Abstract
To explore potential biomarkers for amoxicillin/clavulanate‐induced liver injury (AC‐DILI), we conducted a clinical trial in 32 healthy subjects based on multi‐omics approaches. Every subject was administered amoxicillin/clavulanate for 14 days. The liver‐specific microRNA‐122 (miR‐122) level increased prior to and correlated well with the observed alanine aminotransferase (ALT) level increase. This result indicates its potential as a sensitive early marker for AC‐DILI. We also identified urinary metabolites, such as azelaic acid and 7‐methylxanthine, with levels that significantly differed among the groups classified by ALT elevation level on day 8 after drug administration (P < 0.05). Lymphocyte proliferation in response to the drug was also observed. These findings demonstrate sequential changes in the process of AC‐DILI, including metabolic changes, increased miR‐122 level, increased liver enzyme activity, and enhanced lymphocyte proliferation after drug administration. In conclusion, this study provides potential biomarkers for AC‐DILI based on currently known mechanisms using comprehensive multi‐omics approaches.
Study Highlights
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
✓Aminotransferases such as ALT and AST are used as gold standards for the evaluation of DILI. However, their elevation may not be specific to liver injury and may not reflect the mechanisms of DILI.
WHAT QUESTION DID THIS STUDY ADDRESS?
✓To identify sensitive biomarkers for AC‐DILI, we evaluated changes in the levels of miR‐122, urinary metabolites, and lymphocyte proliferation both in response to drug administration and with respect to risk genotypes.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE
✓This study identifies potential biomarkers for AC‐DILI based on currently known mechanisms and suggests that drug‐induced hepatocellular injury, oxidative stress, and the adaptive immune response are underlying mechanisms of AC‐DILI.
HOW THIS MIGHT CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE
✓The biomarkers identified in this study by integrating omics data could enable more sensitive and earlier predictions of liver injury. These biomarkers could also provide mechanistic insight that is currently only available from liver biopsies.
Drug‐induced liver injury (DILI) is a major challenge for both clinical care and drug development.1 Monitoring the activity of the enzymes alanine aminotransferase (ALT) and aspartate aminotransferase (AST), which reflect hepatic function, is the primary screening tool for DILI.2 However, elevated levels of these enzymes may not be specific to liver disease and could represent an asymptomatic response.3, 4, 5 Thus, there is a medical need for biomarkers that are more sensitive and specific for the early detection of DILI, and the rapidly evolving and high‐throughput “‐omics” technologies have been applied to identify prognostic biomarkers of DILI.
Pharmacogenomic studies have demonstrated specific human leukocyte antigen (HLA) alleles that are associated with DILI that is caused by the following drugs: flucloxacillin (B*57:01), ximelagatran (DRB1*07:01 and HLA‐DQA1*02), co‐amoxiclav (DRB1*15:01), and antituberculosis drugs (HLA‐DQB1*0502).6, 7, 8, 9 Glutathione s‐transferase (GST) is an essential phase II metabolic enzyme that is related to drug detoxification, and the mu‐1 and theta‐1 (GSTM1 and GSTT1) null genotypes have also been identified as genetic risk factors for DILI.6, 10
Numerous studies have identified microRNAs, which are small, single‐stranded noncoding regulatory RNA molecules, as possible sensitive biomarkers for DILI.11, 12, 13, 14 Notably, microRNA‐122 (miR‐122) is elevated earlier and demonstrates increased sensitivity in patients with acetaminophen‐induced liver injury compared with ALT.11, 15 Although the mechanism of miR‐122 elevation is not yet known, hepatocyte damage may induce the release of cellular miR‐122 into the circulation, which leads to increased miR‐122 levels in the peripheral blood.15 In a recent study, differences in the circulating serum levels of exosomal miR‐122 were observed between hepatocyte injury and inflammation.12
Pharmacometabolomic approaches have been used to identify novel DILI biomarkers.16, 17 The results of a clinical trial that was performed to analyze the pharmacometabolomics patterns in urine samples from healthy subjects before and after the administration of acetaminophen indicated that N‐acetyl‐p‐benzoquinone imine may be a useful biomarker of DILI.18 Another clinical study demonstrated that acetaminophen‐induced DILI was inversely correlated with p‐cresol sulfate, which competes with acetaminophen entry into the portal circulation from the liver.19
Amoxicillin/clavulanate was selected as study drug in this study because it is the frequently prescribed antibiotic worldwide and is often associated with hepatotoxicity.20, 21 The delayed nature and HLA association of DILI indicate the involvement of the adaptive immune system, and immunological idiosyncrasies have been proposed as a possible mechanism for amoxicillin/clavulanate‐induced DILI (AC‐DILI).8, 22, 23 In a recent study, drug‐responsive T cells that were specific for either amoxicillin or clavulanate were isolated from a patient with AC‐DILI.24 The mechanism that underlies AC‐DILI has not yet been clearly identified; however, the activation of T cells by xenobiotics is likely to mediate the immune response that is involved in DILI.
Although several promising biomarkers have been identified in patients with DILI, including HLA,25 miR‐122,26 and endogenous metabolites,27 well‐controlled prospective clinical trials have not been conducted to validate these biomarkers. Based on the multidirectional approaches described above, we evaluated potential biomarkers of DILI in healthy subjects after multiple administrations of amoxicillin/clavulanate.
MATERIALS AND METHODS
Study design
A total of 32 healthy Korean male volunteers were enrolled and grouped according to four GSTT1/M1 genotypes (eight subjects per group): wild/wild, null/wild, wild/null, and null/null. Subjects were included in this study if they were in good health as indicated by their previous medical history, physical examination, vital signs, 12‐lead electrocardiography (ECG), serology, and urinary drug screening. Subjects who had used any prescription drugs, including antibiotics such as amoxicillin/clavulanate or herbals agents, within 2 weeks before the drug administration of the study were excluded. (ClinicalTrials.gov registry number: NCT02143323).
This study had an open‐label, single‐sequence, three‐period design. The workflow of the study design is shown in Figure 1. Each subject received three tablets of amoxicillin/clavulanate (375/125 mg) twice daily for 14 days. Blood and urine samples were obtained for laboratory tests, pharmacokinetic (PK) evaluations, and multi‐omics analyses such as miRNA‐122, pharmacometabolites, HLA typing, and LTT.
Figure 1.

Study design. GSTT1/M1, blood collection for genotyping of glutathione s‐transferase mu‐1/theta‐1; LTT, blood collection for lymphocyte transformation test; HLA, blood collection for genotyping human leukocyte antigen (HLA); PK, blood collection for pharmacokinetic analyses of amoxicillin and clavulanate; Metabolomics, 12‐h interval urine collection for metabolomics analysis; miRNA, blood collection for microRNA analyses.
The Institutional Review Board of Seoul National University Hospital, Seoul, Korea, approved this clinical study, which was conducted in compliance with the ethical principles of the Declaration of Helsinki. All of the subjects provided written informed consent before being screened for eligibility.
PK analyses
The plasma concentrations of amoxicillin and clavulanate were determined using validated high‐performance liquid chromatography‐tandem mass spectrometry with an Agilent 1260 series chromatography system (Agilent, Santa Clara, CA) coupled to an API 4000 mass spectrometer (Sciex, Toronto, Canada). Chromatographic separation of amoxicillin was conducted using a Synergi 4U HydroRP column (100 × 2.0 mm, 4 μm) (Phenomenex, Torrance, CA). A Luna column (100 × 2.0 mm, 5 μm) (Phenomenex) was used for the chromatographic separation of clavulanate. The lower limit of quantitation was 10 ng/mL for both amoxicillin and clavulanate. The intraday and interday precision data are described in Supplementary Table S1.
The PK parameters were derived using noncompartmental analyses in Phoenix WinNonlin (v. 6.3., Certara, St. Louis, MO). The Cmax (maximum plasma concentration at steady state), AUC0‐12h (area under the plasma concentration–time curves from zero to 12 h after a single administration), and AUCss,τ (area under the plasma concentration–time curves from zero to 12 h at steady state) were calculated using the linear‐up and log‐down method.
GSTT1/GSTM1 and HLA typing
A multiplex polymerase chain reaction method was used to simultaneously analyze the GSTM1 and GSTT1 genotypes using blood samples that were obtained during screening (DNA Link, Seoul, Korea).42 The primers and specific methods are available in the Supplemental Methods.
High‐resolution sequence‐based HLA typing was performed at the following loci: HLA‐A, HLA‐B, HLA‐C, HLA‐DRB1, HLA‐DQB1, and HLA‐DPB1 (Histogenetics, Ossining, NY). The HLA types of the study subjects are shown in Supplementary Table S2.
Measurement of serum miRNAs
Total RNA was extracted for quantitative analyses of serum microRNAs using a miRNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. A total volume of 100 μL of serum was used for the RNA extraction. A synthetic miRNA from Caenorhabditis elegans was included in the sample after homogenization in QIAzol Lysis Reagent (Qiagen) to validate the RNA extraction efficiency. Complementary DNA was prepared using a miScript Reverse Transcription Kit II (Qiagen). The 20 μL PCR reaction contained 10 μL of miScript SYBR Green PCR Master Mix (Qiagen), 4 μL of nuclease‐free water, 2 μL of 10× miScript primer assay, 2 μL of 10X miScript Universal Primer, and 2 μL of cDNA template. Amplification was performed using a CFX96 Real‐Time System (Bio‐Rad, Hercules, CA) under the following conditions: initial denaturation at 95ºC for 15 min followed by 40 cycles of denaturation for 15 s at 94ºC, annealing for 30 s at 55ºC, and elongation for 30 s at 70ºC. A total exosome isolation reagent (Invitrogen, Carlsbad, CA) was used for exosome isolation from serum according to the manufacturer's instructions.
Pharmacometabolomic analyses
Untargeted metabolomics profiling was performed using urine samples that were collected at 12‐h intervals before and after the administration of the study drug. The collected urine samples were centrifuged at 14,000 rpm for 20 min at 4ºC to remove any solid debris, and the supernatant was diluted with distilled water. A 4‐μL aliquot of the prepared urine sample was injected into a 2.1 × 100 mm ACQUITY 1.8 μm HSS T3 column using an ACQUITY UPLC system (Waters, Milford, MA) coupled to a Waters Xevo Q‐TOF. The gradient mobile phase condition consisted of phase A (water with 0.1% formic acid) and phase B (methanol containing 0.1% formic acid). Each sample was resolved for 20 min at a flow rate of 0.4 mL/min under the following gradient conditions: 0–1 min, 5% B; 1–4 min, 5–20% B; 4–7.5 min, 20–60% B; 7.5–11.5 min, 60–95% B; 11.5–15.5 min, 95% B; 15.5–16.2 min, 95‐5% B; and 16.2–20 min, 5% B. The data were collected in centroid mode over the range of 50–1000 m/z with a scan time of 0.4 s.
The multivariate data were analyzed using the EZinfo software (Waters). The stability and reproducibility of the unsupervised principal components analysis were checked using the pareto‐scaled data by clustering quality controls (QCs).
Lymphocyte transformation test (LTT)
Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation from blood that was collected on day 1 before the study drug administration, day 20–22, and day 60. The proliferation of PBMCs (0.15 × 106 cells) in response to amoxicillin (0.25–2 mM), clavulanate (0.0625–0.5 mM), and PHA (5 μg/mL; positive control) was measured using the LTT as previously described.24 The proliferative response was calculated as a stimulation index (SI; cpm in drug‐treated cells/cpm in mock‐treated cells).
PBMCs secreting interferon‐gamma were visualized using ELISpot (MabTech, Nacka Strand, Sweden) by culturing the PBMCs with each drug for 48 h.
Statistical analysis was done with SPSS (v. 23.0; IBM, Armonk, NY)
The arithmetic means, standard deviations (SDs), medians, and maximum and minimum values of the continuous data were calculated. The subjects were classified into subgroups based on the extent of the ALT elevations, HLA and GSTT1/M1 genotypes, and LTT results to evaluate the associations between the biomarkers. The genotypic frequencies of the GSTM1 and T1 polymorphic variants and the groups that were classified by ALT elevation were compared. The LTT results were also used to classify subjects as positive type, recovery type, or negative type using a cutoff value of 3 for the stimulation index (SI).43 An analysis of variance was conducted to compare the maximum ALT change among the groups.
A t‐test was conducted to compare the changes in ALT and endogenous metabolites between the groups. The correlations between the potential biomarkers and liver function parameters were evaluated using Pearson's correlation test.
RESULTS
Study subject demographics and group classification
A total of 31 subjects completed the study. The mean age was 30 years, and all of the subjects were male. The means ± standard deviations (SDs) of the weights and heights of the subjects were 71.9 ± 7.5 kg and 174.3 ± 5.2 cm, respectively. One subject (AN103) dropped out because of adverse events, including headache and vomiting on day 4. A total of 48 adverse events were reported in 24 subjects during the entire study period. No serious adverse events occurred, and the most frequent adverse events were nausea, dyspepsia, and diarrhea, which have commonly been associated with the use of amoxicillin and clavulanate. The demographics, biochemical variables, and lymphocyte transformation test (LTT) results are provided in Table 1.
Table 1.
Study demographics of all subjects
| Demographics | Biochemical variables | LTT a results | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Peak levels fold change | ||||||||||
| ID | GSTT/GSTM | Age (yr) | Weight (kg) | Height (cm) | T. bilirubin | ALT | AST | ALP | Type | Drug |
| AN001 | Wild/Wild | 27 | 67.2 | 171.3 | 1.57 | 1.31 | 1.15 | 1.04 | Nc | |
| AN002 | 25 | 71.5 | 177.8 | 1.33 | 1.15 | 1.09 | 1.10 | N | ||
| AN003 | 32 | 71.9 | 167.5 | 1.25 | 1.43 | 1.42 | 1.24 | N | ||
| AN004 | 40 | 71.3 | 170.6 | 1.00 | 3.29 | 4.26 | 1.23 | Pb | Ce | |
| AN005 | 30 | 77.5 | 179.4 | 1.10 | 1.38 | 1.13 | 1.11 | N | ||
| AN006 | 30 | 73.3 | 180.4 | 0.90 | 1.73 | 1.73 | 1.14 | Rd | A | |
| AN007 | 28 | 71.9 | 178.1 | 1.31 | 1.36 | 1.09 | 1.08 | P | ||
| AN008 | 26 | 69.0 | 174.8 | 1.71 | 2.25 | 2.21 | 1.07 | R | A | |
| AN101 | Wild/Null | 31 | 77.6 | 177.7 | 1.00 | 3.89 | 2.17 | 1.23 | N | |
| AN102 | 30 | 80.0 | 178.5 | 1.11 | 1.42 | 1.22 | 1.05 | R | A | |
| AN103* | 20 | 82.6 | 178.6 | 1.60 | 1.27 | 1.19 | 1.23 | NA | ||
| AN104 | 26 | 75.9 | 174.7 | 1.18 | 1.61 | 1.29 | 1.14 | N | ||
| AN105 | 30 | 73.8 | 171.3 | 1.10 | 1.68 | 1.16 | 1.10 | N | ||
| AN106 | 32 | 82.5 | 182.5 | 1.22 | 2.00 | 1.00 | 1.12 | N | ||
| AN107 | 29 | 59.9 | 175.7 | 1.20 | 1.54 | 1.82 | 1.14 | N | ||
| AN108 | 35 | 64.1 | 171.1 | 1.33 | 0.88 | 0.94 | 1.00 | N | ||
| AN201 | Null/Wild | 37 | 69.3 | 173.1 | 1.80 | 1.38 | 1.18 | 1.23 | N | |
| AN202 | 23 | 88.4 | 183.4 | 1.00 | 2.33 | 1.38 | 1.29 | N | ||
| AN203 | 39 | 72.5 | 176.5 | 0.83 | 3.00 | 1.67 | 1.02 | P | C, Af | |
| AN204 | 36 | 71.4 | 167.4 | 1.25 | 1.08 | 1.09 | 1.18 | N | ||
| AN205 | 26 | 65.2 | 168.1 | 1.50 | 1.00 | 1.13 | 1.14 | N | ||
| AN206 | 30 | 59.5 | 168.4 | 1.00 | 3.00 | 1.86 | 1.03 | N | ||
| AN207 | 27 | 62.1 | 173.4 | 1.25 | 0.75 | 1.12 | 1.17 | R | A | |
| AN208 | 40 | 60.0 | 161.5 | 1.50 | 3.00 | 2.79 | 1.25 | N | ||
| AN301 | Null/Null | 38 | 62.9 | 168.2 | 1.33 | 0.91 | 1.50 | 1.10 | N | |
| AN302 | 25 | 81.5 | 178.7 | 1.30 | 1.17 | 1.10 | 1.14 | N | ||
| AN303 | 30 | 77.3 | 172.6 | 1.60 | 2.92 | 1.64 | 1.13 | N | ||
| AN304 | 26 | 68.2 | 172.2 | 1.10 | 1.64 | 1.16 | 1.08 | N | ||
| AN305 | 32 | 65.3 | 171.7 | 1.11 | 1.23 | 1.38 | 1.08 | N | ||
| AN306 | 32 | 83.3 | 179.8 | 1.10 | 1.00 | 0.92 | 1.04 | N | ||
| AN307 | 23 | 71.8 | 174.8 | 1.70 | 2.00 | 4.00 | 1.11 | N | ||
| AN308 | 25 | 81.6 | 182.8 | 1.00 | 1.45 | 1.24 | 1.01 | N | ||
LTT, Lymphocyte transformation test;bP, positive type (stimulation index > 3.0);cN, negative type (stimulation index ≤ 2.0);dR, recovery type (stimulation index > 3.0 at day 20–22 but decreased below 3.0 at day 60);eC, clavulanate;fA, amoxicillin *Dropped out due to adverse events (headache, nausea). NA, not applicable.
Based on the extent of their ALT elevations relative to the baseline, the subjects were classified into three groups (Figure 2): (1) Responder (n = 6), who had greater than a twofold change in ALT at more than two sampling points; (2) Nonresponder (n = 17), who had less than a 1.5‐fold change in ALT at all of the sampling points; and (3) Intermediate (n = 8), who had a 1.5‐ to 2.0‐fold change in ALT at all of the sampling points. Most of the ALT levels were within the normal range (0–40 IU/L). However, in 19.35% (6/31) of the subjects, the ALT levels after drug administration showed more than twofold changes above the baseline values (i.e., ALT before drug administration on day 1) at two or more sampling points.
Figure 2.
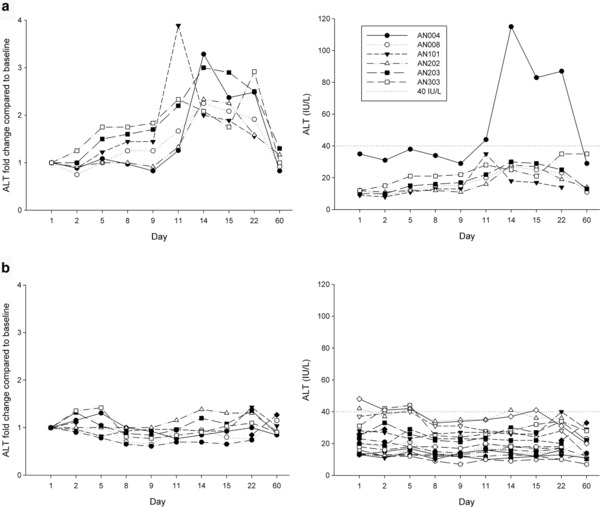
ALT fold changes relative to baseline levels (IU/L) in the subjects over the course of the 60‐day study in the (a) Responder group (left: ALT fold change, right: ALT levels) and the (b) Nonresponder group (left: ALT fold change, right: ALT levels). The baseline ALT levels were defined as the value of the first day predose sample.
Correlation of PK parameters with ALT elevations
Comparative analyses between the Responder and Nonresponder groups revealed no statistically significant differences in the primary PK parameters of amoxicillin or clavulanate, including AUC0‐12h, AUClast,ss, and Cmax (Supplementary Table S3), which indicates that the nature of idiosyncratic DILI that is induced by amoxicillin and clavulanate may not be related to the dose or the drug pharmacokinetics. There were also no significant differences in the mean maximum ALT fold change among the groups that were classified by GSTT1/GSTM1 genotype (Supplementary Figure S1).
Correlation between miR‐122 and ALT elevation
We measured the serum levels of miR‐122, which has been proposed as an alternative biomarker of DILI. After drug administration, the miR‐122 levels exhibited a similar increasing trend to that of the ALT levels (Pearson's r of 0.613–0.923, P < 0.05) but to a larger extent. Figure 3 shows these results for both the Responder and Nonresponder groups. The miR‐122 levels tended to increase on day 8, which was earlier than the increase in the ALT levels that were first noted on day 14. We found significant correlations between the serum miR‐122 and ALT fold changes (Pearson's r of 0.613–0.923, P < 0.05, Supplementary Table S4).
Figure 3.
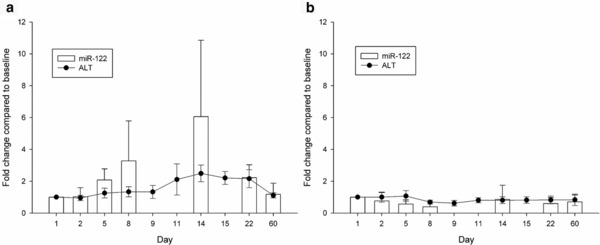
Time courses depicting the serum miR‐122 and ALT fold changes in the (a) Responder and (b) Nonresponder groups. The data represent the mean ± standard deviation.
Pharmacometabolomic analyses
A total of 5,169 peaks in electrospray ionization in the positive ion mode (ESI+) and 4261 peaks in the negative ion mode (ESI‐) were detected after peak alignment and were subsequently imported into the EZinfo software. According to the OPLS‐DA model (Supplementary Figure S2), the concentrations of 5 urinary metabolites (variable influence on the projection > 5) were significantly different in the Responder and Nonresponder groups (Table 2). The identified urinary markers 7‐methylxanthine (7MX), 7‐methyluric acid (7MU), 3‐methylxanthine (3MX), acetylcarnitine (ACar), and azelaic acid (AzA) were quantified using QuanLynx (Waters) and normalized to the creatinine concentration.
Table 2.
Urinary metabolites that were significantly different between the Responder and Nonresponder groups at day 8 after amoxicillin/clavulanate administration
| Responder | Nonresponder | |||||||
|---|---|---|---|---|---|---|---|---|
| Metabolite | VIP | Baseline | Day 8 | Day 14 | Baseline | Day 8 | Day 14 | Ratio of metabolites |
| Azelaic acid | 5.68 | 1,227.9 ± 468.4 | 530.3 ± 409.3 | 971.5 ± 762.1 | 1,415.2 ± 1,298.1 | 1,191.7 ± 706.2 | 1,084.9 ± 983.6 | 0.4* |
| 7‐Methylxanthine | 11.78 | 5,020.6 ± 7,165.8 | 12,159.2 ± 6,520.7 | 8,352.7 ± 6,863.8 | 2,441.2 ± 2,203.0 | 3,703.3 ± 4,612.0 | 4,277.7 ± 5,458.8 | 3.3* |
| 3‐Methylxanthine | 7.67 | 1,631.5 ± 955.3 | 2,802.6 ± 1,096.9 | 2,343.3 ± 1,691.5 | 968.0 ± 876.2 | 1,137.1 ± 1,226.6 | 1,494.5 ± 2,132.8 | 2.5* |
| 7‐Methyluric acid | 8.71 | 944.5 ± 076.4 | 2,759.1 ± 1,799.7 | 2,489.4 ± 2,122.3 | 650.3 ± 681.0 | 712.3 ± 905.9 | 897.0 ± 1,176.2 | 3.9* |
| Acetylcarnitine | 5.93 | 5,490.0 ± 221.6 | 2,570.5 ± 1,654.6 | 6,299.0 ± 4,098.6 | 5,593.9 ± 4,417.5 | 7,724.3 ± 9,887.3 | 6,502.1 ± 10,705.5 | 0.3 |
*
P < 0.05 by t‐test statistics using an independent two‐sample t‐test between the Responder and Nonresponder averages on day 8; Ratio of endogenous metabolites, ratio for a given metabolite (Responder/Nonresponder) assayed on day 8; VIP, variable influence on the projection.
Four of the urinary metabolites showed significant changes between days 1 and 8 after amoxicillin/clavulanate administration in the Responder group but not in the Nonresponder group (Figure 4). Although the urinary metabolite levels showed large interindividual variations, the levels of 7MX, 7MU, and 3MX in the Responder group on day 8 were significantly higher (3.3, 3.9, and 2.5 times, respectively) than in the Nonresponder group, whereas the AzA level was significantly lower (0.4 times; Figure 4).
Figure 4.
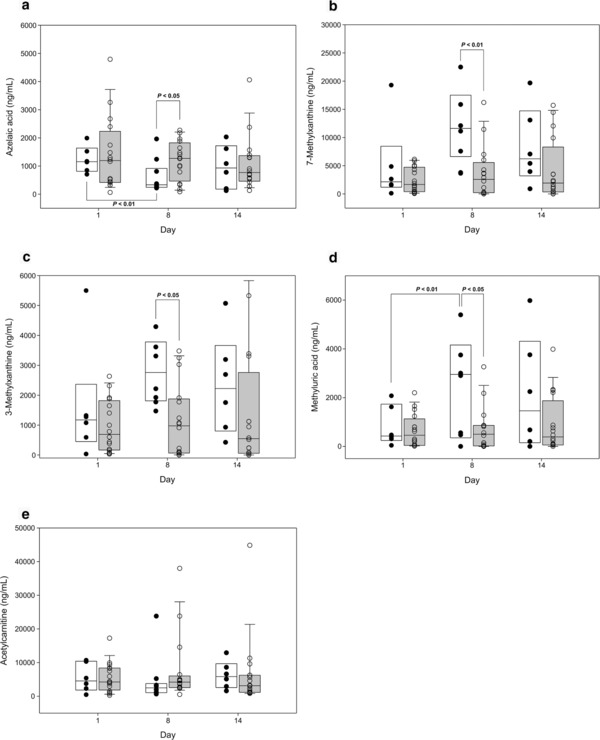
Box‐whisker plots for significantly regulated endogenous metabolites. (a) Azelaic acid, (b) 7‐methyluric acid, (c) 3‐methylxanthine, (d) 7‐methylxanthine, and (e) acetylcarnitine on days 1, 8, and 14 (white box: Responder; gray box: Nonresponder). The box plot shows the median (line) and 25–75% interquartile range for endogenous metabolites. Circles represent outliers.
Lymphocyte proliferation against amoxicillin and clavulanate
The LTT was performed on three different visits (day 1, days 20–22, and day 60) to determine the lymphocyte response against amoxicillin and clavulanate. Based on the LTT results, for which stimulation index values above 3 were considered positive, amoxicillin appeared to transiently induce lymphocyte proliferation at 20–22 days after administration in four subjects (AN006, AN008, AN102, AN207), although the proliferation levels had returned to a normal range on day 60. After clavulanate stimulation, a strong proliferative response was observed in two subjects (AN004 and AN203) on day 60. The lymphocyte response of one subject (AN004) appeared to follow a similar process to that of a patient with AC‐DILI (Figure 5), with a very strong proliferative response to clavulanate and enhanced interferon gamma (IFN‐γ) release from lymphocytes after clavulanate stimulation. The biomarkers showed that there were metabolic changes on day 8, increased miR‐122 levels on day 9, increased enzyme activity, and then an enhanced proliferative response to clavulanate.
Figure 5.

Time courses depicting changes in (a) urinary metabolites, (b) miR‐122, (c) ALT and AST, (d) lymphocyte proliferation, and (e) INF‐γ release in subject AN004.
DISCUSSION
Given the heterogeneous and complex nature of DILI, we used multidirectional approaches to identify and evaluate biomarkers in a well‐controlled clinical trial in healthy subjects.
Consistent with previous findings,11 the liver‐specific miR‐122 response was highly correlated with ALT changes. Our results demonstrated that the miR‐122 levels changed more dramatically than ALT, which indicates that miR‐122 is more sensitive to liver injury. The miR‐122 also began to increase earlier than ALT, which demonstrates the potential of miR‐122 as an early marker of AC‐DILI.
We used a pharmacogenomics approach combined with immunologic concepts to evaluate GSTT1/M1 genotypes and HLA types as risk factors for DILI. We hypothesized that there would be a higher incidence of ALT elevation in subjects with the GSTT1/M1 null/null genotype28 or with specific HLA types that are known risk factors for DILI.28 However, we did not find an association between ALT changes and these genotypes. This result may have been related to the limited sample size or to slight elevations in ALT.
We also used a metabolomics platform to investigate the metabolomic signatures of amoxicillin/clavulanate exposure in healthy volunteers as predictors of AC‐DILI. Although the levels of urinary metabolites showed large interindividual variations, we identified four urinary markers with the potential to predict ALT elevation: AzA was significantly downregulated, whereas 7MX, 3MX, and 7MU were significantly upregulated.
AzA is known to scavenge radicals and modulate the inflammatory response in human keratinocytes.29 A recent study in mice demonstrated that exposure to 2,3,7,8‐tetrachlorodibenzo‐p‐dioxin (TCDD) induced oxidative stress in the liver and decreased AzA levels.30 The AzA levels changed even with low doses of TCDD, which did not significantly increase the serum ALT or AST levels. Therefore, AzA was suggested to be an early indicator of liver damage that is related to mitochondrial oxidative stress. Interestingly, 7MX also appears to be related to oxidative stress. It is metabolized in the liver by xanthine oxidase to yield 7MU, which generates reactive oxygen species (ROS) in the liver,31 which indicates that these metabolites are involved in drug‐induced oxidative stress in the liver.32, 33 The metabolomics results thus support the conclusion that mitochondrial oxidative stress could be another mechanism that underlies AC‐DILI.
The immune basis of AC‐DILI has been supported by the delayed onset, individual susceptibility, HLA association, and presence of drug‐specific T cells in patients with AC‐DILI.23, 24 The technologically demanding LTT assay was used here as an in vitro testing method for detecting drug‐responsive T cells.34 As expected, because of the low incidence of DILI, we observed that a small number of subjects showed a positive response to the drug. The lymphocyte response to amoxicillin appears to be transient and unrelated to changes in ALT, miR‐122, or other metabolic markers, which indicates that it may be an asymptomatic response. However, the positive lymphocyte response to clavulanate in one subject (AN004) appears to be related to AC‐DILI because we found a similar sequence of metabolic, biochemical, and biological changes with early metabolic changes detected on day 8 and increased miR‐122 levels on day 9 followed by increased enzyme activity (ALT/AST), and finally by an enhanced proliferative response to clavulanate. Overall, this finding indicates that one biomarker alone cannot distinguish between the various etiologies of DILI, which supports the need for multi‐omics approaches to identify a prognostic biomarker for DILI.
We also attempted to define the DILI mechanism by examining the levels of circulating miR‐122. MicroRNAs can circulate in the blood either bound to proteins (e.g., argonaute, lipoprotein) or packaged in extracellular vesicles such as exosomes and microvesicles.35, 36 A recent study reported that circulating miRNAs are associated with either the exosome‐rich or protein‐rich fraction, depending on the type of liver injury.12, 37 Circulating miR‐122 in the exosome‐rich fraction is more related to inflammation. On the other hand, miR‐122 in protein‐rich fraction is known to be associated with hepatocyte injury induced by necrosis.12 Our results showed that miR‐122 increased by 7.2‐ and 5.2‐fold in the exosome‐rich and protein‐rich fractions, respectively (Supplementary Figure S3), which indicates that amoxicillin and clavulanate induce a mixed‐type liver injury.
A major limitation of this study is the relatively small number of subjects, which decreases the statistical power. Another limitation is in the study subjects themselves. In general, DILI occurs more frequently in elders and females,20 while this study was performed with healthy male volunteers, not with DILI patients. Realistically, applying amoxicillin/clavulanate to relatively vulnerable groups, such as elders and females, may be problematic because of subject safety and ethical issues. It has also been reported that while the frequency of DILI differs depending on gender and age, the manifestations of DILI do not differ significantly.38 Considering these factors, we assumed that potential biomarkers that are identified in males could represent those present in all populations. Finally, our study was limited by the fact that the levels of transaminases such as ALT, which we used as an evaluation marker, may change depending on diet or alcohol intake.39, 40 However, we limited alcohol consumption during the study period by performing alcohol air breathing tests upon hospitalization to exclude the possibility of ALT elevation by alcohol. We also attempted to minimize interfering factors other than drugs that could affect liver function by limiting the intake of any prescribed drugs and high‐carbonate or high‐lipid diets beginning 2 weeks before the test.
Nevertheless, to the best of our knowledge, this study is the first prospective clinical trial that has been performed on healthy subjects with antibiotic‐induced DILI that is based on multi‐omics tools. Previously, DILI biomarker explorations have typically been retrospective studies on DILI patients. Considering this, the significance of this study is the detection of altered biomarker levels upon slight changes in liver function under well‐controlled normal conditions or subclinical conditions.41 However, a control group of either the same subjects or different subjects not taking the antibiotic would have added validity to changes in transaminases.
In conclusion, we evaluated and identified potential DILI biomarkers based on currently known AC‐DILI mechanisms. Our results demonstrate that drug‐induced hepatocellular injury, oxidative stress, and adaptive immune response may represent the main mechanisms that underlie DILI. Further confirmatory studies that demonstrate and validate the clinical benefits of these markers for use in bedside applications should be performed.
Supporting information
Supplementary Table S1. Accuracy and precision of (a) amoxicillin and (b) clavulanic acid concentration measurements using liquid chromatography‐tandem mass spectrometry (n = 5).
Supplementary Table S2. The HLA types of the study subjects.
Supplementary Table S3. PK parameters (Cmax, AUC0‐12h, and AUCss,tau) of amoxicillin and clavulanate in the Responder and Nonresponder groups.
Supplementary Table S4. Correlations between ALT changes and microRNA‐122 (miR‐122) changes throughout the study.
Supplementary Figure S1. Mean of maximum ALT fold change classified by GSTT1/M1 genotype. GSTT, glutathione s‐transferase T; GSTM, glutathione s‐transferase mu; W/W, wild/wild; W/N, wild/null; N/W, null/wild; N/N, null/null.
Supplementary Figure S2. S‐plot corresponding to the OPLS‐DA analysis for the new model built on samples in (A) positive ion mode and (B) negative ion mode.
Supplementary Figure S3. Changes in miR‐122 levels in exosome‐rich and protein‐rich fractions separated from Nonresponder serum (a) and Responder serum (b). In the Responder group, miR‐122 levels massively increased in both fractions at peak ALT time points. However, in the Nonresponder group, miR‐122 levels in both fractions were unchanged at peak ALT time points. The data represent the means ± standard deviation.
Acknowledgments
We thank Jingshun Li and Soyoung Kim for coordinating the clinical trials. We acknowledge Min Chang Kim and Jaejin Lee for the quantitative analyses of the study drug. ClinicalTrials.gov registry number:NCT02143323
Author Contributions
I‐J.J., J.L., S.Y., J‐Y.C., and S‐H.K. wrote the manuscript; I‐J.J., J.L., S.C.J., K‐H.S., J‐Y.C., K.S.L., S.L., J‐Y.C., and K‐S.Y. designed the research; I‐J.J., J.L., S.C.J., B.K., S.Y., K.S.L., and H‐S.P. performed the research; J.L., S.C.J., B.K., K‐H.S., S.L., S.H.Y., K‐S.Y., and H‐S.P. analyzed the data; S.C.J., J‐Y.C., S.H.Y., and S‐H.K. contributed new reagents/analytical tools.
Conflict of Interest
The authors declared no conflict of interest.
Financial Support
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, which is funded by the Ministry of Health & Welfare, Republic of Korea (HI14C0065 and HI14C2770).
Contributor Information
SH Kim, Email: kimsh@ajou.ac.kr.
IJ Jang, Email: ijjang@snu.ac.kr.
References
- 1. Qureshi, Z.P. , Seoane‐Vazquez, E. , Rodriguez‐Monguio, R. , Stevenson, K.B. & Szeinbach, S.L. Market withdrawal of new molecular entities approved in the United States from 1980 to 2009. Pharmacoepidemiol. Drug Saf. 20, 772–777 (2011). [DOI] [PubMed] [Google Scholar]
- 2. Regev, A. & Bjornsson, E.S. Drug‐induced liver injury: morbidity, mortality, and Hy's law. Gastroenterology 147, 20–24 (2014). [DOI] [PubMed] [Google Scholar]
- 3. Kong, A.P. et al Independent associations of alanine aminotransferase (ALT) levels with cardiovascular risk factor clustering in Chinese adolescents. J. Hepatol. 49, 115–122 (2008). [DOI] [PubMed] [Google Scholar]
- 4. Lee, J.K. et al Estimation of the healthy upper limits for serum alanine aminotransferase in Asian populations with normal liver histology. Hepatology 51, 1577–1583 (2010). [DOI] [PubMed] [Google Scholar]
- 5. Dufour, D.R. Alanine aminotransferase: is it healthy to be “normal”? Hepatology 50, 1699–1701 (2009). [DOI] [PubMed] [Google Scholar]
- 6. Daly, A.K. et al HLA‐B*5701 genotype is a major determinant of drug‐induced liver injury due to flucloxacillin. Nat. Genet. 41, 816–819 (2009). [DOI] [PubMed] [Google Scholar]
- 7. Kindmark, A. et al Genome‐wide pharmacogenetic investigation of a hepatic adverse event without clinical signs of immunopathology suggests an underlying immune pathogenesis. Pharmacogenomics J. 8, 186–195 (2008). [DOI] [PubMed] [Google Scholar]
- 8. Lucena, M.I. et al Susceptibility to amoxicillin‐clavulanate‐induced liver injury is influenced by multiple HLA class I and II alleles. Gastroenterology 141, 338–347 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen, R. et al The association between HLA‐DQB1 polymorphism and antituberculosis drug‐induced liver injury: a case‐control study. J. Clin. Pharm. Ther. 40, 110–115 (2015). [DOI] [PubMed] [Google Scholar]
- 10. Huang, Y.‐S. et al Genetic polymorphisms of manganese superoxide dismutase, NAD (P) H: quinone oxidoreductase, glutathione S‐transferase M1 and T1, and the susceptibility to drug‐induced liver injury. J. Hepatol. 47, 128–134 (2007). [DOI] [PubMed] [Google Scholar]
- 11. Starkey Lewis, P.J. et al Circulating microRNAs as potential markers of human drug‐induced liver injury. Hepatology 54, 1767–1776 (2011). [DOI] [PubMed] [Google Scholar]
- 12. Bala, S. et al Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug‐induced, and inflammatory liver diseases. Hepatology 56, 1946–1957 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagano, T. et al Liver‐specific microRNAs as biomarkers of nanomaterial‐induced liver damage. Nanotechnology 24, 405102 (2013). [DOI] [PubMed] [Google Scholar]
- 14. Shah, N. , Nelson, J.E. & Kowdley, K.V. MicroRNAs in liver disease: bench to bedside. J. Clin. Exp. Hepatol. 3, 231–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Starkey Lewis, P.J. et al Serum microRNA biomarkers for drug‐induced liver injury. Clin. Pharmacol. Ther. 92, 291–293 (2012). [DOI] [PubMed] [Google Scholar]
- 16. O'Connell, T.M. & Watkins, P.B. The application of metabonomics to predict drug‐induced liver injury. Clin. Pharmacol. Ther. 88, 394–399 (2010). [DOI] [PubMed] [Google Scholar]
- 17. Mattes, W. et al Detection of hepatotoxicity potential with metabolite profiling (metabolomics) of rat plasma. Toxicol. Lett. 230, 467–478 (2014). [DOI] [PubMed] [Google Scholar]
- 18. Winnike, J.H. , Li, Z. , Wright, F.A. , Macdonald, J.M. , O'Connell, T.M. & Watkins, P.B. Use of pharmaco‐metabonomics for early prediction of acetaminophen‐induced hepatotoxicity in humans. Clin. Pharmacol. Ther. 88, 45–51 (2010). [DOI] [PubMed] [Google Scholar]
- 19. Clayton, T.A. , Baker, D. , Lindon, J.C. , Everett, J.R. & Nicholson, J.K. Pharmacometabonomic identification of a significant host‐microbiome metabolic interaction affecting human drug metabolism. Proc. Natl. Acad. Sci. U S A 106, 14728–14733 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robles, M. , Toscano, E. , Cotta, J. , Lucena, M.I. & Andrade, R.J. Antibiotic‐induced liver toxicity: mechanisms, clinical features and causality assessment. Curr. Drug Saf. 5, 212–222 (2010). [DOI] [PubMed] [Google Scholar]
- 21. Andrade, R.J. et al Drug‐induced liver injury: an analysis of 461 incidences submitted to the Spanish registry over a 10‐year period. Gastroenterology 129, 512–521 (2005). [DOI] [PubMed] [Google Scholar]
- 22. Invernizzi, P. Drug‐induced liver injury: is it time for genetics to change our clinical practice? J. Hepatol. 53, 993–994 (2010). [DOI] [PubMed] [Google Scholar]
- 23. Stephens, C. et al HLA alleles influence the clinical signature of amoxicillin‐clavulanate hepatotoxicity. PLoS One 8, e68111 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim, S.H. et al Characterization of amoxicillin‐ and clavulanic acid‐specific T‐cells in patients with amoxicillin‐clavulanate‐induced liver injury. Hepatology 62, 887–899 (2015). [DOI] [PubMed] [Google Scholar]
- 25. Tujios, S. & Fontana, R.J. Mechanisms of drug‐induced liver injury: from bedside to bench. Nat. Rev. Gastroenterol. Hepatol. 8, 202–211 (2011). [DOI] [PubMed] [Google Scholar]
- 26. Hornby, R.J. , Starkey Lewis, P. , Dear, J. , Goldring, C. & Park, B.K. MicroRNAs as potential circulating biomarkers of drug‐induced liver injury: key current and future issues for translation to humans. Expert Rev. Clin. Pharmacol. 7, 349–362 (2014). [DOI] [PubMed] [Google Scholar]
- 27. Zhang, A. , Sun, H. , Wang, P. , Han, Y. & Wang, X. Metabonomics for discovering biomarkers of hepatotoxicity and nephrotoxicity. Pharmazie 67, 99–105 (2012). [PubMed] [Google Scholar]
- 28. Lucena, M.I. et al Glutathione S‐transferase m1 and t1 null genotypes increase susceptibility to idiosyncratic drug‐induced liver injury. Hepatology 48, 588–596 (2008). [DOI] [PubMed] [Google Scholar]
- 29. Mastrofrancesco, A. et al Azelaic acid modulates the inflammatory response in normal human keratinocytes through PPARgamma activation. Exp. Dermatol. 19, 813–820 (2010). [DOI] [PubMed] [Google Scholar]
- 30. Matsubara, T. et al Metabolomics identifies an inflammatory cascade involved in dioxin‐ and diet‐induced steatohepatitis. Cell Metab. 16, 634–644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frederiks, W.M. & Bosch, K.S. The proportion of xanthine oxidase activity of total xanthine oxidoreductase activity in situ remains constant in rat liver under various (patho)physiological conditions. Hepatology 24, 1179–1184 (1996). [DOI] [PubMed] [Google Scholar]
- 32. Kellogg, E.W. 3rd & Fridovich, I. Superoxide, hydrogen peroxide, and singlet oxygen in lipid peroxidation by a xanthine oxidase system. J. Biol. Chem. 250, 8812–8817 (1975). [PubMed] [Google Scholar]
- 33. Chowdhury, A. et al Induction of oxidative stress in antitubercular drug‐induced hepatotoxicity. Indian J. Gastroenterol. 20, 97–100 (2001). [PubMed] [Google Scholar]
- 34. Pichler, W.J. & Tilch, J. The lymphocyte transformation test in the diagnosis of drug hypersensitivity. Allergy 59, 809–820 (2004). [DOI] [PubMed] [Google Scholar]
- 35. Arroyo, J.D. et al Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U S A 108, 5003–5008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vickers, K.C. , Palmisano, B.T. , Shoucri, B.M. , Shamburek, R.D. & Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high‐density lipoproteins. Nat. Cell Biol. 13, 423–433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hunter, M.P. et al Detection of microRNA expression in human peripheral blood microvesicles. PLoS One 3, e3694 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lucena, M.I. et al Determinants of the clinical expression of amoxicillin‐clavulanate hepatotoxicity: a prospective series from Spain. Hepatology 44, 850–856 (2006). [DOI] [PubMed] [Google Scholar]
- 39. Kechagias, S. et al Fast‐food‐based hyper‐alimentation can induce rapid and profound elevation of serum alanine aminotransferase in healthy subjects. Gut 57, 649–654 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johnston, D.E. Special considerations in interpreting liver function tests. Am. Fam. Physician 59, 2223–2230 (1999). [PubMed] [Google Scholar]
- 41. Giboney, P.T. Mildly elevated liver transaminase levels in the asymptomatic patient. Am. Fam. Physician 71, 1105–1110 (2005). [PubMed] [Google Scholar]
- 42. Abdel‐Rahman, S.Z. , el‐Zein, R.A. , Anwar, W.A. & Au, W.W. A multiplex PCR procedure for polymorphic analysis of GSTM1 and GSTT1 genes in population studies. Cancer Lett. 107, 229–233 (1996). [DOI] [PubMed] [Google Scholar]
- 43. Schreiber, J. , Zissel, G. , Greinert, U. , Schlaak, M. & Muller‐Quernheim, J. Lymphocyte transformation test for the evaluation of adverse effects of antituberculous drugs. Eur. J. Med. Res. 4, 67–71 (1999). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Accuracy and precision of (a) amoxicillin and (b) clavulanic acid concentration measurements using liquid chromatography‐tandem mass spectrometry (n = 5).
Supplementary Table S2. The HLA types of the study subjects.
Supplementary Table S3. PK parameters (Cmax, AUC0‐12h, and AUCss,tau) of amoxicillin and clavulanate in the Responder and Nonresponder groups.
Supplementary Table S4. Correlations between ALT changes and microRNA‐122 (miR‐122) changes throughout the study.
Supplementary Figure S1. Mean of maximum ALT fold change classified by GSTT1/M1 genotype. GSTT, glutathione s‐transferase T; GSTM, glutathione s‐transferase mu; W/W, wild/wild; W/N, wild/null; N/W, null/wild; N/N, null/null.
Supplementary Figure S2. S‐plot corresponding to the OPLS‐DA analysis for the new model built on samples in (A) positive ion mode and (B) negative ion mode.
Supplementary Figure S3. Changes in miR‐122 levels in exosome‐rich and protein‐rich fractions separated from Nonresponder serum (a) and Responder serum (b). In the Responder group, miR‐122 levels massively increased in both fractions at peak ALT time points. However, in the Nonresponder group, miR‐122 levels in both fractions were unchanged at peak ALT time points. The data represent the means ± standard deviation.


