Abstract
Lipids and nucleic acids (NAs) can hierarchically self-organize into a variety of nanostructures of increasingly complex geometries such as the 1D lamellar, 2D hexagonal, and 3D bicontinuous cubic phases. The diversity and complexity of those lipid–NA assemblies are interesting from a fundamental perspective as well as being relevant to the performance in gene delivery and gene silencing applications. The finding that not only the chemical make of the lipid-NA constructs, but their actual supramolecular organization, affects their gene transfection and silencing efficiencies has inspired physicists, chemists, and engineers to this field of research. At the moment it remains an open question how exactly the different lipid-NA structures interact with cells and organelles in order to output an optimal response. This article reviews our current understanding of the structures of different lipid–NA complexes and the corresponding cellular interaction mechanisms. The recent advances in designing optimal lipid–based NA carriers will be introduced with an emphasis on the structure–function relations.
Keywords: self-assembly, gene delivery, lipid-DNA, lipid-siRNA
Graphical abstract
1. Introduction
Lipids are a class of organic compounds possessing both hydrophilic and hydrophobic moieties. The amphiphilic nature of lipids leads to molecular self-assembly behavior in solutions, forming various nanostructures of liquid crystalline nature. The characteristics of such assemblies that they are nano-sized and structured have led to numerous practical applications in pharmaceutical and biomedical technologies. Especially, the ability of lipid self-assembled aggregates to encapsulate nucleic acids (NAs) have earned researcher’s attention to exploit them as gene delivery vehicles (vectors) as early as in 1979. The advantages of these vectors include the ease of preparation, low immunogenicity, and ability to transfer a large amount of NA. However, low gene delivery efficiency of those vectors has limited their therapeutic applications, provoking the improvements of the vectors by rational design. The rational design of gene delivery vectors requires a coherent understanding of their structures and interactions with cells. The step-by-step mechanisms of actions are still not clearly understood but we know from substantial research efforts that the structure of lipid-NA complexes is one of the important parameters determining the gene delivery efficiency. In addition, such systems have attracted the attention of a broad range of disciplines as the assembly of lipid-NA constructs is orchestrated by a balance of forces of different natures such as long range electrostatic attraction/repulsion and short range repulsions, many of which remain poorly understood at a very fundamental level.1–4 The current research efforts are directed towards clarifying the interplay between distinct nanoscale structures of lipid-NA complexes and their interactions with cells. In line with this research trend, we provide an overview of the development of the lipid-NA complexes focusing on the structural aspect, and deliver insights for the future research in this field.
In the first part of this review, we present an introduction to the general principles governing lipid self-assembly behavior. This section will help the reader to make the link between the molecular structure of lipids and the resulting self-assembly morphology of lipid-NA complexes. A chronological account of the various structures of lipid-NA constructs is provided as well as how they correlate to cellular interactions and gene delivery/silencing efficacy. Finally, we propose a number of future research directions and applications for lipid-NA complex materials.
2. Lipid self-assembly
Lipid molecules are mostly known to self-organize into bilayer structures resembling that of plasma membranes. However, depending on a number of intrinsic (e.g. molecular packing and membrane curvature) or extrinsic (salinity, solvent properties, or external cues) properties, lipids offer a rather diverse design space of nanostructures. Examples include 2D hexagonal phases and 3D cubic phases of multiple symmetries.5
The pioneering work on fundamental theory of self-assembly of amphiphiles was proposed by Tanford6 who quantitatively modeled the standard free energy change associated with the transfer of singly dispersed free amphiphiles in solution to self-assembled aggregates. He introduced the concept of opposing energy contributions; a negative free energy contribution arising from the hydrophobic effect and a positive free energy contribution due to the repulsive interactions emanating from lipid head groups. The hydrophobic interaction drives the transfer of the hydrophobic lipid tail that disrupts the H-bonding water network to the aggregate core which possess the hydrocarbon-like environment. While the hydrophobic effect favors the formation of aggregates, the assembly of lipid molecules entail the repulsive interactions such as steric interactions for all types of lipid head groups, dipole-dipole interactions for zwitterionic head groups, and ion-ion repulsions for ionic head groups. The competition between the hydrophobic effect and repulsive interactions, therefore, explains why the self-assembly process happens, how it is highly tunable by environmental conditions, and why the resulting aggregates have a finite size. Although Tanford’s model explains the basic features of self-assembly, it could not predict the final shape of the equilibrium aggregates.
Israelachvili et al.7 further developed the framework of the Tanford free energy model by combining the concept of a molecular packing parameter with thermodynamics to predict the shape of the self-assembly aggregates in equilibrium. To parameterize the geometric constraints imposed by molecular packing, a specific packing parameter P was introduced. A molecular packing parameter is defined as P = v/(alc), where v = the volume of the hydrocarbon, a = the effective area of the head group, and lc = the effective length of the lipid tail.
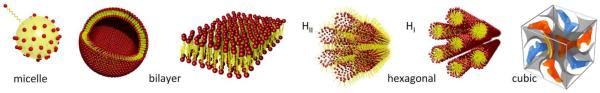
The magnitude of packing parameter is related to the shape of aggregate formed. Cone-shaped lipids with bulky head group (e.g., lysophosphocholine or LPC) with small P (0-1/3) tend to form spherical aggregates such as micelles. Truncated cone-shaped lipids with P value between 1/3 and 1/2 organize into cylindrical micelles, and those with packing parameter between 1/2 and 1 prefers to form flexible bilayers or vesicles. Cylinder-shaped lipids (e.g., dioleylphosphocholine or DOPC) tend to form the flat bilayer with nearly zero curvature, i.e. lamellar phase (P ≈ 1). For lipids with small effective head group area such as dioleylphosphatidylethanolamine or DOPE, the relative tail splay becomes wide (P > 1), leading to non-bilayer lipid phases such as an inverted hexagonal phase. Figure 1 schematically represents typical phases obtained with lipid systems.
Figure 1.
Schematic representation of various lipid self-assembled structures. Lipids in the presence of water can form micelle, liposome, inverse and normal hexagonal phases, bicontinuous cubic phase, respectively from left to right.
Although the general principles of lipid self-assembly could be well understood from the pioneering work by Tanford and Israelachvili, it could not capture the complete morphology of all lipid structures. As an alternative to the packing parameter, Gruner8 explained nonlamellar phase formation in terms of spontaneous curvature. If a molecule has a preferred average geometry, the deviations from the preferred geometry yield a bending energy cost. Often, it is necessary to think about the stabilization of the different lipid phases in terms of lipid membrane elasticity that was appropriately described by Helfrich.9,10 The Helfrich equation is expressed as the following: the Helfrich energy (E) per area (A) = 0.5κ(C-C0)2+κGC1C2 where κ and κG are the bending and the Gaussian modulus respectively. The spontaneous curvature C0 represents a tendency of a flat monolayer to bend spontaneously. Practically, it means a stress-free state of a monolayer at given geometry. The first term thus describes the energetic cost of bending a lipid layer from its spontaneous curvature C0 to the deformed curvature C. The second term accounts for the topology of the lipid layer where C1 and C2 are two principal curvatures of the lipid layer plane. The Helfrich equation becomes useful when describing for example lipid bicontinuous cubic phases that were firstly introduced by Luzzati.11 The failure of explaining the formation of biocontinuous cubic phases by the packing parameter concept arises from the negligence of membrane curvature arguments with global packing constraints. While the packing parameter P treats geometrical constraints locally, i.e. at the single-molecule level, the concept of a membrane curvature extends the packing constraints to be macroscopic. In general, the shape of the resulting self-assembly aggregate can be predicted as a function of the spontaneous curvature C0 of membranes.
3. Interactions between nucleic acids and lipid aggregates
To successfully deliver nucleic acid-lipid complexes to the target site of delivery, stable loading of nucleic acids into lipid formulations should be achieved first. DNA-lipid binding and DNA condensation are the first stage of the gene delivery process. In this context, we will take a look into electrostatic interactions that mainly govern the DNA-lipid binding process and DNA condensation. The hydration of the nucleic acid-lipid complexes also comes into play in determining the self-assembled structure of nucleic acid-lipid complexes. The finite amount of water included in the complexes exerts repulsive forces preventing the collapse of the complexes. As the interactions among nucleic acids, lipid headgroup, lipid tail, and surrounding water molecules occur in a highly cooperative manner, it is hard to separate out the individual component of such interactions. However, the examination of two main interactions between nucleic acids and lipid aggregates will help us to gain a better understanding of the structure of lipid-NA complexes.
3.1 Electrostatic interaction
DNA is a negatively charged polyelectrolyte whose conformation is influenced by the ionization of phosphate groups in its backbone. When cationic lipid such as 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) or 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA) binds to the extended DNA backbone, the positive charge of the lipid headgroup neutralizes the phosphate groups on the DNA leading to DNA condensation. During such lipid-DNA binding process via electrostatic interactions, counterions that were both bound to DNA and charged lipids become released in bulk solution, resulting in a large increase in entropy. According to a mathematical model of lipid-DNA binding thermodynamics,12 lipid-DNA interaction is mainly mediated by two contributions: 1) a translational entropic gain due to counterion release and 2) an increase in entropy due to additional disorder of the lipid aliphatic chains. The direct evidence of counterion release upon CL-DNA condensation was demonstrated by Wagner et al.13 with the conductivity measurement of the solution before and after the CL-DNA complex formation.
The electrostatic interaction is strongest in the absence of salt ions, i.e. when there is no electrostatic screening effects. Thus use of salt ions with different charge and concentration can modulate the strength of the electrostatic interaction bringing additional changes to the complex structure. For example, addition of salt ions to DNA-surfactant complexes has been reported to increase the uptake of water where the driving force of swelling is based on the higher contribution of salt to the osmotic pressure accompanied with decreased electrostatic attraction.14
The electrostatic interactions between DNA and cationic/zwitterionic lipid membranes are significant when it comes to the molecular mobility and the packing of lipids. In DNA-lipid complexes reported by Leal et al., the preferential binding of DNA to cationic lipids (dimethyldioctadecylammonium or DDA) over zwitterionic lipids (1,2-dilauroyl-sn-glycero-3-phosphocholine or DLPC) resulted in retarded mobility of DNA and decreased diffusion rate of the associated cationic lipids (DDA) with increased local mobility of alkyl chains.15
The binding of DNA to zwitterionic lipids also occurs in the presence of divalent cations such as Mg2+ or Ca2+.16,17 The divalent cations are predicted to neutralize the phosphate groups of the zwitterionic lipids causing the reorientation of lipid headgroup and leaving net positive charge in lipid membrane.17,18 The driving force of DNA and zwitterionic lipid interaction is electrostatic in nature but the mechanism differs from that of cationic lipid-DNA analogues as the redistribution of divalent cations from solution to zwitterionic lipid headgroups plays a key role in complex formation.18 Binding salt ions to the zwitterionic lipid membranes induces electrostatic repulsion of membranes, suggesting the possibility of modulating DNA and lipid membrane interactions using salt ions.19
Additionally, it was shown that the phase of lipids affects electrostatic binding of nucleic acids to zwitterionic lipid membrane in the presence of divalent cations. In heterogeneous lipid membranes composed of lipid ordered (Lo) and lipid disordered phases (Ld), coiled DNA selectively adsorbed onto the Lo phase.20 This experimental finding is in line with the ion adsorption mechanism explaining the interactions among DNA, zwitterionic lipids, and divalent cations given that closely packed Lo phases have higher surface charge density than the parent Lo phases.21 Higher surface charge density would make the binding of DNA on the membrane more stable and thus more preferable.
Besides cationic and zwitterionic lipids, anionic lipids also have been examined as an alternative gene delivery vector in an effort to circumvent the potential toxicity in cells. To condense DNA in anionic lipid systems, divalent cations are essentially required as a mediator of DNA and anionic lipid interactions. Divalent cations such as Zn2+ are assumed to “bridge” anionic lipid headgroups and anionic DNA through direct electrostatic interactions.22,23 The membrane charge density and ion concentration were shown to affect the DNA-anionic lipid complex formation.23
3.2 Hydration of nucleic acid-lipid aggregates
The hydration force is often described as a short range repulsive interaction acting between hydrated bilayers approaching each other.24 The origin of this force is still under debate with the two main interpretations being either i) the structure of liquid water chemically or physically attached onto the membrane surface or ii) the result of confinements of surface excitations.25 Even though the mechanism is not yet clearly defined, there seems to be strong experimental evidence that this short range repulsion force is of entropic nature.26
At this stage of research, we recognize that 1) water penetrates and distributes in the lipid membrane with well controlled and finite amounts and 2) water mediates the lipid-lipid membrane interaction and lipid-nucleic acid complex formation, however the exact role and the extent of involvement of water on the complex structure are still under investigation.27
The study on hydration of a DNA-Cetrimonium bromide (CTAB) complex revealed that the amount of water uptake by the complex is lower than that by pure DNA, suggesting that the presence of cationic surfactant CTAB and its interaction with DNA influenced the equilibrium water layer of the complex. The hydration fluctuations at the water-lipid interface were monitored during short DNA(oligonucelotide)-lipid complex formation. The observed hydration level at different DNA to lipid ratio suggests that water is excluded from the lipid headgroup region while electrostatic pinning occurs, possibly replaced by short DNA molecules.28
Of note, it is known that the certain salts affect the level of hydration in lipid membranes. According to molecular dynamics simulation studies by Gurtovenko et al., the addition of Na+ ions to 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)/ 1,2-dipalmitoyl-3-trimethylammonium-propane (DPTAP) lipid membranes decreased the hydration level of lipids as Na+ ions held water molecules instead of lipids, hence required 1.5 fold increased number of water molecules to reach the hydration equilibrum.29 Interesting experimental results obtained by infrared spectroscopy indicated that divalent ions differ in their binding ability to lipids, affecting the local environment of water molecules near lipid headgroups. For instance, Zn2+ ions interact with lipid headgroups strongly, thereby reducing the number of water molecules near headgroups. Likewise Cu2+ ions have a partial dehydration effect while Mg2+ ions have a minimal effect on lipid headgroup dehydration.30 Such ion-specific interactions with lipid membranes are important when one wants to control the hydration level, lipid headgroup structure, and the phase behavior.
4. Structure of nucleic acid-lipid complexes
Ever since the pioneering work on the use of cationic lipids (CL) as non-viral vectors of nucleic acids (NA) by Felgner et al.,31 CL-NA complexes have been extensively investigated for gene therapy applications. The advantages of CL-NA complexes for gene delivery and gene silencing include low toxicity, low immunogenicity, ease of production, and capacity to store large NA amounts.32 Despite these advantages, the low transfection/silencing efficiency retards the clinical applications of the CL-NA complexes, giving rise to a need for better understanding of their nanoscale structures which have been shown to influence the transfection/silencing pathways and mechanisms. In this section, we will present an overview of different structures of CL-DNA complexes focusing on the nanoscale assembly features and their implications on the transfection/silencing performance. The recent studies on the CL-NA complex structures provide insight into the possibility of controlling the structure upon external stimuli that can be advantageous for actuated gene delivery.
4.1 Lamellar phase
The equilibrium supramolecular structure of CL-DNA complexes was first elucidated by means of synchrotron small angle X-ray scattering (SAXS) experiments by Rädler et al. in 1997.33
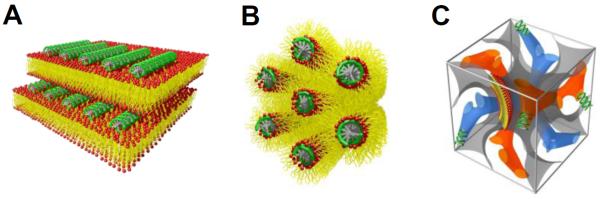
In the absence of DNA, the binary mixture of neutral zwitterionic DOPC and univalent cationic DOTAP lipids forms liposomes which are closed vesicles. Upon addition of DNA, a liposome undergoes a topological transition into an open layered structure while adapting DNA molecules within the structure. Such layered structure, so called a lamellar phase () is composed of sheets of bilayers that are separated by bulk water. SAXS studies revealed that a single layer of DNA is located between lipid bilayers constituting a sandwiched lamellar structure as shown in Figure 2A. The driving force of the CL-DNA self-assembly is the entropic gain due to counterion release upon DNA binding to cationic lipids as stated in the section 3.1.
Figure 2.
(A) The lamellar phase of CL-DNA complexes (). DNA monolayers (green) are intercalated between lipid bilayers. (B) The inverted hexagonal phase of CL-DNA complexes (). DNA molecules (green) are inserted within water channels of the inverse lipid tubules forming a hexagonal array. (C) The unit cell of the double gyroid lipid cubic phase of CL-siRNA complexes (). siRNAs (green) are incorporated within two intertwined but independent water channels (blue and orange). The lipid bilayer is represented as a gray surface.
In SAXS scans, a broader and weaker peak that does not belong to lamellar peaks appears at different locations depending on the average bilayer charge density and cationic lipid to DNA charge ratio. The appearance of such peak indicates the existence of DNA-DNA correlations. The line shape analysis of the DNA peak demonstrates that the alignment of DNA molecules adsorbed onto lipid bilayers is governed by electrostatic repulsions rather than pure hard-core interactions, steric repulsion, or hydration forces.34 Those DNA correlations are short-range, but strong enough to guide DNA chains to constitute a 1D lattice with a regular interaxial spacing (i.e., 2D smectic phase). Additionally, the linewidth of the DNA peak conveys information regarding the domain size of correlated DNA which was found to consist of ~10 neighboring chains. Altogether, the SAXS data analysis demonstrates that the supramolecular structure of CL-DNA complex exhibits a lamellar structure where parallel DNA chains are intercalated between lipid bilayers with a positional correlation between adjacent layers.
CL-siRNA (short interfering RNA) complexes show a similar structure to that of CL-DNA complexes. Alternating layers of lipid bilayers and siRNA single layers are periodically spaced comprising a lamellar structure. However, much shorter length of siRNA molecules (typically between 19 and 25 bp) than that of DNA (~48 000 bp for λ–DNA and 5000 bp for plasmid DNA) brings different features in the formation of complex structure. First, siRNA molecules possess greater rotational and translational degrees of freedom than DNA. This leads to weaker correlations among siRNA molecules when adsorbed to lipid membranes compared to DNA-DNA correlations. Thus in SAXS scans, the siRNA correlation peak is absent or appears very broad and weak (the former observed in DOPC/DOTAP-siRNA systems and the latter observed using multivalent cationic lipid materials, N1-[2-((1S)-1-[(3-aminopropyl)amino]-4-[di(3-amino-propyl)amino]butylcarboxamido)ethyl]-3,4-di[oleyloxy]-benzamide (MVL5)/DOTAP-siRNA).35 This loss of orientational order of siRNA is in accordance with the observation that siRNA layers exist as a liquid-like phase. Second, siRNA molecules have a larger diffusion constant than DNA, thereby shortening the time required to reach the equilibrium state of CL-siRNA complexes. According to time-resolved XRD studies, CL-siRNA complexes were equilibrated in a matter of hours35 in contrast to CL-DNA complexes which required days.33
The correlation among DNAs that are confined to a two-dimensional lipid plane was systemically studied by Bouxsein et al. in 2011.36 Short DNA rods (11, 24, and 48 bp) with nonpairing overhangs exhibited rich liquid crystal phase behavior in CL-sDNA complexes from an isotropic phase to 2D nematic (N) and 2D columnar nematic (NC) phases. Such rich phase behavior of sDNA originates from the end-to-end stacking interactions37 between sDNA molecules which could be modulated by the length of the nonpairing overhangs.36 This work implicates the possibility of modulating end-to-end interactions to guide the stacking of short DNA or other rod-like molecules to form a certain self-assembled phase. The application of such end-to-end pairing has not yet been reported in CL-siRNA complexes, but it is expected to improve the packing efficiency of siRNAs in the complexes.
Recent studies on the lamellar structure of NA-lipid complexes take novel design approaches to induce a stimuli-activated structural reorganization so as to exert control over the release and activity of nucleic acids. For example, the inclusion of redox-active lipids in CL-siRNA complexes permitted to induce the molecular reorganization of complex structure from an amorphous state to a lamellar structure via controlling the redox state, resulting in the high level of gene silencing.38 Another study employed pH-sensitive lipids in CL-siRNA complexes. The synthesis of pH-cleavable lipids which release fatty alcohols into phospholipid bilayers under a weak acidic condition promoted the destabilization of existing liposomes and transition into a lamellar phase.39 Such phase destabilization and transition at the lower endosomal pH led to a successful release of siRNAs into cellular cytoplasm.39
4.2 Inverted hexagonal phase
Soon after the initial finding of phase in 1997,33 a new phase of CL-DNA complexes has been discovered by Koltover et al. in 1998.40 The new phase appeared upon the inclusion of neutral DOPE lipids in CL-DNA mixtures which are well-known as “helper-lipids” along with DOPC lipids. The DOPE lipids have a much smaller effective headgroup size than DOPC lipids because of the weaker hydration. Such chemical structure drives the resulting self-assembly structures of DOPE lipids to have negative spontaneous curvature. Thus when incorporated to the flat bilayers with zero curvature (Lα phase), DOPE lipids would introduce lipid packing stress into structures and tend to change structures to have more negative curvature.41
The supramolecular structure of CL-DNA complexes containing DOPE lipids was revealed by SAXS measurements.40 The identified diffraction peaks corresponded to a 2D hexagonal lattice with a high degree of regularity which the author referred to as phase. The plausible packing of lipids and DNA molecules in a 2D hexagonal lattice would be extended arrays of long lipid tubes with hydrocarbon tails on the outside and the polar headgroups on the inside generating aqueous channels inwards where DNA molecules are trapped as shown in Figure 2B. The calculated unit cell spacing corresponded to 67.4 Å at ΦDOPE = 0.75 for DOPE/DOTAP-DNA system. The subtraction of an average lipid monolayer thickness from the unit cell spacing leaves an estimated diameter of the cylindrical void that is near 28 Å. This confers enough space for accommodating DNA molecules inside the lipid inverse micelles.
A more detailed structure of phase was obtained from an electron-density reconstruction of the x-ray diffraction data. The electron density calculation along the [10] direction of the hexagonal lattice showed different profiles between HII phase of pure DOPE and phase of the DOPE/DNA/Fe2+ complex.42 The electron density profile of the complex assures the occupation of DNA at the core of cylinders which were filled by only water molecules in the absence of DNA.
Apart from the addition of lipid with negative spontaneous curvature, the addition of membrane cosurfactant molecules such as pentanol and hexanol can also induce the formation of phase, which is attributed to the reduction of the membrane bending rigidity κ. The increase in a membrane fluidity reduced the elastic energy barrier to form the phase, facilitating to phase transitions in DOPC/hexanol/DOTAP-DNA systems.40
The structure of CL-siRNA complexes has been reported in glycerol monooleate (GMO/DOTAP-siRNA and MVL5/DOTAP-siRNA systems.35 The structure of CL-siRNA complexes has the same geometries as the one in CL-DNA complexes. Although there are not much differences between CL-DNA complexes and CL-siRNA complexes in terms of the self-assembly structure, the performance as a gene delivery vector presents unexpected differences, which will be described in section 5.
Worthy of note, some studies exploited to phase transitions as a means of designing better CL-NA complexes that undergo phase transitions upon interactions with cell membranes. This approach has been started based on the hypothesis that the degree of leakiness and release of nucleic acids from CL-NA complexes would be facilitated in the vicinity of phase transitions.43,44
4.3 Normal hexagonal phase
The third phase of CL-DNA complexes was discovered by Ewert et al in 2006.45 A highly charged (16+) dendritic multivalent cationic lipid, gave rise to the appearance of a novel hexagonal phase, denoted , that differs from the phase. The structure of a new phase was elucidated by means of SAXS as previous two phases of CL-DNA complexes ( and ). The SAXS patterns for the phase indicated that lipid and DNA molecules assembled into a hexagonal-close-packed lattice with the lattice constant of a = 81.5 Å, which is larger than that of the phase (67.4 Å). This indirectly implies the different arrangement of DNA and lipids in the new phase . In the phase elongated cylinder-shaped lipid micelles constitute a honeycomb lattice and the DNA rods are located in the interstices of the lipid micelle arrangement. It is interesting to note that a geometric nature of normal hexagonal phase () involves a contiguous packing of DNA in the complex as opposed to an isolated packing of DNA in the inverted hexagonal phase () and lamellar phase ().
The phase was only present at around 25 mol % of dendritic lipid in DOPC/dendritic-lipid/DNA system. A dendritic bulky headgroup of the dendritic lipid makes the molecule cone-shaped, driving the formation of micelles. Higher molar content of MVLBG2 above 25 mol% yielded an undefined structure with lower symmetry. The optimum inclusion of MVLBG2 and the balance with DOPC lipids were the key to drive self-assembly of tubular lipid micelles into a hexagonal array.
The dendritic-lipid/DOPC-DNA complexes showed superior transfection efficiencies in many different cell lines compared to DOTAP/DOPC-DNA complexes in the whole range of cationic lipid molar fraction. As phase is present in a very narrow range of dendritic-lipid, it is difficult to examine the transfection efficiency of sole phase and relate that with the structural uniqueness of phase. Nevertheless, a continuous DNA substructure of phase is intriguing in that such interconnected DNA lattice does not exist in other CL-DNA complexes which raises questions such as — Will the linked substructure of DNA facilitate its release upon exposure to cells? We expect further studies to unveil the role of a continuous DNA substructure in cellular transfection mechanism and efficiency.
4.4 Cubic phases
The bicontinuous lipid cubic phase is another class of thermodynamically stable lipid self-assembled structures. The bicontinuous cubic phase has a complex architecture, and thus its structure became gradually revealed and accepted during the 1980s with earlier pioneering work by Luzzati and Lutton.46,47 The lipid bicontinuous cubic phase can be expressed by infinite series of periodic minimal surfaces, which divide the space into two separated water channels. The cubic phase has several structural advantages including isotropic material exchange and fusogenic properties due to its resemblance of the intermediate states (lipid bilayer pores) in a membrane fusion process. Therefore the cubic phase has been recently recognized as an effective vehicle to deliver nucleic acids, which has a limiting step for endosomal escape process with previously tested vehicles.48
The cellular delivery of nucleic acids using bicontinuous cubic phases was first studied by Leal and coworkers in 2010 using siRNA.49 The authors studied an inverse gyroid bicontinuous cubic phase made of GMO and DOTAP. A combined study of optical microscopy and SAXS on GMO/DOTAP/siRNA systems reveals that there is a composition window where a cubic phase of the gyroid type is preserved with embedded siRNAs in the lipid matrix. Figure 2C shows the schematic of gyroid lipid cubic phase in which siRNAs are incorporated within its two water channels (). The authors hypothesized that highly charged siRNAs are favorably located in hydrophilic water channel regions. They assumed that a positively charged bicontinuous cubic phase has swelled water channels, thereby allowing inclusion of siRNA molecules inside the swelled channel. This lipid cubic phase-siRNA complex showed superior efficiency over conventional cationic lipid-siRNA (CL-siRNA) complex with significantly low cytotoxicity by minimizing cationic charge density in the cubic lipid matrix. This study sheds light on the importance of fundamental understanding on interaction between nucleic acids and lipid with cubic phase.
In 2013, Leal and coworkers studied how DNA length affects the structure of lipid-nucleic acid complexes.50 They used “sticky” adenine (A)–thymine (T) (dAdT) overhangs for long DNA demonstration. As control groups, no overhangs (blunt) and “nonsticky” (dTdT) overhangs were used respectively. The temperature modulation allows control of DNA assemblies which results in controlling the length of linearly stacked DNAs within the cubic phase matrix. From this study, they found that the assembly of short DNAs with sticky ends induces reversible structural transitions from the cubic geometry towards inverse hexagonal phases. This structural change can be understood by considering the highly curved channel structure of the gyroid cubic phase. In other words, the nucleic acids are accommodated within bicontinuous cubic phases only at DNA lengths shorter than the unit cell dimension of the gyroid cubic phase.
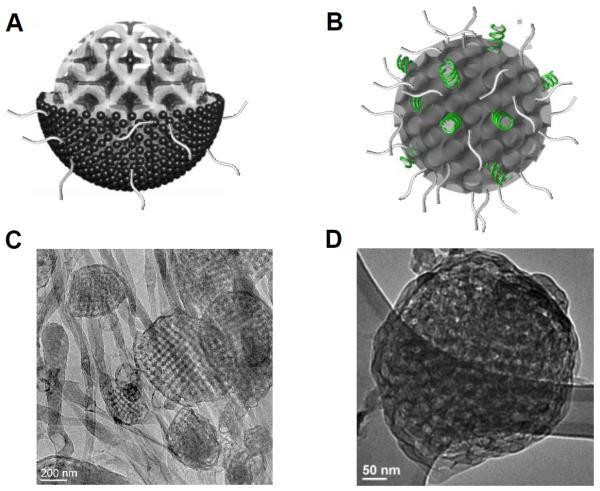
The studies described above carefully characterized how nucleic acids interact with bulk lipid cubic phases. However, for the application of gene delivery it is necessary to translate into a particulate system where colloidal stability can be achieved. For lamellar CL-NA phases it is known that the bulk sandwiched phase translates to a multilayer particulate analogue (lipoplexes) of diameters ranging within a few hundred nanometers.51 However, for other CL-NA phases, little is known about the formation of particles with colloidal stability retaining the structural identity of the mother bulk phase. In 2015, Kim and Leal reported a nanometer scale lipid bicontinuous cubic phase-siRNA complex, which were named as cuboplexes.52 The schematic representation of such system as well as the correspondent cryogenic transmission electron microscopy (Cryo-EM) images are depicted in Figure 3.
Figure 3.
Schematic illustration of (A) lipid-only and (B) lipid-siRNA Im3m cubic phase nanoparticles showing the internal bicontinuous cubic structure. siRNA (green) is incorporated in the water channels. PEG chains are located towards the outside of the particles as represented with gray tails. Cryo-Em images of (C) lipid-only and (D) lipid-siRNA Im3m cubic phase particulate systems obtained from GMO/DOTAP/GMO-PEG (95/4/1)-siRNA mixtures. Before and after siRNA incorporation, the internal cubic symmetry is preserved while maintaining the spherical shape of the particles. Note that water channels in (D) appear darker than that in (C) implying the presence of encapsulated siRNA molecules within the channels.
In this work, the authors report on the fragmentation of the siRNA embedded cubic phase matrix to several hundreds of nanometers through indirect sonication, and successfully imaged individual cubic phase-siRNA complexes through Cryo-EM. This significant advance was achieved by adopting a third lipid component, a modified GMO lipid attached to a polyethylene glycol unit-GMO-polyethylene glycol2k (GMO-PEG). Very small amounts of GMO-PEG impart crucial colloidal stability of the cubic phase without its disassembly. Through in-house SAXS studies a few important findings were revealed in the GMO/DOTAP/GMO-PEG-siRNA system. Firstly, two bicontinous cubic phases of different symmetry were observed that showed distinct responses upon siRNA inclusion. In addition to the gyroid phase previously obtained with the GMO/DOTAP-siRNA system, addition of GMO-PEG enables the stabilization of a primitive bicontinuous cubic phase (QIIP, Im3m).53 Importantly, for these PEG-loaded systems only the primitive cubic phase is preserved during siRNA incorporation while the gyroid cubic phase completely transformed to an hexagonal phase. This interesting behavior is in contrast to previous GMO/DOTAP-siRNA systems where the gyroid maintained its inner structure.49 The authors explained this behavior through the size and the degree of curvature of the water channels in the different bicontinuous cubic phases. Lower degree of curvature and larger water channels are eventually more favorable for the inclusion of rigid, rod-like siRNA molecules. Combined with previous studies involving DNA length and bicontinuous cubic phase stabilization,50 we argue that there is an optimum length and amount of nucleic acids that can be incorporated into cubic phase matrices without changing their internal structure. The Cryo-EM images are consistent with the idea of the GMO-PEG lipid units preferentially locating at the outer rims of cubosomes (lipid only particles) and cuboplexes (lipid-siRNA complex particles).
5. Interactions between CL-NA complexes and cells
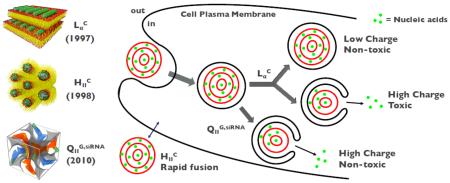
Transfection efficiency (TE) of distinct phases of CL-DNA complexes shows significant differences, implying the possibility of different phases going through phase-specific transfection pathways and mechanisms.
For the phase, the membrane charge density (σM, average charge per unit area of the membrane) was found to be a key parameter affecting TE of CL-DNA complexes. The TE of the complexes is highly dependent on σM values. In contrast, TE of complexes is in general higher than that of complexes and independent of σM. The differences in the dependence of TE on σM between phase and phase were so significant that several models have been proposed to explain such differences. According to the model proposed by Lin et al.,54 the complexes that are entrapped in endosomes escape from the endosomes via fusion. The fusion between the cationic membrane of phase complexes and anionic endosomal membranes is anticipated to facilitate release of DNA into the cytoplasm. At low σM, insufficient electrostatic interactions leave the complexes as trapped in the endosome. At very high σM, the strong electrostatic interactions between cationic lipid membrane and DNA hamper the dissociation of DNA from the complexes. Hence there exists optimal σM that allows highly efficient escape of DNA into the cytoplasm, achieving the highest level of TE. For complexes, it was suggested that the outer layer of the complex contributes to the rapid fusion of the complex membrane with the cell plasma and endosomal membranes.54 The curvature of the outer layer imposes energetic cost, which is independent of σM, driving a facile fusion process. The complex also showed independence of TE on σM,45 but the underlying mechanism remains poorly understood. Further studies should be geared towards elucidating the transfection mechanisms of CL-DNA complexes in connection with their supramolecular structures in order to optimize the TE.
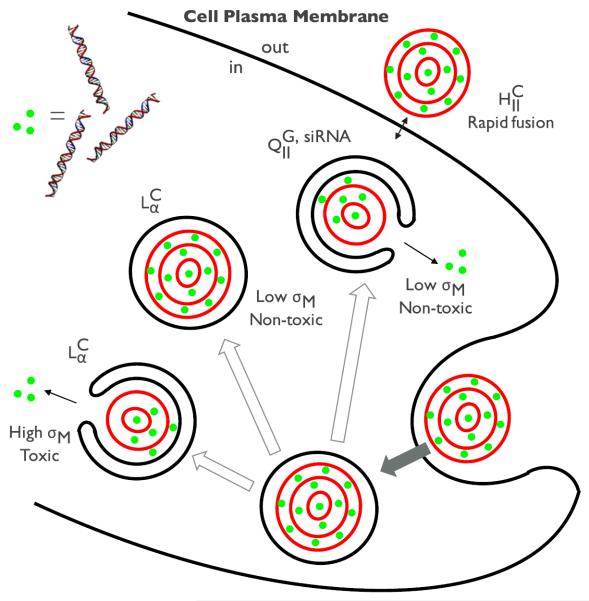
For siRNA gene silencing applications, CL-siRNA complexes of and phases have been firstly explored. Despite their structural similarities to CL-DNA analogues, the delivery of siRNA was not successful with CL-siRNA complexes because of the high level of cell toxicity.35 The discovery of bicontinuous cubic CL-siRNA complexes () solved the toxicity problem as it was found to be efficient in gene silencing even at low σM.49 Such highly efficient gene silencing of the cubic phase results from enhanced membrane fusion between the cubic phase and plasma membrane or endosomal membrane, which is attributed to negative Gaussian curvatures intrinsic to bicontinuous cubic phases. The efficient gene silencing benefitted from the inherent structure of cubic phase indicates the close correlation between the structure of CL-NA complexes and their gene delivery mechanisms. Figure 4 shows a schematic of the different proposed mechanisms for the interaction of each supramolecular structure with cells.
Figure 4.
A schematic representation of the cellular pathway and transfection mechanisms of different CL-NA complexes. The lamellar phases of CL-DNA complexes () enter the cells through endocytosis while the inverted hexagonal phases of CL-DNA complexes () rather directly interacts with the plasma membrane through facilitated fusion process. The fate of entered complexes (i.e. trapped in endosomes) turned out to be dependent on the membrane charge density (σM). High σM facilitates the fusion with the endosomal membrane effectively releasing DNA inside the cell, but increases the cytotoxicity at the same time. Low σM involves the low cytotoxicity but this state lacks the capability to escape out of the endosomes. On the other hand, the cubic phase of CL-siRNA complexes () show the high efficiency of escaping endosomes even at low σM, which arises from the negative Gaussian curvatures intrinsic to bicontinuous cubic structures.
6. Future perspectives
The design of functional lipids such as redox-active lipids,38 pH-sensitive lipids,39 and ionizable lipids55,56 in the recent few years enabled the self-assembly of advanced lipid-based structures that are responsive to external stimuli and/or trigger an efficient endosomal escape of encapsulated nucleic acids. One challenge associated with lipid mediated gene delivery is that lipids are not always amenable to facile chemical modification. We expect that future work on synthesis of new lipids with desired functionalities will facilitate the resulting self-assembly structures to be tailored for specific applications. For example we now have a reliable way to modify lipid molecules with PEG units for liposomal gene delivery.57,58 Also the modification of lipid molecules to include cell-targeting motifs such as Arginylglycylaspartic acid (RGD) and other recognition sequences for integrins59,60 could transform the field in terms of cell specificity of lipid-based gene delivery technologies. Constructing stimuli-responsive complex structures using specifically designed functional lipids with cell surface recognition moieties seem to be a logical next step towards the realization of optimized gene delivery vectors.
The characteristic self-assembled structures of different complexes have been shown to be linked to their transfection mechanisms including cell specific attachment, internalization capabilities, and endosomal escape. The recent finding of cuboplexes is noteworthy in that it does not rely on cationic lipids to efficiently deliver NA cargo as much as lamellar phases. The implication of reduced cell toxicity and non-specific gene knockdown is an important one that needs to be further explored. To deeply understand the important structural parameters relevant to gene delivery/silencing efficiencies, the detailed mechanism of cellular interactions should be clarified on a molecular level. In this respect biotechnology methodologies to isolate endosomes61,62 and new emerging high resolution imaging techniques63,64 will be instrumental.
Also it should be noted that there are lipid self-assembly structures that have not yet been explored as gene delivery vectors such as sponge phases or non-cubic 3D phases (e.g. the hexagonal P63/mmc) that may hold new capabilities that cannot be found in existing CL-NA complexes. As lipid phase behavior for complex 2D phases10 becomes better understood, opportunities will arise to appropriately functionalize those phases with nucleic acids.
Finally, while the various self-assembled structures of CL-NA complexes in equilibrium have been discovered over the past decades, researchers still lack basic understanding of the kinetics of self-assembly. Understanding the dynamics of complex formation will allow us to better unravel the gene transfection/silencing spatiotemporal mechanisms which ultimate govern the performance of all steps involved in gene delivery and silencing.
Acknowledgements
This work was supported by the National Science Foundation under grant no. DMR-1554435.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
8. References
- 1.Bruinsma R. Electrostatics of DNA-cationic lipid complexes: isoelectric instability. Eur. Phys. J. B. 1998;4:75–88. [Google Scholar]
- 2.Golubović L, Golubović M. Fluctuations of Quasi-Two-Dimensional Smectics Intercalated between Membranes in Multilamellar Phases of DNA-Cationic Lipid Complexes. Phys. Rev. Lett. 1998;80:4341–4344. [Google Scholar]
- 3.Harries D, May S, Gelbart WM, Ben-Shaul A. Structure, stability, and thermodynamics of lamellar DNA-lipid complexes. Biophys. J. 1998;75:159–173. doi: 10.1016/S0006-3495(98)77503-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gelbart WM, Bruinsma RF, Pincus PA, Parsegian VA. DNA-Inspired Electrostatics. Phys. Today. 2000;53:38. [Google Scholar]
- 5.Evans DF, Wennerström H. The colloidal domain: where physics, chemistry, biology, and technology meet. Wiley-VCH; 1999. [Google Scholar]
- 6.Tanford C. The hydrophobic effect: formation of micelles and biological membranes. Wiley; 1980. [Google Scholar]
- 7.Israelachvili JN, Marcelja S, Horn RG. Physical principles of membrane organization. Q. Rev. Biophys. 1980;13:121–200. doi: 10.1017/s0033583500001645. [DOI] [PubMed] [Google Scholar]
- 8.Gruner SM. Intrinsic curvature hypothesis for biomembrane lipid composition: a role for nonbilayer lipids. Proc. Natl. Acad. Sci. U. S. A. 1985;82:3665–3669. doi: 10.1073/pnas.82.11.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z. Für Naturforschung Teil C Biochem. Biophys. Biol. Virol. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 10.Seddon JM, Templer RH. Handbook of Biological Physics. Vol. 1. Elsevier; 1995. pp. 97–160. [Google Scholar]
- 11.Luzzati V, Tardieu A, Gulik-Krzywicki T, Rivas E, Reiss-Husson F. Structure of the Cubic Phases of Lipid–Water Systems. Nature. 1968;220:485–488. doi: 10.1038/220485a0. [DOI] [PubMed] [Google Scholar]
- 12.Matulis D, Rouzina I, Bloomfield VA. Thermodynamics of Cationic Lipid Binding to DNA and DNA Condensation: Roles of Electrostatics and Hydrophobicity. J. Am. Chem. Soc. 2002;124:7331–7342. doi: 10.1021/ja0124055. [DOI] [PubMed] [Google Scholar]
- 13.Wagner K, Harries D, May S, Kahl V, Rädler JO, Ben-Shaul A. Direct Evidence for Counterion Release upon Cationic Lipid–DNA Condensation. Langmuir. 2000;16:303–306. [Google Scholar]
- 14.Leal C, Moniri E, Pegado L, Wennerström H. Electrostatic Attraction between DNA and a Cationic Surfactant Aggregate. The Screening Effect of Salt. J. Phys. Chem. B. 2007;111:5999–6005. doi: 10.1021/jp065358h. [DOI] [PubMed] [Google Scholar]
- 15.Leal C, Sandström D, Nevsten P, Topgaard D. Local and translational dynamics in DNA–lipid assemblies monitored by solid-state and diffusion NMR. Biochim. Biophys. Acta BBA - Biomembr. 2008;1778:214–228. doi: 10.1016/j.bbamem.2007.09.035. [DOI] [PubMed] [Google Scholar]
- 16.Gromelski S, Brezesinski G. DNA Condensation and Interaction with Zwitterionic Phospholipids Mediated by Divalent Cations. Langmuir. 2006;22:6293–6301. doi: 10.1021/la0531796. [DOI] [PubMed] [Google Scholar]
- 17.Ainalem M-L, Kristen L, Edler KJ, Höök F, Sparr E, Nylander T. DNA Binding to Zwitterionic Model Membranes. Langmuir. 2010;26:4965–4976. doi: 10.1021/la9036327. [DOI] [PubMed] [Google Scholar]
- 18.Mengistu DH, Bohinc K, May S. Binding of DNA to Zwitterionic Lipid Layers Mediated by Divalent Cations. J. Phys. Chem. B. 2009;113:12277–12282. doi: 10.1021/jp904986j. [DOI] [PubMed] [Google Scholar]
- 19.Petrache HI, Zemb T, Belloni L, Parsegian VA. Salt screening and specific ion adsorption determine neutral-lipid membrane interactions. Proc. Natl. Acad. Sci. 2006;103:7982–7987. doi: 10.1073/pnas.0509967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato A, Tsuji A, Yanagisawa M, Saeki D, Juni K, Morimoto Y, Yoshikawa K. Phase Separation on a Phospholipid Membrane Inducing a Characteristic Localization of DNA Accompanied by Its Structural Transition. J. Phys. Chem. Lett. 2010;1:3391–3395. [Google Scholar]
- 21.Hamada T, Fujimoto R, Shimobayashi SF, Ichikawa M, Takagi M. Molecular behavior of DNA in a cell-sized compartment coated by lipids. Phys. Rev. E. 2015;91 doi: 10.1103/PhysRevE.91.062717. [DOI] [PubMed] [Google Scholar]
- 22.Lau AWC, Pincus P, Levine D, Fertig HA. Electrostatic attraction of coupled Wigner crystals: Finite temperature effects. Phys. Rev. E. 2001;63 doi: 10.1103/PhysRevE.63.051604. [DOI] [PubMed] [Google Scholar]
- 23.Liang H, Harries D, Wong GCL. Polymorphism of DNA-anionic liposome complexes reveals hierarchy of ion-mediated interactions. Proc. Natl. Acad. Sci. 2005;102:11173–11178. doi: 10.1073/pnas.0502416102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leikin S, Parsegian VA, Rau DC, Rand RP. Hydration Forces. Annu. Rev. Phys. Chem. 1993;44:369–395. doi: 10.1146/annurev.pc.44.100193.002101. [DOI] [PubMed] [Google Scholar]
- 25.Israelachvili JN, Wennerstroem H. Hydration or steric forces between amphiphilic surfaces? Langmuir. 1990;6:873–876. [Google Scholar]
- 26.Wennerström H, Sparr E. Thermodynamics of membrane lipid hydration. Pure Appl. Chem. 2003;75 [Google Scholar]
- 27.Disalvo EA. In: Membrane Hydration. Disalvo EA, editor. Vol. 71. Springer International Publishing; 2015. pp. 1–16. [Google Scholar]
- 28.Meidan VM, Cohen JS, Amariglio N, Hirsch-Lerner D, Barenholz Y. Interaction of oligonucleotides with cationic lipids: the relationship between electrostatics, hydration and state of aggregation. Biochim. Biophys. Acta BBA - Biomembr. 2000;1464:251–261. doi: 10.1016/s0005-2736(00)00151-6. [DOI] [PubMed] [Google Scholar]
- 29.Gurtovenko AA, Miettinen M, Karttunen M, Vattulainen I. Effect of Monovalent Salt on Cationic Lipid Membranes As Revealed by Molecular Dynamics Simulations. J. Phys. Chem. B. 2005;109:21126–21134. doi: 10.1021/jp053667m. [DOI] [PubMed] [Google Scholar]
- 30.Binder H, Zschörnig O. The effect of metal cations on the phase behavior and hydration characteristics of phospholipid membranes. Chem. Phys. Lipids. 2002;115:39–61. doi: 10.1016/s0009-3084(02)00005-1. [DOI] [PubMed] [Google Scholar]
- 31.Felgner PL, et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Dosari MS, Gao X. Nonviral Gene Delivery: Principle, Limitations, and Recent Progress. AAPS J. 2009;11:671–681. doi: 10.1208/s12248-009-9143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radler JO. Structure of DNA-Cationic Liposome Complexes: DNA Intercalation in Multilamellar Membranes in Distinct Interhelical Packing Regimes. Science. 1997;275:810–814. doi: 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- 34.Salditt T, Koltover I, Rädler JO, Safinya CR. Self-assembled DNA–cationic-lipid complexes: Two-dimensional smectic ordering, correlations, and interactions. Phys. Rev. E. 1998;58:889–904. [Google Scholar]
- 35.Bouxsein NF, McAllister CS, Ewert KK, Samuel CE, Safinya CR. Structure and Gene Silencing Activities of Monovalent and Pentavalent Cationic Lipid Vectors Complexed with siRNA †. Biochemistry (Mosc.) 2007;46:4785–4792. doi: 10.1021/bi062138l. (*This paper presents the rich phase behavior of DNA packing in lipid self-assembled structures and its implications in gene silencing and chemical delivery.)
- 36.Bouxsein NF, Leal C, McAllister C, Ewert KK, Li Y, Samuel CE, Safinya CR. Two-Dimensional Packing of Short DNA with Nonpairing Overhangs in Cationic Liposome–DNA Complexes: From Onsager Nematics to Columnar Nematics with Finite-Length Columns. J. Am. Chem. Soc. 2011;133:7585–7595. doi: 10.1021/ja202082c. (*This paper reports for the first time the end-to-end adhesion between DNA oligomers. It is important in that the end-to-end adhesion leads to the formation of liquid crystalline phases. )
- 37.Nakata M, Zanchetta G, Chapman BD, Jones CD, Cross JO, Pindak R, Bellini T, Clark NA. End-to-end stacking and liquid crystal condensation of 6 to 20 base pair DNA duplexes. Science. 2007;318:1276–1279. doi: 10.1126/science.1143826. (*This paper introduces an interesting approach to design functional lipids and their applications in siRNA delivery.)
- 38.Aytar BS, Muller JPE, Kondo Y, Talmon Y, Abbott NL, Lynn DM. Redox-Based Control of the Transformation and Activation of siRNA Complexes in Extracellular Environments Using Ferrocenyl Lipids. J. Am. Chem. Soc. 2013;135:9111–9120. doi: 10.1021/ja403546b. (*This paper presents an interesting approach for designing stimuli-responsive lipids which exerts the possibility of controlling the stability of lipid-based complexes.)
- 39.Oumzil K, Benizri S, Tonelli G, Staedel C, Appavoo A, Chaffanet M, Navailles L, Barthélémy P. pH-Cleavable Nucleoside Lipids: A New Paradigm for Controlling the Stability of Lipid-Based Delivery Systems. ChemMedChem. 2015;10:1797–1801. doi: 10.1002/cmdc.201500381. [DOI] [PubMed] [Google Scholar]
- 40.Koltover I. An Inverted Hexagonal Phase of Cationic Liposome-DNA Complexes Related to DNA Release and Delivery. Science. 1998;281:78–81. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- 41.Gruner SM. Stability of lyotropic phases with curved interfaces. J. Phys. Chem. 1989;93:7562–7570. [Google Scholar]
- 42.Francescangeli O, Pisani M, Stanić V, Bruni P, Weiss TM. Evidence of an inverted hexagonal phase in self-assembled phospholipid-DNA-metal complexes. Europhys. Lett. EPL. 2004;67:669–675. [Google Scholar]
- 43.Koynova R, Wang L, MacDonald RC. An intracellular lamellar-nonlamellar phase transition rationalizes the superior performance of some cationic lipid transfection agents. Proc. Natl. Acad. Sci. 2006;103:14373–14378. doi: 10.1073/pnas.0603085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tenchov BG, Wang L, Koynova R, MacDonald RC. Modulation of a membrane lipid lamellar–nonlamellar phase transition by cationic lipids: A measure for transfection efficiency. Biochim. Biophys. Acta BBA - Biomembr. 2008;1778:2405–2412. doi: 10.1016/j.bbamem.2008.07.022. (*This paper is important because the initial finding of a normal hexagonal lipid phase incorporating DNA and the effectiveness of such structure are presented. )
- 45.Ewert KK, Evans HM, Zidovska A, Bouxsein NF, Ahmad A, Safinya CR. A columnar phase of dendritic lipid-based cationic liposome-DNA complexes for gene delivery: hexagonally ordered cylindrical micelles embedded in a DNA honeycomb lattice. J. Am. Chem. Soc. 2006;128:3998–4006. doi: 10.1021/ja055907h. [DOI] [PubMed] [Google Scholar]
- 46.Luzzati V, Gulik-Krzywicki T, Tardieu A. Polymorphism of Lecithins. Nature. 1968;218:1031–1034. doi: 10.1038/2181031a0. [DOI] [PubMed] [Google Scholar]
- 47.Lutton ES. Phase behavior of aqueous systems of monoglycerides. J. Am. Oil Chem. Soc. 1965;42:1068–1070. doi: 10.1007/BF02636909. (*This paper is important in that the obstacles of siRNA delivery were experimentally revealed providing the future research directions of effective siRNA delivery.)
- 48.Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, Karagiannis E, Love K, Chen D, Zoncu R, Buganim Y, Schroeder A, Langer R, Anderson DG. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. (**This paper presents the formation of cubic phase nanoparticle and its application in siRNA delivery. This work is important in that the superiority of cubic structure in siRNA delivery was suggested for the first time. )
- 49.Leal C, Bouxsein NF, Ewert KK, Safinya CR. Highly Efficient Gene Silencing Activity of siRNA Embedded in a Nanostructured Gyroid Cubic Lipid Matrix. J. Am. Chem. Soc. 2010;132:16841–16847. doi: 10.1021/ja1059763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leal C, Ewert KK, Bouxsein NF, Shirazi RS, Li Y, Safinya CR. Stacking of short DNA induces the gyroid cubic-to-inverted hexagonal phase transition in lipid–DNA complexes. Soft Matter. 2013;9:795–804. doi: 10.1039/C2SM27018H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Desigaux L, Sainlos M, Lambert O, Chevre R, Letrou-Bonneval E, Vigneron J-P, Lehn P, Lehn J-M, Pitard B. Self-assembled lamellar complexes of siRNA with lipidic aminoglycoside derivatives promote efficient siRNA delivery and interference. Proc. Natl. Acad. Sci. 2007;104:16534–16539. doi: 10.1073/pnas.0707431104. (**This work is interesting as a nanometer scale lipid bicontinuous cubic phase-siRNA complex (cuboplex) was presented. A well-defined internal structure of cuboplex and its high siRNA delivery efficacy are notable. )
- 52.Kim H, Leal C. Cuboplexes: Topologically Active siRNA Delivery. ACS Nano. 2015;9:10214–10226. doi: 10.1021/acsnano.5b03902. [DOI] [PubMed] [Google Scholar]
- 53.Gupta A, Stait-Gardner T, de Campo L, Waddington LJ, Kirby N, Price WS, Moghaddam MJ. Nanoassemblies of Gd–DTPA–monooleyl and glycerol monooleate amphiphiles as potential MRI contrast agents. J. Mater. Chem. B. 2014;2:1225. doi: 10.1039/c3tb21069c. [DOI] [PubMed] [Google Scholar]
- 54.Lin AJ. Three-dimensional imaging of lipid gene-carriers: membrane charge density controls universal transfection behavior in lamellar cationic liposome-DNA complexes. Biophys. J. 2003;84:3307–3316. doi: 10.1016/S0006-3495(03)70055-1. (*This paper presents an interesting design approach of ionizable lipids which were shown to be ffective in escaping endosomes. )
- 55.Habrant D. Design of Ionizable Lipids To Overcome the Limiting Step of Endosomal Escape: Application in the Intracellular Delivery of mRNA, DNA, and siRNA. J. Med. Chem. 2016;59:3046–3062. doi: 10.1021/acs.jmedchem.5b01679. (*This paper introduces a rational approach to designing cationic lipids for in vivo siRNA delivery. The efficacy of siRNA delivery using such approach is noteworthy. )
- 56.Semple SC. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 57.Zalipsky S. Synthesis of an end-group functionalized polyethylene glycol-lipid conjugate for preparation of polymer-grafted liposomes. Bioconjug. Chem. 1993;4:296–299. doi: 10.1021/bc00022a008. [DOI] [PubMed] [Google Scholar]
- 58.Heyes J, Hall K, Tailor V, Lenz R, MacLachlan I. Synthesis and characterization of novel poly(ethylene glycol)-lipid conjugates suitable for use in drug delivery. J. Controlled Release. 2006;112:280–290. doi: 10.1016/j.jconrel.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 59.Dubey PK, Mishra V, Jain S, Mahor S, Vyas SP. Liposomes Modified with Cyclic RGD Peptide for Tumor Targeting. J. Drug Target. 2004;12:257–264. doi: 10.1080/10611860410001728040. [DOI] [PubMed] [Google Scholar]
- 60.Temming K, Schiffelers RM, Molema G, Kok RJ. RGD-based strategies for selective delivery of therapeutics and imaging agents to the tumour vasculature. Drug Resist. Updat. 2005;8:381–402. doi: 10.1016/j.drup.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 61.Singer-Krüger B, Frank R, Crausaz F, Riezman H. Partial purification and characterization of early and late endosomes from yeast. Identification of four novel proteins. J. Biol. Chem. 1993;268:14376–14386. [PubMed] [Google Scholar]
- 62.Tjelle TE, Brech A, Juvet LK, Griffiths G, Berg T. Isolation and characterization of early endosomes, late endosomes and terminal lysosomes: their role in protein degradation. J. Cell Sci. 1996;109(Pt 12):2905–2914. doi: 10.1242/jcs.109.12.2905. [DOI] [PubMed] [Google Scholar]
- 63.Kirchhausen T. Imaging endocytic clathrin structures in living cells. Trends Cell Biol. 2009;19:596–605. doi: 10.1016/j.tcb.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lidke DS, Lidke KA. Advances in high-resolution imaging - techniques for three-dimensional imaging of cellular structures. J. Cell Sci. 2012;125:2571–2580. doi: 10.1242/jcs.090027. [DOI] [PMC free article] [PubMed] [Google Scholar]