Abstract
The RNA binding protein Lin28 is best known for the critical role in cell development, recent researches also have implied its oncogenic function in various human cancers, including breast cancer. Specifically, aberrant Lin28 participates in multiple pathological processes, such as proliferation, metastasis, radiotherapy and chemotherapy resistance, metabolism, immunity and inflammation as well as stemness. In this review, we summarize the let-7-dependent and let-7-independent mechanism regulated by Lin28, focusing on its relation with tumor hallmarks in breast cancer, and subsequently discuss our present knowledge of Lin28 to develop a molecular-based therapeutic strategy against breast cancer.
Keywords: Lin28, Let-7, breast cancer, metastasis, drug resistance
INTRODUCTION
Breast cancer is a complex and poorly understood disease which affects 1 in 10 females. It is the leading cause of death in women between the ages of 40 and 50 years [1]. Since breast cancer is highly heterogeneous in terms of biological behavior, researchers have made extensive efforts to conquer this challenge. In the last few decades, several novel molecular or cellular factors were identified in breast cancer, including Lin28 and its related factors [2]. Currently, the mainstream opinion is that Lin28 mainly functions as an oncogene. Mechanistically, Lin28 is involved in various pathological processes of cancers via let-7 dependent and let-7 independent pathways [3]. With a deeper understanding of how Lin28 is involved in breast cancer initiation and progression, new findings will be valuable in helping us to explore variably targetable mechanisms of breast cancer. The purpose of this article is to review existing literatures on the molecular mechanisms of Lin28, along with its roles in breast cancer. To our knowledge, this is the first review focusing on Lin28 in breast cancer. We believe that a more comprehensive understanding of Lin28 function will yield preventive, diagnostic, predictive and therapeutic advances for breast cancer.
MOLECULAR MECHANISMS OF LIN28 AND ITS TWO PATHWAYS
Basic knowledge of Lin28
Firstly identified as a regulator of developmental timing in Caenorhabditis elegans [4], Lin28 is now well established as an RNA binding protein and transcription factor implicated in stem cell differentiation, normal development, glucose metabolism, and cancer [5]. Lin28 has two RNA-binding motifs: a cold shock domain (CSD) and a Cys-Cys-His-Cys (CCHC) zinc finger domain [6]. Mammals produce two Lin28 paralogs: Lin28A and Lin28B, which have identical regions in structures and functional similarities but differ in a few respects [7]. For instance, Lin28B has additional sections: a nuclear localization signal (NLS) and a nucleolar localization signal (NoLS), and is therefore primarily located in nucleus and nucleolus. Lin28A on the other hand is predominantly located in the cytosol of cells (Figure 1).
Figure 1. Domains of human Lin28A and Lin28B proteins.

Lin28A and Lin28B share several common domains: the CSD (yellow) and CCHC (orange), while Lin28B contains both NoLS and NLS (gray). Numbers denote amino acids.
The let-7 family of microRNA (miRNA), which was also firstly discovered in Caenorhabditis elegans [8], has been reported to be a key suppressive target of Lin28, and serves as a potent tumor suppressor via post-transcriptional repression of multiple oncogenic messenger RNA (mRNA) [9]. Meanwhile, the convergence of abundant in vitro and in vivo structural studies have discovered the molecular basis of the interaction between Lin28 and other target RNAs, whose Lin28 binding sites vary depending on their sequence and context [10]. It is therefore appropriate to assign the critical functions of Lin28 to one of two classifications: let-7 dependent and let-7 independent.
Let-7 dependent functionality
Lin28 homologs (Lin28A and Lin28B) are small (< 30kDa) proteins which can block the processing of let-7 family members by binding to the terminal loop of the let-7 precursor (pre-let-7) hairpin via a CSD and two retroviral-like CHCC zinc-finger knuckles [11–13]. The GGAG sequences in the terminal loop of let-7 precursors serve as the binding sites for the zinc finger domains critical for let-7 regulation [14]. Subsequent reports have demonstrated that Lin28 blocks the processing of let-7 at primary, precursor, and mature forms of let-7 family members, as Microprocessor complexes (DGCR8 and Drosha) and DICER complexes cannot associate with Lin28-bound let-7 [15–17]. Furthermore, studies revealed that Lin28A and Lin28B inhibited let-7 biogenesis by distinct mechanisms. Lin28B sequesters primary let-7 transcripts and inhibits their processing by the Microprocessor [18], while Lin28A recruits TUTase4 to induce oligo-uridylation of pre-let-7, which blocks DICER processing and facilitates degradation of the RNAs [11–13]. Another study indicated that Lin28 used two different TUTases to control let-7 expression and had important implications for stem cell biology as well as cancer [19]. Taken together, regulation of let-7 expression is controlled by Lin28 proteins through the post-transcriptional blockade of let-7 biogenesis. Interestingly, Lin28A and Lin28B mRNAs themselves have potential let-7 complementary sites (3′UTR of Lin28) and serve as let-7 targets, making let-7 regulate Lin28 expression by cleaving Lin28 mRNAs or inhibiting translation of Lin28 mRNAs [6, 20–25]. Additionally, the let-7 target genes, such as c-MYC [26] and CDC25A [23], could regulate their downstream Lin28 via translational repression, making let-7 an indirect inhibitor of Lin28. Therefore, Lin28/let-7 axis establishes a double-negative feedback loop whereby either let-7 or Lin28 is expressed at high levels, promoting physiological or pathological conditions, respectively (Figure 2).
Figure 2. Lin28′s let-7-dependent functionality.
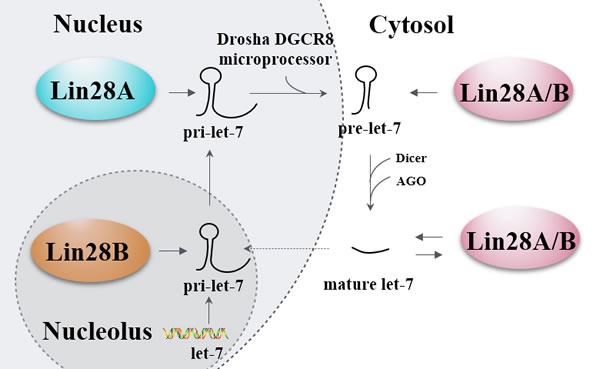
A primary let-7 (pri-let-7) transcript produced by let-7 gene is processed by the Drosha DGCR8 microprocessor in the nucleus. Then the generated precursor let-7 (pre-let-7) is transported to the cytosol and further processed by the Dicer and Argonaute proteins (AGO) to generate the mature let-7. The biogenesis of pri-let-7 is blocked by Lin28A in the nucleus and Lin28B in the nucleolus, the biogenesis of pre-let-7 and mature let-7 are blocked by Lin28A/B in the cytosol, and the mature let-7 can in turn block the biogenesis of Lin28A/B. Solid line arrows refer to induction and promotion, dash line arrows refer to only promotion, while hammerheads refer to inhibition.
Let-7 independent functionality
Interaction of Lin28 and mRNA
As for let-7 independent way, Cho et al. mapped the Lin28A binding sites on the genomic scale by RNA crosslinking-immunoprecipitation-sequencing (CLIP-seq) technology and ribosome footprinting. They observed that Lin28A binds to a large number of spliced mRNAs by recognizing AAGNNG, AAGNG, and less frequently UGUG, which are located in the terminal loop of a small hairpin [27]. In their study, Lin28A was found to be enriched in the peri-endoplasmic reticulum region and bound to the cytosolic surface of rough endoplasmic reticulum (RER) on which endoplasmic reticulum-associated mRNAs were translated in undifferentiated stem cells. Several other studies have reported that Lin28 could activate OCT4 mRNA translation through binding to OCT4 mRNA in complex with RNA helicase A in polysomes [28, 29], and that Lin28 could directly enhance translation efficiency of IGF2 mRNA during murine muscle cell differentiation [30]. It is worth mentioning that besides OCT4 and IGF2, researchers further found that a series of mRNAs bound by Lin28 were enriched in glycolysis and glucose metabolism, e.g., GPAA1 and GNPDA1 [31]. These interactions between Lin28 and mRNAs are based on linear sequence recognition motifs, Lei et al. also reported specific structural determinants for Lin28 binding with its targeted mRNAs, including HMGA1, EEF1G and RPS13 mRNAs [32]. More precisely, the motif is characterized by a small ‘A’ bulge flanked by two G:C base pairs embedded in a larger secondary structure, which mediates Lin28-dependent stimulation of mRNA translation.
Interaction of Lin28 and miRNA
Lin28 also could activate or inhibit other miRNAs besides the let-7 family. For example, Lin28 inhibition has recently been demonstrated to decrease the level of abundance of miR-17∼92 family miRNAs, which may be connected with the molecular basis of GGAG motif in miRNAs [33]. Among aforementioned miRNAs, Peters et al. focused on the putative tumor suppressor miR-363, discovering that miR-363 RNA segment, which contains the GGAG motif, formed a relatively stable complex with the Lin28 protein. Knockdown of Lin28 could lead to decreased level of mature miR-363, implying that Lin28 functioned as a positive regulator of miR-363 biogenesis [34]. Additionally, Lin28 could differentially promote and inhibit specific fates of cells by inhibiting miR-302d with its CSD domain [35], miR-125b could efficiently reduce Lin28 protein levels via directly binding to the 3′-UTR region of the Lin28 mRNA [36]. Intriguingly, Willbert et al. have reported that Lin28 directly enhances its own expression level by binding to sites within its 3′UTR, revealing a mechanism of positive feedback regulation by Lin28 [37]. In fact, Lin28 can regulate multiple tumor-associated progressions in let-7 independent way, including proliferation, chemo-resistance, metabolism, inflammation, stemness and cell development (Figure 3).
Figure 3. Lin28′s let-7-independent functionality.
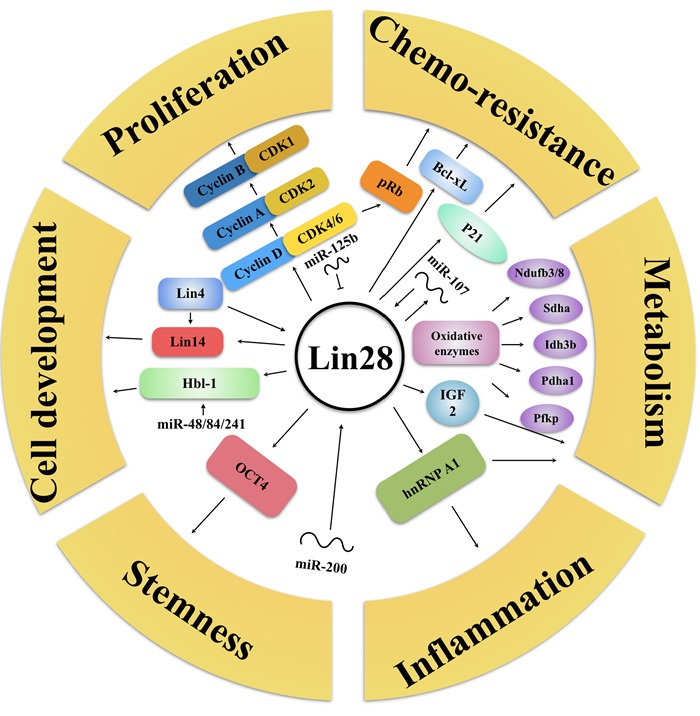
Lin28 can regulate multiple tumor-associated progressions without let-7, but with proliferation (CyclinA/B/D, CDK1/2/4/6, miR-125b), chemoresistance (pRb, p21, Bcl-xL, miR-107), metabolism (IGF2, Oxidative enzymes), inflammation (hnRNP A1), stemness (OCT4, miR-200), cell development (Hbl-1, Lin4/14, miR-48/84/241) related proteins and RNAs. Arrows refer to promotion, while hammerheads refer to inhibition.
MULTIPLE PROCESSES REGULATED BY LIN28 IN BREAST CANCER
In malignancies, a more common phenomenon is that the expression of Lin28 is up-regulated. The correlational studies have shown that Lin28 may be transcriptionally activated by upstream factors, e.g. c-Myc, Src, NF-κB and Wnt [18, 38]. For instance, c-Myc is found to be associated with the genomic locus encoding Lin28 and transactivate the Lin28 promoter, leading Lin28 to become the direct target of MYC [39]. Besides, Chen et al. demonstrated that the C allele of rs3811463, a SNP that located near the let-7 binding-site of Lin28, could weaken the suppression of Lin28 by let-7, which means an increasing level in Lin28 expression along with a reduction of let-7 level, elevating the risk of breast cancer [40]. In fact, Lin28 is reactivated by oncogenes in approximately 15% of all analyzed cancers, and its expression is correlated to advanced stages of various types of cancer, including breast cancer [41]. Using tissue microarrays, researchers have demonstrated that Lin28 is expressed in a collection of human breast cancer cell lines, while cultured normal human breast epithelial cells have no detectable Lin28 expression. Furthermore, Lin28 expression was detected in ductal carcinoma in situ (DCIS) specimens but not in any benign breast tumor specimens [42]. Taken together, these data have shown that expression of Lin28 is frequently up-regulated in breast cancer. Here, we will overview mechanisms and functions of Lin28 as well as multiple cancer-associating progressions regulated by Lin28 (Figure 4).
Figure 4. Lin28 regulates multiple progressions in breast cancer.
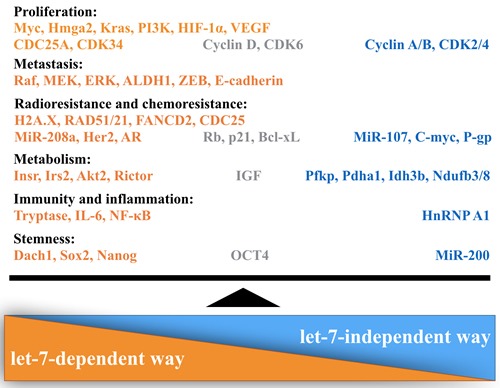
Lin28 exerts its critical role in breast cancer through two distinct ways: let-7 dependent and let-7 independent. Orange refers to factors of let-7-dependent way, blue refers to factors of let-7-independent way, gray means factors involved in both mechanisms.
Lin28 promotes cancer cell proliferation
Arguably, the most fundamental feature of Lin28 in breast cancer cells is it ability to promote and sustain chronic proliferation. Many studies focusing on proliferative signaling in cancers suggest that Lin28 functions as an oncogene by repressing let-7, leading to the dysregulation of multiple genes regulated directly or indirectly by let-7, including MYC, Hmga2, and components of PI3K-mTOR pathway [43–46]. In the aspect of cell cycle, Li et al. have demonstrated that Lin28 promotes proliferation of tumor cells through regulating the G0/G1 transition in cell cycle, namely, increasing the expression of Cyclin D1/D2, CDC25A, CDK34, CDK6, as well as other cell cycle-related factors by depressing let-7 [42]. We should note that Figure 5 in Li's article showed that Lin28A dramatically promoted the G0/G1 transition rather than its role of enhancing G2/M transition in human embryonic stem cells (ESCs), further works are needed to explain the reason why Lin28A controls different cell cycle regulators among different cells. Adding another layer of meaning to the complex downstream effects of Lin28 are some studies in which Lin28 itself binds to many mRNAs and enhances their translation, including IGF2 and Cyclin A/B [30, 35].
Furthermore, Sharma et al. reported that combining the Lin28/let-7a/Kras axis inhibitors (NVP-LDE-225 and NVP-BEZ-235) could inhibit tumor growth by inhibiting Bcl-2 family members and activating caspases and by suppressing PI3K-mTOR pathway, suggesting that Lin28/let-7a may be involved in the apoptosis and proliferation of cancer cells simultaneously [47]. Since several studies indicate that the tumor-associated neovasculature, generated by the process of angiogenesis, could also promote tumor growth, researchers have investigated in the involvement of Lin28/let-7 axis in angiogenesis. For instance, Isanejad et al. found that markers for aggressive breast cancer cells (such as Ki67 and ERα) or for tumor blood vessels (such as HIF-1α, CD31 and VEGF) could be down-regulated by the combination hormone therapy through the let-7a pathway. Taken together, these evidences support the hypothesis of Lin28/let-7 axis contributing to the anti-angiogenesis effects of the breast cancer [48].
Lin28 expression promotes cancer cell metastasis
Abnormal Lin28 activation is found in diverse human malignancies, especially in poorly differentiated and highly aggressive tumors [49]. Rosner et al. have revealed that Lin28 facilitates bone metastasis in Raf kinase inhibitory protein (RKIP)-expressing breast tumors [50]. More specifically, the results support a model in which RKIP suppresses Raf-1/MEK/ERK activity, leading to the inhibition of Lin28 and induction of let-7, finally blocking the induction of Snail transcription and other genes involved in tumor cell invasion and metastatic colonization. Epithelial-to-mesenchymal transition (EMT) is known to accelerate tissue remodeling from epithelial phenotype to mesenchymal phenotype, and Lin28/let-7 axis is also a prerequisite for the process of EMT among some cases [51]. Liu et al. found that over-expression of Lin28 in breast cancer cells remarkably decreased the expression of E-cadherin but increased Vimentin, thus facilitating metastasis [52]. In the Table 3 of the article, they also reported that Lin28 correlated with axillary lymph node, distal metastases and PR. However, the authors found no significant correlation between Lin28 and ER or HER2 expression, therefore large-scale clinical samples are needed to ensure Lin28 to serve as a prognostic marker for patients with breast cancer. Furthermore, Wang et al. demonstrated that in breast cancer cell lines, let-7a could suppress cell migration by significantly blocking the direct binding target of Lin28, which provided evidence for the potential therapeutic role of targeting Lin28 strategies in conquering metastasis in breast cancer [53].
Lin28 contributes to the resistance of radiation treatment and chemotherapy
Resistance to radiotherapy and chemotherapy has long been a great challenge in the treatment of cancer patients, and abundant evidences have shown that over-expression of Lin28 contributed to cancer cell radiotherapy and chemotherapy resistance [54]. For instance, Wang et al. found that Lin28 expression was up-regulated in radiation-resistant breast cancer cells. Lin28 transfection could induce radiation resistance via inhibiting H2A.X pathway while over-expression of let-7 enhanced the sensitivity to radiation [55]. Sun et al. have demonstrated that miR-208a-SOX2/β-catenin-Lin28-let-7a-DICER1 regulatory feedback loop participates in the therapy resistance of breast cancer through promoting the induction of the cancer stem cells [56]. Based on the Figure 5 of their work, miR-208a increased the activity of DICER1 while let-7a directly targeted and degraded DICER1 mRNA, it is necessary to investigate whether the observation could be explained by the competitive endogenous RNA (ceRNA) hypothesis. Since the impact of Lin28/let-7 axis on radio-sensitivity has been confirmed in vitro, increasing evidence has shown that down-expression of Lin28 and over-expression of let-7 could decrease the expression of RAS oncogene and genes associated with DNA like RAD51, RAD21, FANCD2 and CDC25, eventually radiosensitizing the cancer cells [57–59].
In the aspect of drug resistance, Teng et al. reported that over-expression of Lin28 decreased the sensitivity to chemotherapy (e.g. Oxaliplatin, Paclitaxel, Doxorubicin and Fluorouracil) via inhibiting miR-107, as well as the RNA and protein expression of c-Myc and P-gp [60]. Other researchers also have found that Lin28-induced chemotherapy resistance is associated with let-7, Rb, p21 and Bcl-xL, thus unraveling complicated relationship between Lin28 and tumor resistance [61, 62]. As for clinical study, by evaluating the polymorphism of Lin28 gene between breast cancer females and healthy controls, researchers have found that genetic variations in Lin28 might be a possible mechanism underlying susceptibility and clinical features in breast cancer, including the response to neoadjuvant chemotherapy [63].
Lin28 contributes to enhanced cell metabolism
Sixty years ago, Otto Warburg first noted that under normoxic conditions, normal cells metabolize glucose by using mitochondrial oxidative phosphorylations (OxPhos) instead of glycolysis to maximize the production of adenosine triphosphate (ATP), but cancer cells choose to rely on glycolysis even under the sufficient oxygen condition, a phenomenon later known as the Warburg effect [64]. Since then interest in this topic has increased, and major areas of knowledge have been gained, but fundamentally important questions remain unresolved.
Extensive studies have provided the evidence that conditional deletion of Lin28 could lead to insulin resistance and impaired glucose uptake. Let-7 transgenic mice not only reduced body size and growth retardation, but also led to hyperglycemia and glucose intolerance [45]. As for breast cancer, Yang et al. performed RNA-protein immunoprecipitation (RIP) coupled with genome-wide sequencing (RIP-Seq) to identify endogenous Lin28 mRNA targets. Data suggested that Lin28 regulated the expression of a unique set of mRNAs, whose expression is involved in classic cancer metabolism [65]. More accurately, Lin28 down-expression could inhibit insulin sensitivity mostly by suppressing the let-7 targets Insr, IGF1r, Irs2, Akt2, and Rictor, and also in part by directly reducing the ribosomal translation of mRNAs encoding IGF2 and mitochondrial OxPhos enzymes.
Of course, other researches on metabolism have different voices. Zhang et al. found that induction of Lin28 over-expressing mouse ESCs compromised basal and maximal oxygen consumption rate (OCR), but that levels of let-7 did not change from their already suppressed levels [66]. According to Figure S4 in their study, authors not only verified that let-7 mimic in Lin28-overexpressing cells could not reverse the reduced OCR, but also knockdowned Lin28 in Dgcr8−/− ESCs (scant quantities of let-7) to exclude other possibly let-7-related microRNAs function. Both the experimental design and result are quite rigorous and precise, which deserves our respect and learning. Other studies also found that gain or loss of function of let-7 in wild-type cells did not change OCR, suggesting that Lin28 reduced OCR through several let-7-independent mechanisms [67].
Lin28 is involved in immunity and inflammation
In addition to the above-mentioned role of Lin28 in cancer, several frontier researches have suggested that Lin28 may be implicated in the immune system [68]. For example, researchers reported that Lin28 in myeloid could affect mast cell differentiation and promote mast cell malignancy [69]. Mast cells, the critical components of the innate immune system, release a serine protease called Tryptase, which was demonstrated to play a positive role in tumor angiogenesis of breast cancer [70]. Several other investigators have provided evidence that Lin28 promotes tumor growth via enhancing the development of T cells, B cells, and natural killer T (NKT) cells [71–74].
In regards to inflammation, Iliopoulos et al. have observed that Src, an oncoprotein in immortalized breast cells, could activate a rapid inflammatory response mediated by NF-κB. The response then led to Lin28-regulated expression of the anti-inflammatory cytokine interleukin-6 (IL-6) via inhibiting let-7 expression, thus revealing a new mechanism containing cancer cells and immune molecules [75, 76]. They also found that let-7 inhibited IL-6 expression both directly through its 3′UTR and indirectly by interacting with RAS to reduce the NF-κB activity. As for other inflammatory mechanisms, Yang et al. have reported that breast cancer cells lacking Lin28 could increase levels of anti-inflammatory cytokines, and that the regulation of the major cytokine genes is dependent on the expression of hnRNP A1, suggesting a mechanism independent of let-7 [65]. Meanwhile, Yang et al. also found that despite of the cooperation to regulate nuclear processes, depletion of Lin28 and hnRNP A1 proteins yielded nonidentical effects on mRNA splicing (presented in Figure 6 of the article). The phenomenon may be attributed to that RT-PCR assay is not sensitive enough and transcript isoforms are confounded by other cellular factors.
Lin28 is involved in cancer stem cells
In the cancer microenvironment, a fraction of tumor cells, called tumor initiating cells (TICs) or cancer stem cells (CSCs), have been found to propagate in various tumors, including breast cancer [77]. Human mammary epithelial cells transformed by EMT could significantly enhance their self-renewal and tumor-initiating capabilities, and lead to the expression of stem-cell markers typically associated with the stemness of breast cancer cells [78]. Studies in the past decade have shown that the Lin28/let-7 axis plays a significant role in stem cell renewal [79]. For example, Cai et al. have reported that Lin28 upregulation and let-7 posttranscriptional downregulation were identified in the Wnt-β-catenin pathway-stimulated breast CSCs phenotype, while loss of function of Lin28 impaired Wnt-β-catenin-pathway-mediated let-7 inhibition and breast cancer stem cell expansion [80]. The in-depth mechanism revealed that Lin28 could act as a novel direct downstream target of the Wnt-β-catenin pathway, namely, both LEF1 and β-catenin were found to directly bind to the Lin28 promoter. Feng et al. have implicated that over-expression of HER2 in breast cancer increases the population of CSCs, which also correlates with the high Lin28 expression level [81]. Specifically, their discoveries suggested that Lin28 could bind to HER2 mRNA and stimulate its translation via a 200-nucleotide sized Lin28-responsive element (LRE) in HER2 mRNA. But according to the Figure 4 illustrated in Feng's article, targeting HER2 alone by Herceptin (Trastuzumab) is insufficient to inhibit the level of Lin28, thus the study of Lin28/HER2 positive feedback loop still need to be investigated. Moreover, Wu et al. found that cell fate determination factor Dach1 reduced breast tumor formation in serial transplantations in vivo, further unraveling that in breast cancer cells Dach1 directly bonds to the promoter region of Lin28, Sox2 and Nanog genes and inhibits their expression, finally decreases expansion of CSCs population and blocks breast tumor growth [82]. It is worth mentioning that Jolly et al. discovered that independent of let-7, Lin28 was strongly inhibited by miR-200 which pushed epithelial end towards mesenchymal end of CSCs, thus allowing mesenchymal phenotype cells to gain stemness [83] (Table 1).
Table 1. Multiple processes regulated by Lin28 in breast cancer.
| Tumorigenic mechanisms | Key detection methods | Major findings | References |
|---|---|---|---|
| Proliferation | Immunocytochemistry, Microarray analysis, Motility assay |
|
[30, 35, 42–48] |
| Metastasis | Transwell migration assay, Flow cytometry analysis |
|
[49–53] |
| Radiotherapy and chemotherapy resistance | Immunofluorescence,Cell drug-resistance assay, Xenograft Assay |
|
[54–63] |
| Metabolism | Immunoprecipitation, RIP-Seq, Metabolomics Analysis |
|
[45, 64–67] |
| Immunity and inflammation | Flow cytometric analysis, RNA-Seq, Immunoprecipitation,ELISA |
|
[65, 68–76] |
| Stemness | Microarrays, Immunoprecipitation, In vivo stem cell assay |
|
[77–83] |
CURRENT OPINIONS AND FUTURE PERSPECTIVES ON LIN28 IN BREAST CANCER
Breast cancer is a disease plagued by recurrences with progressive drug resistance, ultimately leading to uncontrolled cancer progress. It is now evident that Lin28 does play roles in breast cancer initiation, progression and maintenance. However, the challenge that lies ahead is how to integrate our improved molecular understanding of Lin28 and breast cancer pathogenesis with novel therapeutic options, ultimately improving patient outcomes.
The canonical targets of Lin28, let-7 family members, have been most notably implicated in cancer [84]. The association of let-7 with pathogenesis of breast cancer is supported by studies examining let-7 expression in breast cancer cell lines and clinical samples (Table 2). The majority of studies revealed that most of let-7 family members were down-regulated in breast cancer samples with either lymph node metastasis or higher proliferation index versus normal tissues [85–88], while one study showing up-regulation of let-7b [89]. Subsequently, several functional studies have reported novel mechanisms of let-7 in breast cancer cells [90–97]. As for clinical researches, apart from cases for let-7 as the classical suppressor [98], other studies have suggested that let-7 does not function as a tumor suppressor under all circumstances [99–101].
Table 2. The expression of let-7 in breast cancer.
| let-7 family members | Down-regulated | Up-regulated | Targets | Reference |
|---|---|---|---|---|
| let-7a-1 | √ | [85] | ||
| let-7 family members | √ | [86] | ||
| let-7a-2/let-7a-3/let-7d/let-7f-2/let-7i | √ | [87] | ||
| let-7a-2 | √ | [88] | ||
| let-7b | √ | [89] | ||
| let-7d/let-7f/let-7a/let-7e/let-7c/ let-7g/let-7i | √ | |||
| let-7a | √ | H-RAS, HMGA2 | [90, 91] | |
| let-7a | √ | CCR7 | [92] | |
| let-7d | √ | Wnt | [93] | |
| let-7c | √ | Wnt | [94] | |
| let-7 family members | √ | Critical components of ER/mitogenic/cell cycle signaling pathways | [95] | |
| let-7b | √ | PAK1, DIAPH2, RDX | [96] | |
| let-7b | √ | BSG | [97] | |
| let-7a/let-7b/let-7c/let-7i/let-7g | √ | GAB2, FN1 | [98] | |
| let-7a-2/miR-98 | √ | ADRB2*,CEP164*,CYP19A1*,TARBP2* | [99] | |
| let-7i | √ | [100] | ||
| let-7i | √ | [101] |
*Predicted putative targets
For neoteric findings, studies using 26 breast cancer surgical specimens found Lin28 expression to be positively correlated with ER and PR status but inversely correlated with HER2 status [102, 103]. In addition to the known risk factors like ER, PR and HER2, Shen et al. have found that Lin28 and AR (androgen receptor) co-expression is a strong predictor of poor prognosis in breast cancer [104]. Although the positive feedback mechanism of Lin28 and AR is still not clear, researchers can conduct in-depth studies in this new research domain. Moreover, Liu et al. found that Hepatitis B X-interacting protein (HBXIP), a novel oncoprotein, could promote proliferation of breast cancer cells by up-regulating Lin28B [105]. Segalla et al. reported that the ribonuclease DIS3 could promote let-7 maturation and inhibit Lin28B mRNA levels through recognition of AU-rich elements in the 3′UTR [106]. Therefore, therapeutically canonical and non-canonical key factors may serve as the target to control upstream or downstream miRNAs and direct mRNA regulation of Lin28, which will guide attempts to use Lin28 as a target for treating human breast cancer.
Recent advanced studies also have revealed that long non-coding RNA H19 acts as a sponge to antagonize let-7. For example, the ability of let-7 to repress the expression of an array of metastasis-promoting genes is compromised when H19 expression is high, leading to decreased bio-availability of let-7, increased expression of c-Myc, Hmga2 and Imp3, and activation of cell migration and invasion [107], thus prompting us to study the relationship among H19, let-7 and Lin28 during carcinogenesis. Researchers’ findings also included that metformin, a drug used for treating type-2 diabetes, could down-regulate H19 at least in part by altering methylation of the H19 promoter, thus presenting anti-tumor effects on tumor cells. In addition, Lin28 has been found to participate in other popular research fields, including exosomes, autophagy and cells polarity. For instance, high Lin28A expressing exosomes could induce EMT-related gene expression and promote non-metastatic target cells to migrate and invade [108], Lin28 suppressor let-7 could activate autophagy by repressing the mTOR signaling pathway [109], human induced pluripotent stem cells (iPSCs) generated by Lin28 could spontaneously differentiate into polarized retinal pigmented epithelium [110]. Taken together, it is necessary for us to further investigate the concrete mechanism of Lin28, which may open up new research avenues for cancer diagnostics and treatment.
In fact, aberrant expression of Lin28 stimulates tumorigenesis in many other kinds of cancers, including lung [111], hepatocellular [112], gastric [113], colon [114], kidney [115], and ovarian carcinoma [116]. More precisely, novel findings involve that Lin28 interacts with a protein expressed by Hepatitis B virus to enhance the proliferation of hepatoma cells [112], Lin28 activates genes that simultaneously enhance both metastasis and cell differentiation in colon cancer [114], higher Lin28 expression is associated with worse pathologic tumor responses under the neoadjuvant chemotherapy [113], Lin28 expression in kidney cells promotes metabolic switching (the increasing rate of glycolysis) to a phenotype related with cancer [115]. IL-1β-miR-101-Lin28 is certified as a novel pathogenic inflammatory signaling in non-small cell lung cancer [111]. Lin28 together with OCT4 can be used to identify subgroups of stem cell-like cells in ovarian cancer [116]. Additionally, except for researches that Lin28/let-7 loop is involved in ten hallmarks of cancer [117], the aberrant loop also regulates cellular senescence and has connection with various oncogenes and signaling pathways, including MYC, RAS, MAPK signaling and PI3K/AKT signaling [118]. Although above mechanisms have not fully been verified in breast cancer, they could provide us new thoughts and references, based on similarity of traits of Lin28 and relative factors, we may propose a series of bold experiments to promote new discoveries.
Nowadays, available data suggest that inhibition of Lin28 in breast cancer holds great therapeutic potential for the treatment of breast cancer. However, it remains to be determined when and to what extent, Lin28 might be involved in preventive, predictive or prognostic functions. Further endeavors will unravel its precise roles in the pathogenesis of breast cancer. It is not difficult to predict that exciting times are still ahead for researches studying Lin28.
CONCLUSIONS
In summary, Lin28 has been observed to be frequently up-regulated in breast cancer. Through canonical and non-canonical pathways, high levels of Lin28 could promote cellular proliferation, metastasis, radio- and chemo-resistance, metabolism reprogramming, immunity and inflammation, and tumor-associated stemness in breast cancer. Although the Lin28 related regulatory network remains unclear, it would certainly be an intriguing frontier to study and a valuable hotspot for future breast cancer therapeutic strategies.
Acknowledgments
We thank reviewers for their critical reading and informative advice during the revision process. We apologize to all researchers whose relevant contributions were not cited due to space limitations.
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed. None of the contents of this manuscript has been previously published or is under consideration elsewhere. All the authors read and approved the final version of the manuscript prior to submission.
FUNDING
This work is supported by National Natural Science Foundation of China (No. 81602471, No. 81672729) and grant from sub-project of China National Program on Key Basic Research Project (973 Program) (No. 2014CB744505).
REFERENCES
- 1.Licznerska B, Baer-Dubowska W. Estrogen intracrinology: therapy and chemoprevention of breast cancer. Postepy Hig Med Dosw (Online) 2010;64:220–30. [PubMed] [Google Scholar]
- 2.Huang Y. A mirror of two faces: Lin28 as a master regulator of both miRNA and mRNA. Wiley Interdiscip Rev RNA. 2012;3:483–94. doi: 10.1002/wrna.1112. [DOI] [PubMed] [Google Scholar]
- 3.Mayr F, Heinemann U. Mechanisms of Lin28-mediated miRNA and mRNA regulation--a structural and functional perspective. Int J Mol Sci. 2013;14:16532–53. doi: 10.3390/ijms140816532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–46. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 5.Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22:474–82. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rehfeld F, Rohde AM, Nguyen DT, Wulczyn FG. Lin28 and let-7: ancient milestones on the road from pluripotency to neurogenesis. Cell Tissue Res. 2015;359:145–60. doi: 10.1007/s00441-014-1872-2. [DOI] [PubMed] [Google Scholar]
- 7.Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 9.Powers JT, Tsanov KM, Pearson DS, Roels F, Spina CS, Ebright R, Seligson M, de Soysa Y, Cahan P, Theissen J, Tu HC, Han A, Kurek KC, et al. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature. 2016;535:246–51. doi: 10.1038/nature18632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Zhao Q, Deng K, Guo X, Xia J. Lin28: an emerging important oncogene connecting several aspects of cancer. Tumour Biol. 2016;37:2841–8. doi: 10.1007/s13277-015-4759-2. [DOI] [PubMed] [Google Scholar]
- 11.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283:21310–4. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 12.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–5. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–91. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desjardins A, Yang A, Bouvette J, Omichinski JG, Legault P. Importance of the NCp7-like domain in the recognition of pre-let-7g by the pluripotency factor Lin28. Nucleic Acids Res. 2012;40:1767–77. doi: 10.1093/nar/gkr808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–79. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) Rna. 2012;18:1875–85. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–93. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 21.Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258:432–42. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- 22.Wu L, Belasco JG. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol. 2005;25:9198–208. doi: 10.1128/mcb.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–91. doi: 10.1158/0008-5472.can-08-0264. [DOI] [PubMed] [Google Scholar]
- 24.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–78. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su B, Zhao W, Shi B, Zhang Z, Yu X, Xie F, Guo Z, Zhang X, Liu J, Shen Q, Wang J, Li X, Zhang Z, et al. Let-7d suppresses growth, metastasis, and tumor macrophage infiltration in renal cell carcinoma by targeting COL3A1 and CCL7. Mol Cancer. 2014;13:206. doi: 10.1186/1476-4598-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ioannidis P, Mahaira LG, Perez SA, Gritzapis AD, Sotiropoulou PA, Kavalakis GJ, Antsaklis AI, Baxevanis CN, Papamichail M. CRD-BP/IMP1 expression characterizes cord blood CD34+ stem cells and affects c-myc and IGF-II expression in MCF-7 cancer cells. J Biol Chem. 2005;280:20086–93. doi: 10.1074/jbc.M410036200. [DOI] [PubMed] [Google Scholar]
- 27.Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, Kim VN. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151:765–77. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38:1240–8. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin J, Jing W, Lei XX, Feng C, Peng S, Boris-Lawrie K, Huang Y. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res. 2011;39:3724–34. doi: 10.1093/nar/gkq1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–38. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng S, Chen LL, Lei XX, Yang L, Lin H, Carmichael GG, Huang Y. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29:496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- 32.Lei XX, Xu J, Ma W, Qiao C, Newman MA, Hammond SM, Huang Y. Determinants of mRNA recognition and translation regulation by Lin28. Nucleic Acids Res. 2012;40:3574–84. doi: 10.1093/nar/gkr1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warrander F, Faas L, Kovalevskiy O, Peters D, Coles M, Antson AA, Genever P, Isaacs HV. lin28 proteins promote expression of 17∼92 family miRNAs during amphibian development. Dev Dyn. 2016;245:34–46. doi: 10.1002/dvdy.24358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters DT, Fung HK, Levdikov VM, Irmscher T, Warrander FC, Greive SJ, Kovalevskiy O, Isaacs HV, Coles M, Antson AA. Human Lin28 Forms a High-Affinity 1: 1 Complex with the 106∼363 Cluster miRNA miR-363. Biochemistry. 2016;55:5021–7. doi: 10.1021/acs.biochem.6b00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- 36.Wan Y, Sun G, Wang Z, Guo J, Shi L. miR-125b promotes cell proliferation by directly targeting Lin28 in glioblastoma stem cells with low expression levels of miR-125b. Neuroreport. 2014;25:289–96. doi: 10.1097/wnr.0000000000000085. [DOI] [PubMed] [Google Scholar]
- 37.Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX, Yan BY, Nathanson JL, Hutt KR, Lovci MT, Kazan H, Vu AQ, Massirer KB, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao K, Qiu S, Tian L, Snider WD, Flannery JG, Schaffer DV, Chen B. Wnt Regulates Proliferation and Neurogenic Potential of Muller Glial Cells via a Lin28/let-7 miRNA-Dependent Pathway in Adult Mammalian Retinas. Cell Rep. 2016;17:165–78. doi: 10.1016/j.celrep.2016.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, Shah SP, Tanwar PS, Mermel CH, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–8. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen AX, Yu KD, Fan L, Li JY, Yang C, Huang AJ, Shao ZM. Germline genetic variants disturbing the Let-7/LIN28 double-negative feedback loop alter breast cancer susceptibility. PLoS Genet. 2011;7:e1002259. doi: 10.1371/journal.pgen.1002259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–9. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Li N, Zhong X, Lin X, Guo J, Zou L, Tanyi JL, Shao Z, Liang S, Wang LP, Hwang WT, Katsaros D, Montone K, Zhao X, et al. Lin-28 homologue A (LIN28A) promotes cell cycle progression via regulation of cyclin-dependent kinase 2 (CDK2), cyclin D1 (CCND1), and cell division cycle 25 homolog A (CDC25A) expression in cancer. J Biol Chem. 2012;287:17386–97. doi: 10.1074/jbc.M111.321158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.can-07-2462. [DOI] [PubMed] [Google Scholar]
- 44.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–9. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108:21075–80. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma N, Nanta R, Sharma J, Gunewardena S, Singh KP, Shankar S, Srivastava RK. PI3K/AKT/mTOR and sonic hedgehog pathways cooperate together to inhibit human pancreatic cancer stem cell characteristics and tumor growth. Oncotarget. 2015;6:32039–60. doi: 10.18632/oncotarget.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Isanejad A, Alizadeh AM, Amani Shalamzari S, Khodayari H, Khodayari S, Khori V, Khojastehnjad N. MicroRNA-206, let-7a and microRNA-21 pathways involved in the anti-angiogenesis effects of the interval exercise training and hormone therapy in breast cancer. Life Sci. 2016;151:30–40. doi: 10.1016/j.lfs.2016.02.090. [DOI] [PubMed] [Google Scholar]
- 49.Murray MJ, Saini HK, Siegler CA, Hanning JE, Barker EM, van Dongen S, Ward DM, Raby KL, Groves IJ, Scarpini CG, Pett MR, Thornton CM, Enright AJ, et al. LIN28 Expression in malignant germ cell tumors downregulates let-7 and increases oncogene levels. Cancer Res. 2013;73:4872–84. doi: 10.1158/0008-5472.can-12-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dangi-Garimella S, Yun J, Eves EM, Newman M, Erkeland SJ, Hammond SM, Minn AJ, Rosner MR. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. Embo j. 2009;28:347–58. doi: 10.1038/emboj.2008.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang CJ, Hsu CC, Chang CH, Tsai LL, Chang YC, Lu SW, Yu CH, Huang HS, Wang JJ, Tsai CH, Chou MY, Yu CC, Hu FW. Let-7d functions as novel regulator of epithelial-mesenchymal transition and chemoresistant property in oral cancer. Oncol Rep. 2011;26:1003–10. doi: 10.3892/or.2011.1360. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Li H, Feng J, Cui X, Huang W, Li Y, Su F, Liu Q, Zhu J, Lv X, Chen J, Huang D, Yu F. Lin28 induces epithelial-to-mesenchymal transition and stemness via downregulation of let-7a in breast cancer cells. PLoS One. 2013;8:e83083. doi: 10.1371/journal.pone.0083083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Wang YX, Zhang DZ, Fang XJ, Sun PS, Xue HC. Let-7a mimic attenuates CCL18 induced breast cancer cell metastasis through Lin 28 pathway. Biomed Pharmacother. 2016;78:301–7. doi: 10.1016/j.biopha.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 54.Hummel R, Hussey DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer. 2010;46:298–311. doi: 10.1016/j.ejca.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 55.Wang L, Yuan C, Lv K, Xie S, Fu P, Liu X, Chen Y, Qin C, Deng W, Hu W. Lin28 mediates radiation resistance of breast cancer cells via regulation of caspase, H2A.X and Let-7 signaling. PLoS One. 2013;8:e67373. doi: 10.1371/journal.pone.0067373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun X, Jiang S, Liu J, Wang H, Zhang Y, Tang SC, Wang J, Du N, Xu C, Wang C, Qin S, Zhang J, Liu D, et al. MiR-208a stimulates the cocktail of SOX2 and beta-catenin to inhibit the let-7 induction of self-renewal repression of breast cancer stem cells and formed miR208a/let-7 feedback loop via LIN28 and DICER1. Oncotarget. 2015;6:32944–54. doi: 10.18632/oncotarget.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collis SJ, Barber LJ, Clark AJ, Martin JS, Ward JD, Boulton SJ. HCLK2 is essential for the mammalian S-phase checkpoint and impacts on Chk1 stability. Nat Cell Biol. 2007;9:391–401. doi: 10.1038/ncb1555. [DOI] [PubMed] [Google Scholar]
- 58.Oh JS, Kim JJ, Byun JY, Kim IA. Lin28-let7 modulates radiosensitivity of human cancer cells with activation of K-Ras. Int J Radiat Oncol Biol Phys. 2010;76:5–8. doi: 10.1016/j.ijrobp.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 59.Estrada-Bernal A, Chatterjee M, Haque SJ, Yang L, Morgan MA, Kotian S, Morrell D, Chakravarti A, Williams TM. MEK inhibitor GSK1120212-mediated radiosensitization of pancreatic cancer cells involves inhibition of DNA double-strand break repair pathways. Cell Cycle. 2015;14:3713–24. doi: 10.1080/15384101.2015.1104437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teng R, Hu Y, Zhou J, Seifer B, Chen Y, Shen J, Wang L. Overexpression of Lin28 Decreases the Chemosensitivity of Gastric Cancer Cells to Oxaliplatin, Paclitaxel, Doxorubicin, and Fluorouracil in Part via microRNA-107. PLoS One. 2015;10:e0143716. doi: 10.1371/journal.pone.0143716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lv K, Liu L, Wang L, Yu J, Liu X, Cheng Y, Dong M, Teng R, Wu L, Fu P, Deng W, Hu W, Teng L. Lin28 mediates paclitaxel resistance by modulating p21, Rb and Let-7a miRNA in breast cancer cells. PLoS One. 2012;7:e40008. doi: 10.1371/journal.pone.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian N, Han Z, Li Z, Zhou M, Fan C. Lin28/let-7/Bcl-xL pathway: the underlying mechanism of drug resistance in Hep3B cells. Oncol Rep. 2014;32:1050–6. doi: 10.3892/or.2014.3292. [DOI] [PubMed] [Google Scholar]
- 63.Tulsyan S, Agarwal G, Lal P, Mittal B. Significant association of combination of OCT4, NANOG, and SOX2 gene polymorphisms in susceptibility and response to treatment in North Indian breast cancer patients. Cancer Chemother Pharmacol. 2014;74:1065–78. doi: 10.1007/s00280-014-2588-4. [DOI] [PubMed] [Google Scholar]
- 64.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 65.Yang J, Bennett BD, Luo S, Inoue K, Grimm SA, Schroth GP, Bushel PR, Kinyamu HK, Archer TK. LIN28A Modulates Splicing and Gene Expression Programs in Breast Cancer Cells. Mol Cell Biol. 2015;35:3225–43. doi: 10.1128/mcb.00426-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Ratanasirintrawoot S, Chandrasekaran S, Wu Z, Ficarro SB, Yu C, Ross CA, Cacchiarelli D, Xia Q, Seligson M, Shinoda G, Xie W, Cahan P, et al. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell. 2016;19:66–80. doi: 10.1016/j.stem.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 67.Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, Daley GQ. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 2013;155:778–92. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang S, Baltimore D. RNA-binding protein Lin28 in cancer and immunity. Cancer Lett. 2016;375:108–13. doi: 10.1016/j.canlet.2016.02.050. [DOI] [PubMed] [Google Scholar]
- 69.Wang LD, Rao TN, Rowe RG, Nguyen PT, Sullivan JL, Pearson DS, Doulatov S, Wu L, Lindsley RC, Zhu H, DeAngelo DJ, Daley GQ, Wagers AJ. The role of Lin28b in myeloid and mast cell differentiation and mast cell malignancy. Leukemia. 2015;29:1320–30. doi: 10.1038/leu.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marech I, Ammendola M, Sacco R, Capriuolo GS, Patruno R, Rubini R, Luposella M, Zuccala V, Savino E, Gadaleta CD, Ribatti D, Ranieri G. Serum tryptase, mast cells positive to tryptase and microvascular density evaluation in early breast cancer patients: possible translational significance. BMC Cancer. 2014;14:534. doi: 10.1186/1471-2407-14-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335:1195–200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Y, Li YS, Bandi SR, Tang L, Shinton SA, Hayakawa K, Hardy RR. Lin28b promotes fetal B lymphopoiesis through the transcription factor Arid3a. J Exp Med. 2015;212:569–80. doi: 10.1084/jem.20141510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beachy SH, Onozawa M, Chung YJ, Slape C, Bilke S, Francis P, Pineda M, Walker RL, Meltzer P, Aplan PD. Enforced expression of Lin28b leads to impaired T-cell development, release of inflammatory cytokines, and peripheral T-cell lymphoma. Blood. 2012;120:1048–59. doi: 10.1182/blood-2012-01-401760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rao S, Lee SY, Gutierrez A, Perrigoue J, Thapa RJ, Tu Z, Jeffers JR, Rhodes M, Anderson S, Oravecz T, Hunger SP, Timakhov RA, Zhang R, et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor. Lin28B. Blood. 2012;120:3764–73. doi: 10.1182/blood-2012-03-415349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cabodi S, Taverna D. Interfering with inflammation: a new strategy to block breast cancer self-renewal and progression? Breast Cancer Res. 2010;12:305. doi: 10.1186/bcr2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 78.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 80.Cai WY, Wei TZ, Luo QC, Wu QW, Liu QF, Yang M, Ye GD, Wu JF, Chen YY, Sun GB, Liu YJ, Zhao WX, Zhang ZM, et al. The Wnt-beta-catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J Cell Sci. 2013;126:2877–89. doi: 10.1242/jcs.123810. [DOI] [PubMed] [Google Scholar]
- 81.Feng C, Neumeister V, Ma W, Xu J, Lu L, Bordeaux J, Maihle NJ, Rimm DL, Huang Y. Lin28 regulates HER2 and promotes malignancy through multiple mechanisms. Cell Cycle. 2012;11:2486–94. doi: 10.4161/cc.20893. [DOI] [PubMed] [Google Scholar]
- 82.Wu K, Jiao X, Li Z, Katiyar S, Casimiro MC, Yang W, Zhang Q, Willmarth NE, Chepelev I, Crosariol M, Wei Z, Hu J, Zhao K, et al. Cell fate determination factor Dachshund reprograms breast cancer stem cell function. J Biol Chem. 2011;286:2132–42. doi: 10.1074/jbc.M110.148395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jolly MK, Jia D, Boareto M, Mani SA, Pienta KJ, Ben-Jacob E, Levine H. Coupling the modules of EMT and stemness: A tunable ‘stemness window’ model. Oncotarget. 2015;6:25161–74. doi: 10.18632/oncotarget.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 85.Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 86.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 87.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.can-05-1783. [DOI] [PubMed] [Google Scholar]
- 88.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nam S, Kim B, Shin S, Lee S. miRGator: an integrated system for functional annotation of microRNAs. Nucleic Acids Res. 2008;36:D159–64. doi: 10.1093/nar/gkm829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20. doi: 10.1158/0008-5472.can-07-5019. [DOI] [PubMed] [Google Scholar]
- 91.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 92.Kim SJ, Shin JY, Lee KD, Bae YK, Sung KW, Nam SJ, Chun KH. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C-C chemokine receptor type 7. Breast Cancer Res. 2012;14:R14. doi: 10.1186/bcr3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun H, Ding C, Zhang H, Gao J. Let7 miRNAs sensitize breast cancer stem cells to radiationinduced repression through inhibition of the cyclin D1/Akt1/Wnt1 signaling pathway. Mol Med Rep. 2016;14:3285–92. doi: 10.3892/mmr.2016.5656. [DOI] [PubMed] [Google Scholar]
- 94.Sun X, Xu C, Tang SC, Wang J, Wang H, Wang P, Du N, Qin S, Li G, Xu S, Tao Z, Liu D, Ren H. Let-7c blocks estrogen-activated Wnt signaling in induction of self-renewal of breast cancer stem cells. Cancer Gene Ther. 2016;23:83–9. doi: 10.1038/cgt.2016.3. [DOI] [PubMed] [Google Scholar]
- 95.Barh D, Parida S, Parida B, Viswanathan G. Let-7, miR-125, miR-205, and miR-296 are prospective therapeutic agents in breast cancer molecular medicine. Gene Ther Mol Biol. 2008;12:189–206. [Google Scholar]
- 96.Hu X, Guo J, Zheng L, Li C, Zheng TM, Tanyi JL, Liang S, Benedetto C, Mitidieri M, Katsaros D, Zhao X, Zhang Y, Huang Q, et al. The heterochronic microRNA let-7 inhibits cell motility by regulating the genes in the actin cytoskeleton pathway in breast cancer. Mol Cancer Res. 2013;11:240–50. doi: 10.1158/1541-7786.mcr-12-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ma L, Li GZ, Wu ZS, Meng G. Prognostic significance of let-7b expression in breast cancer and correlation to its target gene of BSG expression. Med Oncol. 2014;31:773. doi: 10.1007/s12032-013-0773-7. [DOI] [PubMed] [Google Scholar]
- 98.Qian P, Zuo Z, Wu Z, Meng X, Li G, Wu Z, Zhang W, Tan S, Pandey V, Yao Y, Wang P, Zhao L, Wang J, et al. Pivotal role of reduced let-7g expression in breast cancer invasion and metastasis. Cancer Res. 2011;71:6463–74. doi: 10.1158/0008-5472.can-11-1322. [DOI] [PubMed] [Google Scholar]
- 99.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. Rna. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen GQ, Zhao ZW, Zhou HY, Liu YJ, Yang HJ. Systematic analysis of microRNA involved in resistance of the MCF-7 human breast cancer cell to doxorubicin. Med Oncol. 2010;27:406–15. doi: 10.1007/s12032-009-9225-9. [DOI] [PubMed] [Google Scholar]
- 101.Xin F, Li M, Balch C, Thomson M, Fan M, Liu Y, Hammond SM, Kim S, Nephew KP. Computational analysis of microRNA profiles and their target genes suggests significant involvement in breast cancer antiestrogen resistance. Bioinformatics. 2009;25:430–4. doi: 10.1093/bioinformatics/btn646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sakurai M, Miki Y, Masuda M, Hata S, Shibahara Y, Hirakawa H, Suzuki T, Sasano H. LIN28: a regulator of tumor-suppressing activity of let-7 microRNA in human breast cancer. J Steroid Biochem Mol Biol. 2012;131:101–6. doi: 10.1016/j.jsbmb.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 103.Xie R, Wang Y, Nie W, Huang W, Song W, Wang Z, Guan X. Lin28B expression correlates with aggressive clinicopathological characteristics in breast invasive ductal carcinoma. Cancer Biother Radiopharm. 2014;29:215–20. doi: 10.1089/cbr.2014.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shen H, Yang Y, Zhao L, Yuan J, Niu Y. Lin28A and androgen receptor expression in ER-/Her2+ breast cancer. Breast Cancer Res Treat. 2016;156:135–47. doi: 10.1007/s10549-016-3744-9. [DOI] [PubMed] [Google Scholar]
- 105.Liu Q, Bai X, Li H, Zhang Y, Zhao Y, Zhang X, Ye L. The oncoprotein HBXIP upregulates Lin28B via activating TF II D to promote proliferation of breast cancer cells. Int J Cancer. 2013;133:1310–22. doi: 10.1002/ijc.28154. [DOI] [PubMed] [Google Scholar]
- 106.Segalla S, Pivetti S, Todoerti K, Chudzik MA, Giuliani EC, Lazzaro F, Volta V, Lazarevic D, Musco G, Muzi-Falconi M, Neri A, Biffo S, Tonon G. The ribonuclease DIS3 promotes let-7 miRNA maturation by degrading the pluripotency factor LIN28B mRNA. 2015;43:5182–93. doi: 10.1093/nar/gkv387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yan L, Zhou J, Gao Y, Ghazal S, Lu L, Bellone S, Yang Y, Liu N, Zhao X, Santin AD, Taylor H, Huang Y. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2015;34:3076–84. doi: 10.1038/onc.2014.236. [DOI] [PubMed] [Google Scholar]
- 108.Enriquez VA, Cleys ER, Da Silveira JC, Spillman MA, Winger QA, Bouma GJ. High LIN28A Expressing Ovarian Cancer Cells Secrete Exosomes That Induce Invasion and Migration in HEK293 Cells. Biomed Res Int. 20152015 doi: 10.1155/2015/701390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dubinsky AN, Dastidar SG, Hsu CL, Zahra R, Djakovic S, Duarte S, Esau CC, Spencer B, Ashe TD, Fischer KM, MacKenna DA, Sopher BL, Masliah E, et al. Let-7 coordinately suppresses components of the amino acid sensing pathway to repress mTORC1 and induce autophagy. Cell Metab. 2014;20:626–38. doi: 10.1016/j.cmet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Buchholz DE, Hikita ST, Rowland TJ, Friedrich AM, Hinman CR, Johnson LV, Clegg DO. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009;27:2427–34. doi: 10.1002/stem.189. [DOI] [PubMed] [Google Scholar]
- 111.Wang L, Zhang LF, Wu J, Xu SJ, Xu YY, Li D, Lou JT, Liu MF. IL-1beta-mediated repression of microRNA-101 is crucial for inflammation-promoted lung tumorigenesis. Cancer Res. 2014;74:4720–30. doi: 10.1158/0008-5472.can-14-0960. [DOI] [PubMed] [Google Scholar]
- 112.You X, Liu F, Zhang T, Lv N, Liu Q, Shan C, Du Y, Kong G, Wang T, Ye L, Zhang X. Hepatitis B virus X protein upregulates Lin28A/Lin28B through Sp-1/c-Myc to enhance the proliferation of hepatoma cells. Oncogene. 2014;33:449–60. doi: 10.1038/onc.2012.618. [DOI] [PubMed] [Google Scholar]
- 113.Teng RY, Zhou JC, Jiang ZN, Xu CY, Li ZD, Wang QC, Xu CP, Guo JF, Shen JG, Wang LB. The relationship between Lin28 and the chemotherapy response of gastric cancer. Onco Targets Ther. 2013;6:1341–5. doi: 10.2147/ott.s45705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS, Rustgi AK. LIN28B promotes colon cancer progression and metastasis. Cancer Res. 2011;71:4260–8. doi: 10.1158/0008-5472.can-10-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Docherty CK, Salt IP, Mercer JR. Lin28A induces energetic switching to glycolytic metabolism in human embryonic kidney cells. Stem Cell Res Ther. 2016;7:78. doi: 10.1186/s13287-016-0323-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene. 2010;29:2153–9. doi: 10.1038/onc.2009.500. [DOI] [PubMed] [Google Scholar]
- 117.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 118.Wang T, Wang G, Hao D, Liu X, Wang D, Ning N, Li X. Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Mol Cancer. 2015;14:125. doi: 10.1186/s12943-015-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


