Abstract
Tumor-infiltrating CD8+ T cells are associated with improved survival of patients with Merkel cell carcinoma (MCC), an aggressive skin cancer causally linked to Merkel cell polyomavirus (MCPyV). However, CD8+ T-cell infiltration is robust in only 4%–18% of MCC tumors. We characterized the T-cell receptor (TCR) repertoire restricted to one prominent epitope of MCPyV (KLLEIAPNC, “KLL”) and assessed whether TCR diversity, tumor infiltration, or T-cell avidity correlated with clinical outcome. HLA-A*02:01/KLL tetramer+ CD8+ T cells from MCC patient peripheral blood mononuclear cells (PBMC) and tumor-infiltrating lymphocytes (TIL) were isolated via flow cytometry. TCRβ (TRB) sequencing was performed on tetramer+ cells from PBMC or TIL (n = 14) and matched tumors (n = 12). Functional avidity of T-cell clones was determined by IFNγ production. We identified KLL tetramer+ T cells in 14% of PBMC and 21% of TIL from MCC patients. TRB repertoires were diverse (mean of 12 and 29 clonotypes/patient in PBMC and TIL, respectively) and mostly private. An increased fraction of KLL-specific TIL (> 1.9%) was associated with significantly increased MCC-specific survival P = 0.0009). Forty-two distinct KLL-specific TCRα/β pairs were identified. T-cell clones from patients with improved MCC-specific outcomes were more avid (P < 0.05) and recognized an HLA-appropriate MCC cell line. T cells specific for a single MCPyV epitope display marked TCR diversity within and between patients. Intratumoral infiltration by MCPyV-specific T cells was associated with significantly improved MCC-specific survival, suggesting that augmenting the number or avidity of virus-specific T cells may have therapeutic benefit.
Keywords: skin cancer, Merkel cell carcinoma, CD8+ T cells, T-cell receptors, transgenic T-cell therapy
INTRODUCTION
Merkel cell carcinoma (MCC) is a highly aggressive skin cancer associated with UV exposure, advanced age, immune suppression, and in approximately 80% of cases, the Merkel cell polyomavirus (MCPyV) (1–3). MCC incidence is approximately 2000 cases per year in the US (4, 5). It has not been possible to effectively treat advanced disease, leading to a 5-year survival rate of 0–18% for patients with distant metastatic disease and a median survival of only 9.6 months from diagnosis of initial metastasis to death (5–7). Although approximately half of patients initially respond to chemotherapy, responses are not durable, with a median time to progression of only 94 days (8), highlighting the need for improved therapies to treat MCC.
MCPyV is a prevalent, chronic virus that normally does not cause disease. Rarely, it clonally integrates into the host chromosomal DNA, and when paired with UV-induced mutations, can cause MCC (1, 9, 10). Virus-positive MCC is characterized by persistent expression of two MCPyV oncoproteins (the small T- and truncated large T-Antigens) (10), which share a common N-terminus (common T-Ag). Multiple studies have linked the immune system with the incidence and prognosis of MCC. Although < 10% of MCC patients have systemic immune suppression, these patients have significantly poorer MCC-specific survival (11, 12). The adaptive immune system can recognize and mount protective responses to MCPyV. Specifically, increased intratumoral CD3+ cell counts are an independent prognostic factor of increased MCC-specific survival (13). Robust intratumoral CD8+ lymphocyte infiltration, though present in < 20% of MCCs, has been associated in two independent cohorts with 100% MCC-specific survival, independent of tumor stage at diagnosis (14, 15). Recent clinical trials of T cell–activating therapies, such as PD-1 axis blockade, have shown impressive initial response rates and durability (16), which provide further impetus for the study of T cell–based therapies for MCC.
CD8+ T cells recognizing at least 17 unique epitopes of the persistently expressed T-antigens of MCPyV can be isolated and tracked in blood and tumors from MCC patients using HLA class I tetramers (17–19 and our unpublished observations). MCPyV-specific CD8+ T cells have been harnessed for adoptive T-cell therapy and yet resulted in a durable response in just one of four patients treated (20, 21). One strategy to increase the efficacy of adoptive T-cell therapy, and/or offer T cell–based therapies to patients who lack endogenous MCPyV-specific T cells, would be to engineer T cells to express highly avid MCPyV-specific T-cell receptors (TCRs).
To better characterize the MCPyV-specific CD8+ TCR repertoire and measure correlations between TCR clonotype repertoire, intratumoral infiltration, and patient outcomes, we studied primary CD8+ T cells that recognize the epitope KLLEIAPNC (derived from the MCPyV common T-Ag), restricted to human leukocyte antigen (HLA) A*02:01, an HLA class I type present in ~50% of our patient cohort (hereafter, KLL-specific T cells). Similar to other infections and malignancies (22–26), we hypothesized that diversity or functional avidity of the TCR repertoire recognizing this epitope may be correlated with the effectiveness of the T-cell response to MCC in vivo.
Using next-generation TCRβ sequencing, we found significant genetic diversity among TCRs recognizing the KLL epitope. Using paired blood and tumor specimens, we can now extend previous findings of positive correlations between MCC tumor T-cell infiltration and effector gene signatures (14) to the level of tumor antigen-specific CD8+ T cells. In addition, patients with greater KLL-specific clonotype diversity in their tumors have significantly improved MCC-specific and recurrence-free survival. We studied the functional avidity of CD8+ clones expressing unique KLL-specific TCRs, and found that clones generally expressed a narrow range of functional avidities that were largely similar within each patient. Only 5 of 28 clonotypes tested from one of four patients recognized a MCPyV+ MCC cell line. Tumor cell recognition by T-cell clones correlated with high functional avidity. These studies have elucidated the genetic diversity of CD8+ T cells restricted to KLL and support continued investigation of methods to increase intratumoral CD8+ T-cell infiltration. The avid T-cell clones we identified could potentially lend their effector function through transgenic TCR therapy for the ~80% of HLA-A*02+ MCC patients who lack endogenous KLL-specific T cells.
MATERIALS & METHODS
Human subjects and samples
This study was approved by the Fred Hutchinson Cancer Research Center (FHCRC) Institutional Review Board and conducted according to Declaration of Helsinki principles. Informed consent was received from all participants. Subjects were HLA class I typed via polymerase chain reaction (PCR) at Bloodworks Northwest (Seattle, WA). All samples were clinically annotated with long-term patient follow-up data. PBMC: Heparinized blood was obtained from MCC patients and peripheral blood mononuclear cells (PBMCs) were cryopreserved after routine Ficoll preparation at a dedicated specimen processing facility at FHCRC. Patient Tumors: When available, fresh MCC tumor material from core and/or punch biopsy samples were processed and TIL cultured for two weeks before analysis as described (17). For excised tumors of larger volume (> 1 cm3), the remaining tissue was digested as described (18), and single cell suspensions were cryopreserved.
T-cell receptor β sequencing and analysis
Tetramer+ Cells: At least 2 million PBMC or TIL were stained with PE-conjugated monoclonal antibody (mAb) to CD8 (Clone 3B5, Life Technologies), A*02/KLL-APC tetramer (Immune Monitoring Lab, FHCRC), and 7-AAD viability dye (BioLegend). Tetramer+, CD8high cells were sorted via FACSAriaII (BD) and flash frozen (average of 710 cells from PBMC (n = 9), 5776 cells from TIL (n = 5), range 350–8,000 and 1844–12799, respectively). Samples were submitted to Adaptive Biotechnologies (Seattle, WA) for genomic DNA extraction, TRB sequencing and normalization. All TRB sequences detected in ≥ 2 cells (estimated number of genomes ≥ 2) were categorized as tetramer+ clonotypes. Whole tumor sequencing: Primary tumors were used for analysis, except when patients presented with unknown primaries and nodal disease (n = 2), primaries with limited material but abundant nodal disease available for analysis (n = 1) or metastatic disease (n = 1). Tumor samples consisted of molecular curls of 25 microns from formalin-fixed, paraffin embedded (FFPE) tissue blocks (n = 10), nodal tumor digest frozen ex vivo (n = 1) or flash frozen core biopsy of a metastatic lesion (n = 1). Samples were submitted to Adaptive Biotechnologies as described above. T cell receptor clonality: For tetramer-sorted cells, Shannon entropy was calculated on the estimated number of genomes (≥ 2) of all productive TRB and normalized by dividing by the log2 of unique productive sequences in each sample. Clonality was calculated as 1 – normalized entropy. For whole tumors, clonality was calculated in the same method, using all TRB sequences in the sample to calculate normalized entropy. Tetramer+ cell infiltration: KLL-specific clonotypes within tumors (n = 12 tumors) were identified based on TCRβ CDR3 amino acid sequences from the tetramer-sorted samples. The frequency of all KLL-specific T cells within each tumor is reported as the cumulative percentage of productive sequencing reads of clonotypes detected in both the tetramer-sorted sample and the tumor.
Immunohistochemistry
FFPE-embedded tumor tissue was stained (Experimental Histopathology at FHCRC) and slides scored by a dermatopathologist who was blinded to patient characteristics. Samples were stained with anti-CD8 (Dako, clone 144B at 1:100) and intratumoral CD8+ T cells (completely surrounded by tumor without neighboring stroma) on a scale from 0 (absent CD8+ cells) to 5 (> 732 intratumoral CD8+ cells/mm2) as described by Paulson et. al (14). In addition, tumors were stained with anti-MHC class I (27) (MBL, clone EMR8–5) and CM2B4 to measure MCPyV T-antigen expression (28) (Santa Cruz, 1:50). Tumors were stained with anti-CD4 (Cell Marque clone SP35, 1:25) and anti-FoxP3 (eBiosciences clone FJK-16s, 1:25) and reported as the number of positive cells/mm2.
Survival and recurrence analysis
Statistical analyses were performed on Stata software version 14.0 for Macintosh (StataCorp, College Station, TX) and Prism 6 for Mac OS X (Graph Pad Software, Inc). MCC-specific survival is defined as the interval from the diagnostic biopsy date to death by MCC. Recurrence-free survival was defined as the interval from the diagnostic biopsy date to the date of MCC recurrence, last follow up or death by MCC. Log-rank analysis was performed and a p-value of .05 was considered statistically significant. Kaplan-Meier survival curves were created to visualize MCC-specific survival and recurrence-free survival data; groupings of patients were based on percentage of tetramer+ T cells in the tumor (Higher = 1.9%–18%, n = 9 versus Lower = 0%–0.14%, n = 2) as well as number of T-cell clonotypes (Many = 5–108, n = 7; versus Few = 0–3, n = 4) were selected a priori. Patients who were alive at the last time of follow-up were censored on their last day of follow-up and patients who died of unknown causes were censored on their day of death.
Creation of KLL-specific T-cell clones
PBMC or lymphocytes from tumor digest were stained and sorted as described above into T-cell medium (TCM) containing RPMI, 8% human serum, 200 nM L-glutamine and 100 U/ml Penicillin-Streptomycin, and cloned at 0.25 to 3 cells per well with allogeneic irradiated feeders, IL2 (Hemagen Diagnostics) and PHA (Remel) as described (29) with addition of rIL15 (20 ng/mL, R&D Systems) after day 2. After 2 w, microcultures with visible growth were screened for specificity via tetramer; TCRβ variable chain (TCRVβ) expression was assessed by staining clones with fluorescent mAbs toTCRVβ (IOTest Beta Mark, Beckman Coulter). Wells selected for screening, expansion, and TCR analysis came from plates with < 37% of cultures having visual growth, yielding a 95% chance of clonality per the Poisson distribution (30). Cultures with tetramer+ cells, reactivity to peptide and dissimilar TCRVβ chains were further expanded with IL2 and the OKT3 mAb clone to CD3 (Miltenyi Biotec) as described (17), plus rIL15 (20 ng/mL). Prior to harvesting RNA for TCR analysis, cultures were held at least 2 weeks to minimize persistent feeder cell-derived RNA. CD8-independent Tetramer Staining: Clones were stained with a HLA-A*02:01/KLL tetramer containing D227K/T228A mutations in HLA-A*02:01, using methods as above. These mutations abrogate HLA class I:CD8 binding to identify clones expressing TCRs with the ability to bind independent of CD8 stabilization, which may indicate high TCR avidity (31, 32).
Sequencint of TCRα & TCRβ clones
Simultaneous sequencing of TCRα and TCRβ repertoires was performed as described (33). Briefly, total RNA was isolated from clonally expanded populations using Qiagen RNeasy Plus, followed by One Step RT/PCR (Qiagen) primed with multiplexed TCR primers. This reaction was used as template with a second set of nested TCRα and TCRβ primers, followed by PCR to add barcoding and paired end primers. Templates were purified using AMPure (Agencourt Biosciences) then normalized prior to running on Illumina MiSeq v2-300. Pairs of 150 nucleotide sequences were merged into contigs using PandaSeq (34). Merged sequences were then separated according to inline barcodes identifying the plate and well of origin, generating one file of derived sequences for each clone of interest. Files for each clone were processed with MiXCR (35) to identify and quantify clonotypes and assign VDJ allele usage. Cultures in which the dominant TCRβ nucleotide sequence was present at < 97% of productive sequence reads were classified as possibly polyclonal and excluded from further analysis.
T-cell functional assays
T-cell clones were tested for specificity and functional avidity via cytokine release assays. Cytokine Release with Peptide-pulsed Targets: Secreted IFNγ was measured after co-incubating 2 x 104 clonal KLL-specific T cells with 5 x 104 T2 cells (ATCC, 2015, validated by ATCC via short tandem repeat (STR) analysis) plus antigenic peptide at log10 dilutions to final concentration from 10−6 to 10−12 molar in 200 μl TCM for 36 h. Due to possible oxidation and dimerization of cysteine residues in the antigenic KLLEIAPNC peptide, the homolog KLLEIAPNA was used to allow for efficient HLA class I presentation; similar substitution has been shown to not alter recognition of HLA-peptide complex by TCRs raised against the native peptide (36). IFNγ in cell culture supernatants was assayed by ELISA according to manufacturer’s recommendations (Human IFN gamma ELISA Ready-SET-Go Kit, affymetrix). To estimate EC50 (the amount of peptide leading to 50% of maximum IFNγ secretion), IFNγ secretion by each T-cell clone was analyzed via nonlinear regression using Prism version 6.0 (GraphPad). In addition, IFNγ release by KLL-specific clonotypes was measured after incubation with three MCPyV+, HLA-A*02+ MCC cell lines (WaGa and MKL-2 [gift of Dr. Becker, German Cancer Research Center, 2015. Authenticated by Becker lab via STR analysis in 2014, and MS-1 [gift of Dr. Shuda, University of Pittsburg, 2015. Validated by STR and deposited into European Collection of Authenticated Cell Cultures]. Cell lines were early passage and authenticated with independent STR profiling (ATCC). Cell lines were stimulated with IFNβ (Betaseron, BayerHealthCare; 3,000 U/mL) for 24 h to induce expression of HLA class I, followed by 24 h of culture after IFNβ washout. 2 x 104 clonal KLL-specific T cells were incubated with 5 x 104 cells from each MCC cell line, ± IFNβ treatment, and incubated for 36 h. Supernatants were assayed by ELISA as described above. Cytokine Release with Large T-Ag transfected Targets: T-cell clones were incubated with antigen presenting cells transiently transfected with plasmids encoding HLA-A*02:01 and GFP-truncated Large T-Ag (tLTAg) fusion protein (pDEST103-GFP-tLTAg). pDEST103-GFP-tLTAg was created using Gateway recombination cloning technology (Gateway) to insert tLTAg from pCMV-MCV156 (37) into pDEST103-GFP. 3 x 104 COS-7 cells (ATCC, CRL-1651, 2005) were plated in flat-bottom 96-well plates in DMEM + 10% FBS, 200 nM L-glutamine and 100 U/ml Penicillin-Streptomycin. 24 h later, wells were transfected using FuGENE HD (Promega) at a 6:1 ratio of transfection reagent to DNA with 25 ng HLA-A*02:01 and limiting dilution of pDEST103-GFP-tLTAg (25-0.08 ng) plus irrelevant DNA (pcDNA-6/myc-His C, Gateway) to a total of 25 ng. 48 h after transfection, 104 viable KLL-specific T cells in TCM were added to target wells in duplicate. After 36 h, supernatants were assayed by ELISA for IFNγ secretion and EC50 calculated as above. Transfected COS-7 cells were harvested at 48 and 72 h post-transfection to quantitate transfection efficiency by flow cytometry.
RESULTS
Minority of MCC patients have detectable A02/KLL-specific CD8+ T cells
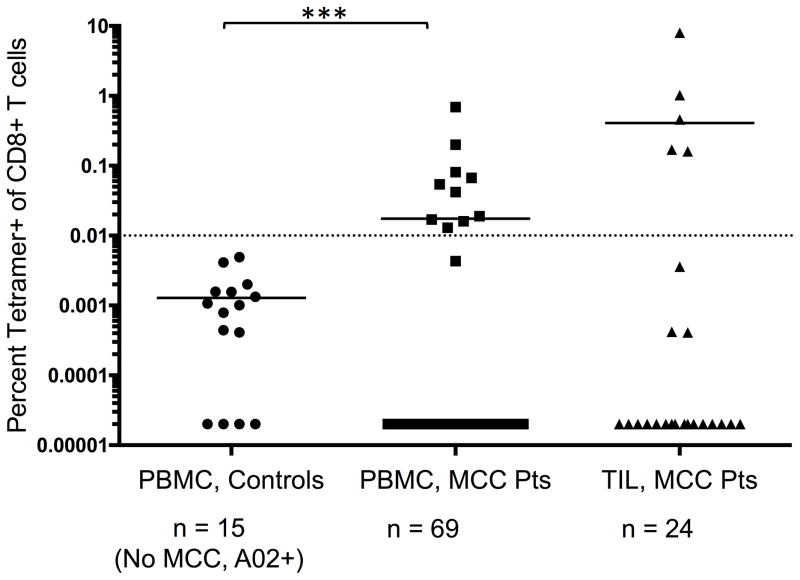
HLA-A*02 is a prevalent HLA-type present in ~55% of our MCC cohort (n = 97 low-resolution HLA class I typed patients; HLA-A*02:01 is the dominant A02 allele). We detected A*02-restricted T-cell responses in MCC patients to an epitope of the common T-Ag (aa 15–23) in 14% of PBMC (10 of 69) and 21% of cultured TIL (5 of 24; TIL were expanded with mitogen/cytokine for 2 weeks; ref 17) from HLA-A*02+ patients. No tetramer+ cells were detected in PBMC from healthy HLA-matched controls (0 of 15, Fig. 1). Among HLA-A*02+ patients, neither MCC-specific survival nor recurrence-free survival were significantly different between patients with or without detectable KLL-specific tetramer+ T cells (P = 0.593 and P = 0.643, data not shown). We believe that the detected KLL-specific T cells were primed by MCPyV due to the limited homology between this epitope and the homologous region of other polyomaviruses known to infect humans (Supplementary Table 1). Moreover, this epitope is predicted to bind to HLA-A*02 ~100x better than these homologous peptides (IC50 for the KLL MCPyV peptide is 299 nM versus 6950–25799 nM for all other homologs as determined by the Immune Epitope Database; ref 38).
Figure 1. Frequencies of KLL tetramer+ CD8+ T cells in PBMC and TIL from MCC patients and controls.
MCPyV-specific T-cell frequencies among HLA-A*02+ patients (n = 69 for PBMC, 24 for TIL) or PBMC from control subjects (n = 15). PBMC acquired when patients had evidence of disease was used in all analyses. Mean for each group is depicted, with dashed line at threshold for credible responses. The mean frequency of tetramer+ CD8+ cells was significantly different between MCC patient PBMC and control subjects (P = 0.0004 by Mann Whitney test) but not significantly different between MCC patient TIL and control PBMC (P = 0.11).
Characteristics of patients with KLL-specific T cells
Twelve patients had robust populations of KLL tetramer+ cells (> 0.04% of CD8+ T cells) in their PBMC and/or cultured TIL. Patient demographics, relevant disease metrics, and frequency of tetramer+ populations are summarized in Supplementary Table 2. All patients were Caucasian, with a median age of 65 (range 50–77). The patients presented at varying stages of disease. Some developed progressive disease and others showed no evidence of disease after definitive treatment during a median follow up period of 2.7 years (range 1.1 – 6.0) years.
Significant clonotypic diversity of KLL tetramer+ T cells within and between patients
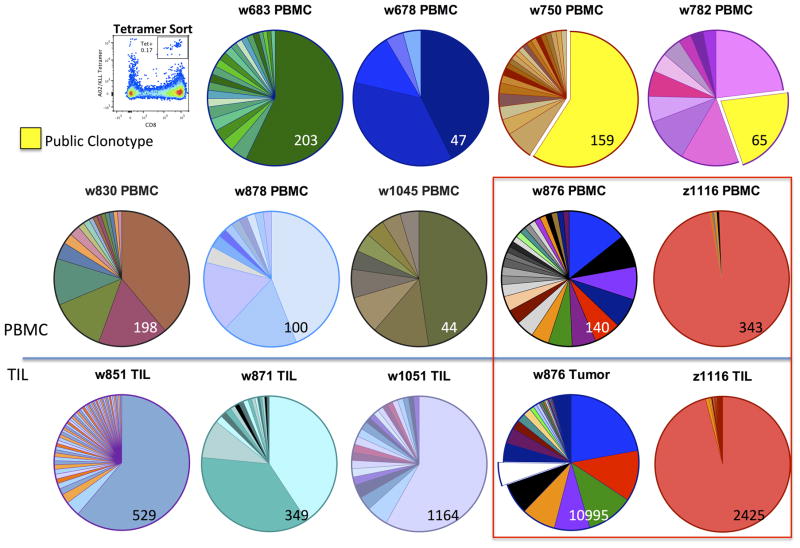
We sequenced the complementarity determining region 3 (CDR3) region of TRB of KLL tetramer-sorted cells from PBMC (n = 9) and/or TIL (n = 5) from 12 patients (Fig. 2). Out of 397 unique TRB sequences, only one public TCRβ clonotype was detected and shared between just two patients. This clonotype dominated the KLL-specific repertoire of these patients (59.1 or 21.5% of KLL-specific TRB sequencing reads). Complete TCRβ sequence results for each patient, in order of decreasing frequency, are in Supplementary Table 3. Paired KLL tetramer+ T cells from both PBMC and TIL were available for two patients (boxed). The diversity of the tetramer+ TRB repertoire varied greatly between patients. The overall TRB diversity in a sample was not correlated with the frequency of tetramer+ T cells among total CD8+ cells in PBMC (Fig. 1). We determined the clonality of each tetramer+ sample from PBMC (range: 0–1 with a completely clonal sample = 1; see Methods for details) and found no significant difference in MCC-specific survival or recurrence-free survival between patients with a less clonal (clonality < 0.3, n = 6) or more clonal (clonality >0.3, n = 3) KLL-specific repertoire in their PBMC (Supplementary Fig. S2, P = 0.52 and P = 0.81 by log-rank test).
Figure 2. TRB CDR3 clonotype diversity among KLL tetramer+ CD8+ cells from PBMC and TIL of 12 patients.
KLL tetramer+ CD8+ T cells were sorted by flow cytometry (a representative plot is shown) and the CDR3 region from TRB was sequenced. All productive TRB clonotypes with an estimated number of genomes ≥ 2 within each sample are indicated in proportion to their prevalence with a pie chart, with the total number of T cells sequenced indicated at bottom right in each pie. Patients are identified by unique “w” or “z” number. Among 397 total TRB clonotypes, only one shared clonotype was detected among two patients (highlighted in yellow). Paired tumor and PBMC samples were available for two patients (boxed).
T-cell repertoire within matched tumor samples
Archival tumor samples were analyzed from 11 of 12 patients; tumor from w750 was unavailable. When possible, primary tumors were acquired (n = 6). For four patients with an unknown primary who presented with nodal disease, lymph nodes were analyzed. Primary tumor from w878 had insufficient material for study and we therefore analyzed a metastasis corresponding to the time of PBMC collection. The primary tumor sample from w782 was small and therefore to ensure adequate sampling we also analyzed a nodal tumor present at time of diagnosis from w782. Tumors were assessed via immunohistochemistry (IHC) for viral status; HLA-I expression; and CD8+, CD4+ and FoxP3+ T-cell infiltration (Supplementary Fig. S3A). All patients were robustly positive for MCPyV Large T-Ag protein by IHC. CD8+ cells were more predominant than CD4+ or FoxP3+ T cells in the majority of samples. TRB CDR3 of all T cells in each tumor sample were sequenced and total unique TCRβ clonotypes/tumor were plotted in Supplementary Fig. S3A (n = 12, range = 16–41, with 645 unique clonotypes/tumor).
We measured whether having a greater number of total T cells was associated with a survival benefit. A priori, patients were binned by whether their tumors had many infiltrating T cells (≥ 0.8 T cells/ng tumor DNA, n = 7) or few T cells < 0.3 T cells/ng tumor DNA, n = 3); these two groups of patients had no survival difference (Supplementary Fig. S3B, P = 0.59 by log-rank test). In addition, we calculated the TRB clonality of each tumor analyzed. Increased clonality of the immune infiltrate within tumors is thought to represent an enrichment of cancer antigen-specific T cells and has been associated with improved response to immunotherapy (39). However, MCC-specific survival or recurrence-free survival was similar in patients whose tumors had a less clonal repertoire (clonality <0.1, n = 7) to those with a more clonal repertoire (clonality > 0.1, n = 4; Supplementary Fig. S4A and B, P = 0.50 and P = 0.64 by log-rank test).
Intratumoral A02/KLL-specific T cells are associated with survival
We next assessed how frequently KLL-specific T cells infiltrated MCC tumors. KLL-specific clonotypes within tumors were identified by determining the intersection between TCRβ CDR3 amino-acid sequences in the tetramer-sorted sample (from Fig. 2) and whole tumor samples from each patient. KLL-specific T cells constituted between 0–18% of the T-cell repertoire of each tumor based on the total number of T-cell genomes sequenced (n = 12, mean 6.3%, SD = 5.8, Supplementary Fig. S5A). Tumors contained between 0–108 unique KLL-specific TCRβ clonotypes (mean = 19.4, SD = 32, Supplementary Fig. S5B). The rank (based on frequency) of each KLL-specific clonotype within each tumor was plotted; individual clonotypes ranged between being the most prevalent clonotype to rare within each autologous tumor. KLL-specific clonotypes appeared to be more abundant (based on total percentage of all KLL-specific T cells in tumor) and predominant (based on percentage of individual KLL-specific clonotypes) in patients that were alive at last follow up (Fig. 3). Patients were binned a priori based on percentage of tumor with KLL-specific T cells. MCC-specific survival was significantly increased for patients who had a higher (1.9–18%; n = 7) versus lower (0–0.14%; n = 2) percentage of KLL-associated T cells in tumor (Fig. 4A, P = 0.0009 by log-rank test).
Figure 3. Frequency and number of KLL-specific clonotypes in tumors from patients with KLL-specific T cells in PBMC or cultured TIL.
A wedge (the length of which represents the total number of productive unique clonotypes in each tumor) is indicated for each tumor on a log scale. Each tumor is identified by patient “w” or “z” number and type of tumor. Tumors from 11 of 12 patients were analyzed; no tumor could be acquired for patient w750. Primary tumor from w782 was small and LN was analyzed to ensure adequate sampling. KLL-specific clonotypes (yellow) are depicted within each tumor with a width approximately proportional to their frequency within each tumor. More predominant clonotypes are located to the left for each tumor. The number of KLL-specific clonotypes out of the total number of unique clonotypes is tabulated at far right. Wedges for tumors from patients alive at time of censoring are in green, and wedges for tumors in grey are from patients who have died of MCC.
Figure 4. Increased frequency and number of KLL-specific clonotypes in tumors is associated with improved MCC-specific survival.
A) MCC-specific survival was significantly increased for patients who had higher (n = 9) versus lower (n = 2) percentage of KLL-specific T cells in tumor (1.9%–18% versus 0%–0.14%, P = 0.0009 by log-rank test). (B) MCC-specific survival was increased for patients who had many unique KLL-specific clonotypes (5–108 clonotypes, n = 7 patients) in their tumors, compared to patients with few KLL-specific clonotypes (0–3, n = 4; P = 0.0051 by log-rank test). (C) Patients were grouped by whether they developed metastatic disease (n = 7) or remained disease-free after definitive treatment of first presentation of disease (n = 3). The percentage of KLL-specific T cells tended to be higher in patients without recurrence (range 4.3%–18%) compared to those who developed metastatic disease (range 0%–10.8%, P =0.11). (D) The number of KLL-specific clonotypes was significantly higher in patients without recurrence (median 38, range 9–108) compared to those who developed metastatic disease (median 2, range 0–17, P = 0.02).
In addition, we asked whether the number of unique KLL-specific TCRβ CDR3 clonotypes infiltrating tumors was associated with survival. Indeed, patients who had more unique KLL-specific clonotypes in their tumors (5–108 clonotypes, n = 7 patients) had a significant survival advantage, compared to patients with few KLL-specific clonotypes (0–3, n = 4; Fig. 4B, P = 0.0051). When patients were separated into those who did and did not have a recurrence, the frequency of KLL-specific T cells was higher in tumors from patients without disease recurrence (median 10.4%) compared to patients who did recur (median of 3.2%, Fig. 4C, P = 0.11). In addition, the diversity of unique KLL-specific clonotypes was significantly higher in patients who did not have recurrences (median of 38 clonotypes) compared to patients who did (median of 2 clonotypes; Fig. 4D, P = 0.02). Collectively, these data show a significant survival advantage for patients whose tumors contain a higher relative percentage or a greater clonotypic diversity of KLL-specific T cells.
TCRα/β sequence diversity among KLL-specific CD8+ T-cell clones
To gain insight into functional differences of unique KLL-specific TCRs, we generated KLL-specific T-cell clones from MCC patients’ PBMC (n = 4) and/or ex vivo tumor digest (n = 1) by sorting KLL-tetramer+ cells followed by limiting dilution cloning. Diversity of the TCRVβ of several KLL-reactive clones per patient was studied with fluorescent anti-TCRVβ antibodies via flow cytometry, and clones encompassing the spectrum of TCRVβ usage were expanded for further study. We determined the V, J, and CDR3 sequences of both TCRα and β chains for 120 clones and identified 71 monoclonal cultures, 42 of which were comprised of distinct TCRs, recognizing this epitope among 4 patients (Table 1). Among many private TCRα chains sequenced, one public TCRα chain using TRAV12-1*01 and encoding the CDR3 “CVLNNNDMRF” was found among clones from three of four patients.
Table 1.
TCRα/β sequences of HLA-KLL tetramer+ clones from four MCC patients
| Alpha Chain | Beta Chain | Functional Assays | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pt | V gene | CDR3 region | J gene | V gene | CDR3 region | J gene | EC50 (ng/mL peptide) |
EC50 (ng/uL DNA) |
Recog. MS-1? |
Mutant Tet+? |
| w830 Clonotypes | ||||||||||
| 1 | TRAV24*01 | CAFNTDKLIF | TRAJ34*01 | TRBV12-4*01 | CASSLAGFRFF | TRBJ2-1*01 | 4.6 0.4 |
No | Lower | |
| 2 | TRAV38-1*01 | CALTSGSRLTF | TRAJ27*01 | TRBV19*01 | CASSIMLYSNQPQHF | TRBJ1-5*01 | 12 1.6 |
140 | No | Equal |
| 3 | TRAV38-1*01 | CAYPSTDKLIF | TRAJ34*01 | TRBV19*01 | CASSILGASNQPQHF | TRBJ1-5*01 | 270 | Equal | ||
| 4 | TRAV12-1*01 | CVLNNNDMRF | TRAJ43*01 | TRBV19*01 | CASSILGASNQPQHF | TRBJ1-5*01 | 1.1 | No | Equal | |
| 5 | TRAV12-1*01 | CVVNANDMRF | TRAJ43*01 | TRBV10-3*01 | CAIRARDQNTGELFF | TRBJ2-2*01 | 5.1 0.89 |
No | Equal | |
| w683 Clonotypes | ||||||||||
| 1 | TRAV12-1*01 | CVVALYSGGGADGLTF | TRAJ45*01 | TRBV6-5*01 | CASRSQNYYGYTF | TRBJ1-2*01 | 1.9 0.36 0.26 |
No | Lower | |
| 2 | TRAV12-1*01 | CVLNNNDMRF | TRAJ43*01 | TRBV6-5*01 | CASRSQNYYGYTF | TRBJ1-2*01 | 0.21 | No | Lower | |
| w678 Clonotypes | ||||||||||
| 1 | TRAV12-1*01 | CVLNNNDMRF | TRAJ43*01 | TRBV10-3*01 | CAIRQFDANTGELFF | TRBJ2-2*01 | 0.43 0.50 0.47 |
0.32 | Yes | Equal |
| 1.5 | UNKNOWN* | TRBV10-3*01 | CAIRQFDANTGELFF | TRBJ2-2*01 | 0.84 | Yes | Equal | |||
| 2 | TRAV38-1*01 | CAFRVSHDMRF | TRAJ43*01 | TRBV19*01 | CASSIIAGSSYNEQFF | TRBJ2-1*01 | 0.017 0.012 |
Yes | Lower | |
| 3 | TRAV12-1*01 | CVVATYSGGGADGLTF | TRAJ45*01 | TRBV19*01 | CASSIIAGSSYNEQFF | TRBJ2-1*01 | 0.022 0.013 |
1.0 | Yes | Equal |
| 4 | TRAV12-1*01 | CVVATYSGGGADGLTF | TRAJ45*01 | TRBV10-2*01 | CASSSGNPSTDTQYF | TRBJ2-3*01 | 0.0094 0.028 |
Yes | Equal | |
| 5 | TRAV38-1*01 | CAFRVSHDMRF | TRAJ43*01 | TRBV10-2*01 | CASSSGNPSTDTQYF | TRBJ2-3*01 | 0.11 0.054 |
Yes | Lower | |
| UNKNOWN* | UNKNOWN* | 0.060 0.058 |
Yes | Equal | ||||||
| w876 Clonotypes | ||||||||||
| 1 | TRAV12-1*01 | CVVGEYSGGGADGLTF | TRAJ45*01 | TRBV28*01 | CAIRAGASYNEQFF | TRBJ2-1*01 | 0.31 | 5.6 | Lower | |
| 2 | TRAV12-1*01 | CVVTEYSGGGADGLTF | TRAJ45*01 | TRBV10-3*01 | CASRGQNTGELFF | TRBJ2-2*01 | 1.2 | No | Lower | |
| 3 | TRAV19*01 | CALGGGTFTSGTYKYIF | TRAJ40*01 | TRBV9*02 | CASSVEDYTGELFF | TRBJ2-2*01 | 0.12 | 11 | No | Equal |
| 4 | TRAV12-1*01 | CVVYTGYSGGGADGLTF | TRAJ45*01 | TRBV10-2*01 | CASSVLNTGELFF | TRBJ2-2*01 | 0.31 | 14 | No | Equal |
| 5 | TRAV38-1*01 | CAYNQGGKLIF | TRAJ23*01 | TRBV10-2*01 | CASSVLNTGELFF | TRBJ2-2*01 | 0.11 | No | Equal | |
| 6 | TRAV12-1*01 | CVVPLYSSASKIIF | TRAJ3*01 | TRBV6-1*01 | CASSDTPDLNTEAFF | TRBJ1-1*01 | 0.015 0.035 |
No | Lower | |
| 7 | TRAV12-1*01 | CVLNNNDRF | TRAJ43*01 | TRBV6-1*01 | CASSDTPDLNTEAFF | TRBJ1-1*01 | 0.14 | 3.6 | No | Lower |
| 8 | TRAV12-1*01 | CVVYASKIIF | TRAJ3*01 | TRBV6-1*01 | CASSDTPDLNTEAFF | TRBJ1-1*01 | 4.2 | Lower | ||
| 9 | TRAV12-1*01 | CVGNNNDMRF | TRAJ43*01 | TRBV10-3*01 | CAISARDQNTGELFF | TRBJ2-2*01 | 0.12 0.24 |
No | Lower | |
| 10 | TRAV12-1*01 | CVVYGSSNTGKLIF | TRAJ37*02 | TRBV10-3*01 | CAIRRQDQNTGELFF | TRBJ2-2*01 | 0.70 | No | Lower | |
| 11 | TRAV12-1*01 | CVVYTGYSGGGADGLTF | TRAJ45*01 | TRBV10-3*01 | CAIHEGDSNTGELFF | TRBJ2-2*01 | Equal | |||
| 12 | TRAV3*01 | CAVRDNSGTYKYIF | TRAJ40*01 | TRBV7-2*04 | CASSLAGLAGTDTQYF | TRBJ2-3*01 | Lower | |||
| 13 | TRAV12-1*01 | CVVTDTSGGGADGLTF | TRAJ45*01 | TRBV7-2*04 | CASSLAGLAGTDTQYF | TRBJ2-3*01 | 3.9 | |||
| 14 | TRAV12-1*01 | CVVPSAGKSTF | TRAJ27*01 | TRBV2*03 | CASSEFAGQETQYF | TRBJ2-5*01 | 5.4 | Lower | ||
| 15 | UNKNOWN* | TRBV6-5*01 | CASRASNTYGYTF | TRBJ1-2*01 | 80 | No | ||||
| 16 | TRAV38-1*01 | CAYNQGGKLIF | TRAJ23*01 | TRBV19*01 | CASSTLSGTHNEQFF | TRBJ2-1*01 | 0.12 | No | Lower | |
| 17 | TRAV12-1*01 | CVVYGSSNTGKLIF | TRAJ37*02 | TRBV7-2*04 | CASSLAGLANNEQFF | TRBJ2-1*01 | Lower | |||
| 18 | TRAV14 | CAMREAQSGGYQKVTF | TRAJ13*01 | TRBV12-4*01 | CASSFGSGTKDTQYF | TRBJ2-3*01 | Equal | |||
| 19 | TRAV12-1*01 | CVVYTGYSGGGADGLTF | TRAJ45*01 | TRBV10-2*01 | CASSGQNTGELFF | TRBJ2-2*01 | 0.92 | No | Lower | |
| 20 | TRAV10*01 | CVVSAGINGADGLTF | TRAJ45*01 | TRBV12-4*01 | CASSPWDEQFF | TRBJ2-1*01 | Lower | |||
| 21 | TRAV3*01 | CAVRDNSGTYKYIF | TRAJ40*01 | TRBV13*02 | CASSSGTSGGLTYNEQFF | TRBJ2-1*01 | ||||
| 22 | UNKNOWN* | TRBV13*02 | CASSSRTKAYEQYF | TRBJ2-7*01 | ||||||
| 23 | TRAV12-3*01 | CAMSVAQGGSEKLVF | TRAJ57*01 | TRBV6-6*01 | CASSYQIGLSYEQYF | TRBJ2-7*01 | ||||
| 24 | UNKNOWN* | TRBV28*01 | CASSFDSKGSNTGELFF | TRBJ2-2*01 | ||||||
| 25 | TRAV27*01 | CAGDQGGSEKLVF | TRAJ57*01 | TRBV16*01 | CASSQLRTGDEYEQYF | TRBJ2-7*01 | ||||
| 26 | TRAV12-1*01 | CVVYTGYSGGGADGLTF | TRAJ45*01 | TRBV13*02 | CASSSGTSGGLNYNEQFF | TRBJ2-1*01 | ||||
| 27 | TRAV3*01 | CALTGYSTLTF | TRAJ11*01 | TRBV19*01 | CASRSQLAVLNEQFF | TRBJ2-1*01 | ||||
| 28 | UNKNOWN* | TRBV28*01 | CASRGGSSYNEQFF | TRBJ2-1*01 | ||||||
| 29 | TRAV12-1*01 | CVVPLYSSASKIIF | TRAJ3*01 | TRBV10-2*01 | CASSVLNTGELFF | TRBJ2-2*01 | 0.12 0.16 |
3.3 | No | Equal |
| 30 | UNKNOWN* | TRBV10-3*01 | CATRDINTGELFF | TRBJ2-2*01 | ||||||
| 31 | UNKNOWN* | TRBV6-1*01 | CASSDTPDLNTEAFF | TRBJ1-1*01 | 24 | |||||
select TRA or TRB sequences were unresolved with next-generation sequencing.
KLL-specific clones display a hierarchy of functional avidity
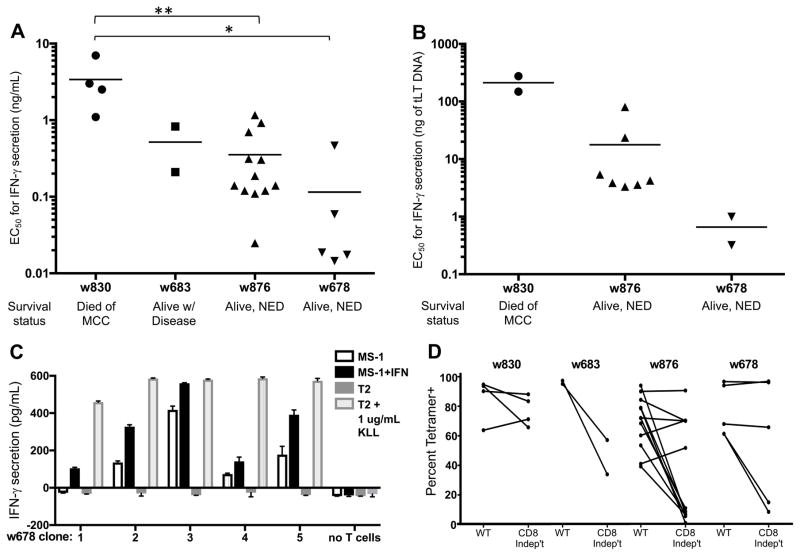
To investigate functional differences among MCPyV-specific T-cell clones, we measured secretion of IFNγ, a canonical type 1 helper T cell cytokine, after stimulation with T2 target cells pulsed with limiting dilution of an alanine-substituted variant of the peptide (KLLEIAPNA; this peptide is antigenic but less susceptible to oxidation, allowing direct comparison of T-cell clones to each other. See Methods for details). Clones displayed narrow ranges of intra-patient variability for functional avidity (Table 1 Fig. 5A). Concordant results were obtained in a separate but analogous assay using targets transfected with limiting dilution of plasmid encoding truncated Large T-Ag (Fig. 5B). Importantly, patients with improved MCC-specific survival had more functionally avid T-cell clonotypes (P < 0.05). To further interrogate the effector function of these clonotypes, we tested the ability of 28 unique KLL-specific clonotypes to recognize the MCPyV+, HLA-A*02+ MCC cell lines (WaGa, MS-1 and MKL-2) ± IFNβ treatment. Five unique clonotypes secreted IFNγ when incubated with MS-1; this response was generally lower than that to T2 cells pulsed with a maximal concentration of peptide. No clones recognized WaGa or MKL-2 (Table 1 and Fig. 5C). Reactive clones were derived from patient w678 who had the most functionally avid clonotypes in the IFNγ release assay. Lastly, we compared the ability of KLL-specific clonotypes to bind both wild-type and ‘CD8-independent’ tetramers that contain mutations in HLA-A*02:01 to abrogate CD8 stabilization of the TCR:pMHC interaction, which may select for more avid TCRs (31, 32). More functionally avid clonotypes in the IFNγ assays (Fig. 5A & B) often bound both wild-type (WT) and CD8-independent tetramers equivalently; however, there were many clonotypes that did not follow this trend (Table 1 and Fig. 5D). Indeed, when clones from each patient were binned by whether they bound the CD8-independent tetramer ‘equally’ or ‘lower’, the mean EC50 of these two groups did not differ significantly (P = 0.57 for w678 by Mann-Whitney test, P = 0.30 for w830, insufficient data for w830 and w683). No significant correlations between clonotype avidity and enrichment within tumors were identified.
Figure 5. Functional avidity of 28 KLL-specific clonotypes from 4 patients.
EC50 values for IFNγ secretion by KLL-specific clones in response to peptide concentration (A) or concentration of tLT-Ag DNA transfected into Cos7 cells (B) are plotted for each patient, with mean of all clones/patient depicted by the horizontal bar. For replicate experiments of clones with the same TCR, a single point representing the mean EC50 is plotted. Clonotypes from the same patient generally had similar functional avidities; more avid clonotypes are detected among patients with better MCC-specific survival. Statistical comparisons were made between patients; *, P < 0.05; **, P < 0.01, Mann Whitney test. (C) Clonotypes from one patient respond to the MCPyV+, HLA-A02+ MCC cell line MS-1 ± IFNβ treatment to upregulate HLA-I; responses of each clone to T2 cells ± KLL peptide are shown for comparison. Mean of duplicates + SEM are shown after subtracting background IFNγ secretion by T cells without targets; representative results from one of at least two separate experiments with each clone are shown. (D) Select clonotypes are able to bind a ‘CD8-independent’ KLL-tetramer.
DISCUSSION
The purpose of our study was to characterize the TCR repertoire restricted to a naturally processed epitope of MCPyV in the context of the prevalent HLA-A*02 allele, and to assess whether differences in either the breadth or avidity of the TCRs correlated with the effectiveness of the T-cell response in vivo. We identified KLL tetramer+ CD8+ T cells in a minority of HLA-matched MCC patients. Presence of circulating KLL-specific T cells was not associated with differences in survival or recurrence among HLA-A*02+ The TCRβ repertoires of the KLL-specific T cells were strikingly diverse. When we examined the T cells within tumors, higher frequencies of KLL-specific T cells, as identified by their signature TRB CDR3 sequences, correlated with a significant MCC-specific survival advantage. These findings identify the diversity of the CD8+ T-cell response to MCC, and suggest therapeutic maneuvers to boost tumor immunity. Overall, our findings provide a rationale for active or passive immunization to increase MCPyV-specific CD8+ T-cell diversity and avidity, and for manipulations of the immunosuppressive tumor microenvironment to promote the infiltration by these T cells.
This study is unique in its focus on the TCR repertoire specific for particular epitopes of MCPyV, using a high-throughput TCR sequencing approach to study tumor antigen–specific TCRs, which has revealed diversity that may have been missed using earlier methods. TCR sequencing was accompanied by the generation of KLL-specific clones and functional avidity characterization in a physiologically relevant system, allowing for paired analysis of unique TCRs and matched T-cell clones. Similar approaches have been utilized in other virally associated cancers to select the ‘best’ TCRs for transgenic T-cell therapy (40), a therapeutic modality that has merit in MCC.
The frequency of KLL-specific T cells among A*02+ MCC patients was lower than expected (only 14% of A*02+ MCC patients have detectable KLL-specific T cells), given our finding that T cells restricted to an HLA-A*2402-epitope were found in 64% of HLA-matched PBMC (18). These findings could be the result of poor processing and presentation of the KLL epitope (for instance, the A*2402 restricted epitope is two-fold more avid for cognate HLA compared to the KLL epitope by IEDB; ref 36), which would prevent patients from being able to mount an immune response. Indeed, this hypothesis is supported by our finding that functionally avid KLL-specific clonotypes were only able to recognize one of three tested MCPyV+ A*02+ cell lines, even after upregulation of MHC-I on all three cell lines.
Alternatively, there may be adequate antigen presentation and yet patients are unable to mount their own endogenous effector response to this epitope (due to local immunosuppression or a host of other factors). These patients in particular may benefit from therapy with T cells expressing transgenic TCRs (tTCRs) restricted to this epitope. The TCR sequences generated from this study could be used to create useful reagents (i.e., tTCR clones) to detect and quantify KLLEIAPNC on the surface of primary MCCs to help distinguish between these hypotheses.
In this in-depth examination of MCPyV-specific TCRs, we found that out of 397 KLL-specific TCRs detected among 12 patients, the vast majority (396) were private, with only one public TCR observed across two individuals. Public TCRs have been observed in multiple species in response to many viral infections and tumor antigens (41), and might be expected to be particularly prevalent in the response to a DNA virus thought to have low mutational capacity such as MCPyV. However, that idea is discordant with our observation in this study of a predominantly private repertoire, although the small sample size of our study (n = 12) is a limitation. Increased diversity of an antigen-specific T-cell response led to improved outcomes in models of chronic infections such as CMV (42) and herpes (43). In our study, the diversity of the circulating KLL-specific T-cell repertoire and MCC outcomes were not associated; however, within tumors an increased number of KLL-specific clonotypes was associated with improved MCC-specific survival. Although these studies elucidate TCR diversity restricted to a single epitope of MCPyV, we now have validated tetramers for 5 other peptide/HLA combinations (unpublished observations) and can replicate these studies to assess whether this striking diversity in immune response is specific to the KLL epitope or more broadly observed in the CD8+ T-cell response to MCPyV.
Our finding that a higher frequency of KLL-specific T cells within primary tumors is associated with a significant MCC-specific survival advantage builds on previously published work that CD8+ infiltration into tumors is associated with improved survival (14, 15), but is the first to confirm the importance of MCPyV-specific T cells in the infiltrate. In contrast, the presence of detectable circulating KLL-specific T cells was not associated with improved MCC-specific survival. Therefore, efforts should be focused on improving the tumor homing and infiltration of both endogenous and therapeutic MCPyV-specific T cells. Several candidate agents in are currently in preclinical development. In a mouse model of ovarian cancer, modification of histone methylation was associated with restored secretion of Th-1 cytokines by tumor cells, promoting T-cell infiltration and improved outcomes with T cell-based therapies such as anti-PD-L1 (44).
In parallel to other infections and malignancies (22–26), we hypothesized that increased avidity of the TCR repertoire may be correlated with the effectiveness of the T-cell response in vivo. Forty-two distinct clonotypes recognizing this epitope were identified by creating clones from PBMC or tumor of four patients. Clones generally expressed a narrow intra-patient range of functional avidities with more avid clones isolated from patients with better MCC-specific outcomes. We detected specific IFNγ responses by the most avid clones from a patient with a favorable outcome, but only with 1 of 3 HLA-appropriate MCPyV cell lines. Antigen presentation by MCC cell lines is expected to be relatively minimal, because in vivo these cells survived by their ability to escape immune detection. There are numerous known mechanisms by which such immune evasion could be minimized, such as proteasome, TAP, and HLA class I/β2-microglobulin expression and function. We hypothesize that variation in these factors in vivo may correlate with T-cell infiltration and immune checkpoint inhibitor responses and are investigating these areas in ongoing research.
In a trial of anti-PD-1 (pembrolizumab) in patients with metastatic MCC (16), objective responses were observed in 56% of patients overall and in 62% of patients with MCPyV-positive tumors. Among the 38% of patients with MCPyV+ MCCs that had stable or progressive disease (16), it is possible that these nonresponders, similar to the patients in this study with poor survival outcomes, lacked high avidity and/or tumor infiltrating MCPyV-specific T cells. Analyses similar to those outlined in this study may therefore elucidate prognostic markers for response to PD-1 axis blockade.
There are several limitations to our study. Our sample size was limited by the number of subjects with this rare cancer in our research cohort with detectable KLL-tetramer+ T cells (n = 12). Because there are no durable treatments for advanced disease, patients received a variety of therapies including chemotherapy, radiation, and immunotherapies that were not standardized between patients. All patients in this study who received no further therapy since definitive excision of their presenting lesion (n = 4) are currently alive with no evidence of disease (median follow up time of 2.9 years; average recurrence for most MCC patients is 9 months), supporting the importance of their immune system in fighting MCC. Another limitation is that we studied only a single MCPyV epitope. It is almost certain that MCPyV-specific T cells recognizing other epitopes besides KLL contributed to the antitumor immune response. However, KLL-specific T cells were among the top 10 most frequent clonotypes within tumors of 7 of 9 patients studied, strongly suggesting that these T cells are a predominant factor in the effector response to MCC.
In summary, MCPyV-specific CD8+ T cells are detectable ex vivo in a substantial portion of HLA A*02+ MCC patients and have considerable TCR diversity that corresponds to several orders of magnitude of functional avidity. Our hypothesis that infiltration of MCPyV-specific T cells leads to superior tumor control is supported by our findings of increased MCC-specific survival in patients with a higher frequency of intratumoral KLL-tetramer+ T cells. Our findings support further investigation of agents that improve T-cell tumor homing and infiltration, as well as use of avid TCRs for transgenic T-cell therapy in advanced MCC.
Supplementary Material
Acknowledgments
Funding: Supported by NIH F30-CA192475-02, NIH K24-CA139052, NIH R01-CA162522-05, NIH R01-CA CA176841, NIH R01-AI094109, Adaptive Biotechnologies Young Investigator Award, Kelsey Dickson Team Science Courage Research Team Award, Prostate Cancer Foundation Award #15CHAS04, ARCS Fellowship, Environmental Pathology/Toxicology Training Grant T32ES007032-36, NIH Center for Aids Research (CFAR) AI027757, National Cancer Institute (NCI) Cancer Target Discovery and Development (CTD2) U01 CA176270, the David & Rosalind Bloom Endowment for MCC Research, the Michael Piepkorn Endowment Fund, and the UW MCC Patient Gift Fund, and the Deutsche Forschungsgemeinschaft (SFB TR36).
We thank the patients who made this research possible. In addition, we thank Dr. Juergen Becker for MCC cell lines WaGa and MKL-2, and Dr. Masahiro Shuda for MCC cell line MS-1.
Footnotes
Authors’ disclosures of potential conflicts of interest:
PN serves as a paid consultant for EMD Serono.
References
- 1.Feng H, Shuda M, Chang Y, Moore PS. Clonal Integration of a Polyomavirus in Human Merkel Cell Carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heath M, Jaimes N, Lemos B, Mostaghimi A, Wang LC, Peñas PF, et al. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: the AEIOU features. J Am Acad Dermatol. 2008;58:375–381. doi: 10.1016/j.jaad.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodig SJ, Cheng J, Wardzala J, DoRosario A, Scanlon JJ, Laga AC, et al. Improved detection suggests all Merkel cell carcinomas harbor Merkel polyomavirus. J Clin Invest. 2012;122:4645–4653. doi: 10.1172/JCI64116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen PJ. Merkel Cell Carcinoma: Prognosis and Treatment of Patients From a Single Institution. J Clin Oncol. 2005;23:2300–2309. doi: 10.1200/JCO.2005.02.329. [DOI] [PubMed] [Google Scholar]
- 5.Lemos BD, Storer BE, Iyer JG, Phillips JL, Bichakjian CK, Fang LC, et al. Pathologic nodal evaluation improves prognostic accuracy in Merkel cell carcinoma: analysis of 5823 cases as the basis of the first consensus staging system. J Am Acad Dermatol. 2010;63:751–761. doi: 10.1016/j.jaad.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santamaria-Barria JA, Boland GM, Yeap BY, Nardi V, Dias-Santagata D, Cusack JC., Jr Merkel cell carcinoma: 30-year experience from a single institution. Ann Surg Oncol. 2013;20:1365–1373. doi: 10.1245/s10434-012-2779-3. [DOI] [PubMed] [Google Scholar]
- 7.Miller NJ, Bhatia S, Parvathaneni U, Iyer JG, Nghiem P. Emerging and mechanism-based therapies for recurrent or metastatic Merkel cell carcinoma. Curr Treat Options Oncol. 2013;14:249–263. doi: 10.1007/s11864-013-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer JG, Blom A, Doumani R, Lewis C, Anderson AC, Ma C, et al. Response rate and durability of chemotherapy for metastatic Merkel cell carcinoma among 62 patients. 2014 ASCO Annual Meeting; Chicago, IL. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int J Cancer. 2012;130:847–856. doi: 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- 10.Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, et al. Merkel Cell Polyomavirus-Infected Merkel Cell Carcinoma Cells Require Expression of Viral T Antigens. J Virol. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulson KG, Iyer JG, Blom A, Warton EM, Sokil M, Yelistratova L, et al. Systemic immune suppression predicts diminished Merkel cell carcinoma-specific survival independent of stage. J Invest Dermatol. 2013;133:642–646. doi: 10.1038/jid.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarantola TI, Vallow LA, Halyard MY, Weenig RH, Warschaw KE, Grotz TE, et al. Prognostic factors in Merkel cell carcinoma: analysis of 240 cases. J Am Acad Dermatol. 2013;68:425–432. doi: 10.1016/j.jaad.2012.09.036. [DOI] [PubMed] [Google Scholar]
- 13.Sihto H, Joensuu H. Tumor-infiltrating lymphocytes and outcome in Merkel cell carcinoma, a virus-associated cancer. OncoImmunology. 2012;1:1420–1421. doi: 10.4161/onci.21120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulson KG, Iyer JG, Tegeder AR, Thibodeau R, Schelter J, Koba S, et al. Transcriptome-Wide Studies of Merkel Cell Carcinoma and Validation of Intratumoral CD8+ Lymphocyte Invasion As an Independent Predictor of Survival. J Clin Oncol. 2011;29:1539–1546. doi: 10.1200/JCO.2010.30.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulson KG, Iyer JG, Simonson WT, Blom A, Thibodeau RM, Schmidt M, et al. CD8+ lymphocyte intratumoral infiltration as a stage-independent predictor of Merkel cell carcinoma survival: a population-based study. Am J Clin Pathol. 2014;142:452–458. doi: 10.1309/AJCPIKDZM39CRPNC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nghiem PT, Bhatia S, Lipson EJ, Kudchadkar RR, Miller NJ, Annamalai L, et al. PD-1 Blockade with Pembrolizumab in Advanced Merkel-Cell Carcinoma. N Engl J Med. 2016;374:2542–2552. doi: 10.1056/NEJMoa1603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer JG, Afanasiev OK, McClurkan C, Paulson K, Nagase K, Jing L, et al. Merkel Cell Polyomavirus-Specific CD8+ and CD4+ T-cell Responses Identified in Merkel Cell Carcinomas and Blood. Clin Cancer Res. 2011;17:6671–6680. doi: 10.1158/1078-0432.CCR-11-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Afanasiev OK, Yelistratova L, Miller N, Nagase K, Paulson K, Iyer JG, et al. Merkel polyomavirus-specific T cells fluctuate with Merkel cell carcinoma burden and express therapeutically targetable PD-1 and Tim-3 exhaustion markers. Clin Cancer Res. 2013;19:5351–5360. doi: 10.1158/1078-0432.CCR-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyngaa R, Pedersen NW, Schrama D, Thrue CA, Ibrani D, Met O, et al. T-cell responses to oncogenic merkel cell polyomavirus proteins distinguish patients with merkel cell carcinoma from healthy donors. Clin Cancer Res. 2014;20:1768–1778. doi: 10.1158/1078-0432.CCR-13-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapuis AG, Afanasiev OK, Iyer JG, Paulson KG, Parvathaneni U, Hwang JH, et al. Regression of metastatic Merkel cell carcinoma following transfer of polyomavirus-specific T cells and therapies capable of re-inducing HLA class-I. Cancer Immunol Res. 2014;2:27–36. doi: 10.1158/2326-6066.CIR-13-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Institutes of Health. [Accessed March 2014];Viral Oncoprotein Targeted Autologous T Cell Therapy for Merkel Cell Carcinoma. Available at http://www.clinicaltrials.gov/ct2/show/NCT01758458?term=Merkel+cell+carcinoma&rank=7.
- 22.Nauerth M, Weissbrich B, Knall R, Franz T, Dossinger G, Bet J, et al. TCR-ligand koff rate correlates with the protective capacity of antigen-specific CD8+ T cells for adoptive transfer. Sci Transl Med. 2013;5:192ra187. doi: 10.1126/scitranslmed.3005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derby M, Alexander-Miller M, Tse R, Berzofsky J. High-avidity CTL exploit two complementary mechanisms to provide better protection against viral infection than low-avidity CTL. J Immunol. 2001;166:1690–1697. doi: 10.4049/jimmunol.166.3.1690. [DOI] [PubMed] [Google Scholar]
- 24.Dutoit V, Rubio-Godoy V, Dietrich PY, Quiqueres AL, Schnuriger V, Rimoldi D, et al. Heterogeneous T-cell response to MAGE-A10(254–262): high avidity-specific cytolytic T lymphocytes show superior antitumor activity. Cancer Res. 2001;61:5850–5856. [PubMed] [Google Scholar]
- 25.Zeh HJ, 3rd, Perry-Lalley D, Dudley ME, Rosenberg SA, Yang JC. High avidity CTLs for two self-antigens demonstrate superior in vitro and in vivo antitumor efficacy. J Immunol. 1999;162:989–994. [PubMed] [Google Scholar]
- 26.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paulson KG, Tegeder A, Willmes C, Iyer JG, Afanasiev OK, Schrama D, et al. Downregulation of MHC-I expression is prevalent but reversible in Merkel cell carcinoma. Cancer Immunol Res. 2014;2:1071–1079. doi: 10.1158/2326-6066.CIR-14-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernandez-Figueras MT, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int J Cancer. 2009;125:1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koelle DM, Corey L, Burke RL, Eisenberg RJ, Cohen GH, Pichyangkura R, et al. Antigenic specificities of human CD4+ T-cell clones recovered from recurrent genital herpes simplex virus type 2 lesions. J Virol. 1994;68:2803–2810. doi: 10.1128/jvi.68.5.2803-2810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen WF, Wilson A, Scollay R, Shortman K. Limit-dilution assay and clonal expansion of all T cells capable of proliferation. J Immunol Methods. 1982;52:307–322. doi: 10.1016/0022-1759(82)90003-5. [DOI] [PubMed] [Google Scholar]
- 31.Choi EM, Chen JL, Wooldridge L, Salio M, Lissina A, Lissin N, et al. High avidity antigen-specific CTL identified by CD8-independent tetramer staining. J Immunol. 2003;171:5116–5123. doi: 10.4049/jimmunol.171.10.5116. [DOI] [PubMed] [Google Scholar]
- 32.Laugel B, van den Berg HA, Gostick E, Cole DK, Wooldridge L, Boulter J, et al. Different T-cell receptor affinity thresholds and CD8 coreceptor dependence govern cytotoxic T lymphocyte activation and tetramer binding properties. J Biol Chem. 2007;282:23799–23810. doi: 10.1074/jbc.M700976200. [DOI] [PubMed] [Google Scholar]
- 33.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods. 2015;12:380–381. doi: 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 36.Webb AI, Dunstone MA, Chen W, Aguilar MI, Chen Q, Jackson H, et al. Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue. J Biol Chem. 2004;279:23438–23446. doi: 10.1074/jbc.M314066200. [DOI] [PubMed] [Google Scholar]
- 37.Paulson KG, Carter JJ, Johnson LG, Cahill KW, Iyer JG, Schrama D, et al. Antibodies to Merkel Cell Polyomavirus T Antigen Oncoproteins Reflect Tumor Burden in Merkel Cell Carcinoma Patients. Cancer Res. 2010;70:8388–8397. doi: 10.1158/0008-5472.CAN-10-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y, Ponomarenko J, Zhu Z, Tamang D, Wang P, Greenbaum J, et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40:W525–530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Draper LM, Kwong ML, Gros A, Stevanovic S, Tran E, Kerkar S, et al. Targeting of HPV-16+ Epithelial Cancer Cells by TCR Gene Engineered T Cells Directed against E6. Clin Cancer Res. 2015;21:4431–4439. doi: 10.1158/1078-0432.CCR-14-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Ye C, Ji G, Han J. Determinants of public T-cell responses. Cell Res. 2012;22:33–42. doi: 10.1038/cr.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang GC, Dash P, McCullers JA, Doherty PC, Thomas PG. T-cell receptor alphabeta diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci Transl Med. 2012;4:128ra142. doi: 10.1126/scitranslmed.3003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Messaoudi I, Guevara Patino JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T-cell avidity, and diversity in immune defense. Science. 2002;298:1797–1800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 44.Peng D, Kryczek I, Nagarsheth N, Zhao L, Wei S, Wang W, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.