Abstract
Benzaldehyde dehydrogenase from Pseudomonas putida (PpBADH) belongs to the Class 3 aldehyde dehydrogenase (ALDH) family. The Class 3 ALDHs are unusual in that they are generally dimeric (rather than tetrameric), relatively non-specific and utilize both NAD+ and NADP+. To date, X-ray structures of three Class 3 ALDHs have been determined, of which only two have cofactor bound, both in the NAD+ form. Here we report the crystal structure of PpBADH in complex with NADP+ and a thioacyl intermediate adduct. The overall architecture of PpBADH resembles that of most other members of the ALDH superfamily, and the cofactor binding residues are well conserved. Conversely, the pattern of cofactor binding for the rat Class 3 ALDH differs from that of PpBADH and other ALDHs. This has been interpreted in terms of a different mechanism for the rat enzyme. Comparison with the PpBADH structure, as well as multiple sequence alignments, suggest that one of two conserved glutamates, at positions 215 (209 in rat) and 337 (333 in rat), would act as the general base necessary to hydrolyze the thioacyl intermediate. While the latter is the general base in the rat Class 3 ALDH, site-specific mutagenesis indicates that Glu215 is the likely candidate for PpBADH, a result more typical of the Class 1 and 2 ALDH families. Finally, this study shows that hydride transfer is not rate limiting, lending further credence to the suggestion that PpBADH is more similar to the Class 1 and 2 ALDHs than it is to other Class 3 ALDHs.
Keywords: hydride transfer, isotope effect, mutagenesis, NAD(P)+, X-ray
Introduction
Benzaldehyde dehydrogenase (PpBADH; EC 1.2.1.7), the terminal enzyme in the mandelamide/mandelate pathway of Pseudomonas putida ATCC 12633, catalyzes the conversion of benzaldehyde to benzoic acid with the concomitant reduction of NAD+ or NADP+ to NADH or NADPH, respectively. Benzoic acid subsequently enters the β-ketoadipate pathway and the citric acid cycle (Stanier et al., 1953). PpBADH has been identified as a Class 3 aldehyde dehydrogenase (ALDH) (McLeish et al., 2003) and a member of the large superfamily of ALDHs which is composed of NAD(P)+-dependent enzymes that catalyze aldehyde oxidation (Lindahl, 1992).
The ALDH superfamily was originally defined by activity as NAD(P)-dependent aldehyde oxidases with broad or narrow specificity. Broad specificity ALDHs were divided based on localization within the cell into Class 1 (cytosolic), Class 2 (mitochondrial) and Class 3 (found in tumors and often utilizing both NAD+ and NADP+). Others were defined by their substrate specificity. As protein sequence data for ALDHs became available (Lindahl, 1992), the superfamily was redefined on the basis of sequence identity rather than activity. Genome studies showed that Class 1 and 2 genes were homologous, sharing much of their intron/exon structure. The genomic organization for Class 3 genes was not yet known, but careful analysis of the Class 1 and 2 genes suggested that Class 3 genes had been extensively modified from a shared ancestor. To date more than 10 ALDH gene families have been identified. More information can be found on the ALDH web site (http://www.aldh.org).
ALDHs are found in organisms from bacteria and archaea to yeast, plants and mammals. Of these, the Class 3 ALDHs are unusual in that they are generally dimeric (rather than tetrameric), and have the unique ability to utilize both NAD+ and NADP+ equally well in vitro (Perozich et al., 1999). The structures of many members of the ALDH superfamily are known, and show striking similarity in tertiary structure even when sequence identity is low (Steinmetz et al., 1997; Moore et al., 1998; D'Ambrosio et al., 2006; Tsybovsky et al., 2007). However, to date the structures for only three Class 3 ALDHs have been determined, the ALDH3 from rat liver (rALDH3) (Liu et al., 1997), the human ALDH3A1 (Khanna et al., 2011; Parajuli et al., 2011, 2014) and the human fatty ALDH (Keller et al., 2014). Of those, there are only two examples with bound cofactor, both NAD+ (Liu et al., 1997; Parajuli et al., 2014), and it is not known how the extra phosphate of NADP+ is accommodated. Thus the ability of the ALDH3 family to use both NAD+ and NADP+ remains to be explained. Furthermore, the rALDH3 structure displays a highly unusual form of NAD+ binding which may not represent a productive complex. In fact, this unusual binding mode relative to Class 1 and 2 enzymes has been interpreted in terms of a differing mechanism for the Class 3 enzymes (Liu et al., 1997).
Recently PpBADH, the product of the chromosomal mdlD gene of P. putida ATCC 12633, was isolated, purified and subjected to spectrophotometric and kinetic analysis (McLeish et al., 2003; Yeung et al., 2008). Testing with a variety of substrates showed that aliphatic aldehydes provided 1–2 times the activity of benzaldehyde, but that reactivity decreased rapidly as the chain length was reduced until no activity was detected with acetaldehyde. This, combined with the fact that cyclohexanal showed nearly as much activity as benzaldehyde, clearly showed that PpBADH is by no means specific for aromatic aldehydes (Yeung et al., 2008). The study also found that PpBADH had very similar Km values for benzaldehyde in the presence of either NAD+ or NADP+ and that the Km value for NAD+ (110 μM) was only ~3-fold lower than that for NADP+ (290 μM). By comparison, despite 86% sequence identity with PpBADH and a similar Km value for benzaldehyde (~7 μM), the benzaldehyde dehydrogenase from Pseudomonas stutzeri has cofactor Km values in the low millimolar range (1 mM and 6.1 mM for NAD+ and NADP+, respectively), i.e. an order of magnitude higher than for PpBADH, although the preference for NAD+ remains (Saehuan et al., 2007). Conversely, the rat Class 3 ALDH has strong (>30-fold) preference for NAD+ (Perozich et al., 2000, 2001). Clearly, there is some variation among the Class 3 ALDHs.
In an attempt to understand some of these apparent inconsistencies, we have determined the structure of PpBADH crystallized in the presence both NADP+ and its product, benzoic acid. Intriguingly the binding mode for NAD+ observed in the rALDH3 structure (Liu et al., 1997) was not found for the NADP+ in the PpBADH structure, suggesting that the alternate mechanism proposed for ALDH3 may not apply to PpBADH. To clarify that issue, we report the use of the structure to guide mutagenesis studies aimed at identifying the catalytic glutamic acid residue and determining whether hydride transfer or deacylation is the rate-limiting step for PpBADH.
Materials and methods
Chemicals
Buffers were purchased from Thermo-Fisher and Sigma-Aldrich and were of the highest quality available. The benzaldehyde and α-[2H]benzaldehyde used in activity assays was purchased from Sigma, redistilled and used within a week of opening to limit potential oxidation on exposure to air. Luria Broth (LB) and LB-agar powder were from Thermo-Fisher while ampicillin, NAD+, NADP+, sodium benzoate and sodium octanoate were purchased from Sigma. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was purchased from Gold Bioscience. Nickel-NTA resin used for the purification of PpBADH was purchased from Qiagen. Bradford reagent and BSA standards for protein quantitation were purchased from BioRad. Sitting drop crystal trays and Crystal Screen II was purchased from Hampton Research.
Expression and purification of PpBADH
The expression plasmid for PpBADH, pET19BADH, was available from a previous study (Yeung et al., 2008). This vector expresses a protein with a N-terminal tag that includes 10 histidine residues and an enterokinase cleavage site. The presence of the tag did not appear to affect the kinetic parameters (Yeung et al., 2008) so the tag was left intact during all experiments described herein. PpBADH was expressed in Escherichia coli BL21(DE3) and purified by affinity chromatography using Nickel-NTA resin as described (Yeung et al., 2008). The purified enzyme was desalted using a BioRad DE 10 column and exchanged into storage buffer (100 mM HEPES, 100 mM KCl, 2 mM DTT, 2 mM NADP+, pH 7.5). Protein concentration was determined by the Bradford method (Bradford, 1976) using BSA as protein standard. The final solution contained PpBADH at ~20 mg/mL and was stored at −80°C. Under these conditions the purified protein could be stored with or without cofactor (NAD+ or NADP+) without any apparent ill effect.
Protein crystallization
Crystallization screening was initiated for PpBADH at 10 mg/mL (in 50 mM HEPES pH 7.5, 50 mM KCl, 1 mM DTT, 1 mM NADP+) mixed 1:1 with the conditions of Hampton Crystal Screen II. Condition 5 (2.0 M ammonium sulfate and 5% by volume isopropanol) gave suitable crystals. This condition was used throughout without modification. Fragile, diamond-shaped crystals appeared after 5–7 days. Crystals were soaked briefly in 3 M sodium malonate as cryoprotectant and flash frozen by immersion in liquid nitrogen. Crystals of the benzoate complex were obtained from a protein solution containing 10 mg/mL PpBADH and 1 mM benzoate. Crystals were cryoprotected as previously described. All efforts to obtain crystals in the presence of NAD+ were unsuccessful.
Data collection
A native X-ray data set of PpBADH with NADP+ and benzoate was collected to a resolution of 2.28 Å at the GM/CA-CAT Sector 23 beamline ID-B at the Advanced Photon Source (APS) at the Argonne National Lab (ANL), Argonne, IL. Data were indexed and scaled in space group I4122 using the HKL2000 package (Otwinowski and Minor, 1997).
Phasing and refinement
Phases for the PpBADH/NADP+/benzoate complex were obtained by molecular replacement (MR) using Phaser (McCoy et al., 2007) in Phenix (Adams et al., 2010). The search model was based on the structure of the rALDH3 dimer (38% identical; PDB accession code 1AD3) in which all residues were truncated to alanine, and residues 83–99 and 425–447, which form a beta sheet at the dimer interface, were removed from the coordinates in order to obtain a MR solution. The resulting electron density was clear and the residues of the PpBADH protein were fitted by hand using Refmac (Murshudov et al., 1997) for refinement and Coot (Emsley et al., 2010) for model building. Simulated annealing and additional refinement were performed with Phenix (Adams et al., 2010). Data collection and refinement statistics are found in Table I.
Table I.
PpBADH data collection and refinement statisticsa
| Data set | NADP+ and benzoate |
|---|---|
| Space group | I4122 |
| Unit cell dimensions | |
| a, b, c (Å) | 195.0, 195.0, 129.1 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution range (Å) | 34.86–2.28 (2.33–2.28) |
| No. reflections | 804 671 |
| No. unique reflections | 56 492 |
| Completeness (%)a | 99.5 (97.2) |
| Redundancya | 14.2 (11.1) |
| Linear Rmergeb | 17.9 |
| CC1/2 (highest-resolution bin) | 0.785 |
| Mean I/σ (I) | 16.9 (1.9) |
| Refinement statistics | |
| Rworkc | 0.190 |
| Rfreed | 0.234 |
| No. of atoms | 7017 |
| Protein | 6664 |
| Cofactor, NAP | 96 |
| Modified cysteine, ZBZ | 28 |
| Water | 229 |
| B-factors (Å2) | |
| Wilson B | 34.6 |
| Protein | 29.3 |
| Cofactor, NAP | 44.1 |
| Modified cysteine, ZBZ | 39.1 |
| Water | 30.5 |
| RMSD (bonds, Å) | 0.004 |
| RMSD (angles, °) | 0.80 |
| Ramachandran favored (%) | 96.88 |
| Ramachandran allowed (%) | 2.89 |
| Ramachandran outliers (%) | 0.23 |
| Clashscore | 3.62 |
| MolProbity score | 1.56 |
aValues in parentheses refer to statistics in the highest-resolution shell.
bLinear Rmerge = Σ|Iobs–Iavg|/ΣIavg.
cRwork = Σ|Fobs–Fcalc|/ΣFobs.
dRfree was calculated as Rwork where 5% of the reflection data was selected at random as a test set and excluded from refinement.
RMSD, root mean square deviation.
Figure production and structure analysis
Reaction schemes were drawn in ChemBioDraw (CambridgeSoft 2009). PyMOL (Schrödinger Inc.) was used to perform structure alignments and to prepare the figures presented in this paper. Sequence alignments were carried out using Clustal Omega (Sievers et al., 2011).
Site-directed mutagenesis
Polymerase chain reaction (PCR) primers for mutagenesis experiments (Table S1) were obtained from Integrated DNA Technologies (IDT). The PpBADH variants were generated by the QuikChange mutagenesis protocol (Stratagene) using pET19BADH (Yeung et al., 2008) as template. Initial screening for mutations was done by restriction analysis. The presence of the mutation and the fidelity of the PCR amplification were confirmed by sequencing (University of Michigan Sequencing Core).
Purification of PpBADH variants
The variants were all expressed and purified to homogeneity using the method described for the wild-type (WT) enzyme. All expressed as soluble protein with no evidence of aggregation. Circular dichroism spectropolarimetry indicated that there were no gross structural changes.
Kinetic analysis of PpBADH variants
Activity assays were carried out at 30°C in a reaction mixture containing TAPS buffer, pH 8.5 (100 mM), KCl (100 mM), DTT (1 mM), NAD+ (1 mM) with varying concentrations of benzaldehyde or α-[2H]benzaldehyde, as appropriate. The final volume was 1 mL. The reactions were followed spectrophotometrically, monitoring the production of NADH at 340 nm. Reaction rates were determined using a molar extinction coefficient of 6220 M−1 for NADH at 340 nm. Initial rate data (triplicates) were fitted to the Michaelis–Menten equation.
Results and discussion
PpBADH was expressed and purified as described previously (Yeung et al., 2008). The protein expresses well and readily purifies to apparent homogeneity. Using the vapor diffusion method in sitting drops, crystals of PpBADH were obtained in the presence of both NADP+ and sodium benzoate. The crystals belong to the I4122 space group and diffracted to a resolution of 2.28 Å.
Overall structure of PpBADH
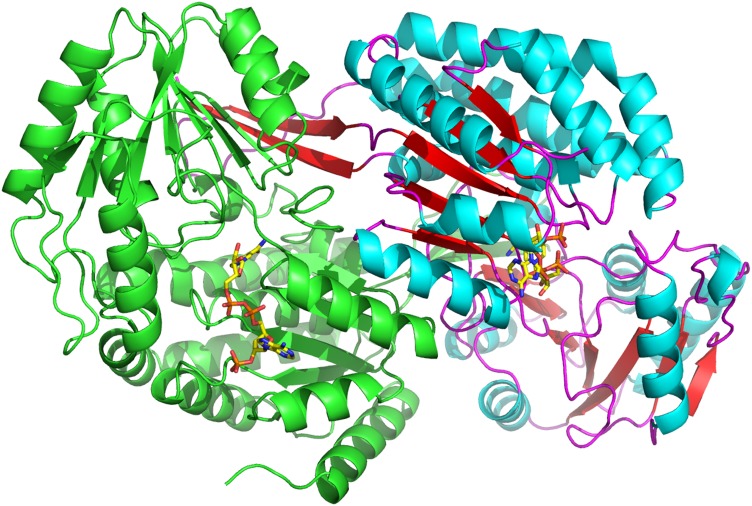
PpBADH occurs as a dimer in solution and crystallizes as a dimer (Fig. 1). The electron density was clear and all residues except the N-terminal His-tag and the first residue of the protein were modeled. Given the 38% sequence identity the overall PpBADH structure is, not surprisingly, very similar to that of rALDH3, with an RMSD of <1.2 Å for the backbone atoms. The two monomers are similar to each other at the level of <0.3 Å RMSD for all atoms.
Fig. 1.
Cartoon representation of the PpBADH dimer in the asymmetric unit. Secondary structural elements are highlighted in the second monomer, alpha-helices in blue, beta strands in red. Both monomers show the bound NADP+ (yellow).
Domains and secondary structure
As with other members of the ALDH superfamily, the PpBADH monomer comprises a nucleotide-binding domain and a catalytic (or substrate binding) domain, plus an oligomerization arm sometimes referred to as the oligomerization or bridging domain. However, the oligomerization domain is not an independent domain, rather it is an integral part of the catalytic domain of the opposite monomer of the dimer and forms part of the opening of the substrate tunnel (Fig. 2).
Fig. 2.
‘Front’ and ‘back’ view of the PpBADH dimer, showing the oligomerization arm (or bridging domain) that helps to hold the dimer together. The bridging domain forms an integral part of the catalytic domain of the other monomer. The cofactor can be seen in yellow.
A summary of the secondary structure elements is provided in Fig. S1. The nucleotide-binding domain is a modified Rossmann fold containing the nucleotide-binding motif. The phosphate binding motif (GXGXXG) is found in the β4-αD loop while Cys249, the catalytic cysteine residue (Yeung et al., 2008), is found in the catalytic domain on a flexible loop between α9 and β7. The section between β12 and α14 has been identified as the conserved motif 10 (Perozich et al., 1999) and consists of a short helix-like segment continuing in the same direction as β12, followed by a long loop going sideways along the interface between the two domains. This structure has been referred to as a ‘U-turn’ (Liu et al., 1997).
Structure of the PpBADH/NADP+/benzoate complex
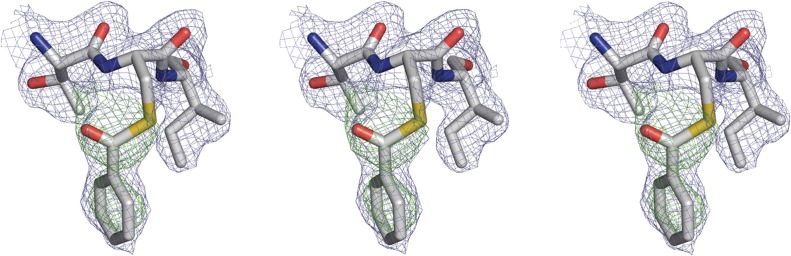
This experiment was designed to trap the unproductive ternary complex between PpBADH, oxidized cofactor (NADP+) and oxidized product (benzoate). Instead, the electron density was found to be best modeled as a covalently bound thioacyl intermediate adduct (Fig. 3). How this was formed is not obvious. It is conceivable that the cofactor was reduced in the X-ray beam or that the NADP+ contained a small amount of NADPH, both of which would permit reaction with benzoate. Less likely is that the commercial benzoate contained a small amount of benzaldehyde that reacted with NADP+, forming the trapped intermediate. Yet another possibility is that the thioacyl intermediate arose from the reversible addition of benzoate to the PpBADH:NADP+ complex. Regardless of how the intermediate was formed, the electron density in this region is clear and supports the modification of Cys249. It is notable that a similar enzyme-bound thioacyl intermediate was observed when the NADP+ form of the E268A variant of glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans was briefly soaked with glyderaldehyde-3-phosphate (D'Ambrosio et al., 2006). In that case the carbonyl oxygen of the thioacyl intermediate was stabilized by interactions with the amide nitrogen of Cys302 and the side chain amide of Asn169, two groups postulated to comprise an oxyanion hole. In the PpBADH/NADP+/benzoate adduct, it appears that the carbonyl is similarly stabilized in an oxyanion hole provided by the backbone amide of Cys249 and the side chain amide of Asn120. The phenyl ring of the benzoate is located in a hydrophobic pocket made up of residues Tyr121, Leu125, Thr248, Ile250, Ala399, Phe400 and Phe406. His418, the outlier in the Ramachandran plot, is adjacent to this hydrophobic pocket. However, the electron density for this residue is well defined in both subunits, indicating correct modeling of this residue.
Fig. 3.
Electron density map for the benzoate adduct at Cys249 contoured at 1.0σ. The electron density (2Fo−Fc) is shown in blue for the ternary complex. A difference electron density (Fo−Fc contoured at 3σ) shown in green was calculated prior to addition of the adduct to the model. The figure shows the adduct in wall-eyed stereo for the left two images and cross-eyed stereo for the right two images.
Orientation of the cofactor
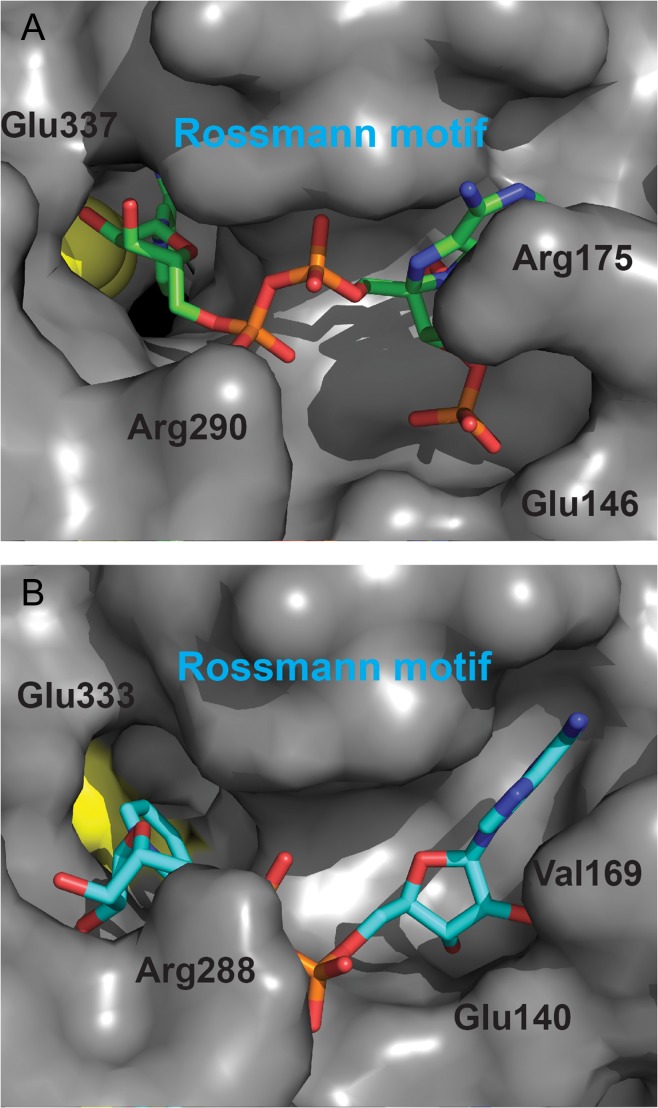
The electron density for the NADP+ cofactor is contiguous. However, based on the electron density, the cofactor appears to be about three quarters occupied in this structure. The nicotinamide ring is held in place by interactions between the NH of nicotinamide with the CO of Leu216 and by hydrophobic interactions with Asn120, Leu125, Thr192, Glu215, Leu216, Cys249, Phe339 and Leu366; the ribose ring is held in place by interactions between the hydroxyl oxygens and Glu337, and with Gly193 and Phe339; the pyrophosphate is held in place by interactions with the backbone of Phe119 and backbone and side chain of Ser194; the 3′-hydroxyl group of the second ribose ring interacts with Lys143 and the backbone carbonyl of Gly117, while the pyrimidine ring is held in place by interactions with Arg175, Asn178 and Val197. The terminal phosphate is held by interactions with Ser145, Glu146 and Thr147.
It is notable that several distinct positions have been observed for the cofactor in structures of Class 2 ALDHs. These have been interpreted in terms of a hydride transfer position associated with the presence of bound NAD+ and a deacylation (hydrolysis) position associated with the presence of bound NADH (Steinmetz et al., 1997; Perez-Miller and Hurley, 2003; Inagaki et al., 2006; Muñoz-Clares et al., 2011). In the PpBADH, structure the nicotinamide ring is oriented in the hydride transfer position (Fig. 4A). Intriguingly, the orientation of the cofactor does not match that of NAD+ in the rALDH3 structure (Fig. 4B). The latter showed NAD+ in an unusual orientation in which, based on the known stereospecificity of the reaction, the nicotinamide ring was not positioned properly for hydride transfer (Liu et al., 1997). This observation has been used to support an alternative mechanism for Class 3 ALDHs in which the reaction occurs on the cofactor side of the active site (Hempel et al., 1999). However, the position of the cofactor in PpBADH does not fit with this hypothesis. Rather it suggests that the mechanism for this enzyme is likely to be similar to that proposed for all other ALDHs.
Fig. 4.
Comparison of the cofactor binding region of (A) PpBADH (PDB ID 5UCD) and (B) rat ALDH3 (PDB ID 1AD3). In both cases, the catalytic cysteine is in yellow and the cofactors are clearly oriented differently relative to the active site. In addition, the conformations of the two cofactors from the two enzymes are different.
Origin of NAD+/NADP+ specificity
PpBADH is able to utilize both NAD+ and NADP+ (Yeung et al., 2008). This is relatively unusual among ALDHs, as most are specific for NAD+ or, less frequently, NADP+ (Perozich et al., 2000, 2001; González-Segura et al., 2015). The former typically has a glutamic acid residue (Glu195 using the standardized hALDH2 numbering, Fig. S1) that coordinates the 2′ and 3′ hydroxyls of the NAD adenine ribose, thereby blocking the 2′-phosphate of NADP+ both sterically and electrostatically. In NADP+ specific enzymes, Glu195 has generally been replaced by threonine or serine, whose side chains are too short to interact with the ribose oxygens, although replacements such as alanine, glycine, glutamine, isoleucine, leucine and valine have been observed (González-Segura et al., 2015). There is a small group of ALDHs that retain Glu195 but are able to accommodate the additional phosphate moiety. They achieve this by changes in positions not specifically related to coenzyme binding, but which allow sufficient movement of Glu195 to accommodate the additional 2′-phosphate of NADP+ (González-Segura et al., 2015). PpBADH has solved this problem in a manner seen before only in betaine aldehyde dehydrogenase from Pseudomonasaeruginosa (PaBALDH), another enzyme that uses both NAD+ and NADP+ (Velasco-Garcia et al., 2000). PaBALDH accomplishes this by pulling the glutamate out of the way with a salt bridge to an arginine residue (González-Segura et al., 2009). In PpBADH Glu146 (Glu195, hALDH2 numbering) is pulled out of the way by a hydrogen bond with Tyr3 (Fig. S2). The fact that a similar mechanism is employed in both cases, combined with the fact that Class 3 ALDHs and BALDHs are evolutionarily related (Perozich et al., 1999) suggests that this mechanism may have evolved from a shared ancestor.
Substrate specificity
The majority of Class 3 ALDHs identified to date are of mammalian origin and act on medium and long chain aliphatic aldehydes, as well as aromatic aldehydes (Muzio et al., 2012). This observation is consistent with specificity data for PpBADH, which show that long chain aliphatics containing 5–8 carbons have ca. twice the relative rate of reactivity than benzaldehyde. As the chain length decreases, the activity reduced quickly, with <0.1% activity being observed with acetaldehyde (Yeung et al., 2008). This same trend of higher activity for longer straight-chain aliphatic substrates is also seen in human Class 1 and 2 ALDH as well as yeast Class 2 ALDH, although the Class 2 enzymes have significantly more activity with acetaldehyde than does PpBADH (Wang et al., 2009). Several substituted benzaldehydes were also tested on PpBADH, and while some had no activity (suggesting steric problems), substrates containing electron-withdrawing groups showed less activity than benzaldehyde, consistent with previously published results on Class 3 ALDHs (Evces and Lindahl, 1989; Yeung et al., 2008). Recently there has been a comprehensive study of the structural determinants for the various classes of ALDHs (Riveros-Rosas et al., 2013). It was found that four residues (170, 177, 296 and 465, hALDH2 numbering) contribute to an ‘aromatic box’ likely to be involved in substrate binding, while another four residues (121, 124, 301 and 303, hALDH2 numbering) may also be involved in aldehyde binding (Riveros-Rosas et al., 2013). The structure of the PpBADH/NADP+/benzoate complex provides an opportunity to test that hypothesis. Residues lying within 4 Å of the aromatic ring of the benzoate adduct include Tyr121, Asn124, Ala399 and Phe406. Two of these, Tyr121 and Phe406, correspond to the ‘aromatic box’ hALDH2 residues Tyr170 and Phe465 (Riveros-Rosas et al., 2013). However, with the exception of Glu68 (Asp121) there seems to be little conservation in the putative aldehyde binding residues.
Relating structure to mechanism
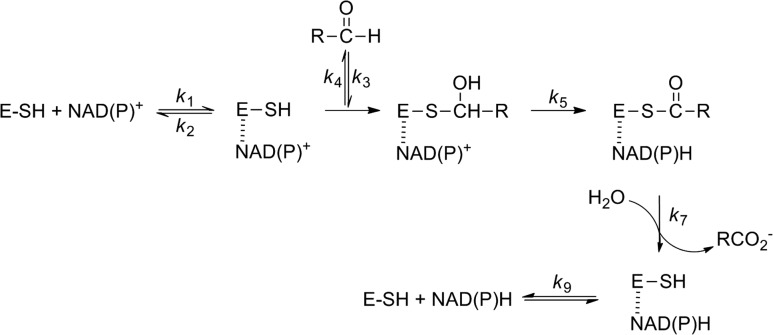
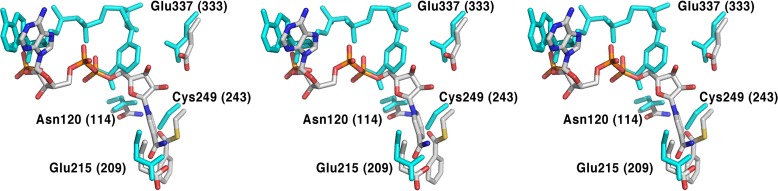
The basic mechanism of ALDHs is straightforward: the active site cysteine acts as a nucleophile reacting with the aldehyde substrate to form a covalently bound thiohemiacetal intermediate (Feldman and Weiner, 1972; Mann and Weiner, 1999). Oxidation is achieved by hydride transfer to the pyridine ring of NAD(P)+ producing NAD(P)H and a thioacyl-enzyme intermediate, which is rapidly hydrolyzed to release the oxidized product, concomitantly regenerating the free cysteine (Scheme 1). The active site of PpBADH contains all the residues considered essential for the ALDH reaction (Fig. 5), Cys249, Asn120, Glu215 and Glu337. The catalytic cysteine, Cys249 (Cys302 hALDH2 numbering), together with Asn120 (Asn169), form part of an oxyanion hole, thereby assisting in stabilizing covalently bound reaction intermediates (Steinmetz et al., 1997; Muñoz-Clares et al., 2010, 2011). One glutamic acid residue, Glu215 (Glu268), is considered to be the base that activates the water necessary for hydrolysis of the thioacyl-enzyme intermediate (Wang and Weiner, 1995; Marchal et al., 2000; D'Ambrosio et al., 2006). Glu337 (Glu399) interacts with the nicotinamide ribose of the cofactor (Ni et al., 1997; Steinmetz et al., 1997) and has been shown to play a role in the hydride transfer reaction (Ni et al., 1997).
Scheme 1.
Basic reaction scheme for a NAD(P)+ ALDH (adapted from Mann and Weiner (1999)). Dashed line indicates a non-covalent interaction.
Fig. 5.
Comparison of active site residues and cofactor in PpBADH (5UCD, gray) and rat ALDH3 (1AD3, cyan). Residue numbers in parentheses refer to the rat structure. Structures were aligned using Coot. Note that the view in Fig. 4 is seen from the outside of the protein looking into the active site. For clarity, the view in this figure is seen from the inside of the protein, i.e. the opposite side as Fig. 4. The figure shows the adduct in wall-eyed stereo for the left two images and cross-eyed stereo for the right two images.
Kinetic studies performed on PpBADH revealed a sequential mechanism and a bell-shaped pH-rate profile with apparent pKas around 6.7 and 9.9 using both NAD+ and NADP+ (Yeung et al., 2008). In the absence of cofactor, the pKa of Cys249 was found to be 8.4 (Yeung et al., 2008), consistent with the apparent pKa of 8.5 found for the active site cysteine of non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) (Marchal and Branlant, 1999). The apparent pKa of 9.9 may represent a lysine, such as Lys219 in PpBADH (Lys272 in hALDH numbering), which is thought to be structurally important in Class 3 enzymes (Hempel et al., 2001). Another possibility would be Lys143 (Lys192), which interacts with the 3′-hydroxyl of the second ribose of NADP+. Of the two, Lys219 would seem to be more likely as its pKa, calculated by PROPKA (Olsson et al., 2011; Sondergaard et al., 2011), is 9.9 in the PpBADH ternary complex. The pKa around 6.7 is likely to be the glutamate general base, either Glu215 or Glu337 (see below).
Based on the unusual conformation of NAD+ in the X-ray structure of the rat enzyme (Liu et al., 1997), it was proposed that Class 3 ALDHs may have a different mechanism than other members of the superfamily (Hempel et al., 1999, 2001). Specifically, it was proposed that Glu333 rather than Glu209 would act as the general base for activation of the water necessary to hydrolyze the thioacyl-enzyme intermediate (Hempel et al., 1999). Initially identified on the basis of chemical modification studies (Abriola et al., 1990), with later support from sequence analysis (Hempel et al., 1993), even prior to the availability of X-ray structures, both Glu268 (hALDH2 numbering) (Wang and Weiner, 1995) and Glu399 (Ni et al., 1997; Sheikh et al., 1997) were mutated to investigate their roles in catalysis. Based on the observation that its mutation had a significantly greater effect on kcat values (Table II), Glu268 was assigned the role of the general base for the Class 2 ALDHs. Mutation of Glu399 to glutamine had only a minor effect on reaction rate but did convert the rate-limiting step from deacylation to hydride transfer (Ni et al., 1997). The structure of the bovine ALDH2 lent credence to this analysis when Glu399 was found to interact with the ribose moiety of NAD+. Conversely, the rALDH3 structure did not show this interaction, and the analogous mutation (E333Q) resulted in a 1000-fold decrease in kcat/Km (Table II), a result that seemed to support the alternative mechanism for Class 3 ALDHs (Hempel et al., 2001). Unfortunately, the E209Q variant of rALDH3 was not prepared, so a true comparison of the two glutamates was not achieved. Subsequently, the E209Q and E333Q variants were prepared for hALDH3 (Mann and Weiner, 1999). In this instance the E209Q variant showed only a 3-fold decrease in kcat/Km (Table III) but, unfortunately, the E333Q variant was unstable and therefore the experiment was not definitive. The issue was further complicated by MM simulations which showed that Glu333 is likely to be the general base for rALDH3, although it was admitted that small changes in the active site could easily result in Glu209 being the general base (Wymore et al., 2004). Nonetheless, if all experiments are taken at face value, it would seem that Glu268 is the catalytic base for Class 1 and Class 2 ALDHs, while Glu399 is the base for Class 3 enzymes.
Table II.
Comparison of kinetic parameters of PpBADH variants and selected other ALDHs
| Variant | Km (µM) | kcat (s−1) | kcat/Km (M−1s−1) |
|---|---|---|---|
| PpBADHa,b | |||
| WT | 4.4 ± 2.2 | 156 ± 29 | 3.5 × 107 |
| E215D | 28 ± 3.9 (6.4) | 1.5 ± 0.2 (104) | 5.4 × 104 (640) |
| E215Q | n.a.c | n.a. | n.a. |
| E215L | n.a. | n.a. | n.a. |
| E337D | 6.6 ± 0.7 (1.5) | 45 ± 1 (3.5) | 6.8 × 106 (5.1) |
| E337Q | 20 ± 1.6 (4.5) | 0.8 ± 0.1 (195) | 4.0 × 104 (875) |
| E337L | 20 ± 1.5 (4.5) | 1.3 ± 0.1 (120) | 6.5 × 104 (540) |
| hALDH2d | |||
| WT | 0.5 | 3.2 | 6.4 × 106 |
| E268Q | 0.6 (1.2) | 6.7 × 10−4 (4800) | 6.2 × 102 (10 000) |
| E399Q | 0.3 (0.6) | 0.33 (10) | 1.1 × 106 (5.8) |
| rALDH3e | |||
| WT | 0.2 ± 0.02 | 79 | 3.9 × 108 |
| E333Q | 4.2 (21) | 1.6 (3.0) | 3.8 × 105 (1000) |
| hALDH3f | |||
| WT | 220 | 0.9 | 4.1 × 103 |
| E209Q | 580 (2.6) | 0.8 (0.9) | 1.4 × 103 (2.9) |
| E333Q | n.a. | n.a. | n.a. |
aReactions were carried out in TAPS buffer (0.1 M, pH 8.5) containing 0.1 M KCl, 1 mM NAD+ and 1 mM DTT at 30°C. Data were treated as described in Materials and methods.
bIn parentheses is the fold increase in Km value or decrease in kcat or kcat/Km values.
cn.a. = no enzyme activity detected within the limits of the assay.
dData for reaction of hALDH2 with propionaldehyde from Wang and Weiner (1995) and Sheikh et al. (1997).
eData from Hempel et al. (2001).
fData from Mann and Weiner (1999).
Table III.
Reaction of PpBADH with substituted benzaldehydesa
| Substrate | Km (µM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| Benzaldehyde | 4.4 ± 2.2 | 156 ± 29 | 3.5 × 107 |
| Benzaldehyde-α-d | 3.9 ± 0.5 | 192 ± 11 | 4.9 × 107 |
| p-methoxybenzaldehyde | 2.0 ± 0.3 | 116 ± 4 | 5.8 × 107 |
| p-chlorobenzaldehyde | 1.2 ± 0.2 | 135 ± 2 | 11 × 107 |
| p-nitrobenzaldehyde | 9.3 ± 0.8 | 95 ± 3 | 1.0 × 107 |
aReactions were carried out in TAPS buffer (0.1 M, pH 8.5) containing 0.1 M KCl, 1 mM NAD+ and 1 mM DTT at 30°C. Data were treated as described in Materials and methods.
The position of the cofactor in the PpBADH/NADP+/benzoate complex is consistent with those observed for the majority of ALDHs, suggesting that a ‘normal’ mechanism would be in operation. Surprisingly, but shown clearly in Fig. 5, the positions of the two glutamic acid residues in rALDH and PpBADH, are essentially identical. Making the situation more complicated is the recently determined structure of hALDH3 (PDB 4L2O). That structure too shows the two glutamic acid residues in the same positions as those in Fig. 5, but the NAD+ occupies its ‘normal’ site with Glu333 strongly interacting with the ribose hydroxyls (i.e. as in the PpBADH structure). In an attempt to clarify the roles of the two glutamic acid residues in PpBADH, we carried out site-directed mutagenesis and determined the kinetic parameters for the reaction of several Glu215 and Glu337 variants with benzaldehyde. The results of that study are provided in Table II. While the E215D variant shows a 100-fold decrease in activity, no activity could be detected with the E215Q and E215L. By comparison, all the E337 variants retained some level of activity, all of which implies that Glu215, not Glu337, acts as the general base, i.e. PpBADH acts more like an ALDH2 than an ALDH3.
The rate-limiting step was found to provide another significant difference between Class 2 and Class 3 ALDHs. Based on a combination of primary isotope and substituent effects (Wang and Weiner, 1995; Ni et al., 1997), the rate-limiting step in hALDH2 was shown to be deacylation (k7, Scheme 1). Conversely, a similar study showed that hydride transfer (k5, Scheme 1) is rate limiting in hALDH3 (Mann and Weiner, 1999). To identify the rate-limiting step in the PpBADH reaction, we first determined the isotope effect on this reaction with α-[2H]benzaldehyde. As was observed for hALDH2 (Wang and Weiner, 1995; Ni et al., 1997), the kcat values were within experimental error, i.e. kH/kD = ~1 (Table III). This indicates that hydride transfer must not be rate limiting. By contrast, hALDH3 provided a vH/vD of 2.1 (Mann and Weiner, 1999).
If, on the other hand, deacylation were rate limiting, the reaction would be expected to be favored by electron-withdrawing substituents. This was observed for hALDH2 (Ni et al., 1997) but, in variants where hydride transfer was rate limiting, the reaction was strongly favored by electron donating substituents (Ni et al., 1997; Mann and Weiner, 1999). Unfortunately, while the primary isotope effect was unambiguously in favor of rate-limiting deacylation, the results for the substituent effects on the PpBADH were more confusing (Table III). It seemed that kcat values were marginally reduced in the presence of both electron donating and withdrawing substituents. While formally this is at odds with deacylation being the rate-limiting step, it is not unreasonable to suggest that other substituent effects such as hydrophobicity and steric effects may also be playing a role, thereby masking the expected electronic effects. While not strictly analogous, a study on the substituent effects on the reduction of aromatic aldehydes by xylose reductase from Candida tenuis showed that ring substituents can differentially perturb enzyme interactions at different points of the reaction coordinate, leading to unexpected changes in Hammett ρ values (Mayr and Nidetzky, 2002). Another possibility is that release of NADH, i.e. k9 (Scheme 1) is rate limiting as has been suggested for Class 1 ALDHs (MacGibbon et al., 1977; Blackwell et al., 1987). Both of these possibilities warrant further investigation.
Summary
The structure presented here represents the first structure of a Class 3 ALDH with a ternary complex containing both NADP+ and a covalently bound thioacyl intermediate. The cofactor was bound in the typical hydride transfer orientation observed in many other ALDHs. This orientation is clearly different from that found in the rat enzyme and mitigates against a novel mechanism for Class 3 ALDHs. Mutagenesis studies indicate that Glu215 acts as the general base, and isotope effects suggest that deacylation rather than hydride transfer is rate limiting. Taken together, the data indicate that, although it can utilize both NAD+ and NADP+, PpBADH is structurally and mechanistically more similar to Class 1 and 2 ALDHs than it is to the rat Class 3 ALDH.
Accession numbers
Coordinates and structure factors for the PpBADH/ NADP+/benzoate structures have been deposited in the Protein Data Bank (Berman et al., 2000; pdb.org) with accession code 5UCD.
Supplementary Material
Supplementary data
Supplementary data are available at Protein Engineering, Design & Selection online.
Funding
This work was supported by the National Science Foundation (EF-0425719) to G.A.P., D.R. and M.J.M. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract DE-AC02-06CH11357. GM/CA @ APS has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104).
References
- Abriola D.P., MacKerell A.D. Jr. and Pietruszko R. (1990) Biochem. J., 266, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkoczi G., et al. (2010) Acta. Crystallogr. Sect D-Biol. Crystallogr., 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N. and Bourne P.E. (2000) Nucleic. Acids. Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell L.F., Motion R.L., MacGibbon A.K., Hardman M.J. and Buckley P.D. (1987) Biochem. J., 242, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976) Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio K., Pailot A., Talfournier F., Didierjeau C., Benedetti E., Aubry A., Branlant G. and Corbier C. (2006) Biochemistry., 45, 2978–2986. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G. and Cowtan K. (2010) Acta. Crystallogr. D Biol. Crystallogr., 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evces S. and Lindahl R. (1989) Arch. Biochem. Biophys., 274, 518–524. [DOI] [PubMed] [Google Scholar]
- Feldman R.I. and Weiner H. (1972) J. Biol. Chem., 247, 267–272. [PubMed] [Google Scholar]
- González-Segura L., Riveros-Rosas H., Julián-Sánchez A. and Muñoz-Clares R.A. (2015) Chem. Biol. Interact., 234, 59–74. [DOI] [PubMed] [Google Scholar]
- González-Segura L., Rudiño-Piñera E., Muñoz-Clares R.A. and Horjales E. (2009) J. Mol. Biol., 385, 542–557. [DOI] [PubMed] [Google Scholar]
- Hempel J., Kuo I., Perozich J., Wang B.C., Lindahl R. and Nicholas H. (2001) Eur. J. Biochem., 268, 722–726. [DOI] [PubMed] [Google Scholar]
- Hempel J., Nicholas H. and Lindahl R. (1993) Protein. Sci., 2, 1890–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel J., Perozich J., Chapman T., Rose J., Boesch J.S., Liu Z.J., Lindahl R. and Wang B.C. (1999) Adv. Exper. Med. Biol., 463, 53–59. [DOI] [PubMed] [Google Scholar]
- Inagaki E., Ohshima N., Takahashi H., Kuroishi C., Yokoyama S. and Tahirov T.H. (2006) J. Mol. Biol., 362, 490–501. [DOI] [PubMed] [Google Scholar]
- Keller M.A., Zander U., Fuchs J.E., et al. (2014) Nat. Commun., 5, 4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna M., Chen C.H., Kimble-Hill A., et al. (2011) J. Biol. Chem., 286, 43486–43494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl R. (1992) Crit. Rev. Biochem. Mol. Biol., 27, 283–335. [DOI] [PubMed] [Google Scholar]
- Liu Z.J., Sun Y.J., Rose J., et al. (1997) Nat. Struct. Biol., 4, 317–326. [DOI] [PubMed] [Google Scholar]
- MacGibbon A.K., Buckley P.D. and Blackwell L.F. (1977) Biochem. J., 165, 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann C.J. and Weiner H. (1999) Protein. Sci., 8, 1922–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal S. and Branlant G. (1999) Biochemistry., 38, 12950–12958. [DOI] [PubMed] [Google Scholar]
- Marchal S., Rahuel-Clermont S. and Branlant G. (2000) Biochemistry., 39, 3327–3335. [DOI] [PubMed] [Google Scholar]
- Mayr P. and Nidetzky B. (2002) Biochem. J., 366, 889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C. and Read R.J. (2007) J. Appl. Crystallogr., 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeish M.J., Kneen M.M., Gopalakrishna K.N., Koo C.W., Babbitt P.C., Gerlt J.A. and Kenyon G.L. (2003) J. Bacteriol., 185, 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S.A., Baker H.M., Blythe T.J., Kitson K.E., Kitson T.M. and Baker E.N. (1998) Structure., 6, 1541–1551. [DOI] [PubMed] [Google Scholar]
- Muñoz-Clares R.A., Diaz-Sánchez A.G., González-Segura L. and Montiel C. (2010) Arch. Biochem. Biophys., 493, 71–81. [DOI] [PubMed] [Google Scholar]
- Muñoz-Clares R.A., González-Segura L. and Diaz-Sánchez A.G. (2011) Chem. Biol. Interact., 191, 137–146. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A. and Dodson E.J. (1997) Acta. Crystallogr. D Biol. Crystallogr., 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Muzio G., Maggiora M., Paiuzzi E., Oraldi M. and Canuto R.A. (2012) Free. Radic. Biol. Med., 52, 735–746. [DOI] [PubMed] [Google Scholar]
- Ni L., Sheikh S. and Weiner H. (1997) J. Biol. Chem., 272, 18823–18826. [DOI] [PubMed] [Google Scholar]
- Olsson M.H., Sondergaard C.R., Rostkowski M. and Jensen J.H. (2011) J. Chem. Theory. Comput., 7, 525–537. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor W. (1997) Macromol. Crystallogr. Part. A, 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parajuli B., Fishel M.L. and Hurley T.D. (2014) J. Med. Chem., 57, 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parajuli B., Kimble-Hill A.C., Khanna M., Ivanova Y., Meroueh S. and Hurley T.D. (2011) Chem. Biol. Interact., 191, 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Miller S.J. and Hurley T.D. (2003) Biochemistry., 42, 7100–7109. [DOI] [PubMed] [Google Scholar]
- Perozich J., Kuo I., Lindahl R. and Hempel J. (2001) Chem. Biol. Interact., 130-132, 115–124. [DOI] [PubMed] [Google Scholar]
- Perozich J., Kuo I., Wang B.C., Boesch J.S., Lindahl R. and Hempel J. (2000) Eur. J. Biochem., 267, 6197–6203. [DOI] [PubMed] [Google Scholar]
- Perozich J., Nicholas H., Wang B.C., Lindahl R. and Hempel J. (1999) Protein. Sci., 8, 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveros-Rosas H., González-Segura L., Julián-Sánchez A., Diaz-Sánchez A.G. and Muñoz-Clares R.A. (2013) Chem. Biol. Interact., 202, 51–61. [DOI] [PubMed] [Google Scholar]
- Saehuan C., Rojanarata T., Wiyakrutta S., McLeish M.J. and Meevootisom V. (2007) Biochim. Biophys. Acta., 1770, 1585–1592. [DOI] [PubMed] [Google Scholar]
- Sheikh S., Ni L., Hurley T.D. and Weiner H. (1997) J. Biol. Chem., 272, 18817–18822. [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., et al. (2011) Mol. Syst. Biol., 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondergaard C.R., Olsson M.H., Rostkowski M. and Jensen J.H. (2011) J. Chem. Theory. Comput., 7, 2284–2295. [DOI] [PubMed] [Google Scholar]
- Stanier R.Y., Gunsalus C.F. and Gunsalus I.C. (1953) J. Bacteriol., 66, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz C.G., Xie P., Weiner H. and Hurley T.D. (1997) Structure., 5, 701–711. [DOI] [PubMed] [Google Scholar]
- Tsybovsky Y., Donato H., Krupenko N.I., Davies C. and Krupenko S.A. (2007) Biochemistry., 46, 2917–2929. [DOI] [PubMed] [Google Scholar]
- Velasco-Garcia R., González-Segura L. and Muñoz-Clares R.A. (2000) Biochem. J., 352, 675–683. [PMC free article] [PubMed] [Google Scholar]
- Wang M.F., Han C.L. and Yin S.J. (2009) Chem. Biol. Interact., 178, 36–39. [DOI] [PubMed] [Google Scholar]
- Wang X.P. and Weiner H. (1995) Biochemistry., 34, 237–243. [DOI] [PubMed] [Google Scholar]
- Wymore T., Hempel J., Cho S.S., Mackerell A.D. Jr, Nicholas H.B. Jr. and Deerfield D.W. (2004) Proteins., 57, 758–771. [DOI] [PubMed] [Google Scholar]
- Yeung C.K., Kenyon G.L. and McLeish M.J. (2008) Biochim. Biophys. Acta., 1784, 1248–1255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.