Abstract
Elevated glucose levels in cancer cells can be attributed to increased levels of glucose transporter (GLUT) proteins. Glut1 expression is increased in human malignant cells. To investigate alternative roles of Glut1 in breast cancer, we silenced Glut1 in triple-negative breast-cancer cell lines using a short hairpin RNA (shRNA) system. Glut1 silencing was verified by Western blotting and qRT-PCR. Knockdown of Glut1 resulted in decreased cell proliferation, glucose uptake, migration, and invasion through modulation of the EGFR/MAPK signaling pathway and integrin β1/Src/FAK signaling pathways. These results suggest that Glut1 not only plays a role as a glucose transporter, but also acts as a regulator of signaling cascades in the tumorigenesis of breast cancer.
Keywords: Breast cancer cell, EGFR, Glut, Integrin β1, Triple-negative breast cancer
INTRODUCTION
Some cancer cells depend on glycolysis instead of oxidative phosphorylation for energy production. This phenomenon is known as the Warburg effect (1, 2). As a result, cancer cells take up glucose at an elevated rate to meet their increased energy demands. The most widely expressed glucose transporter is Glut1, which is responsible for basal glucose uptake (3). High expression of Glut1 was correlated with poor prognosis in several cancer types, including breast cancer (4, 5).
There are five distinct subtypes of breast cancer with different clinical outcomes: luminal A, luminal B, HER2-positive, basal-like, and normal-like (6, 7). Basal-like breast cancers generally lack hormone receptors and HER2, and the majority of these cancers are also called triple-negative breast cancer (TNBC) (8). It was previously demonstrated that expression of Glut1 is significantly associated with high histologic grade, ER negativity, PR negativity, CK5/6 negativity, EGFR expression, and high p53 expression (9). Although Glut1 is expressed in TNBCs at a high level (9), the signaling pathways regulated by Glut1 remain poorly understood.
In this study we investigated the effects of Glut1 silencing in two TNBC cell lines, MDA-MB-231 and Hs578T, using a short hairpin RNA (shRNA) system. Glut1 knockdown (Glut1 shRNA) cells were compared with control knockdown (Control shRNA) cells with respect to cell proliferation, colony formation, cell-cycle distribution, glycolytic phenotypes, wound-healing ability, migration, and invasion. We also showed that Glut1 regulated expression of EGFR and integrin β1, and modulated the EGFR/mitogen-activated protein kinase (MAPK) signaling pathway and integrin β1/Src/focal adhesion kinase (FAK) signaling pathway in TNBC cell lines.
RESULTS
Effects of Glut1 silencing on proliferation, colony formation, and cell-cycle distribution
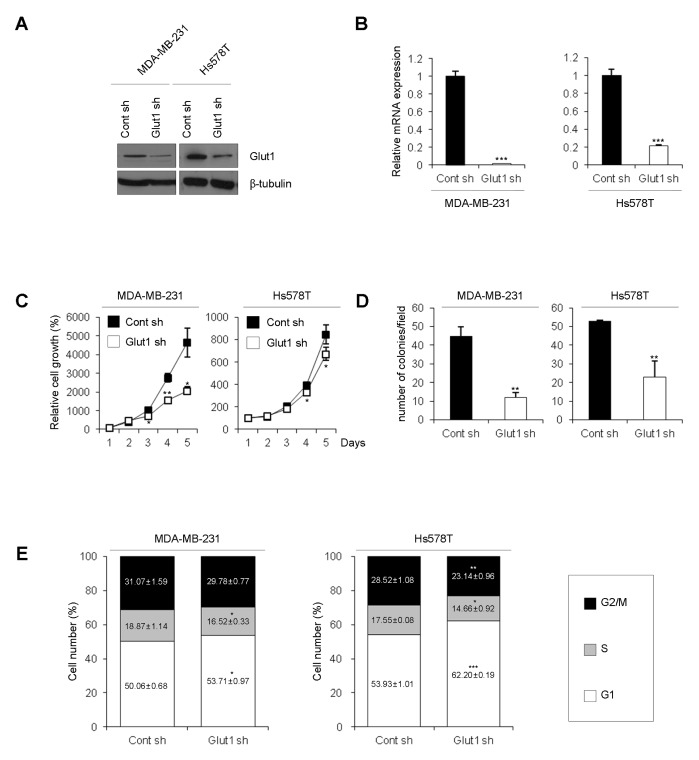
To investigate the role of Glut1 in TNBC cells, we silenced Glut1 in TNBC cells using a shRNA system. Glut1 silencing was verified by Western blot analysis and qRT-PCR (Fig. 1A and 1B). First, we compared the proliferation rates of Glut1 shRNA cells (MDA-MB-231 Glut1 sh and Hs578T Glut1 sh) and Control shRNA cells (MDA-MB-231 Cont sh and Hs578T Cont sh). The growth rate of Glut1 shRNA cells was lower than that of Control shRNA cells (Fig. 1C). Moreover, silencing of Glut1 significantly decreased the rate of colony formation (Fig. 1D). To identify the mechanisms responsible for the reduced cell proliferation in Glut1 shRNA cells, we analyzed the cell-cycle distribution by flow cytometry. Glut1 shRNA cells displayed accumulation of cells in G1 phase with a decrease in the S phase fraction (Fig. 1E).
Fig. 1.
Effects of Glut1 silencing on proliferation, colony formation, and cell-cycle distribution. (A) Glut1 silencing was verified by Western blot analysis using anti-Glut1 antibody in MDA-MB-231 and Hs578T breast cancer cell lines. β-tubulin was used as a loading control. (B) Ablation of Glut1 was confirmed by qRT-PCR using Glut1-specific primers. The values were normalized to GAPDH mRNA (***P < 0.0005). (C) Cont shRNA (Cont sh) cells and Glut1 shRNA (Glut1 sh) cells were seeded at 1 × 104 cells/well in 12-well plates and counted with a hemocytometer over 4 days (*P < 0.05, **P < 0.005). (D) Cells were seeded at 200 cells/well in 6-well plates. The number of colonies (> 20 μm diameter) was counted at 12 days (**P < 0.005, ***P < 0.0005). (E) Cells were seeded at 1 × 106 cells/100-mm dish. After 24 h, cells were harvested, fixed in methanol, and incubated in PBS containing 40 μg/ml propidium iodide and 100 μg/ml RNase A. Propidium iodide-labeled nuclei were analyzed by flow cytometry.
Reduction of glycolytic phenotypes by Glut1 knockdown
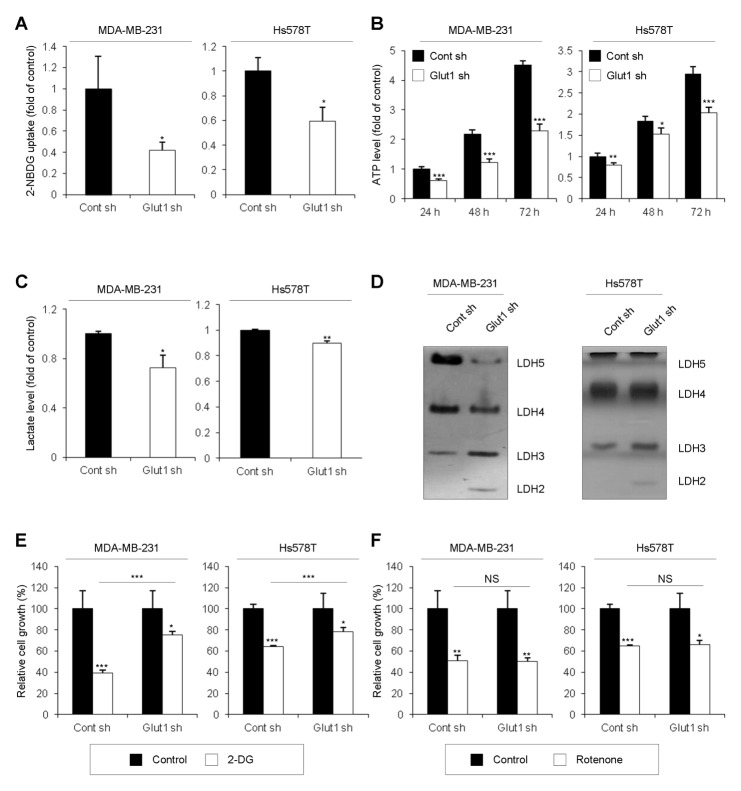
Next, we examined metabolic phenotypes, including glucose-uptake levels, ATP levels, and lactate-production levels. As expected, since Glut1 is responsible for basal glucose uptake, the level of glucose uptake was decreased in Glut1 shRNA cells (Fig. 2A). Moreover, levels of cellular ATP and lactate production were decreased in Glut1 shRNA cells (Fig. 2B and 2C). Lactate dehydrogenase (LDH) is a tetrameric enzyme composed of two major subunits A and/or B, resulting in five isozymes (A4, A3B1, A2B2, A1B3, and B4) that catalyze the interconversion of lactate and pyruvate. LDHA (LDH-5, LDH-M, or A4) favors the conversion of pyruvate to lactate, whereas LDHB (LDH-1, LDH-H, or B4) converts lactate to pyruvate. Visualization of the stained zymography gel showed decreased expression of LDH5 and LDH4 in Glut1 shRNA cells (Fig. 2D). In contrast, expression of LDH3 and LDH2 was increased in Glut1 shRNA cells (Fig. 2D). These results suggest that LDHB is more dominant than LDHA and that LDH mainly converts lactate to pyruvate in Glut1 shRNA cells; correspondingly, the data in Fig. 2C shows that lactate production was decreased in Glut1 shRNA cells. To find out whether the transition of glycolysis to oxidative phosphorylation occurred in Glut1 shRNA cells, the cells were treated with 2-deoxyglucose to inhibit glycolysis. After 24 h, inhibition of cell proliferation in Glut1 shRNA cells was significantly attenuated compared to that in Control shRNA cells (Fig. 2E), indicating that Glut1 shRNA cells were less dependent on glycolysis than Control shRNA cells. Cells were also treated with rotenone for 24 h to inhibit the electron-transport chain in mitochondria. No difference in sensitivity to rotenone was observed between Control shRNA cells and Glut1 shRNA cells (Fig. 2F), suggesting that oxidative phosphorylation occurred at a similar level regardless of Glut1 knockdown. Thus, Glut1 facilitates glycolytic phenotypes such as glucose uptake levels, cellular ATP levels, and lactate production levels.
Fig. 2.
Reduction of glycolytic phenotypes by Glut1 knockdown. (A) For glucose uptake assays, Cont sh and Glut1 sh cells were seeded at 5 × 103 cells/well in 96-well plates, and glucose uptake was measured by 2-NBDG fluorescence using a plate reader (*P < 0.05). (B) For ATP assays, Cont sh and Glut1 sh cells were seeded at 5 × 103 cells/well in 96-well plates. After 24, 48, or 72 h, 100 μl of CellTiter-Glo® Reagent was added to each well. Cellular ATP levels were measured by luminescence (*P < 0.05, **P < 0.005, ***P < 0.0005). (C) For lactate production assays, Cont sh and Glut1 sh cells were seeded at 1 × 106 cells/100-mm dish. After 24 h, the culture medium was replaced with FBS-free DMEM. After a further 8 h, culture medium was harvested and lactate levels were analyzed by colorimetric assay (*P < 0.05, **P < 0.005). (D) For LDH zymography assays, 30-μg protein samples from each cell line were subjected to native gel electrophoresis. (E, F) Cont sh and Glut1 sh cells were seeded at 2 × 103 cells/well in 96-well plates and treated with 20 mM 2-deoxyglucose (2-DG) (E) or 20 μM rotenone (F) for 24 h before analysis by MTT assay (*P < 0.05, **P < 0.005, ***P < 0.0005).
Effects of Glut1 downregulation on migration and invasion
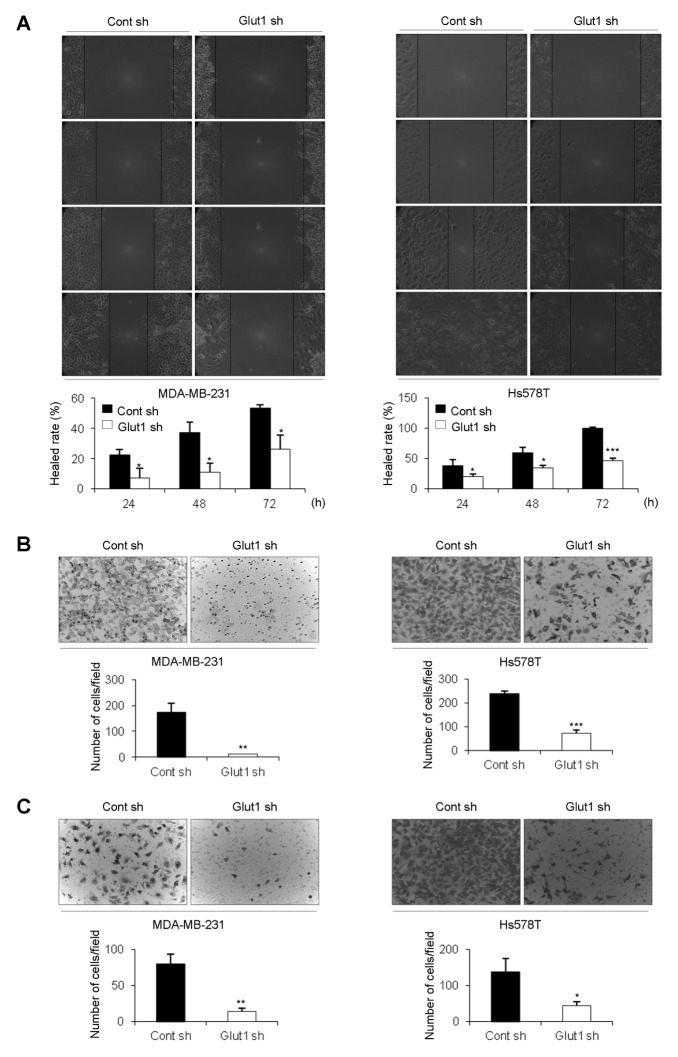
To investigate the effect of Glut1 on cell migration and invasion, we first carried out wound-healing assays. Results of these assays showed that migration of Glut1 shRNA cells was decreased compared with that of Control shRNA cells (Fig. 3A). Similarly, transwell-migration assays showed that Glut1 shRNA cells migrated more slowly than Control shRNA cells (Fig. 3B). Cell invasion ability was also examined using transwell-invasion assays. Glut1 shRNA cells showed reduced invasion ability compared to Cont shRNA cells (Fig. 3C). Therefore, we concluded that Glut1 promotes cell migration and invasion.
Fig. 3.
Effects of Glut1 downregulation on migration and invasion. (A) For the wound-healing assay, cells were seeded at a density of 4 × 105 cells/well in 6-well plates to achieve 90–95% confluence. A wound was created using a 10-μl pipette tip and wound closure was monitored microscopically at the indicated time points. Assays were carried out in triplicate (*P < 0.05, ***P < 0.0005). (B, C) Cells were seeded at a density of 5 × 104 cells/well in transwell plates for (B) migration assay or (C) invasion assay. The number of cells on the bottom of the membrane was counted at 24 h (**P < 0.005, ***P < 0.0005).
Modulation of signaling pathways by Glut1 ablation
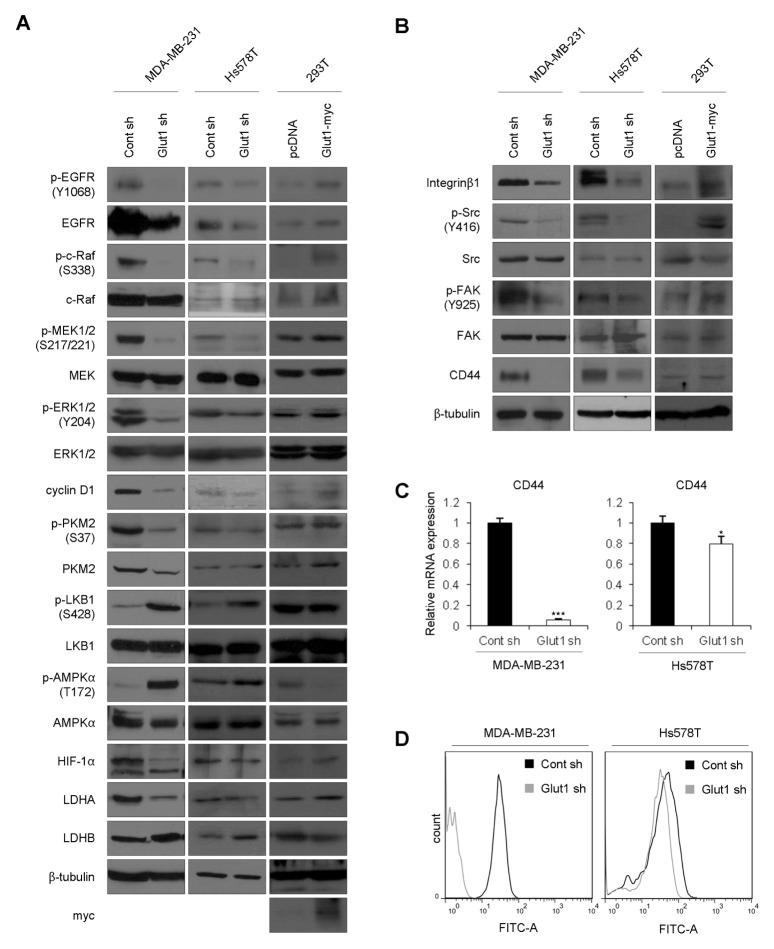
To investigate which signaling pathways were affected by ablation of Glut1, we examined intracellular signaling molecules. First, we measured levels of EGFR expression and EGFR activation, because high levels of EGFR are a common feature of TNBCs (10). Surprisingly, expression and activation of EGFR was decreased in Glut1 shRNA cells (Fig. 4A). Activation of EGFR stimulates several different signaling pathways, including the Ras/MAPK pathway (11). Activation of the MAPK pathway causes transcription of genes that induce cellular proliferation, as well as genes involved in migration and differentiation (12). When Glut1 was silenced in MDA-MB-231 and Hs578T cells, c-Raf/MEK/ERK activity was downregulated (Fig. 4A). Expression of cyclin D1 is up-regulated by ERK activation (13), and cyclin D1 induces the transition from G1 to S-phase (14). Cyclin D1 expression was downregulated in line with decreased ERK activity in Glut1 shRNA cells, in agreement with the cell-cycle analysis (Fig. 1D). A high expression level of the M2 isoform of pyruvate kinase (PKM2) has been detected in several types of cancer cells (15), indicating its involvement in mediating the Warburg effect. It was previously reported that ERK-dependent phosphorylation of PKM2 promotes the Warburg effect (16). Decreased phosphorylation and expression of PKM2 were observed in Glut1 shRNA cells (Fig. 4A) and resulted in reduced glycolytic phenotypes as depicted in Fig. 2. Moreover, serine/threonine kinase LKB1 (liver kinase B1), a known tumor suppressor, directly phosphorylates and activates AMP-activated protein kinase (AMPK), a central metabolic sensor, under conditions of energy stress (17). The decreased glucose uptake (Fig. 2A) and ATP levels (Fig. 2B) in Glut1 shRNA cells might have an influence on activation of LKB1 and AMPKα (Fig. 4A). The expression of proteins involved in glycolysis, including hypoxia-inducible factor 1α (HIF-1α) and LDHA, was down-regulated by Glut1 silencing, whereas LDHB was upregulated in Glut1 shRNA cells (Fig. 4A). These results are consistent with data in Fig. 2C and 2D, which show a decreased level of lactate production and a shift in LDH isoform expression.
Fig. 4.
Modulation of signaling pathways by Glut1 ablation. (A, B) Total cell lysates were prepared from MDA-MB-231 Cont sh, MDA-MB-231 Glut1 sh, Hs578T Cont sh, Hs578T Glut1 sh cells, and 293T cells and subjected to Western blot analyses with the indicated antibodies. Transient transfection experiments were performed using the 293T cell line and pcDNA3.1/myc-His control vector or pcDNA3.1-Glut1/myc-His vector. After 48 h, cell lysates were analyzed by Western blotting using the indicated antibodies. (C) mRNA from each cell line was analyzed by qRT-PCR using primers specific for CD44. The values were normalized to GAPDH mRNA (*P < 0.05, ***P < 0.0005). (D) For cell surface labeling, cells were seeded at a density of 1 × 106 cells/100-mm dish. After 24 h, cells were incubated with anti-CD44 antibody in PBS for 15 min and then with FITC antibody in PBS for 15 min. Cells were washed with PBS and analyzed by flow cytometry. These experiments were performed in triplicate and a representative histogram is shown.
Integrins regulate various biological processes, including cell proliferation, survival, and migration (18, 19). Integrins do not possess intrinsic catalytic activity, but transduce biochemical signals through the recruitment and activation of signaling proteins, such as non-receptor tyrosine kinases, e.g., FAK and c-Src (20, 21). When Glut1 was downregulated in TNBC cells, expression of integrin β1 and phosphorylation of Src/FAK was decreased (Fig. 4B). CD44, which plays an important role in cell migration and invasion, was also downregulated (22). Moreover, mRNA level and cell surface expression of CD44 were decreased in Glut1 shRNA cells (Fig. 4C and 4D). These results are consistent with those of wound healing, migration, and invasion assays (Fig. 3). Furthermore, when Glut1 was introduced into 293T cells, we observed an increase in phosphorylation of EGFR/MAPK and Src/FAK and an increase in EGFR, integrin β1, and CD44, concordant with the effects of Glut1 knockdown (Fig. 4A and 4B).
Taken together, these data indicate that Glut1 promotes proliferation, glycolytic phenotypes, migration, and invasion of TNBC cells by regulating the expression of EGFR and integrin β1 and thereby modulating EGFR/MAPK and integrin β1/Src/FAK signaling pathways.
DISCUSSION
Glut1 is expressed at high levels in several types of cancer, including breast cancers, and correlates with poor survival (4, 5). In particular, Glut1 expression is related to basal-like breast cancers that are negative for ER, PR, HER2, and CK5/6 and express EGFR (9). Grover-McKay et al. demonstrated a strong association between Glut1 expression and the invasive ability of breast-cancer cells (23) and showed that Glut1 was intensely expressed in highly invasive MDA-MB-231 cells. In addition, many reports indicated that Glut1 contributes to tumor aggressiveness, but little is known about the signaling pathways that are regulated by Glut1.
To investigate the effects of Glut1 in TNBC cells, we silenced its expression in MDA-MB-231 and Hs578T cells using a shRNA system. Our results indicate that silencing of Glut1 decreases cell growth, cell cycle, glycolytic phenotype, migration, and invasion in TNBC cells.
Interestingly, the expression of Glut1 was also significantly high in non-TNBC MCF7 and T47D cells (Supplementary Fig. S1). Glut1 might be responsible for basal glucose uptake and have a role as a major glucose transporter in these cells as well. We also generated Glut1 knockdown cells in MCF7 and T47D cells (Supplementary Fig. S2). We compared the proliferation rate of Glut1 shRNA cells and Control shRNA cells. The growth rate of Glut1 shRNA cells was lower than that of Control shRNA cells. Therefore, Glut1 facilitates cell proliferation in non-TNBC cells.
To find out which signaling pathways are affected by downregulation of Glut1, we examined intracellular signaling molecules. Glut1 downregulation decreased phosphorylation of EGFR and c-Raf/MEK/ERK (MAPK cascades) (Fig. 4A). Moreover, expression of EGFR was reduced in Glut1 shRNA cells, although the precise underlying mechanism remains to be clarified. Integrins can transmit signals through co-clustering with FAK and c-Src (21). FAK is recruited to the β-integrin cytoplasmic domain in part by binding with the integrin binding proteins. Src associates with FAK phosphorylated at Tyr397 (autophosphorylation site) and phosphorylates FAK at Tyr576, Tyr577, Tyr861, and Tyr925 for maximal FAK activation (24). We discovered that silencing of Glut1 downregulated expression of integrin β1 and subsequently reduced phosphorylation of c-Src at Tyr416, followed by decreased phosphorylation of FAK at Tyr925 (Fig. 4B). The mechanism for reduced expression of integrin β1 in Glut1 shRNA cells has also yet to be found. In particular, we examined CD44, which is more highly expressed in basal-like breast cancers than in all other subtypes and correlates with epithelial-mesenchymal transition (EMT) and cancer stem cell gene profiles (25, 26). We observed that ablation of Glut1 downregulated expression of CD44 (Fig. 4B–D). However, EMT associated gene expression profiles, twist, nanog, and N-cadherin were increased rather than decreased in Glut1 shRNA cells (data not shown). From these results, we propose that Glut1 might be involved in EMT via regulation of CD44.
In conclusion, Glut1 not only serves as a glucose transporter, but also plays an important role in tumorigenesis in TNBCs. Our results indicate that Glut1 may be a potential therapeutic target for TNBCs, a notion that warrants further investigation.
MATERIALS AND METHODS
Generation of cell lines
Control knockdown cells and Glut1 knockdown cells were generated by transfection with a Control shRNA and Glut1-specific shRNA respectively (Santa Cruz Biotechnology, CA, USA) using Viafect (Promega, WI, USA) according to the manufacturer’s protocol. Expression of Glut1 was confirmed by Western blotting and qRT-PCR.
Cell culture and transfection
MDA-MB-231 Control shRNA (Cont sh), MDA-MB-231 Glut1 shRNA (Glut1 sh), Hs578T Cont sh, Hs578T Glut1 sh human breast cancer cells, and 293T human embryonic kidney cells were maintained in Dulbecco’s modified Eagle’s medium (Corning, NY, USA) containing 10% fetal bovine serum (Youngin Frontier, Seoul, Korea), 100 U/ml penicillin (Corning), and 100 mg/ml streptomycin (Corning). All cell lines were incubated at 37°C in 5% CO2 in a humidified atmosphere. Calcium phosphate transfection was carried out in 293T cells as described previously (27).
Plasmids
The pcDNA3.1-Glut1/myc-His A vector was generated by PCR amplification using the following primers: Fwd 5′-CTA GGA TCC AT ATG GAG CCC AGC AGC AAG-3′; Rv 5′-CGC CTC GAG CAC TTG GGA ATC AGC-3′. The amplicon was cloned into the pcDNA3.1/myc-His A vector (Invitrogen, Carlsbad, CA, USA) between the BamHI and XhoI restriction sites to generate the pcDNA3.1-Glut1/myc-His A vector.
Antibodies
Antibodies against p-EGFR (Tyr1068), p-c-Raf (Ser338), c-Raf, p-MEK1/2 (Ser217/221), MEK1/2, PKM2, p-LKB1 (Ser428), p-AMPKα (Thr172), Myc-Tag, p-Src family (Tyr416), Src, p-FAK (Tyr925), and FAK were from Cell Signaling Technology (Beverly, MA, USA). Antibodies against Glut1, p-ERK1/2 (Tyr204), ERK1, cyclin D1, LKB1, AMPKα, HIF-1α, LDHA, LDHB, integrin β1, CD44, and β-tubulin were from Santa Cruz Biotechnology. EGFR was from Abcam (Cambridge, UK). p-PKM2 (Ser37) was from Signalway Antibody (College Park, MD, USA).
Proliferation assays
Cells were seeded at 1 × 104 cells/well in 12-well plates. Every 24 h for 4 days, cells were harvested by trypsinization, resuspended in 1 ml of medium, and counted in triplicate using a hemocytometer.
Two-dimensional colony formation
For 2D colony-formation assays, cells were seeded at 200 cells/well in 6-well plates. After 12 days, the cells were washed with PBS and stained with 0.5% crystal violet for 30 min. Cells were washed twice with PBS and observed with an optical microscope. The number of colonies (> 20 μm diameter) was counted in triplicate.
Cell-cycle analyses by flow cytometry
Cells were seeded in 100-mm dishes at a density of 1 × 106 cells/dish. After 24 h, cells were harvested and fixed in methanol overnight at −20°C and then incubated in PBS containing 40 μg/ml propidium iodide and 100 μg/ml RNase A for 10 min at RT. Cell cycle analyses were performed on a FACS CantoII (BD Biosciences, Franklin Lakes, NJ, USA) and data were analyzed using FlowJo software. Experiments were repeated three times.
MTT assays
Cell viability was measured using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) thiazolyl blue assay. Cells were seeded in 96-well culture plates at 2 × 103 cells/well and treated with 2-deoxyglucose (2-DG; Sigma, St. Louis, MO, USA) or rotenone (Sigma). After 24 h, 10 mg/ml MTT (Sigma) was added to 100 μl of culture medium and incubated for 3 h at 37°C. After removal of the medium, formazan was dissolved in dimethyl sulfoxide (Sigma), and optical density was measured at 590 nm using a Multiskan EX.
Wound-healing assays
Cells were seeded in 6-well plates and incubated until approximately 90–95% confluence. Wounds were generated by scraping the monolayer with a micropipette tip, and wound closure was monitored microscopically at the indicated time points. Assays were carried out in triplicate.
Transwell migration and invasion assays
Cells (5 × 104) were added to the top chambers of 24-well Transwell plates (Corning) and a medium containing serum was added to the bottom chambers. After 24 h, cells that migrated through the chamber were fixed and stained with 0.5% crystal violet for 30 min. The cells were washed twice with PBS and observed with an optical microscope. The number of cells was counted in triplicate. Cell-invasion assays were performed by the same procedure, except that the upper chamber was filled with Matrigel.
Western blots
Cells were washed once with PBS and lysed with lysis buffer (20 mM Tris-HCl, pH 7.4, 0.1 mM EDTA, 150 mM NaCl, 1% NP-40, 0.1% Triton X-100, 0.1% SDS, 20 mM NaF, 1 mM Na3VO4, 1× protease inhibitor [Roche, Basel, Switzerland]). Equal amounts of protein in cell lysates were measured by standardization with the BCA Protein Assay Kit (Thermo). Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Whatman, Dassel, Germany). After blocking with 5% skim milk in TBST, membranes were incubated with primary antibodies overnight, followed by 2-h incubation with HRP-conjugated secondary antibodies. Protein bands were visualized with Dyne ECL (Dyne, Seongnam, Korea).
Quantitative real-time polymerase chain reaction (qPCR)
RNA was isolated using Trizol reagent (MRC, Cincinnati, OH, USA) and RT-PCR was carried out using an RT-PCR kit (Toyobo). Quantitative real-time PCR was performed with SYBR FAST qPCR kit (KAPA) in a Thermal Cycler Dice (Takara, Otsu, Shiga, Japan) according to the manufacturer’s protocols. The C(t) value was normalized using GAPDH. Transcripts were assessed using the following primers: Glut1 Fwd 5′-AAG CTG ACG GGT CGC CTC ATG-3′, Rev 5′-CTC TCC CCA TAG CGG TGG ACC-3′; GAPDH Fwd 5′-GGC AAA TTC CAT GGC ACC GTC AAG G-3′, Rev 5′-GCC AGC ATC GCC CCA CTT GAT TTT G-3′; CD44 Fwd 5′-GCC TGG GGA CTC TGC CTC GT-3′, Rev 5′-GCC TTG CAG AGG TCA GCG GC-3′.
Cell-surface labeling
Cells were seeded in 100-mm dishes at a density of 1 × 106 cells/dish. After 24 h, cells were treated with trypsin/EDTA solution, washed with PBS, and resuspended with anti-CD44 antibody in PBS for 15 min. After washing with PBS, cells were incubated with FITC-labeled antibody in PBS for 15 min, washed again with PBS, and filtered through a cell strainer for analysis by FACS CantoII.
Supplementary Information
ACKNOWLEDGEMENTS
This work was supported by an NRF grant (2016R1A2B4011196) from the Korean Research Foundation.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 3.Wood IS, Trayhurn P. Glucose transporters (GLUT and SGLT): expanded families of sugar transport proteins. Br J Nutr. 2003;89:3–9. doi: 10.1079/BJN2002763. [DOI] [PubMed] [Google Scholar]
- 4.Kunkel M, Reichert TE, Benz P, et al. Overexpression of Glut-1 and increased glucose metabolism in tumors are associated with a poor prognosis in patients with oral squamous cell carcinoma. Cancer. 2003;97:1015–1024. doi: 10.1002/cncr.11159. [DOI] [PubMed] [Google Scholar]
- 5.Kang SS, Chun YK, Hur MH, et al. Clinical significance of glucose transporter 1 (GLUT1) expression in human breast carcinoma. Jpn J Cancer Res. 2002;93:1123–1128. doi: 10.1111/j.1349-7006.2002.tb01214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005;23:7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 7.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 8.Badve S, Dabbs DJ, Schnitt SJ, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–167. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- 9.Hussein YR, Bandyopadhyay S, Semaan A, et al. Glut-1 Expression Correlates with Basal-like Breast Cancer. Transl Oncol. 2011;4:321–327. doi: 10.1593/tlo.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 11.Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 13.Lee CH, Yun HJ, Kang HS, Kim HD. ERK/MAPK pathway is required for changes of cyclin D1 and B1 during phorbol 12-myristate 13-acetate-induced differentiation of K562 cells. IUBMB Life. 1999;48:585–591. doi: 10.1080/713803574. [DOI] [PubMed] [Google Scholar]
- 14.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 15.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Yang W, Zheng Y, Xia Y, et al. ERK1/2-dependent phosphorylation and nuclear translocation of KM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juliano RL, Haskill S. Signal transduction from the extracellular matrix. J Cell Biol. 1993;120:577–585. doi: 10.1083/jcb.120.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark EA, Brugge JS. Integrins and signal transduction pathways: the road taken. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 20.Guan JL. Role of focal adhesion kinase in integrin signaling. Int J Biochem Cell Biol. 1997;29:1085–1096. doi: 10.1016/S1357-2725(97)00051-4. [DOI] [PubMed] [Google Scholar]
- 21.Mitra SK, Schlaepfer DD. Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol. 2006;18:516–523. doi: 10.1016/j.ceb.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Nagano O, Saya H. Mechanism and biological significance of CD44 cleavage. Cancer Sci. 2004;95:930–935. doi: 10.1111/j.1349-7006.2004.tb03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grover-McKay M, Walsh SA, Seftor EA, Thomas PA, Hendrix MJ. Role for glucose transporter 1 protein in human breast cancer. Pathol Oncol Res. 1998;4:115–120. doi: 10.1007/BF02904704. [DOI] [PubMed] [Google Scholar]
- 24.Schlaepfer DD, Mitra SK, Ilic D. Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim Biophys Acta. 2004;1692:77–102. doi: 10.1016/j.bbamcr.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 25.Klingbeil P, Natrajan R, Everitt G, et al. CD44 is overexpressed in basal-like breast cancers but is not a driver of 11p13 amplification. Breast Cancer Res Treat. 2010;120:95–109. doi: 10.1007/s10549-009-0380-7. [DOI] [PubMed] [Google Scholar]
- 26.Xu H, Tian Y, Yuan X, et al. Enrichment of CD44 in basal-type breast cancer correlates with EMT, cancer stem cell gene profile, and prognosis. Onco Targets Ther. 2016;9:431–444. doi: 10.2147/OTT.S97192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan M, Wurm F. Transfection of adherent and suspended cells by calcium phosphate. Methods. 2004;33:136–143. doi: 10.1016/j.ymeth.2003.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.