Abstract
Acireductone dioxygenase (ARD) from the methionine salvage pathway of Klebsiella oxytoca is the only known naturally occurring metalloenzyme that catalyzes different reactions in vivo based solely on the identity of the divalent transition metal ion (Fe2+ or Ni2+) bound in the active site. The iron-containing isozyme catalyzes the cleavage of substrate 1,2-dihydroxy-3-keto-5-(thiomethyl)pent-1-ene (acireductone) by O2 to formate and the ketoacid precursor of methionine, whereas the nickel-containing isozyme uses the same substrates to catalyze an off-pathway shunt to form methylthiopropionate, carbon monoxide and formate. This dual chemistry was recently demonstrated in vitro by ARD from Mus musculus (MmARD), providing the first example of a mammalian ARD exhibiting metal-dependent catalysis. We now show that human ARD (HsARD) is also capable of metal-dependent dual chemistry. Recombinant HsARD was expressed and purified to obtain a homogeneous enzyme with a single transition metal ion bound. As with MmARD, the Fe2+-bound HsARD shows the highest activity and catalyzes on-pathway chemistry, whereas Ni2+, Co2+ or Mn2+ forms catalyze off-pathway chemistry. The thermal stability of the HsARD isozymes is a function of the metal ion identity, with Ni2+-bound HsARD being the most stable followed by Co2+ and Fe2+, and Mn2+-bound HsARD being the least stable. As with the bacterial ARD, solution NMR data suggest that HsARD isozymes can have significant structural differences depending upon the metal ion bound.
Keywords: acireductone, dual chemistry, Klebsiella oxytoca, mammalian, metal
Introduction
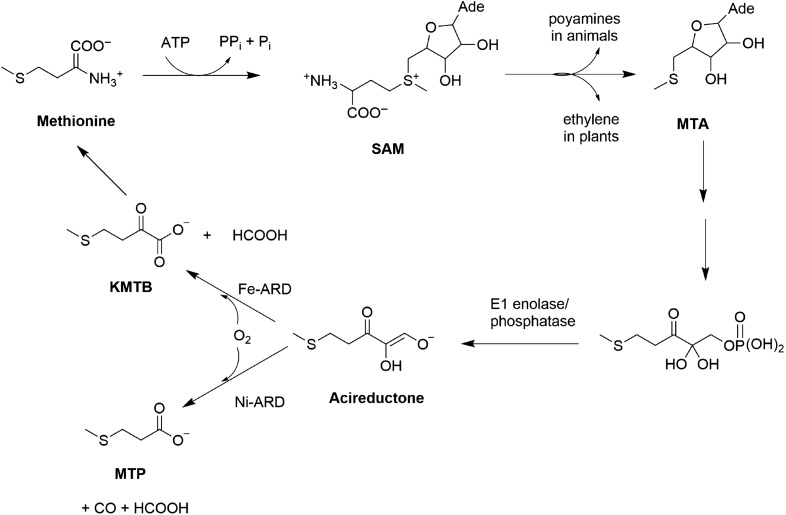
The methionine salvage pathway (MSP) is a ubiquitous pathway found in bacteria, plants and animals. The MSP is essential, recycling the methylthio group of methionine and regulating the concentration of several intermediates in the pathway. The first step in the MSP involves enzymatic synthesis of S-adenosylmethionine (SAM) from methionine and ATP (Fig. 1) (Pochapsky et al., 2007). SAM is involved in the synthesis of spermine and spermidine in animals and ethylene in plants. Polyamines are required for cell growth and proliferation (William-Ashman and Pegg, 1981) while ethylene is required for fruit and vegetable ripening (Taiz, 1991). Defects in polyamine regulation are associated with defects in cell cycle, cell growth and DNA replication (Oredsson, 2003). Methylthioadenosine (MTA) is an intermediate formed from SAM during polyamine and ethylene synthesis, and is an inhibitor of polyamine synthesis (William-Ashman and Pegg, 1981; Williams-Ashman et al., 1982). Inhibition of polyamine synthesis arrests DNA replication whereas elevated polyamine levels are associated with suppression of apoptosis and tumor formation (Pegg, 1988; Marton and Pegg, 1995). Hence the intracellular levels of MTA are tightly regulated, and the MSP is critical in maintaining MTA homeostasis (Shapiro and Barrett, 1981).
Fig. 1.
The MSP in Klebsiella oxytoca.
Acireductone dioxygenase (ARD) catalyzes the penultimate step in the pathway, the oxidative decomposition of substrate acireductone (1, 2-dihydoxy-3-keto-5-(thiomethyl) pent-1-ene) to formate and 2-keto-4-(thiomethyl) butyrate (KMTB). It represents a branch point in the MSP of the bacterium Klebsiella oxytoca: when Fe2+ is bound, ARD catalyzes on-pathway chemistry, but when Ni2+ is bound, it catalyzes an off-pathway shunt leading to production of carbon monoxide, formate and 3-methylthiopropionate (Wray and Abeles, 1995; Dai et al., 1999). Both these forms of ARD have been isolated from native K. oxytoca (Myers et al., 1993), and are also obtained upon overexpression in Escherichia coli. The activities of the two enzymes can be interconverted by replacement and reconstitution with the appropriate metal (Chai et al., 2008). These represent the only known pair of naturally occurring metalloenzymes with distinct functions determined solely by the metal ion identity in the active site. The purpose of the off-pathway chemistry catalyzed by Ni2+-ARD in K. oxytoca is unknown. The solution structures of the two isoforms of ARD from K. oxytoca have been characterized by NMR. In the Ni2+-bound form (Ni-KoARD), the active site was modeled by X-ray absorption spectroscopy due to paramagnetism of the bound metal ion (Pochapsky et al., 2002). The structure of the Fe2+-bound form (Fe-KoARD) is a model based on the structure of a stable metal-free mutant of H98S ARD (Ju et al., 2006).
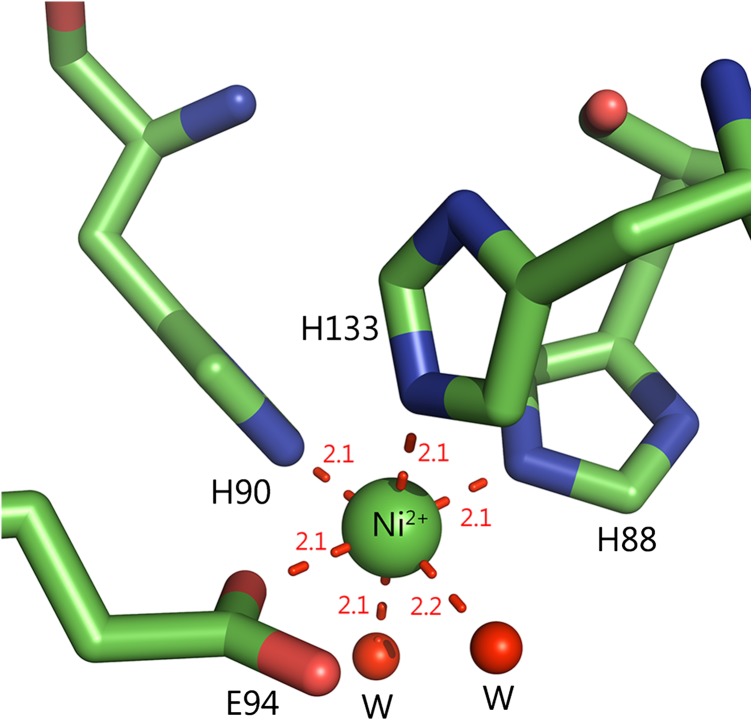
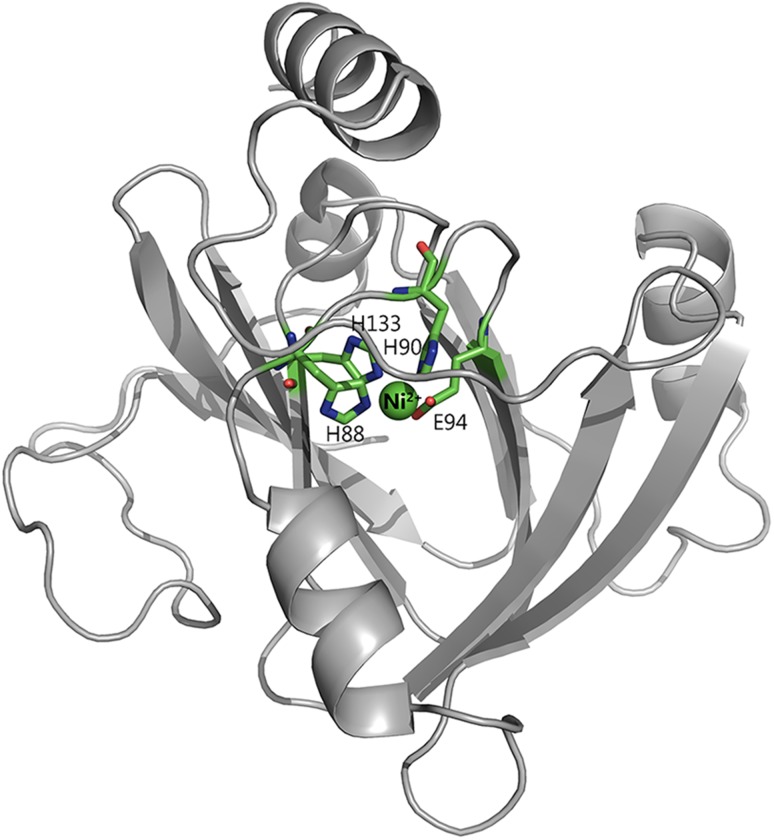
Recently, a mammalian ARD from Mus musculus (MmARD) was shown to exhibit dual chemistry similar to ARD from K. oxytoca (Deshpande et al., 2016), the activity with Fe2+-bound MmARD showing ~10 times higher enzymatic activity than Ni2+, Co2+ or Mn2+-bound MmARD. Crystallographic data indicate that Ni2+ and Co2+-bound forms of MmARD are structurally similar. The metal in both cases is present in a distorted octahedral coordination geometry in the active site bound to protein ligands H88, H90, H133 and E94 (Fig. 2). In addition to the protein ligands, there are two distinct water ligands bound to the metal ion center. Structures of Ni2+-bound MmARD bound to product and substrate analogs provided insight into substrate binding in the active site, metal coordination and catalytic mechanism (Xu et al., 2006; Deshpande et al., 2016).
Fig. 2.
Active site metal coordination geometry. The active site of MmARD is shown with the Ni atom indicated as a green sphere and its protein ligands H88, H90, H133, E94 represented as sticks and two water molecules denoted as ‘W’ shown as red spheres forming a distorted octahedral coordination geometry. The metal coordination distances are shown as red dotted lines with distances measured in Å.
Several studies have shown ARD to serve other functions in mammals in addition to its enzymatic function. An N-terminal truncated version of human ARD (HsARD), referred to as SipL, is implicated in the replication of hepatitis C virus in non-permissive cell lines (Yeh et al., 2001). SipL lacks the first 63 amino acids of full-length HsARD. HsARD is also known to bind the cytoplasmic tail of membrane-type 1 matrix metalloproteinase (MT1-MMP/MMP14) and act as a negative regulator by inhibiting MT1-MMP-mediated cellular invasion (Uekita et al., 2004). The gene ADI1 encoding ARD is downregulated in rat prostate and human prostate cancer cell lines and enforced expression of ADI1 induces apoptosis (Oram et al., 2007). Cultured gastric carcinoma cells and fibrosarcoma cells also have downregulated ADI1 (Oram et al., 2004). A recent study examining the impact of genetic polymorphism on the relationship between exposure to environmental carcinogens and gene expression has shown that the gene ADI1 has a direct link with cancer development (Espin-Perez et al., 2015). These studies suggest that ARD plays an inhibitory role in tumor progression.
In order to understand the molecular mechanisms involved in determining the role of HsARD in cancer, we deemed it important to biochemically and structurally characterize this important enzyme. HsARD is a 21 498 Da protein which shares an 84% sequence identity to MmARD and 28% sequence identity with K. oxytoca ARD (KoARD) (Fig. 3). Herein we describe expression, purification and biochemical characterization of recombinant full-length HsARD bound to different metal ions. We show that, as with MmARD, Fe2+-bound HsARD exhibits on-pathway enzymatic activity, while Ni2+, Co2+ and Mn2+-bound HsARDs catalyze off-pathway chemistry. Solution NMR data provide the first structural evidence of significant tertiary structure differences between Mn2+, Fe2+, Co2+ and Ni2+-bound HsARD isozymes.
Fig. 3.
Sequence alignment of ARD homologs. The metal-binding residues are marked by arrows and the diamond represents the first methionine residue of N-terminal truncated HsARD denoted as SiPL which is implicated in replication of hepatitis C virus in non-permissive cell lines. Sequence alignment was performed using ClustalOmega.
Materials and Methods
Cloning
The HsARD gene was obtained from the Pochapsky lab at Brandeis University. The gene was amplified using the primers 5′-CCATGGCAAGTTGGAGCCACCCGCAGTTCGAGAAG-3′ and 5′- GGCACAGACCGCATAGGAATTC-3′ and cloned into pET28a (Merck Millipore, Darmstadt, Germany) via the restriction sites NcoI and XhoI. Using the appropriate primers, an eight amino acid Strep II affinity tag (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) was placed on the N-terminus of the protein. This tag binds with high affinity to Strep-Tactin, an engineered streptavidin (IBA Life Sciences). All sequences were confirmed by standard methods.
Single-metal expression and purification of HsARD
The plasmid encoding Strep-HsARD was transformed into E. coli RIPL (Agilent Technologies) strain cells and plated onto minimal media without metal supplements. The different metal ion-bound ARD proteins were expressed in M9 minimal media (6 g/L Na2HPO4, 3 g/L KH2PO4, 1 g/L NH4Cl, 0.5 g/L NaCl, 2 mM MgSO4, 0.1 mM CaCl2) with 0.4% glucose as the carbon source. The appropriate metal was added to the medium at induction and purifications conducted as described in Deshpande et al. (2016). Fe2+-bound HsARD was purified in anaerobic conditions as described below. For the 15N-labeled HsARD isozymes, the M9 minimal media was prepared with 15NH4Cl (Cambridge Isotope Laboratories, Andover, MA) as the sole nitrogen source.
Anaerobic purification of Fe2+-bound HsARD
The buffers used for purification of Fe-HsARD were degassed and argon saturated. The harvested cells were suspended in wash buffer 100 mM HEPES, pH 7.0, 100 mM NaCl with protease inhibitors (Roche protease inhibitor cocktail EDTA-free) and sonicated for 5 min. All the subsequent purification steps were performed in a Coy anaerobic chamber (Coy Manufacturing, Ann Arbor, MI) to maintain anaerobic conditions. The cell extract was collected anaerobically in a centrifuge tube to maintain an anaerobic head space during centrifugation. The supernatant collected after centrifugation was passed through Strep-Tactin resin (IBA Life Sciences) which were pre-equilibrated with the wash buffer. The resin was washed with five column volumes (CVs) of wash buffer and eluted with two CV of elution buffer 100 mM HEPES, pH 7.0, 10 mM D-desthiobiotin. The eluent was buffer-exchanged into the storage buffer (50 mM HEPES, pH 7.0, 100 mM NaCl) using Millipore Amicon concentrators which were pre-equilibrated with degassed and Ar-saturated storage buffer to remove the dissolved oxygen in the membrane. The pooled sample was concentrated and stored anaerobically at −80°C.
Enzymatic activity assay
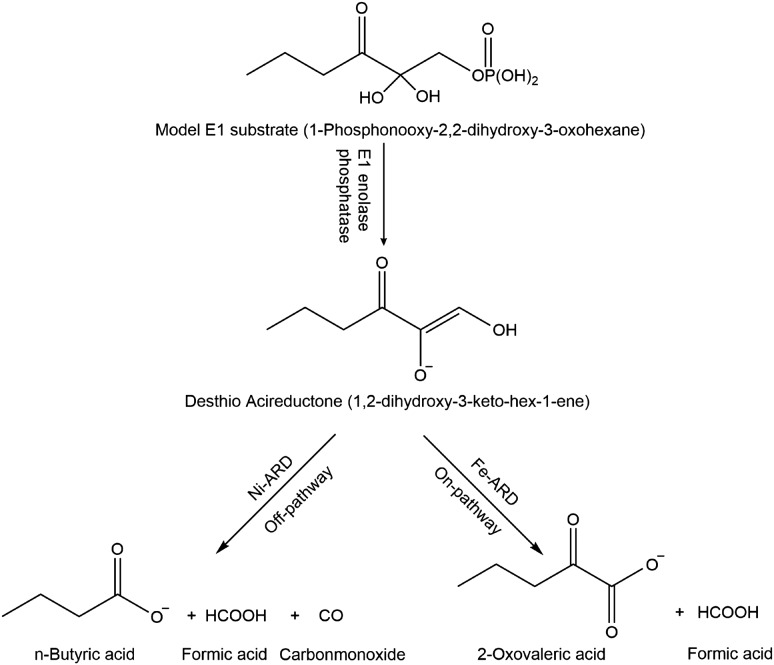
As acireductone is oxidatively unstable and cannot be stored, it must be generated in situ for enzymatic assays by the action of E1 enolase phosphatase on precursor (1-Phosphonooxy-2,2-dihydroxy-3-oxohexane) prior to addition of ARD. E1 and 1-Phosphonooxy-2,2-dihydroxy-3-oxohexane were prepared as described in Zhang et al. (2004) (Fig. 4). The enzymatic assay to measure ARD activity was performed as previously described (Deshpande et al., 2016). All experiments were performed in the assay buffer 50 mM HEPES pH 7.0, 1 mM MgCl2.
Fig. 4.
ARD reactions using desthio E1 substrate to produce desthio-acireductone and the desthio on-pathway and off-pathway ARD chemistry products.
Product quantification assays
The off-pathway reaction product butyric acid derived from desthio-acireductone (1,2-dihydroxy-3-keto-hex-1-ene) substrate was quantified by high pressure liquid chromatography using a Biorad Aminex HPX-87 organic acid column (300 × 7.8 mm). The column was eluted using 5 mM H2SO4 at 0.3 mL/min. Butyric acid elutes at a retention time of ~45 min.
Metal content analysis
The metal content in each protein sample was measured using inductively coupled plasma mass spectrometry (ICP-MS). The measurements were performed at Harvard School of Public Health using a Perkin Elmer Elan DRC-II ICP-MS instrument. The protein sample preparation and ICP-MS measurements were performed as previously described (Deshpande et al., 2016).
Thermal stability
Thermal stability of purified HsARD samples was measured using differential scanning fluorimetry (DSF) performed on a StepOnePlus™ Real-Time PCR System (Applied Biosystems). The samples for analysis were set up in a 96-well plate with each well containing 20 µM protein with 10× SYPRO Orange (Invitrogen) in final volume of 25 µL. The samples were heated at a ramp rate of 0.3°C/min and a temperature range of 25–90°C. The fluorescence data generated was normalized and plotted for each metal ion-bound HsARD.
Oligomeric state
The oligomeric state of HsARD proteins bound to different metal ions was determined using analytical gel filtration. The samples were concentrated to 30–100 µM and loaded onto a Superdex 200 5/150 GL column (GE Healthcare Life Sciences) equilibrated with 50 mM HEPES pH 7.0, 100 mM NaCl. A calibration curve was generated using a set of low-molecular-weight protein standards (GE Biosciences) ribonuclease A (13.7 kDa), carbonic anhydrase (29 kDa), ovalbumin (44 kDa), conalbumin (75 kDa) and aldolase (158 kDa) dissolved in the same buffer.
NMR data collection
15N-labeled Mn2+, Fe2+, Co2+ and Ni2+-bound HsARD samples were each prepared at a final protein concentration of 0.5 mM in 50 mM HEPES pH 7.0, 50 mM KCl, 5% D2O and loaded into a Shigemi tube. The Fe2+-bound HsARD sample was loaded anaerobically in a glove box. NMR spectroscopy experiments were performed on a Bruker Avance 800 NMR spectrometer operating at 800.13 MHz (1H), 81.08 MHz (15N). All experiments were performed at 298 K. The experimental pulse sequences used are standard pulse sequences included in the Bruker Topspin Software (Bruker Biospin Inc.) and were applied without modifications. NMR data were processed using TOPSPIN (Bruker Biospin Inc.).
Results
Expression and purification of HsARD with specific transition metal ion bound
HsARD was overexpressed and purified in minimal media supplemented with appropriate metal salts during induction to express Mn2+, Fe2+, Co2+, Ni2+, Cu2+ and Zn2+-bound HsARDs. The enzyme overexpressed in all cases and was purified to >95% purity using Strep-Tactin beads. Fe-HsARD purification was performed anaerobically, as oxidation to Fe3+ ion leads to metal loss and protein instability. The metal ion content of each isozyme was determined using ICP-MS. The results indicated that each metal ion is occupied to different extent in HsARD. Unlike MmARD, Fe2+ and Ni2+ exhibited lower than 1 mole of metal ion per mole of the protein and Cu2+ was not incorporated at all into HsARD protein. Mn2+ and Zn2+ were also incorporated at lower than 1 mole of metal ion per mole of the protein. Only Co2+ was incorporated close to 1 mole of the desired metal ion per mole of the protein (Table I). Zn2+-bound HsARD was not characterized biochemically due to low metal occupancy.
Table I.
Metal ion content of HsARD proteins determined by inductively coupled plasma mass spectrometry (ICP-MS)
| Protein | Metal ion content (mol of metal ion/mol of protein) | |||||
|---|---|---|---|---|---|---|
| Mn | Fe | Co | Ni | Cu | Zn | |
| Mn-HsARD | 0.64 | – | – | – | – | – |
| Fe-HsARD | – | 0.56 ± 0.06 | – | – | – | – |
| Co-HsARD | – | 0.07 ± 0.03 | 0.96 ± 0.04 | – | – | – |
| Ni-HsARD | – | 0.12 ± 0.12 | – | 0.72 ± 0.05 | – | – |
| Cu-HsARD | – | 0.35 ± 0.16 | – | – | 0.04 | – |
| Zn-HsARD | – | 0.08 ± 0.01 | – | – | – | 0.46 ± 0.02 |
Errors are represented as standard deviations of five replicates. All errors are reported within ± 0.01 mol of metal ion/mol of protein except where noted.
Enzymatic activity
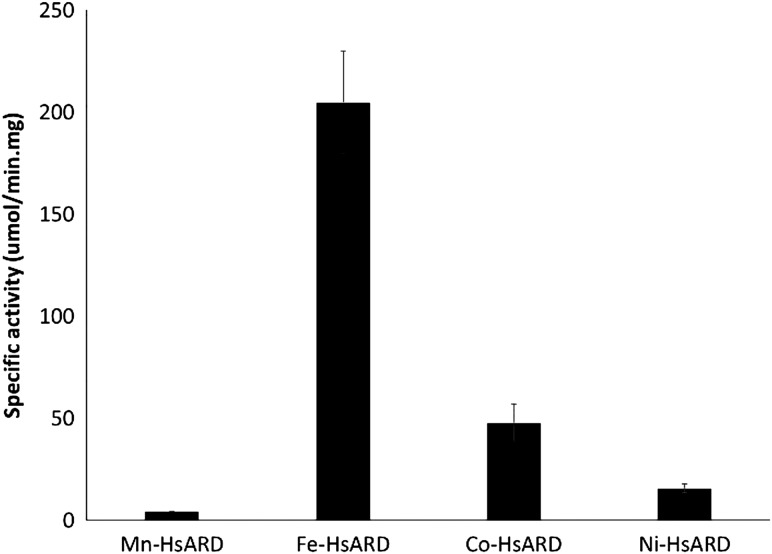
The different metal ion-bound HsARD isozymes were measured for specific activity using the substrate depletion enzymatic assay (Fig. 5). The enzymatic activity of HsARD is metal ion co-factor dependent and the activity trend is similar to MmARD (Fe2+ > Ni2+ = Co2+ > Mn2+). Although the specific activity of Co-HsARD is higher than Ni-HsARD, after correction for metal ion occupancy, the two isozymes exhibit similar activity.
Fig. 5.
Specific activity of HsARD proteins bound to different metal ions. The assay was performed in argon-saturated 50 mM HEPES pH 7.0, 1 mM MgCl2 buffer with 0.6 µM E1 enzyme, 500 µM E1 substrate in a total volume of 1 mL. A controlled amount of ARD was added and the depletion of acireductone was monitored spectrophotometrically at 308 nm. The errors are represented as standard deviations of three replicates.
Enzymatic product analysis
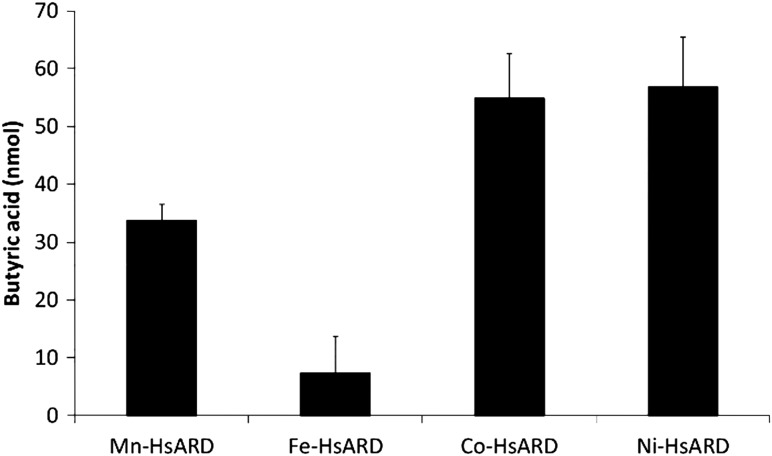
The desthio-product butyric acid from the off-pathway reaction of desthio-acireductone was quantified and the results are summarized in Fig. 6. The bacterial proteins from K. oxytoca Fe-KoARD and Ni-KoARD were used as controls to confirm that the assays are working. The data clearly demonstrate that Fe-HsARD exhibits on-pathway chemistry and Ni-HsARD, Co-HsARD and Mn-HsARD exhibit off-pathway chemistry. HsARD exhibits the same metal ion-dependent dual chemistry as the bacterial enzyme KoARD and mammalian enzyme MmARD.
Fig. 6.
Off-pathway product analysis of different metal ion-bound HsARDs. The enzymatic reaction mixture contained argon-saturated 50 mM HEPES pH 7.0, 1 mM MgCl2 buffer with 0.3 µM E1 enzyme, 250 µM E1 substrate in a total volume of 1 mL. About 100 µL of this reaction mixture was loaded onto the organic acid column. The quantitation is based on a 1 mL reaction mixture. The errors are represented as standard deviations of three replicates.
Relative thermal stability of HsARD bound to specific metal ions
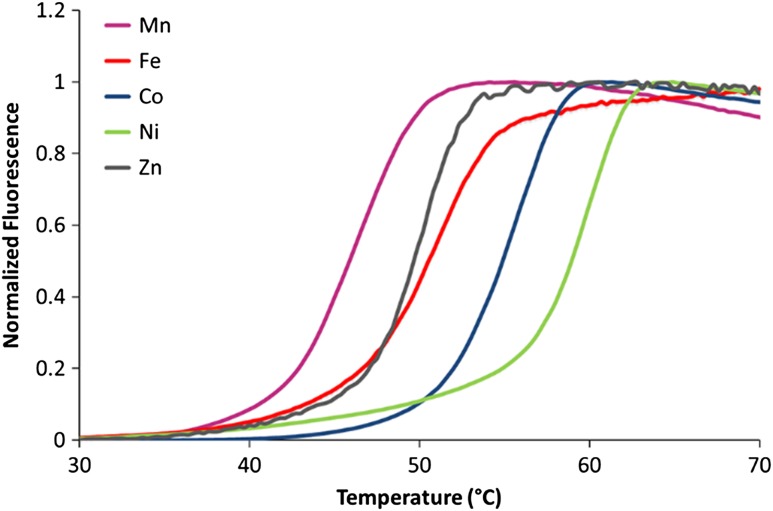
The thermal stability of each metal ion-bound HsARD was established using DSF (Niesen et al., 2007). Similar to MmARD, the data (Fig. 7) indicated that Ni-HsARD is the most thermostable, followed by Co-HsARD, while Mn-HsARD exhibited the least thermal stability. These differences in the melting temperatures likely reflect the binding affinity of a given metal ions for the apo-protein.
Fig. 7.
Thermal stability of HsARD as a function of the bound metal ion. The samples for analysis contained 20 µM protein with 10× SYPRO Orange (Invitrogen) in final volume of 25 µL. The samples were heated at a ramp rate of 0.3°C/min and a temperature range of 25–90°C.
Oligomeric state of HsARD bound to specific metal ions
To investigate whether the oligomeric state of HsARD changes upon binding different metal ions, analytical gel filtration was used to estimate the molecular weights of each of the metal ion-bound HsARD isozymes. All the metal constituted HsARD proteins exhibited a molecular weight of the monomer except Mn-HsARD which eluted with a size of 19 KDa. This difference of 10 KDa in the molecular weight may may reflect the lower stability of the Mn-bound enzyme, with possible unfolding occurring during the experiment.
Solution NMR data of different metal ion-bound HsARD enzymes
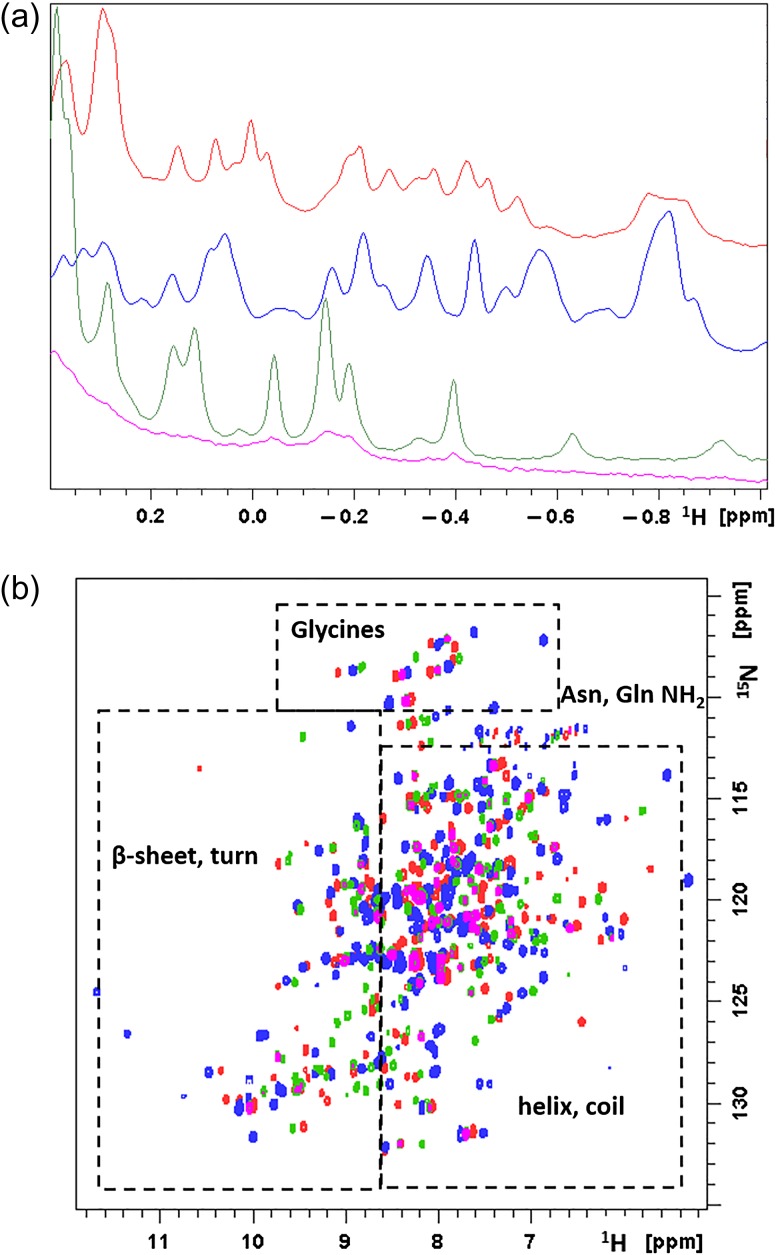
NMR spectroscopy can provide significant insights into the folding and integrity of protein structure. Ideally, a two-dimensional 15N, 1H HSQC spectrum shows a correlation for each backbone amide N–H in the protein (as well as side chain glutamine and asparagine NH2 groups). As the chemical shift dispersion in the spectrum is reflective of the local structural environment, comparison of the HSQC spectra of the different metal ion-bound forms of HsARD provides a rapid evaluation of structural differences (if any) between the isozymes. The dispersion apparent in the Fe-HsARD, Ni-HsARD and Co-HsARD HSQC spectra is typical of well-folded proteins, with peaks downfield of 8 ppm in the 1H dimension (to the left along the horizontal axis) likely representing linearly hydrogen bonded NH groups that occur in β-sheets or tight turns, while those between 6–8 ppm typical of helical or coiled regions. The Mn-HsARD isozyme shows significantly fewer correlations, but whether this is due to the lower stability of the protein (Fig. 6) or the expected high-spin (S = 5/2) of Mn2+ ion in the active site resulting in long-range paramagnetic relaxation will require a more thorough analysis. Differences in magnetic anisotropy of the various metal ions may also contribute to spectral differences near the metal-binding site. Nevertheless, the lack of overlap between many HSQC correlations in the Fe-HsARD and Ni-HsARD isozymes indicates significant differences in the different metal ion-bound HsARD structures (Fig. 8).
Fig. 8.
Solution NMR data of Mn2+, Fe2+, Co2+ and Ni2+-bound HsARD isozymes. (a) Upfield region of 800 MHz 1H NMR spectra of Mn-HsARD (pink), Fe-HsARD (red), Co-HsARD (blue) and Ni-HsARD (green) showing methyl resonances of amino acid side chains ring current-shifted by nearby aromatic residues. The different spectral patterns indicate differences in side chain packing in hydrophobic cores. (b) Overlay of the 2D 1H, 15N HSQC spectra of Mn-HsARD (pink), Co-HsARD (blue), Fe-HsARD (red) and Ni-HsARD (green). Different shift patterns indicate differences local hydrogen bonding and structure. All spectra were obtained at 25°C, pH 7.0 at 800 MHz 1H observe frequency.
Structural differences are further indicated by the upfield shifted regions (<0 ppm) of the one-dimensional 1H spectra of the isozymes. This spectral region typically shows the signals of methyl groups (valine, leucine and isoleucine) that pack against aromatic rings (phenylalanine, tyrosine and tryptophan) in hydrophobic core regions of the fold. In the case of KoARD, these differences were observed due to the metal-dependent repacking of a secondary hydrophobic core on one face of the β-barrel structure (Ju et al., 2006). It is likely that the same regions are responsible for the metal-dependent spectral differences observed for HsARD as well.
Discussion and conclusion
Enzymatic activity of HsARD is metal ion dependent
In this study, we were able to purify one isoform of metal ion-bound HsARD using minimal media expression in the presence of a single metal ion. The metal occupancies were lower than 1 mole metal ion/mole protein except for Co2+. Unlike MmARD, Cu2+ was not incorporated into HsARD. As seen in MmARD, Fe-HsARD has the highest activity compared to Ni-HsARD, Co-HsARD or Mn-HsARD. This is in contrast to ARD in K. oxytoca, where Ni-KoARD has a higher activity than Fe-KoARD (Dai et al., 2001). Biochemical studies on ARD from Oryza sativa L. (OsARD) have shown that Ni-OsARD polymerizes and has a much reduced activity relative to Fe-OsARD (Sauter et al., 2005). Hence we speculate that the off-pathway chemistry may be relevant only in pathology in mammals or eukaryotes, unlike in bacteria (K. oxytoca) where it has been shown that the dual chemistry is seen in the native organism. Under normal conditions, therefore, it is expected that Fe2+ is the native metal ion co-factor in eukaryotes.
Human ARD is capable of performing metal ion-dependent dual chemistry
The data presented here clearly show that, as for MmARD and KoARD, HsARD exhibits metal ion-dependent on- and off-pathway chemistry in vitro. HsARD also exhibits the same metal ion-binding promiscuity seen in KoARD and MmARD, with similar patterns of metal ion-dependent thermostabilites. The dual chemistry of HsARD is interesting since carbon monoxide (CO) which is a product of the off-pathway chemistry is known to be an anti-apoptotic molecule and has been proposed to act as a signaling molecule in mammals (Verma et al., 1993; Thom et al., 2000; Gunther et al., 2002; Liu et al., 2002) and several studies have indicated a role for HsARD in cancer (Uekita et al., 2004; Oram et al., 2007). This leads to the intriguing possibility that the off-pathway chemistry catalyzed by the Ni2+, Co2+ or Mn2+-bound enzymes may be relevant in pathology in mammals. The binding of Ni2+, Co2+ or Mn2+ in HsARD active site could help protect tumor cells from apoptosis by producing carbon monoxide. Ni is known to be toxic and carcinogenic, and no native Ni-binding metalloenzyme has yet been found in mammals. However, since off-pathway chemistry is seen also with Co-HsARD and Mn-HsARD, it will be interesting to test for the presence of off-pathway HsARD chemistry in vivo in methionine dependent cancer cell lines.
Metal-dependent structural perturbations are observed in HsARD
The NMR data presented here provide preliminary evidence of significant structural differences between some of the different metal ion-bound HsARD isoforms. Inspection of both the upfield region of 1D spectra and 2D 1H, 15N HSQC spectra of Mn-HsARD, Fe-HsARD, Co-HsARD and Ni-HsARD isozymes provides evidence for these differences. We are currently performing sequential resonance assignments of Fe-HsARD, Co-HsARD and Ni-HsARD proteins preliminary to obtaining solution structures of the three HsARD isoforms. To date, the only available structure of HsARD is a low-resolution crytstallographic (3.05 Å) structure (PDB ID: 4QGN). The complete details of this structure have not been published but the bound metal ion is proposed to be Fe3+. This is surprising to us, since we have found that the Fe3+ form of the enzyme is unstable and inactive. A homology model of HsARD created using the Ni2+-bound MmARD (PDB code 5I8S) is shown in Fig. 9. The structures of the Ni2+ (PDB ID: 1ZRR) and Fe2+ (PDB ID: 2HJI) forms of ARD from K. oxytoca have been solved by solution NMR spectroscopy (Pochapsky et al., 2002; Ju et al., 2006). While Fe-KoARD was difficult to characterize structurally, as it tended to decompose during long NMR acquisitions,(Ju et al., 2006) Fe-HsARD is relatively stable under anaerobic conditions over the course of the NMR experiments described here. Fe-HsARD is, as such, a promising target for a more complete structural work.
Fig. 9.
Homology model of HsARD. Model is based on the crystal structure of Ni-MmARD (PDB ID: 5I8S). The Ni atom is indicated as a green sphere and the protein ligands H88, H90, H133, E94 are represented as sticks. The remaining two metal coordination sites are shown empty. In the case of MmARD, these are occupied by water molecules as indicated in Fig. 2.
Close inspection of both the 1D and 2D NMR spectra of metal isoforms of HsARD in Fig. 8 shows that, despite the broadness of signals corresponding to Mn2+-bound HsARD (pink in both spectra), they are almost superimposable on the corresponding signals from Ni2+ HsARD (in green). This suggests that, despite differences in electronic relaxation times and resulting coupling with nuclear spin transitions, the structures of the two isozymes are very similar. On the other hand, octahedral complexes of Fe2+ and Ni2+ with N/O ligation both have integer electronic spins (S = 2 and S = 1, respectively) and have similar 1H pseudocontact shift patterns and magnitudes in model complexes (Crans et al., 2003), suggesting that the spectral differences between the Fe2+ and Ni2+ HsARD isoforms reflect genuine structural differences, similar to what was observed in the bacterial ARD enzymes (Ju et al., 2006). Finally, Co2+-HsARD (S = 3/2) will likely exhibit significantly different pseudocontact 1H shifts than the Ni-bound enzyme, based on the results from the model complexes referenced above. So while the similarity between the crystallographic structures of Ni-MmARD and Co-MmARD (RMSD = 0.06 Å) may be to some extent crystallographically artifactual, resolving this issue must await the results of a more complete NMR structural characterization.
Potential ‘moonlighting’ functions of HsARD
HsARD has been shown to serve regulatory functions in addition to its enzymatic function in the MSP. HsARD binds to the cytoplasmic tail of membrane-type 1 matrix metalloproteinase (MT1-MMP) and inhibits MT1-MMP-mediated cellular invasiveness (Uekita et al., 2004). ADI1 encoding ARD is downregulated in rat prostate and human prostate cancer cell lines and its enforced expression induces apoptosis (Oram et al., 2007). An N-terminal truncated version of HsARD called SipL was shown to be implicated in the replication of hepatitis C virus in non-permissive cell lines. This truncated version of HsARD is missing 63 amino acids at the N-terminal end (Yeh et al., 2001). Assuming that the SipL variant folds similar to the full-length HsARD, the intact β-barrel is present in the SipL variant. It is unknown if SipL is a metal-binding protein or if it maintains enzymatic activity. A recent clinical study related these two regulatory functions of HsARD, showing that the physical interaction between MT1-MMP and ADI1 leads to suppression of hepatitis C infection and this inhibitory effect could be reversed by ADI1 overexpression (Yeh et al., 2008).
ADI1 was also shown to be required for normal Drosophila fecundity (Chou et al., 2014). Under conditions of dietary restriction, the egg production of acireductone dioxygenase 1 (DADI1) deficient flies was reduced compared to that of control flies. This fecundity defect in mutant flies was rescued by either supplementing with methionine or reintroduction of the DADI1 gene. A functional homolog of human ADI1 was also able to rescue the fecundity defect. This shows that ARD is multifunctional, acting both as an enzyme and a regulatory protein. This leads to interesting questions such as: Do each of these functions have a different mechanism or are they inter-related? Are these functions performed in different subcellular compartments? Are these non-enzymatic functions dependent on the enzymatic activity of ARD? Future work can be addressed towards delineating some of these functions of ARD and identifying regions of the protein that are involved in the regulatory functions of ARD.
Acknowledgements
The authors thank Nicola Lupoli from Harvard School of Public Health for help with inductively coupled plasma mass spectrometry (ICP-MS) analysis. The methionine salvage pathway and model substrate structures were drawn using ChemDraw Professional 15.0. Figures 2 and 9 were generated using PyMOL (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC).
Funding
This work was supported in part by a grant from the National Institutes of Health (GM26788 to G.A.P. and D.R.).
References
- Chai S.C., Ju T., Dang M., Goldsmith R.B., Maroney M.J. and Pochapsky T.C. (2008) Biochemistry, 47, 2428–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.Y., Lin Y.H., Shiu G.L., Tang H.Y., Cheng M.L., Shiao M.S. and Pai L.M. (2014) J. Biomed. Sci., 21, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crans D.C., Yang L., Gaidamauskas E., Kahn R., Jin W. , Simonis U. (2003) In Paramagnetic Resonance of Metallobiomolecules (Telser, J., Ed.), American Chemical Society, Washington D.C.
- Dai Y., Pochapsky T.C. and Abeles R.H. (2001) Biochemistry, 40, 6379–6387. [DOI] [PubMed] [Google Scholar]
- Dai Y., Wensink P.C. and Abeles R.H. (1999) J. Biol. Chem., 274, 1193–1195. [DOI] [PubMed] [Google Scholar]
- Deshpande A.R., Wagenpfeil K., Pochapsky T.C., Petsko G.A. and Ringe D. (2016) Biochemistry, 55, 1398–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espin-Perez A., de Kok T.M., Jennen D.G., Hendrickx D.M., De Coster S., Schoeters G., Baeyens W., van Larebeke N. and Kleinjans J.C. (2015) Carcinogenesis, 36, 1154–1161. [DOI] [PubMed] [Google Scholar]
- Gunther L., Berberat P.O., Haga M., Brouard S., Smith R.N., Soares M.P., Bach F.H. and Tobiasch E. (2002) Diabetes, 51, 994–999. [DOI] [PubMed] [Google Scholar]
- Ju T., Goldsmith R.B., Chai S.C., Maroney M.J., Pochapsky S.S. and Pochapsky T.C. (2006) J. Mol. Biol., 363, 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.M., Chapman G.B., Wang H. and Durante W. (2002) Circulation, 105, 79–84. [DOI] [PubMed] [Google Scholar]
- Marton L.J. and Pegg A.E. (1995) Annu. Rev. Pharmacol. Toxicol., 35, 55–91. [DOI] [PubMed] [Google Scholar]
- Myers R.W., Wray J.W., Fish S. and Abeles R.H. (1993) J. Biol. Chem., 268, 24785–24791. [PubMed] [Google Scholar]
- Niesen F.H., Berglund H. and Vedadi M. (2007) Nat. Protoc., 2, 2212–2221. [DOI] [PubMed] [Google Scholar]
- Oram S., Jiang F., Cai X., Haleem R., Dincer Z. and Wang Z. (2004) Endocrinology, 145, 1933–1942. [DOI] [PubMed] [Google Scholar]
- Oram S.W., Ai J., Pagani G.M. et al. (2007) Neoplasia, 9, 643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oredsson S.M. (2003) Biochem. Soc. Trans., 31, 366–370. [DOI] [PubMed] [Google Scholar]
- Pegg A.E. (1988) Cancer Res., 48, 759–774. [PubMed] [Google Scholar]
- Pochapsky T.C., Ju T., Dang M., Beaulieu R., Pagani G. and OuYang B. (2007) Sigel A., Sigel H. and Sigel R.K.O. (eds),. Metal Ions in Life Sciences: Nickel and Its Surprising Impact in Nature, 1st edn John Wiley & Sons, Ltd, Chichester, England, pp. 473–500. [Google Scholar]
- Pochapsky T.C., Pochapsky S.S., Ju T., Mo H., Al-Mjeni F. and Maroney M.J. (2002) Nat. Struct. Biol., 9, 966–972. [DOI] [PubMed] [Google Scholar]
- Sauter M., Lorbiecke R., Ouyang B., Pochapsky T.C. and Rzewuski G. (2005) Plant. J., 44, 718–729. [DOI] [PubMed] [Google Scholar]
- Shapiro S.K. and Barrett A. (1981) Biochem. Biophys. Res. Commun., 102, 302–307. [DOI] [PubMed] [Google Scholar]
- Taiz L. (1991) Plant Physiology. Benjamin/Cummins Publishin Co, Redwood City, CA. [Google Scholar]
- Thom S.R., Fisher D., Xu Y.A., Notarfrancesco K. and Ischiropoulos H. (2000) Proc. Natl. Acad. Sci. USA, 97, 1305–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uekita T., Gotoh I., Kinoshita T., Itoh Y., Sato H., Shiomi T., Okada Y. and Seiki M. (2004) J. Biol. Chem., 279, 12734–12743. [DOI] [PubMed] [Google Scholar]
- Verma A., Hirsch D.J., Glatt C.E., Ronnett G.V. and Snyder S.H. (1993) Science, 259, 381–384. [DOI] [PubMed] [Google Scholar]
- William-Ashman H.G. and Pegg A.E. (1981) Polyamines in Biology and Medicine. Marcel Dekker, New York, pp. 43–73. [Google Scholar]
- Williams-Ashman H.G., Seidenfeld J. and Galletti P. (1982) Biochem. Pharmacol., 31, 277–288. [DOI] [PubMed] [Google Scholar]
- Wray J.W. and Abeles R.H. (1995) J. Biol. Chem., 270, 3147–3153. [DOI] [PubMed] [Google Scholar]
- Xu Q., Schwarzenbacher R., Krishna S.S. et al. (2006) Proteins, 64, 808–813. [DOI] [PubMed] [Google Scholar]
- Yeh C.T., Lai H.Y., Chen T.C., Chu C.M. and Liaw Y.F. (2001) J. Virol., 75, 11017–11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C.T., Lai H.Y., Yeh Y.J. and Cheng J.C. (2008) Biochem. Biophys. Res. Commun., 372, 157–161. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Heinsen M.H., Kostic M., Pagani G.M., Riera T.V., Perovic I., Hedstrom L., Snider B.B. and Pochapsky T.C. (2004) Bioorg. Med. Chem., 12, 3847–3855. [DOI] [PubMed] [Google Scholar]