentiated individual SNPs are likely to have been either directly targeted by selection or occur in the vicinity of loci under selection. For each SNP, we calculated the absolute allele frequency difference between wild and domestic rabbits (ΔAF) and sorted these into 5% bins (ΔAF=0-0.05, etc.). The majority of SNPs showed low ΔAF between wild and domestic rabbits (Fig. 2D). We examined exons, introns, UTRs and evolutionarily conserved sites for enrichment of SNPs with high ΔAF, as would be expected under directional selection on many independent mutations (Fig. 2D, Table S6). We observed no consistent enrichment for high ΔAF SNPs in introns, but significant enrichments in exons, UTRs and conserved non-coding sites (χ2 test, P<0.05). Of note, we detected a significant excess of SNPs at conserved non-coding sites for each bin ΔAF>0.45 (χ2 test, P=1.8x10-3 - 7.3x10-17), whereas in coding sequence, a significant excess was only found at ΔAF>0.80 (χ2 test, P=3.0x10-2 - 1.0x10-3). Compared to the relative proportions in the entire dataset, there was an excess of 3,000 SNPs at conserved non-coding sites with ΔAF>0.45, whereas for exonic SNPs with ΔAF>0.80 the excess was only 83 SNPs (Table S6). Thus, changes at regulatory sites have played a much more prominent role in rabbit domestication, at least numerically, than changes in coding sequences.
We selected the 1,635 SNPs at conserved non-coding sites with ΔAF>0.80, which represent 681 non-overlapping 1 Mb blocks of the rabbit genome. In order not to inflate significances due to inclusion of SNPs in strong linkage disequilibrium we selected only one SNP per 50 kb, leaving 1,071 SNPs. More than 60% of the SNPs were located 50 kb or more from the closest transcriptional start site (TSS; Fig. 2E), suggesting that many differentiated SNPs are located in long-range regulatory elements. A gene ontology (GO) overrepresentation analysis (11) examining all genes located within 1 Mb from high ΔAF SNPs showed that the most enriched categories of biological processes involved ‘cell fate commitment’ (Bonferroni P=3.1x10-3-5.4x10-5; Table 1, Database S3), while the statistically most significant categories involved brain and nervous system cell development (Bonferroni P=2.9x10-3-3.7x10-10). Many of the mouse orthologs of genes associated with non-coding high ΔAF SNPs were expressed in brain or sensory organs, and this enrichment was highly significant (Table 1). We also examined phenotypes observed in mouse mutants (http://www.informatics.jax.org) for these genes, revealing a significant (Bonferroni P=3.7x10-2-7.5x10-17) enrichment of genes associated with defects in brain and neuronal development, development of sensory organs (hearing and vision), ectoderm development and respiratory system phenotypes (Fig. S5). These highly significant overrepresentations were obtained because there were many genes in the overrepresented categories (Table 1). For example, we observed high ΔAF SNPs associated with 191 genes (113 expected by chance) from the nervous system development GO category (Bonferroni P=3.7x10-10). Thus, rabbit domestication must have a highly polygenic basis with many loci responding to selection, and where genes affecting brain and neuronal development have been particularly targeted.
Table 1.
Summary of results from enrichment analysis of ΔAF >0.8 SNPs located in conserved non-coding elements. One significantly enriched term was chosen from each group of significantly enriched inter-correlated terms. Full lists of enriched terms and inter-correlations are presented in Database S3 and the most enriched inter-correlated terms are presented in Figure S5
| Enriched term | Number of genes | P1 | Enrich ment | Unique loci O/R)2 | |
|---|---|---|---|---|---|
| Gene Ontology Biological Process | |||||
| GO:0007399 | nervous system development | 191 | 3.7x10-10 | 1.7 | 154/155 |
| GO:0045595 | regulation of cell differentiation | 107 | 4.5x10-6 | 1.8 | 94/91 |
| GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 122 | 2.0x10-5 | 1.7 | 101/100 |
| GO:0045165 | cell fate commitment | 36 | 5.5x10-5 | 2.9 | 31/32 |
| GO:0007389 | pattern specification process | 57 | 1.4x10-4 | 2.2 | 43/44 |
| GO:0009887 | organ morphogenesis | 85 | 2.0x10-3 | 1.8 | 72/73 |
| GO:0048646 | anatomical structure formation involved in morphogenesis | 75 | 2.8x10-3 | 1.8 | 65/64 |
| GO:0045892 | negative regulation of transcription, DNA-dependent | 82 | 1.4x10-2 | 1.7 | 62/62 |
| GO:0034332 | adherens junction organization | 13 | 1.5x10-2 | 4.7 | 11/11 |
| Mouse Genome Informatics gene expression3 | |||||
| 11853 | TS23 diencephalon; lateral wall; mantle layer | 109 | 3.9x10-25 | 3.3 | 86/85 |
| 12449 | TS23 medulla oblongata; lateral wall; basal plate; mantle layer | 115 | 2.6x10-13 | 2.3 | 90/89 |
| 2257 | TS17 sensory organ | 113 | 3.4 x10-13 | 2.3 | 98/99 |
| 1324 | TS15 future brain | 72 | 8.5x10-9 | 2.4 | 61/61 |
| Mouse Genome Informatics phenotype | |||||
| MP:0010832 | lethality during fetal growth through weaning | 240 | 7.5x10-17 | 1.8 | 197/189 |
| MP:0003632 | abnormal nervous system morphology | 237 | 1.2x10-13 | 1.7 | 191/193 |
| MP:0005388 | respiratory system phenotype | 127 | 1.7x10-7 | 1.8 | 101/102 |
| MP:0000428 | abnormal craniofacial morphology | 109 | 1.4x10-6 | 1.9 | 93/92 |
| MP:0002925 | abnormal cardiovascular development | 88 | 3.3x10-5 | 1.9 | 73/73 |
| MP:0005377 | hearing/vestibular/ear phenotype | 73 | 1.8x10-4 | 2.0 | 61/62 |
Bonferroni corrected P-value.
Number of unique non-overlapping 1Mb windows observed (O) and the average number of 1 Mb windows observed in 1000 random (R) samplings of the same number of genes (rounded to the nearest integer).
TS=Thieler stage.
None of the coding SNPs that differed between wild and domestic rabbits was a nonsense or frame-shift mutation - consistent with data from chicken (12) and pigs (13), suggesting that gene loss has not played a major role during animal domestication. This is an important finding as it has been suggested that gene inactivation could be an important mechanism for rapid evolutionary change, for instance during domestication (14). Of 69,985 autosomal missense mutations, there were no fixed differences and only 14 showed a ΔAF above 90%. On the basis of poor sequence conservation, similar chemical properties of the substituted amino acids, and/or the derived state of the domestic allele we assume that most of these result from hitchhiking, rather than being functionally important (Database S4). However, two missense mutations stand out; these may be direct targets of selection because at these two positions the domestic rabbit differs from all other sequenced mammals (>40 species). The first is a Q813R substitution in TTC21B (tetratricopeptide repeat domain 21B protein), which is part of the ciliome and modulates sonic hedgehog signaling during embryonic development (15). The other is a R1627W substitution in KDM6B (lysine-specific demethylase 6B) that encodes a histone H3K27 demethylase involved in HOX gene regulation during development (16).
Deletions unique to domestic rabbits were difficult to identify, because the genome assembly is based on a domestic rabbit, but some convincing duplications were detected with striking frequency differences between wild and domestic rabbits (Database S5). We observed a one bp insertion/deletion polymorphism located within an intron of IMMP2L (inner mitochondrial membrane peptidase-2 like protein), where domestic and wild rabbits were fixed for different alleles. The polymorphism occurs in a sweep region and it is the sequence polymorphism with highest ΔAF in the region (Fig. S6). Mutations in IMMP2L have been associated with Tourette syndrome and autism in humans (17).
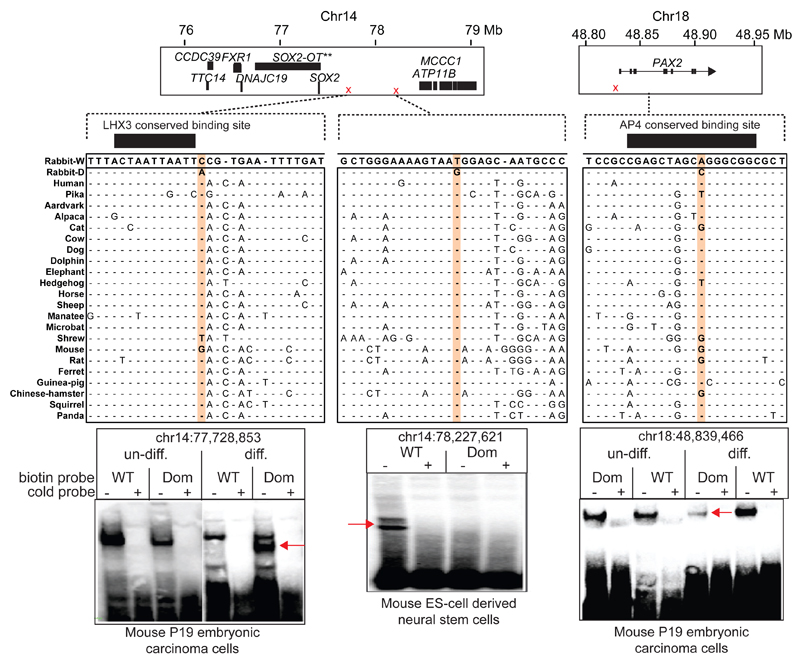
Cell fate determination was a strongly enriched GO category (Database S3) for genes near variants with high ΔAF. We examined the functional significance of twelve SOX2, four KLF4 and one PAX2 high ΔAF SNPs associated with this GO category and where all 17 SNPs were unique to domestic rabbits compared with other sequenced mammals. Electrophoretic mobility shift assay (EMSA) with nuclear extracts from mouse ES-cell derived neural stem cells revealed specific DNA-protein interactions (Figs. 3, S7, Table S7). Four probes, all from the SOX2 region, showed a gel shift difference between wild and domestic alleles. Nuclear extracts from a mouse P19 embryonic carcinoma cell line before and after neuronal differentiation recapitulated these four gel shifts and revealed three additional probes, one in PAX2 and two more in SOX2 that showed gel shift differences between wild-type and mutant probes only after neuronal differentiation. Thus, altered DNA-protein interactions for 7 of the 17 high ΔAF SNPs tested were identified, qualifying them as candidate causal SNPs that may have contributed to rabbit domestication.
Fig. 3. Bioinformatic and functional analysis of candidate causal mutations.
Three examples of SNPs near SOX2 and PAX2 where the domestic allele differs from other mammals. Fourteen such SNPs assessed with electrophoretic mobility shift assays (EMSA) are indicated by red crosses on top. EMSA with nuclear extracts from ES-cell derived neural stem cells or from mouse P19 embryonic carcinoma cells before (un-diff) or after neuronal differentiation (diff) are shown for three SNPs; exact nucleotide positions of polymorphic sites are indicated. Allele-specific gel shifts are indicated by arrows. WT=wild-type allele; Dom=domestic, the most common allele in domestic rabbits. Cold probes at 100-fold excess were used to verify specific DNA-protein interactions.
Our results show that very few loci have gone to complete fixation in domestic rabbits, none at coding sites nor any at non-coding conserved sites. However, allele frequency shifts were detected at many loci spread across the genome and the great majority of ‘domestic’ alleles were also found in wild rabbits, implying that directional selection events associated with rabbit domestication are consistent with polygenic and soft sweep modes of selection (18) that primarily acted on standing genetic variation in regulatory regions of the genome. This stands in contrast with breed-specific traits in many domesticated animals that often show a simple genetic basis with complete fixation of causative alleles (19). Our finding that many genes affecting brain and neuronal development have been targeted during rabbit domestication is fully consistent with the view that the most critical phenotypic changes during the initial steps of animal domestication likely involved behavioral traits that allowed animals to tolerate humans and the environment humans offered. On the basis of these observations, we propose that the reason for the paucity of specific fixed domestication genes in animals is that no single genetic change is either necessary or sufficient for domestication and because of the complex genetic background for tame behavior we propose that domestic animals evolved by means of many mutations of small effect, rather than by critical changes at only a few domestication loci.
Supplementary Material
Acknowledgements
The work was supported by grants from NHGRI (U54 HG003067 to ESL), ERC project BATESON to LA, Wellcome Trust (grant numbers WT095908 and WT098051), the intramural research program of the NIH, NIAID (RGM), the European Molecular Biology Laboratory, POPH-QREN funds from the European Social Fund and Portuguese MCTES [postdoc grants to M.C (SFRH/BPD/72343/2010) and R.C. (SFRH/BPD/64365/2009), and PhD grant to J. Alves (SFRH/BD/72381/2010)], a NSF international postdoctoral fellowship to J.M.G. (OISE- 0754461), by FEDER funds through the COMPETE program and Portuguese national funds through the FCT – Fundação para a Ciência e a Tecnologia – (PTDC/CVT/122943/2010; PTDC/BIA-EVF/115069/2009; PTDC/BIA-BDE/72304/2006; PTDC/BIA-BDE/72277/2006), by the projects “Genomics and Evolutionary Biology” and “Genomics Applied to Genetic Resources” co-financed by North Portugal Regional Operational Programme 2007/2013 (ON.2 – O Novo Norte) under the National Strategic Reference Framework (NSRF) and European Regional Development Fund (ERDF), by travel grants to M.C. (COST Action TD1101) and S.S. was supported by Higher Education Commission in Pakistan. We are grateful to L. Gaffney for assistance with figure preparation, Paulo C. Alves and Scott Mills for providing the snowshoe hare sample and S. Pääbo for hosting M.C., S.A. and R.C. Sequencing was performed by the Broad Institute Genomics Platform. Computer resources were supplied by BITS and UPPNEX at Science for Life Laboratory. The O. cuniculus genome assembly has been deposited in Genbank under the accession number AAGW02000000. The RNA-seq data have been deposited there under the bioproject PRJNA78323, the rabbit genome resequencing data under the bioproject PRJNA242290, and the sequence capture data under the bioproject PRJNA221358.
Footnotes
Author contributions
KLT, FDP, and ESL oversaw genome sequencing, assembly and annotation performed by JA, JTM, JJ, DH, JLC, and SaY. BA, DB, MR, and SS did Ensembl annotations. CRG, VD, LF, RGM, and ZP contributed to the genome project. LA, MC, CJR, NF, and KLT led the domestication study and AMB, NR, and SS contributed with bioinformatic analyses. ShY and GP performed EMSA, AX and KFN developed neural stem cells for EMSA. MC, FWA, JMG, SA, JMA, GB, SB, HAB, RC, HG, GQ, and RV designed, performed and analyzed the sequence capture experiment. LA, MC, CJR, KLT, NF and FDP wrote the paper with input from other authors.
References
- 1.Darwin C. On the origins of species by means of natural selection or the preservation of favoured races in the struggle for life. John Murray; London: 1859. [Google Scholar]
- 2.Clutton-Brock JA. Natural History of Domesticated Mammals. Cambridge University Press; Cambridge: 1999. [Google Scholar]
- 3.Carneiro M, Afonso S, Geraldes A, Garreau H, Bolet G, et al. The genetic structure of domestic rabbits. Mol Biol Evol. 2011;28:1801–1816. doi: 10.1093/molbev/msr003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Materials and methods are available as supplementary material on Science Online
- 5.Carneiro M, Albert FW, Melo-Ferreira J, Galtier N, Gayral P, et al. Evidence for Widespread Positive and Purifying Selection Across the European Rabbit (Oryctolagus cuniculus) Genome. Mol Biol Evol. 2012;29:1837–1849. doi: 10.1093/molbev/mss025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maynard-Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 7.Nielsen R, Williamson S, Kim Y, Hubisz MJ, Clark AG, et al. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Motazacker MM, Rost BR, Hucho T, Garshasbi M, Kahrizi K, et al. A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am J Hum Genet. 2007;81:792–798. doi: 10.1086/521275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 11.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotech. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubin C-J, Zody MC, Eriksson J, Meadows JRS, Sherwood E, et al. Whole genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- 13.Rubin C-J, Megens H, Martinez Barrio A, Maqbool K, Sayyab S, et al. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci USA. 2012;109:19529–19536. doi: 10.1073/pnas.1217149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson MV. When less is more: Gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64:18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran PV, Haycraft CJ, Besschetnova TY, Turbe-Doan A, Stottmann RW, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 17.Deng H, Gao K, Jankovic J. The genetics of Tourette syndrome. Nat Rev Neurol. 2012;8:203–213. doi: 10.1038/nrneurol.2012.26. [DOI] [PubMed] [Google Scholar]
- 18.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20:208–215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson L. Molecular consequences of animal breeding. Curr Opin Genet Dev. 2013;23:295–301. doi: 10.1016/j.gde.2013.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.