Abstract
Nitric Oxide (NO) is a highly-reactive radical gas that can modify a variety of cellular targets in both eukaryotes and bacteria. NO is produced endogenously by a wide variety of organisms: For example, as a cell-signaling molecule in mammals and bacteria via nitric oxide synthase (NOS) enzymes, and as a product of denitrification. As such, it is of great benefit to NO researchers to be able to sensitively detect intracellular NO and stable reactive nitrogen species (RNS) derived from NO. To this end, a protocol for fluorescent detection of intracellular NO/RNS in biofilm cultures of the Gram-positive pathogen Staphylococcus aureus has been optimized using the commercially-available cell-permeable fluorescent stain 4-Amino-5-Methylamino-2’, 7’-Difluorofluorescein Diacetate (DAF-FM diacetate). This compound diffuses into cells and intracellular cleavage by esterase enzymes liberates weakly-fluorescent DAF-FM, which reacts with NO or other specific RNS to become highly fluorescent (Kojima et al., 1999). Although quantification of fluorescence is performed using a fluorescent plate reader, it is envisioned that this protocol could be adapted for intracellular NO/RNS imaging of S. aureus biofilms by confocal microscopy. Likewise, this technique could be optimized for the detection of intracellular NO/RNS in other growth conditions (i.e., planktonic cultures) and/or in other bacteria/archaea.
Materials and Reagents
Wrapping film (Fisher Scientific, Parafilm M™, catalog number: S37440)
Micro-centrifuge tubes (1.7 ml) (Fisher Scientific, catalog number: 14-222-171)
Sterile plastic culture tubes (Fisher Scientific, catalog number: 14-956-6A)
Costar 3524 plates (24-well tissue culture treated) (Fisher Scientific, Corning™ Costar™,catalog number: 07-200-84)
Costar 3904 plates (96-well black tissue-culture treated) (Fisher Scientific, Corning™, catalog number: 07-200-588)
150-ml Nalgene sterile disposable 0.2 μm filter unit (Fisher Scientific, Thermo Scientific™ Nalgene™, catalog number: 09-741-01)
500-ml Nalgene sterile disposable 0.2 μm filter unit (Fisher Scientific, Thermo Scientific™ Nalgene™, catalog number: 09-741-02)
S. aureus stock culture, stored at −80 °C in 25% (v/v) sterile glycerol.
Glycerol (Fisher Scientific, catalog number: G33-1)
Tryptic Soy Agar (TSA) plates (BD, BBL™, catalog number: 221283)
Tryptic Soy Broth (TSB) (BD, Difco™, catalog number: 211825)
Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-1)
Dextrose (Fisher Scientific, catalog number: D16-500)
4-amino-5-methylamino-2’, 7’-difluorofluorescein diacetate (DAF-FM diacetate) (Thermo Fisher Scientific, catalog number: D-23842)
Dimethyl sulfoxide (DMSO) (100 ml) (Sigma-Aldrich, catalog number: 276855)
Human Plasma, lyophilized (5 ml) (Sigma-Aldrich, catalog number: P9523)
Carbonate-Bicarbonate buffer capsules (Sigma-Aldrich, catalog number: C3041)
1× Hank’s buffered salt solution (HBSS) buffer, containing calcium and magnesium (Fisher Scientific, Corning™ cellgro™, catalog number: MT21023CV)
Diethylamine (DEA) (100 ml) (Sigma-Aldrich, catalog number: 471216)
Diethylamine NONOate (DEA/NO) (10 mg vial) (Cayman Chemicals, catalog number: 82100)
2-(4-carboxyphenyl)-4, 5-dihydro-4, 4, 5, 5-tetramethyl-1H-imidazolyl-1-oxy-3-oxide, monopotassium salt (cPTIO) (5 mg vial) (Cayman Chemicals, catalog number: 81540)
Sterile nuclease-free H2O (Fisher Scientific, Invitrogen™ Ambion™, catalog number: AM9930)
NaOH (Thermo Fisher Scientific, catalog number: S318-500)
50% (v/v) Glycerol Stock Solution(see Recipes)
Biofilm Media (see Recipes)
Carbonate-Bicarbonate Buffer (see Recipes)
20% Human Plasma (see Recipes)
DAF-FM diacetate stock solution (see Recipes)
0.01 M NaOH solution (see Recipes)
DEA/NO stock solution (see Recipes)
cPTIO stock solution (see Recipes)
DAF-FM diacetate working solution (see Recipes)
DEA/NO working solution (see Recipes)
DEA working solution (see Recipes)
Equipment
Multi detection microplate reader (Biotek synergy HT, model: SIAFR)
10S Bio UV/Vis Spectrophotometer (Thermo Fisher Scientific, GENESYS™, model: 840-208100)
Plate incubator (VWR International, catalog number: 97058-224, model: 1556)
Shaking incubator (VWR International, Signature™, catalog number: 14004-300, model: 1570)
Pipet-Lite XLS+ single-channel pipettes (2-20 μl, 20-200 μl, 100-1,000 μl) (Rainin, catalog number: 17014406)
Rack LTS tips (“P20” 2-20 μl, “P200” 20-200 μl, “P1000” 100-1,000 μl) (Rainin, Green-Pak™ and SpaceSaver™, catalog number: 17001865, 17001863 and 17001864)
Labconco Class II A2 Biosafety Cabinet (Fisher Scientific, Labconco™Purifier™Logic+™, catalog number: 30-238-1100)
Vortex Mixer (Fisher Scientific, Fisher Scientific™, catalog number: 02-215-365)
Micro-centrifuge (Fisher Scientific, Fisher Scientific™ accuSpin™, catalog number: 13-100-675)
Chemical Fume Hood
Refrigerator (4 °C)
Freezer (−20 °C)
Freezer (−80 °C)
Procedure
General safety notes for entire experiment
When working with BSL-2 organisms such as S. aureus, all potential aerosol-generating steps should be performed in a Class II A2 biosafety cabinet to ensure user safety. An asterick (*) has been placed beside each step below which should be performed in a biosafety cabinet to preserve aseptic technique and user safety.
Please note that although the demonstrations presented in videos 1-4 were filmed at a lab bench (to facilitate clear videography), these steps should be performed under a Class II A2 Biosafety Cabinet when working with BSL-2 bacteria.
Likewise, MSDS sheets for DEA/NO, DEA, and DMSO (available online from the manufacturers) should be read carefully before starting this experiment, and recommended safety precautions be taken to avoid skin exposure and respiratory inhalation as necessary for each chemical. Therefore, perform chemical manipulations as appropriate under a chemical fume hood. Additionally, wear a lab coat, goggles, and nitrile gloves for all steps of the protocol, to comply with both biosafety and chemical safety considerations.
Video 1. Demonstration of biofilm harvesting from 24-well tissue culture plate.
Video 4. Demonstration of sample loading into 96-well plate.
Day 1:
*Streak a TSA plate for isolation from frozen glycerol stock of a S. aureus strain of interest. Incubate in plate incubator at 37 °C for 24 h. In this example protocol the clinical methicillin-susceptible strain UAMS-1 (Gillaspy et al., 1995) was used.
Day 2:
-
2.
*Use a single S. aureus colony (from TSA plate in step 1) to inoculate 3 ml of biofilm media in a sterile plastic culture tube. Grow in shaking incubator at 37 °C, 250 rpm for 16 h.
-
3.
*Add 350 μl 20% (v/v) human plasma solution to 4 wells of a Costar 24-well tissue culture plate. Seal the lid to the plate with wrapping film, and store at 4 °C until use on Day 3.
Day 3:
-
4.
Remove 24 well plate from 4 °C and equilibrate to room temperature.
-
5.
Determine the optical density at 600 nm (OD600) of a 20-fold dilution in sterile TSB of the S. aureus overnight culture using a spectrophotometer. Use sterile TSB as a blank control for the spectrophotometer. Multiply this reading by 20 to calculate the actual OD600/ml of the overnight culture.
-
6.
Calculate how much overnight culture to use to inoculate 1 ml biofilm media to a final OD600/ml = 0.05. Scale-up the volumes as necessary depending on the number of biofilm wells you plan to inoculate, and add 1 ml to account for pipetting error (In this particular sample protocol, 3 biofilm wells will be inoculated, therefore 4 ml of biofilm media containing diluted overnight culture is needed.).
-
7.
Using the P1000 pipette, carefully withdraw all plasma solution from each well of the 24-well plate. Tip the plate at a 15-30° angle and pipette from the corner of each well to ensure complete removal of the plasma solution.
-
8.
*Mix the diluted overnight culture (in this example 4 ml of diluted overnight culture in biofilm medium) by vortexing for 5 sec at top speed, and immediately transfer 1 ml to 3 plasma-coated wells from step 7.
-
9.
*Transfer 1 ml of sterile biofilm media to the forth (empty) plasma-coated well. This will serve as a negative control for aseptic technique.
-
10.
Place 24-well tissue culture plate in 37 °C plate incubator and grow for 7 h.
-
11.
Following growth, visually inspect the negative control well for contamination. The negative control well should look identical to the appearance of sterile media (i.e., no turbid growth or particulate matter should be present). If contamination occurs in the negative control well, do not proceed with the rest of the experiment.
-
12.
*Harvest the total biomass (biofilm + supernatant) from each well by vigorously pipetting, mixing, and scraping the bottom of the well with the P1000 pipette. This step usually takes 30 sec-1 min for each well, depending on the biofilm thickness. Performing work in a sterile environment is not necessary for sample integrity at this and subsequent steps. However, they should be performed under a Biosafety cabinet when working with BSL-2 organisms. Appropriate care should also be used to minimize cross-contamination of samples. Please refer to Video 1 for an example of this step. Transfer contents of each well to a sterile 1.7 ml micro-centrifuge tube.
-
13.
Collect cells by centrifugation at 17,000 × g for 5 min at room temperature.
-
14.
While samples are centrifuging in step 13, dim the lights, remove a 5 μl aliquot of 5 mM DAF-FM diacetate stock solution from the −20 °C freezer, and thaw by vortexing.
Note: DAF-FM is very light sensitive so be sure to perform all subsequent steps with the lights lowered. Dim ambient light (i.e., natural light from windows) is acceptable as long as the DAF-FM is not directly exposed to the light source.
-
15.
Once thawed, prepare the DAF-FM diacetate working solution. The working solution of DAF-FM diacetate should be wrapped in foil until used in step 17 below. This protocol works best if the 5 μM DAF-FM diacetate solution is prepared just prior to use in step 17 below.
-
16.
*Completely remove the culture supernatant from each centrifuged tube (step 13) with the P1000 pipette (Video 2).
-
17.
*Resuspend each cell pellet in 1 ml of 5 μM DAF-FM diacetate (prepared in step 15). S. aureus cell pellets will not resuspend well by vortexing alone, so use the P1000 pipette to break apart the cell pellet by scraping and pipetting (Video 3), then vortex the tube at top speed for 10 sec.
-
18.
Incubate all tubes for 60 min at 37 °C in the plate incubator. Tubes may be covered in foil to reduce exposure to ambient light if this is a concern.
-
19.
While cell suspensions are incubating in step 18, freshly prepare the following solutions: 100 μM DEA in 1× HBSS, 100 μM DEA/NO in 1× HBSS.
-
20.
Collect cell pellets by centrifugation for 5 min, 17,000 × g at room temperature.
-
21.
*Discard supernatants as described in step 16 and Video 2.
-
22.
*To wash residual extracellular stain from cell pellets, resuspend each pellet in 1 ml 1× HBSS buffer as described in step 17 and Video 3.
-
23.
Collect cell pellets by centrifugation for 5 min, 17,000 × g at room temperature.
-
24.
*Discard supernatants as described in step 16 and Video 2.
-
25.*Resuspend each cell pellet (as described in step 17 and Video 3) as follows:
- 1 pellet in 0.65 ml 1× HBSS buffer alone (“Untreated” cells stained with DAF-FM diacetate; baseline level of intracellular NO/RNS)
- 1 pellet in 0.65 ml 1× HBSS containing 100 μM DEA (“DEA” treated cells stained with DAF-FM diacetate; a control for the DEA portion of the chemical NO donor used in step 25c below)
-
1 pellet in 0.65 ml 1× HBSS containing 100 μM DEA/NO (“DEA/NO”-treated cells stained with DAF-FM diacetate; a positive control which should yield high-level intracellular fluorescence relative to the untreated and DEA treated samples)Note: It is important at this step that the sample cell densities are very similar to each other. In our experience, small variations in OD600 (i.e., sample OD600 values that are within 10% of each other) can be accounted for by reporting the data as RFU/OD (as described in step 28 below). However, larger variations in OD600 between samples will lead to inconsistent/difficult to interpret results. It is therefore recommended that the OD600 be checked at this step, and sample volumes adjusted accordingly, when working with different growth conditions or with biological samples that are suspected to grow at different rates during the initial culture incubation (step 10). For S. aureus samples, we typically aim for an OD600 ~1.0 per well. However, we have also observed reproducible results with well OD600 values as high as 2.0, again as long as the OD600 values of all samples across a given experiment are very similar to each other.
-
26.
*Immediately transfer 200 μl aliquots of each cell suspension in triplicate to wells of a Costar 3904 96-well plate (Video 4 and Figure 1). Additionally, transfer 200 μl aliquots of 1× HBSS in triplicate to this 96-well plate (This “buffer only” samples serves as a negative control for any background fluorescence attributable to the buffer itself. 1× HBSS tends to have low auto-fluorescence relative to cell samples.). Plate may be covered in foil to reduce exposure to ambient light at this step if this is a concern.
-
27.
Incubate this 96-well plate in the Biotek Synergy HT fluorescent plate reader. Time-course protocol settings should include the following: 37 °C, 3 sec medium shake prior to each reading, Fluorescence (EX/EM 485 ± 10/516 ± 10) and OD600 measurements recorded every 15 min for up to 60 min total. For S. aureus samples, the peak fluorescence of each sample usually occurs by 30 min.
-
28.
Report data as relative fluorescent units (RFU) per OD600 of each well. Please see (Lewis et al., 2015; Sapp et al., 2014) and Figure 2 for representative data.
Video 2. Demonstration of supernatant removal from centrifuged cell pellet.
Video 3. Demonstration of cell pellet resuspension in DAF-FM diacetate solution.
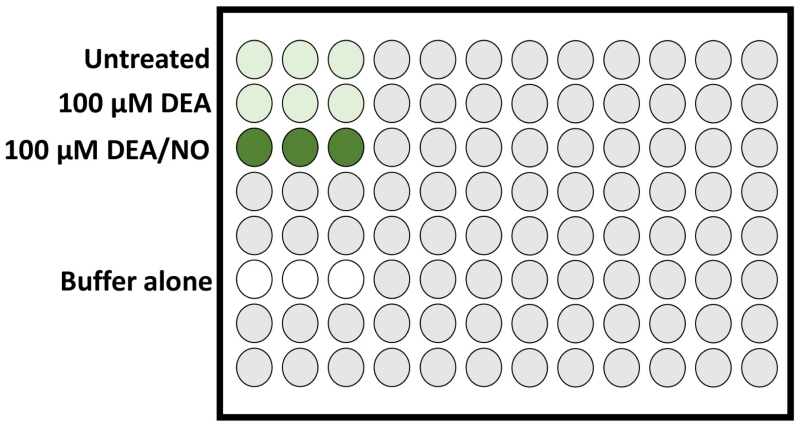
Figure 1. Schematic of DAF-FM stained cell samples loaded in triplicate in a 96-well plate (Step 26).
In this example experiment, higher-level DAF-FM fluorescence is expected in the positive control cell sample treated with DEA/NO (chemical NO donor; dark green wells), whereas detection of endogenous intracellular NO/RNS by DAF-FM fluorescence in the DEA treated and untreated samples (light green wells) are expected to be similar to each other, and both lower than the NO-treated positive control sample. Buffer alone (no DAF-FM stain present; white wells) should have very low levels of auto-fluorescence relative to the DAF-FM stained cells samples.
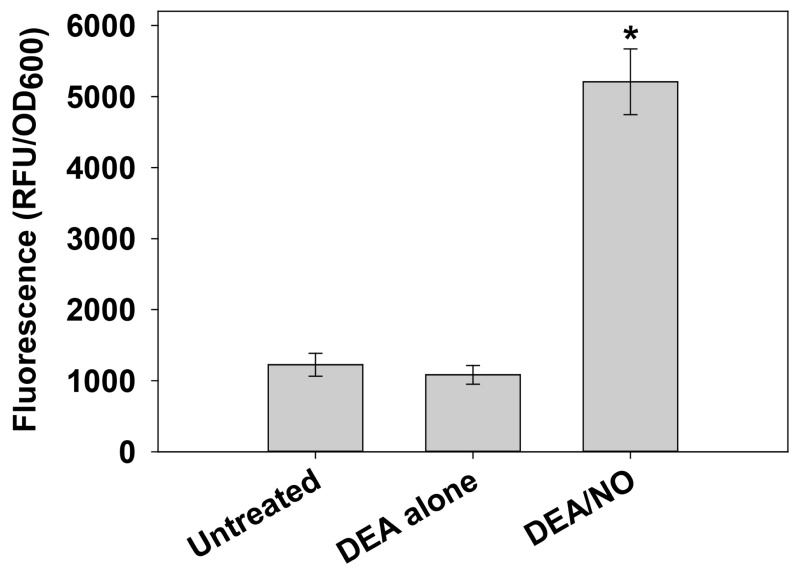
Figure 2. Detection of intracellular NO/RNS in S. aureus with DAF-FM diacetate.
Cells harvested from replicate UAMS-17 h static biofilms were resuspended in 1× HBSS containing 5 μM DAF-FM diacetate. After incubation for 1 h at 37 °C, cells were collected by centrifugation, washed, and resuspended in 1× HBSS alone (“untreated”) or 1× HBSS supplemented with 100 μM DEA or 100 μM DEA/NO. Aliquots (200 μl) of each cell suspension were immediately transferred to a 96-well plate, and incubated at 37 °C in a Synergy HT fluorescent plate reader. Fluorescence and OD600 measurements were recorded after 30 min, and data were reported as relative fluorescent units (RFU) per OD600 of each well. Data represents the average of n=3 independent experiments, error bars = SEM. *statistical significance compared to untreated UAMS-1 (p < 0.05, Tukey Test). The dataset used in this figure was originally published in (Sapp et al., 2014).
Notes
This protocol is most reproducible when analyzing a small set (3-6) of samples per experiment. In our experience, scaling up the experiment to more than 6 samples tends to result in more variability between experiments. Although we have not dissected the reasons for this in detail, it is possible that the increased sample processing time required for more than 6 samples lends to increased variability in the exposure times of the samples to DAF-FM diacetate staining (steps 17-18) and/or to subsequent treatment steps (i.e., exposure of cell samples to DEA/NO as described in step 25), which could in turn influence the timing and level of DAF-FM fluorescence that occurs in the samples in response to NO. It is also possible that increased sample processing times may promote more variability in the exposure of the DAF-FM diacetate to ambient light. Variability between experiments can also minimized by ensuring that other biological variables (i.e., growth time of overnight cultures and biofilms) are kept consistent between experiments.
Experiments should be repeated 3-6 times for adequate power for statistical analysis. In our experience the data tends to follow non-normal distribution, thus non-parametric tests are used to analyze the data for significance.
Although DAF-FM can also react with certain RNS such as nitrosonium ions, the majority of the fluorescent signal has been shown to be due to NO (Kojima, Nakatsubo, et al., 1998, Kojima, Sakurai, et al., 1998). Since the NO radical itself is relatively unstable and may quickly yield other RNS upon exposure to cellular components, intracellular DAF-FM fluorescence should be considered an indirect measurement of NO levels.
As an additional control, 2-(4-carboxyphenyl)-4, 5-dihydro-4, 4, 5, 5-tetramethyl-1H-imidazolyl-1-oxy-3-oxide, monopotassium salt (cPTIO), an NO scavenger, can be added to S. aureus cell suspensions to a concentration of 150 μM during the 1 h DAF-FM diacetate staining step (step 17).
Although this protocol was originally developed to detect intracellular NO in S. aureus biofilm cultures, we have also used it with slight modifications to assess S. aureus intracellular NO levels in cells harvested from planktonic cultures and from agar plates (Sapp et al., 2014). However, when developing this assay for other bacterial or archaeal species, the DAF-FM diacetate concentration used, staining incubation time (step 17), and timing/frequency of fluorescent measurements (step 27) will require careful optimization. Per the manufacturer’s instructions, it is suggested that a concentration range of 1 μM-10 μM DAF-FM diacetate and 15-60 min staining time be initially tested to determine the optimal parameters for the experiment. According to the manufacturer’s protocol, it may take an additional 15-30 min after the initial DAF-FM staining step to allow complete de-esterification of the intra-cellular diacetates and liberation of intracellular DAF-FM. It is also recommended to run the appropriate DEA/NO and cPTIO controls when optimizing this assay. If results under optimized assay conditions are reproducible, these controls may be omitted from future experiments in order to be able to process more “unknown” samples per experiment. As well, the incubation time and/or frequency of RFU/OD600 data collection in the fluorescent plate reader may need to be extended or shortened, depending on the experimental growth conditions, microorganism being tested, and when the earliest “peak” DAF-FM fluorescence occurs.
Recipes
-
50% (v/v) Glycerol Stock Solution
Mix 50 ml of glycerol with 50 ml deionized H2O. Filter-sterilize using a 150-ml Nalgene sterile disposable 0.2 μm filter unit or sterilize by autoclaving (liquid cycle for 30 min).
-
Biofilm Media
Dissolve 3 g TSB, 3 g NaCl, and 0.5 g dextrose in 100 ml deionized H2O. Transfer to a 200 ml glass media bottle and autoclave on liquid cycle for 20-25 min. Prepare freshly-made media for each weekly experiment(s). Biofilm media > 1 week old tends to lead to increased variation in biofilm growth.
-
Carbonate-Bicarbonate Buffer
Empty the contents of one carbonate-bicarbonate capsule into 100 ml of deionized H2O and dissolve. The contents of one capsule yields 100 ml of 0.05 M carbonate-bicarbonate buffer, pH 9.6 at 25 °C. Filter sterilize using a 150-ml Nalgene sterile disposable 0.2 μm filter unit.
-
20% Human Plasma
Add 20 ml carbonate-bicarbonate directly to 5 ml lyophilized human plasma in its original vial. Mix gently for a few minutes to dissolve. This solution will appear as a translucent pale yellow-brown color. Store at 4 °C.
-
DAF-FM diacetate stock solution
Per the manufacturer’s instructions, prepare a 5 mM stock solution of DAF-FM diacetate (MW = 496) by dissolving (vortex to dissolve) the 1 mg packaging (D-23841) in 0.4 ml of high-quality anhydrous DMSO. Once dissolved in DMSO, the DAF-FM diacetate stock solution does not tolerate repeated freeze-thawing. Therefore, aliquot the stock solution into convenient one-time use working volumes (5-10 μl) into sterile 1.7 ml micro-centrifuge tubes pre-chilled on ice. Label tubes and immediately store in a light-tight container at −20 °C.
-
0.01 M NaOH solution
Prepare a 0.01 M NaOH solution by dissolving 200 mg NaOH (FW = 40 g/mol) in 500 ml deionized H2O and dissolve. Filter sterilize using a 500-ml Nalgene sterile disposable 0.2 μm filter unit.
-
DEA/NO stock solution
Per the manufacturer’s instructions, prepare a 100 mM stock solution by dissolving a 10 mg vial of DEA/NO (FW = 206.3) into 323.2 μl of 0.01 M NaOH by vortexing. Once in solution, DEA/NO does not tolerate repeated freeze-thawing. Therefore, immediately aliquot as small working volumes (20-50 μl) into sterile 1.7 ml micro-centrifuge tubes pre-chilled on ice, and store at −80 °C. Do not use DEA/NO stock solution if it has been stored for more than two weeks at −80 °C.
-
cPTIO stock solution
Prepare a 150 mM stock solution of cPTIO (FW = 315.4 g/mol) by dissolving entire 5 mg contents of a cPTIO vial in 105.7 μl sterile nuclease-free H2O. Mix well by vortexing. This should be prepared fresh for each experiment during step 15 of this protocol. Per the manufacturer’s instructions, aqueous solutions of cPTIO are not stable for more than one day.
-
DAF-FM diacetate working solution
Prepare a 5 μM working solution of DAF-FM diacetate by diluting a thawed aliquot of 5 mM DAF-FM stock solution 1000-fold into 1× HBSS buffer. Mix well by vortexing. This should be prepared fresh for each experiment during step 15 of this protocol.
-
DEA/NO working solution
Prepare a 100 μM working solution by adding 3.33 μl DEA/NO stock solution to 5 ml of 1× HBSS. This should be prepared fresh for each experiment during step 19 of this protocol.
-
DEA working solution
First, prepare a 100 mM stock solution of DEA by adding 51.7 μl DEA (0.707 g/ml density, FW = 73.14 g/mol) to 5 ml 1× HBSS. Then, dilute this 1,000-fold in 1× HBSS to a 100 μM working solution. This should be prepared fresh for each experiment during step 19 of this protocol.
Acknowledgments
This work was funded in part by resubmission funding from the University of Florida Emerging Pathogens Institute, a University of Florida IFAS Early Career Award, and NIH grant AI118999, all to KCR. This protocol and the dataset depicted in Figure 2 have both been adapted from (Sapp et al., 2014), and is reproduced here under the Creative Commons Attribution (CC BY) license policy of PLOS ONE.
References
- 1.Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun. 1995;63(9):3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kojima H, Nakatsubo N, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T. Detection and imaging of nitric oxide with novel fluorescent indicators: diaminofluoresceins. Anal Chem. 1998;70(13):2446–2453. doi: 10.1021/ac9801723. [DOI] [PubMed] [Google Scholar]
- 3.Kojima H, Sakurai K, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T. Development of a fluorescent indicator for nitric oxide based on the fluorescein chromophore. Chem Pharm Bull (Tokyo) 1998;46(2):373–375. doi: 10.1248/cpb.46.373. [DOI] [PubMed] [Google Scholar]
- 4.Kojima H, Urano Y, Kikuchi K, Higuchi T, Hirata Y, Nagano T. Fluorescent Indicators for Imaging Nitric Oxide Production. Angew Chem Int Ed Engl. 1999;38(21):3209–3212. doi: 10.1002/(sici)1521-3773(19991102)38:21<3209::aid-anie3209>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 5.Lewis AM, Matzdorf SS, Endres JL, Windham IH, Bayles KW, Rice KC. Examination of the Staphylococcus aureus nitric oxide reductase (saNOR) reveals its contribution to modulating intracellular no levels and cellular respiration. Mol Microbiol. 2015;96:651–69. doi: 10.1111/mmi.12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sapp AM, Mogen AB, Almand EA, Rivera FE, Shaw LN, Richardson AR, Rice KC. Contribution of the nos-pdt operon to virulence phenotypes in methicillin-sensitive Staphylococcus aureus. PLoS One. 2014;9:e108868. doi: 10.1371/journal.pone.0108868. [DOI] [PMC free article] [PubMed] [Google Scholar]