Abstract
Background
Severe fever with thrombocytopenia syndrome bunyavirus (SFTSV) is an emerging tick-borne RNA virus recently identified as the pathogen that causes severe fever with thrombocytopenia syndrome (SFTS) in China. The existing commercial nucleic acid testing (comNAT) assay with a relatively high claimed limit of quantitative detection (LOQD) is not capable of sensitive detection and quantitation of SFTSV. Thus, a new real-time reverse transcriptase (RT)-PCR assay with improved sensitivity is needed for clinical diagnosis; it could also be used to screen blood donors if necessary.
Materials and Methods
We developed a new SFTSV RT-PCR NAT assay (newNAT). 129 plasma samples from 93 suspected SFTS patients with typical clinical symptoms were tested using an anti-SFTSV total antibody ELISA and both comNAT and newNAT. The test performance of the two NAT assays was evaluated and compared.
Results
The newNAT had a lower limit for quantitative testing compared to comNAT.12 samples were comNAT negative but newNAT positive. Out of 35 suspected SFTS patients who were comNAT negative and anti-SFTSV total antibody negative, four tested positive by the newNAT assay And 1 of these 4 seroconverted within two to four days after testing newNAT positive. A high correlation was observed between the Cts of the newNAT and comNAT assays.
Conclusion
The newNAT assay was sensitive for quantitative detection of SFTSV and may be applicable to clinical diagnosis and studies of the need for blood donor screening.
Introduction
Severe fever with thrombocytopenia syndrome bunyavirus (SFTSV) is a newly identified tick-borne pathogen that causes severe fever with thrombocytopenia syndrome (SFTS) initially reported from rural areas in central and eastern China with high initial fatality rates of 10% to 30% [Yu et al., 2011]. The SFTSV epidemic has been expanding in China[Liu et al., 2014], while similar infection cases have been reported in Japan[Takahashi et al., 2014] and Korea[Chang and Woo, 2013; Park et al., 2014], and the Heartland virus with close phylogenetic relationships to SFTSV was detected in the United States,[Savage et al., 2013] indicating possibly globally epidemics.
Effective methods of SFTSV detection are urgently needed for clinical diagnosis and disease control, as well as assessment of risk of SFTSV transmission by blood transfusion. Laboratory testing strategies to detect SFTSV infection have been rapidly provided for clinical diagnosis using serology-based screening by enzyme-linked immunosorbent assay (ELISA) for anti-SFTSV total antibodies[Jiao et al., 2012]. Meanwhile, a commercial NAT (comNAT) assay based on one-step RT-PCR was developed and has been applied in epidemiological investigations [Niu et al., 2013; Wen et al., 2014]. However, over 30% of patients with suspected clinical features of SFTS could not be confirmed by laboratory testing [Wen et al., 2014]. The cut-off Ct for the comNAT assay (the only commercial NAT assay for detection of SFTSV in China) is 35 cycles with a lower limit of quantitative detection (LOQD) of 10 TCID50/ml, according to the assay manual. Thus it is possible that some ELISA negative SFTS infected patients with low viral loads, perhaps resulting in Cts higher than 35 cycles, would not be detected by the comNAT assay. Furthermore, since asymptomatic individuals, such as blood donors, tend to have lower viral loads than clinically ill patients, this assay may not be suitable for SFTSV screening of blood donors. Although currently there is not enough evidence for transfusion–transmitted SFTSV infection to warrant such screening[Zeng et al., 2015], the lack of validated, highly sensitive NAT assays that meet rigorous performance for both clinical diagnosis and blood screening needs to be addressed. In the present study, we sought to develop and validate a sensitive and specific RT-PCR assay for detection and quantitation of SFTSV.
Materials and Methods
Study samples
Xinyang 154 Military Hospital (XMH) is a regional hospital located in one of the concentrated epidemic regions of SFTS in Henan Province, China, with 100–200 SFTS suspected patients receiving treatment annually[Cui et al., 2014]. The average number of days for patients receiving treatment at the hospital was 5 days (1–38 days). Informed consent for testing samples for the purpose of research was collected from the patients during their hospital stay.
For the present study, 129 whole blood samples were collected at XMH between April and August 2013 from 93 suspected SFTS patients during the acute phase of possible SFTS disease. The initial diagnosis for suspected SFTS infection in these 93 patients was based on clinical signs and symptoms which included: fever (100%), malaise (94.6%), myalgia (89.2%), gastrointestinal symptoms (61.3%), thrombocytopenia (81.7%), and leukopenia (78.5%). The collected samples included: 1) 72 paired samples that had been collected from 36 patients, with one initial sample collected from each patient on the 1st day of hospitalization (1DOH) and one follow-up sample collected from the 3rd to 5th day of hospitalization (3–5DOH); 2) single-collection samples were also collected from 57 patients either on 1DOH (n=46) or 3DOH (n=11). Plasma samples were processed from anticoagulated whole blood vacutainer tubes, shipped to the Institute of Blood Transfusion (IBT) via cold chain transportation and stored at −80°C for further ELISA, comNAT and newNAT testing at IBT.
13 clinical samples including: two serotype 1 and eight serotype 2 Dengue virus infections (Guangzhou, Guangdong, China); and three Chikungunya fever virus infections (Dongguan, Guangdong, China) were provided by Guangdong blood center and shipped to IBT to access the analytical specificity of newNAT assay. These clinical samples were all from autochthonous epidemic outbreaks in year 2014 (Dengue fever) and 2010 (Chikungunya fever) in Guangdong province.
Detection of anti-SFTSV total antibody with a commercial ELISA
All clinical samples (n=129) were tested for anti-SFTSV total antibodies (including IgG and IgM) using an ELISA assay (Xin-Lian-Xin, Inc., Wuxi, China) in a 96 well format following the manufacturer’s protocol. Selected ELISA reactive clinical samples were used as external controls. The detailed procedure was reported in a previous study[Zeng et al., 2015]. The assay is a double-antigen sandwich ELISA to detect SFTSV-specific antibodies binding to SFTSV recombinant nucleocapsid protein [Jiao et al., 2012]. The sensitivity and specificity of ELISA assay were both 99.9% given by the manufacturer.
comNAT testing of clinical samples
All clinical samples were tested by comNAT (Daan, Inc., Guangzhou, China) following the manufacturer’s protocol. 140μL plasma sample was extracted for SFTSV RNA using Viral RNA extraction kit (QIAamp, Qiagen, Valencia, CA), per the manufacturer’s instructions. The RNA was diluted in 100μL water. 5μL RNA extract was input per reaction, following the instruction of the kit. The comNAT assay was designed as a one-step real-time RT-PCR based on a Taqman-probe method to detect the S segment of SFTSV. Four quantitative standards as linear range of quantitative detection (1×105, 1×104, 1×103, 1×102TCID50/ml, recombinant pseudotyped virus) were used in the comNAT assay and the cut-off amplification cycle number (Ct) for PCR positive samples was set to 35 cycles with a claimed LOD at 10TCID50/ml. (Table. 1) The SFTSV RNA equivalents of 10TCID50/ml were 4960 copies/ml, given by the manufacturer of comNAT assay.
Table 1.
Detection features of comNAT and newNAT assays.
| PCR procedure | SFTSV RNA extraction | Reaction volume | Limit of detection (LOD) | Quantitative standards | Limit of quantitative detection* | Specificity | |
|---|---|---|---|---|---|---|---|
| comNAT | Taqman-probe one-step RT PCR | 140μL plasma extracted and diluted in 100μL water, 5μL input per RT-PCR reaction | 20μL (one-step including RT and PCR) | 10 TCID50/ml*** | Four standards (1×105, 1×104, 1×103, and 1×102TCID50/ml) | 100TCID50/ml*** | 99.9%*** |
| newNAT | Taqman-probe two-steps RT PCR | 140μL plasma extracted and diluted in 100μL water, 100μL input per RT reaction and 25μL cDNA input per PCR reaction | RT (120μL) and PCR (75μL) | 95% LOD: 5.4 PFU/ml; 50% LOD: 0.8 PFU/ml | Five standards (2×105, 2×104, 2×103, 2×102 and 2×101PFU/ml) | 20 PFU/ml | 99.98% |
10 TCID50/ml (or 4960copies/ml of SFTSV RNA equivalents given by manufacturer) ≈387±27 PFU/ml (From 24 replicates by newNAT); 1 PFU/ml≈13 copies/ml by calculation.
N/A, not available.
Given by manufacturer.
Real-time RT-PCR to detect SFTSV RNA using newNAT
All clinical samples were tested for SFTSV RNA by a new Taqman real-time RT-PCR. The newNAT assay was based on a Taqman-probe method to detect the S segment of SFTSV. RNA from 140μL plasma samples was extracted as described above. All the RNA (100μL) was used for RT-PCR reaction, followed by reverse transcription with RT buffer of 12μL of 10×Solution A + B [Bloch et al., 2013], 1.2μL dNTPs (100 mM - no dUTP; Bioline, Germany), 3μLRNase inhibitor (40U/μL; Roche Diagnostics GmbH, Germany), 3μL reverse transcriptase (50 U/μL, Roche Diagnostics GmbH, Germany) and 0.45μL of downstream primer on S segment (100μM, S-R-3)[Sun et al., 2012], at condition of 42°C for 30 min, 100°C for 10 min and cooling to 4°C to synthesis cDNA. Then, 25μL cDNA was added to 51.4μL of PCR mixture consisting of 50 uL Buffer 55 (patented buffer provided by Blood Systems Research Institute), 0.5μL dNTPs (100 mM-with dUTP; Bioline, Germany), 0.5μL primers (100μM, S-F-3 and S-R-3)[Sun et al., 2012], 0.1μL TaqMan probe (100 μM; S-Probe-3)[Sun et al., 2012] and 0.3μL FastStart Taq (Roche Diagnostics GmbH, Germany). Real-time PCR was performed with conditions of 1 cycle of 95°C for 1 minute followed by 45 cycles of 95°C for 30 seconds and 60°C for 1 minute. The buffers for RT-PCR were optimized for high sensitivity. The RT-PCR method and analytic sensitivity were reported in a previous study focused on application of the assay to blood donor samples [Zeng et al., 2015]. Duplicate testing of each sample was performed and samples were considered to be positive if both wells gave a positive signal at a cycle number (Ct) <40. If the sample was tested to be with one result of Ct<40, the result was considered as negative. The quantitative standards (PFU/ml) from live SFTSV were prepared by the Arbovirus Diseases Branch of the Centers for Disease Control and Prevention (Fort Collins, CO, USA). Five quantitative standards (2×105, 2×104, 2×103, 2×102 and 2×101PFU/ml) were included in each PCR plate. The amplification efficiency of the newNAT assay was calculated based on the slope of the quantitative standard curve (E = 10−1/slope – 1). If the concentration of samples was higher than 2×105PFU/ml upon initial testing, the samples were serially diluted 10-fold in plasma and re-tested to yield results within the range of the quantitative standards. If the concentration was lower than 20 PFU/ml but gave a Ct<40, the viral load was reported as “<20PFU/ml”.
Comparison of LOQD and quantitative and qualitative results of comNAT and newNAT assays
The LOQD of the comNAT assay (package insert claim of 100TCID50/ml) was estimated by amplifying 24 replicates of a 10-fold dilution of the 100TCID50/ml standard to 10TCID50/ml, provided by the comNAT kit using the newNAT assay. 22 clinical samples ranging from 982 to 3.0×105PFU/ml were retested by the two assays to analyze the correlation of the Cts. comNAT negative and newNAT positive samples were retested to confirm the discrepancy between the two assays.
Statistical analysis
Descriptive and correlation analyses were conducted by “XY correlation” analysis using GraphPad Prism Version 6.01 software (GraphPad software Inc., La Jolla, CA, USA). Comparisons between the comNAT and the newNAT assays were assessed using ANOVA (for viral load and Cts) and chi-square or Fisher’s exact tests (for nonparametric outcomes). p<0.05 was considered statistically significant.
Results
comNAT and ELISA testing of clinical samples
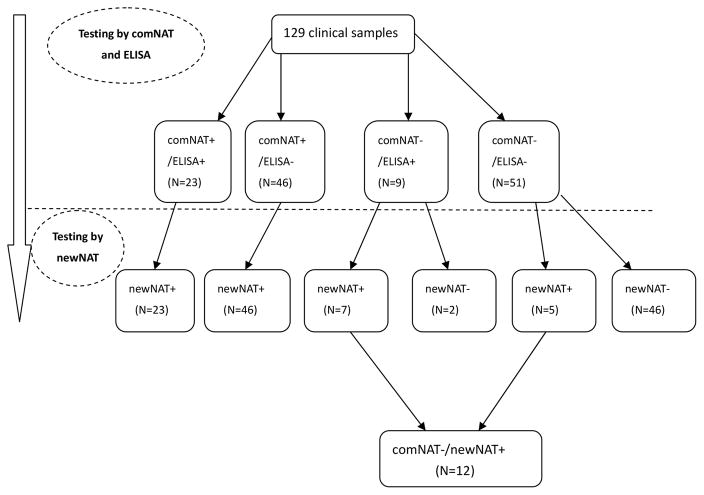
The 129 clinical samples were classified into four groups by ELISA and comNAT testing, as summarized in Table 2: 1) 23 samples were positive by both comNAT and ELISA; 2) 46 samples were comNAT positive/ELISA negative; 3) 9 samples were comNAT negative/ELISA positive; and 4) 51 samples (collected from 35 suspected SFTS patients) were negative by both comNAT and ELISA testing. Overall, 58 suspected patients were tested to be SFTSV positive including: 57 positive by ELISA or/and comNAT and one positive only by ELISA.
Table 2.
ComNAT and ELISA testing on 129 samples*
| Collection times(number of samples) | comNAT+/ELISA+ | comNAT+/ELISA− | comNAT−/ELISA+ | comNAT−/ELISA− |
|---|---|---|---|---|
| 1DOH** (N=82) | 11 | 34 | 5 | 32 |
| 3–5DOH (N=47) | 12 | 12 | 4 | 19 |
| Total (N=129) | 23 | 46 | 9 | 51 |
Out of 93 patients, 58 patients were comNAT and/or ELISA positive on either or/both collection times.
DOH: day of hospitalization.
Performance evaluation of the newNAT assay
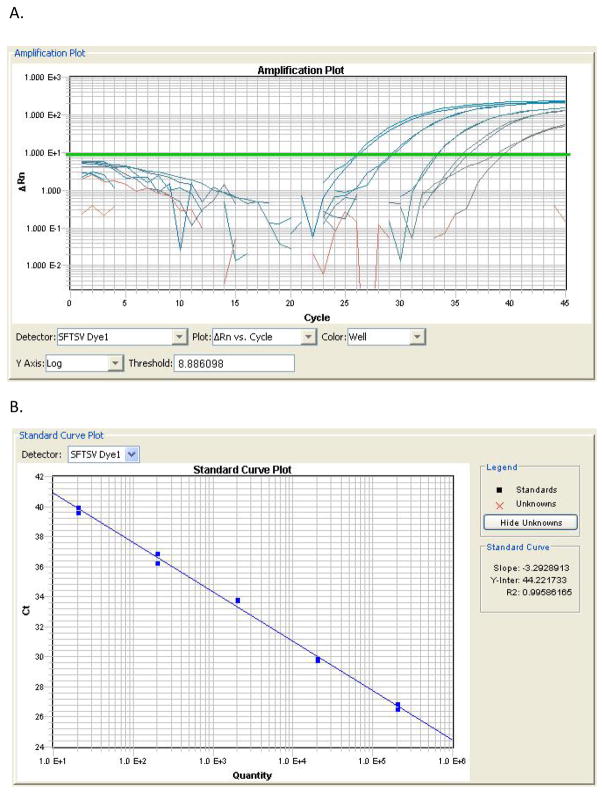
The standard curve and amplification plots of the new real-time PCR assay were evaluated to determine reproducibility, efficiency and dynamic range. The linear correlation (R2>0.99) between Ct value and SFTSV RNA input was high (Figure 1). The assay had 100% amplification efficiency, based on the slope of the standard curve. Furthermore, good reproducibility was observed for the five quantitative standards, with standard deviations of the Cts ranging from 0.23 to 0.53 (Table 3). The lower limit for quantitative testing for the newNAT assay was estimated to be 20 PFU/ml corresponding to the concentration of SFTSV detectable at a Ct of 38.8. The primers and probe of S segment used in study have been systematically proved by group from National Institute for Viral Disease Control and Prevention, China CDC having no cross-react with other bunyaviruses or virus that could cause similar symptoms[Sun et al., 2012]. Further in this study, the newNAT was also used to test against virus RNA extract of Dengue virus serotype 1 and 2 and Chikungunya fever virus, and no positive results were obtained. Assuming that the 2 PCR reactive samples out of 9960 blood donor samples tested in a previous study [Zeng et al., 2015] were false positive, the specificity of the assay was estimated to be 99.98%.
Figure 1.
Table 3.
Reproducibility of the newNAT assay.
| Concentrations of standards (PFU/ml) | PCR amplification cycles (Cts±SD)* |
|---|---|
| 2 x 105 | 26.11±0.23 |
| 2 x 104 | 29.18±0.31 |
| 2 x 103 | 33.01±0.24 |
| 2 x 102 | 35.93±0.37 |
| 2 x 101 | 38.81±0.53 |
Determined from 12 replicates. All replicates were positive.
Testing of clinical samples with the newNAT assay
81 samples collected from 61 patients were SFTSV RNA positive by the newNAT, including 40 paired samples from 20 patients (both positive at 1DOH and 3–5DOH) and 41 single-collection samples (positive either at 1DOH or 3–5 DOH). Viral loads of positive SFTSV clinical samples ranged from 34 to 2.1×107PFU/ml using the newNAT assay. All comNAT positive samples were also positive by the newNAT (N=69) (Figure. 2), while 12 comNAT negative samples from 9 patients were positive using the newNAT; 4 of these 9 patients were initially diagnosed as SFTSV negative due to negative results on ELISA and comNAT. On the newNAT assay, these 12 samples had mean Cts of 36.5±1.3 and viral loads ranging from 34 to 619 PFU/ml (Table. 4).
Figure 2.
Table 4.
comNAT negative and newNAT positive samples (12 samples collected from 9 patients)
| Patient ID* | Anti-SFTSV total antibody | Testing by the comNAT assay** | Testing by the newNAT assay*** | |
|---|---|---|---|---|
|
| ||||
| Mean Ct | Mean Ct | Viral load (PFU/ml) | ||
| 15 | − | 37.2 | 36.7 | 112 |
| 34 | − | 35.3 | 34.3 | 619 |
| 58 | − | 36.7 | 36.1 | 182 |
| 60 | − | 36.3 | 35.5 | 526 |
| 62 | + | 38.8 | 37.1 | 86 |
| 66(1) | + | 38.3 | 37 | 96 |
| 66(2) | + | 36.8 | 36.1 | 172 |
| 68(1) | + | 39.4 | 38.3 | 34 |
| 75(1) | + | 36.4 | 36 | 342 |
| 75(2) | + | 39.7 | 38.5 | 66 |
| 94(1) | − | 35.3 | 35 | 457 |
| 94(2) | + | 37.9 | 37.4 | 84 |
Patient ID (1): The 1st day of hospitalization; Patient ID (2): The 3rd to 5th day of hospitalization.
Determined from 4 replicates; all samples were negative by comNAT (the criteria for comNAT positivity is Cts<35)
Determined from 4 replicates. All replicates were positive by newNAT testing.
For the entire dataset, the viral loads of ELISA positive samples (n=30, 3.1×104±1.0×104PFU/ml) were significantly lower (P<0.05) than those of ELISA negative samples (n=51, 8.1×105±4.4×105PFU/ml).
Of 36 SFTS suspected patients with two collected samples, 20 patients were newNAT positive on both of their samples. (Table. 5) Nine of these 20 patients were ELISA negative on both visits, 4 seroconverted over a 2–4 day interval (i.e., between 1DOH and 3–5DOH), and 7 were ELISA positive on both visits. One patient was ELISA positive/newNAT negative on both visits (Patient ID: 69). For the 4 seroconverted patients, one (Patient ID: 94) tested newNAT positive but comNAT negative. No difference in viral loads was found between 1DOH and 3–5DOH (P=0.57).
Table 5.
20 SFTSV RNA positive patients with two collection times*
| Patient ID | 1DOH* (n=20) | 3–5DOH (n=20) | ||
|---|---|---|---|---|
|
| ||||
| Anti-SFTSV total antibody (Positivity: n=7) | Viral loads by newNAT (PFU/ml)** | Anti-SFTSV total antibody titer (Positivity: n=11) | Viral loads by newNAT (PFU/ml)*** | |
| 63 | − | 2088 | − | 1980 |
| 65 | − | 2530000 | − | 900000 |
| 66 | + | 96 | + | 172 |
| 68 | + | 34 | + | 179500 |
| 70 | + | 1193 | + | 18460 |
| 71 | − | 76800 | − | 100500 |
| 74 | − | 2350 | + | 5480 |
| 75 | + | 342 | + | 66 |
| 77 | − | 26200 | − | 13100 |
| 82 | − | 9500 | − | 5490 |
| 84 | + | 163500 | + | 171800 |
| 86 | − | 1645 | + | 6210 |
| 89 | + | 19440 | + | 9770 |
| 90 | − | 172500 | + | 130500 |
| 91 | − | 6800 | − | 7150 |
| 92 | − | 38500 | − | 12600 |
| 93 | − | 13820 | − | 1006 |
| 94 | − | 457 | + | 84 |
| 97 | − | 20770 | − | 6230 |
| 98 | + | 3210 | + | 2900 |
Out of 36 patients with two collection times, 20 were positive by SFTSV RNA by newNAT; 9 of these were ELISA negative on both visits, 4 seroconverted over a 2–4 day interval and 7 were ELISA positive on both visits.; 1DOH: The 1st day of hospitalization; 3–5DOH: The 3rd to 5th day of hospitalization
Viral loads for 1DOH: 1.5×105±1.3×105PFU/ml
Viral loads for 3–5DOH:7.9×104±4.5×104PFU/ml; No significant difference between 1DOH and 3–5DOH (P=0.57)
Comparison of quantitative assay performance of comNAT and newNAT
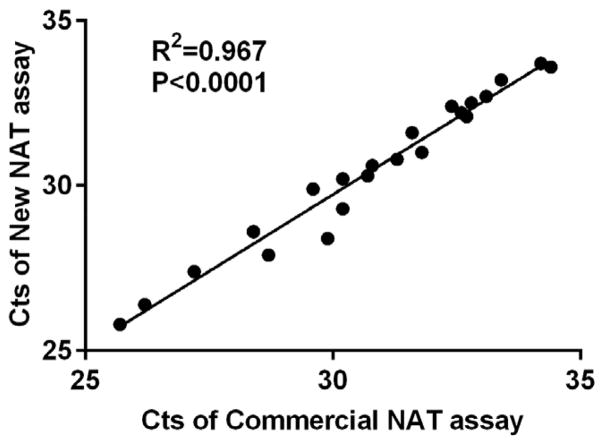
The correlation of the Cts between comNAT and newNAT is displayed in Figure 3. The Cts for selected clinical samples (n=22) ranged from 25 to 34. Significant correlation between the two assays was observed (P<0.0001). To evaluate the sensitivity of the comNAT assay relative to the newNAT assay, the claimed LOQD of quantitative standard of the comNAT assay was diluted to 10 TCID50/ml and amplified using the newNAT assay with its corresponding five quantitative standards (ranging from 2x105 to 20 PFU/ml). Based on this analysis, the claimed LOD of the comNAT assay (10 TCID50/ml or 4960 copies/ml of SFTSV RNA) was estimated to be 387±27 PFU/ml, the equivalent relation of PFU/ml and SFTSV RNA copies/ml can be calculated as: 1 PFU/ml≈13 copies/ml.
Figure 3.
Discussion
A new NAT assay based on real-time RT-PCR was developed to quantitatively detect SFTSV RNA. The assay had a lower limit for quantitative testing of 20 PFU/ml and improved clinical sensitivity relative to a commercially available real-time RT-PCR assay. This newNAT assay can be applied to clinical diagnosis on blood samples from symptomatic SFTS patients, and may also be applicable to studies of SFTSV RNA screening among blood donors. At present, there is still no China Food and Drug Administration (CFDA)-approved assay for either clinical diagnosis or blood screening. For SFTS clinical diagnosis, in addition to typical clinical features that are also common among many other diseases, results of NAT and ELISA testing on suspected patients’ samples may help confirm a diagnosis.
Four comNAT and ELISA negative patients tested positive by the newNAT, although 33% (31/93) of suspected SFTS patients remained negative by both the commercial and new NAT assays and ELISA testing. We speculate that the 31 anti-SFTSV total antibody and RNA negative patients might be infected by other pathogens, since the pathogens in Xinyang and its neighboring regions, such as Anaplasma phagocytophilum[He JG, 2006] and Orientia tsutsugamushi[Xu BL, 2005], result in very similar symptoms such as fever, malaise and thrombocytopenia [Xu et al., 2011]. We also found that most of the newNAT positive samples (63%, 51/81) from the acute phase (within the 5th day of hospitalization) of SFTSV infection were serologically negative. Two ELISA positive/newNAT negative paired samples (from one patient at 1DOH and 3DOH) were from a patient who might have cleared the virus. The results were compatible with findings from other studies indicating that the viremic, seronegative phase (e.g. SFTSV RNA positive but anti-SFTSV total antibody negative) is expected to last about one week (which is longer than the follow-up time in this study) while a few SFTS patients may seroconvert within 5 days of their hospitalization for clinical disease.[Sun et al., 2014; Wen et al., 2014].
High correlation of Cts was found between the two NAT assays by testing on samples with different concentrations of SFTSV. The performance of the two assays was consistent on the samples with Cts<35 cycles indicating similar quantitative performance of the assays over a range of viral loads (Figure 3). However, the interpretive criteria for the comNAT assay testing positive with LOD at 10TCID50/ml or Cts lower than 35 cycles may result in missing low-level viremic SFTS patients, since we have demonstrated that the comNAT assay has lower sensitivity compared to the newNAT assay. While direct comparison of quantitative results was not possible due to the use of assay standards quantified in different units (TCID50/ml for comNAT and PFU/ml for newNAT), we were able to perform a direct comparison by serially diluting the comNAT standard and assuring it using the newNAT assay. Given that we observed good correlation between the Cts of the two NAT assays when we tested the samples with lower viral load (e.g. Ct>35) (Table 4), it is clear that a higher Ct cutoff for the comNAT assay would have resulted in more cases scoring positive. However, we did not further explore a higher Ct cutoff for the comNAT for two reasons: i) the more constrained quantitative range listed in the comNAT product insert and verified in this study implies that detection on low viral load samples might not be as reliable as the newNAT and ii) when we formerly applied the comNAT assays for SFTSV RNA screening in the low risk blood donor population, we found that abnormal non-specific amplifications were commonly observed when the comNAT Ct was >35 (i.e., some samples were amplified with Ct of 38 to 39 in the comNAT, but were negative by newNAT). Thus, further evaluation would be needed to determine the impact of increasing the comNAT cutoff value on that assay’s specificity.
A probit analysis of the analytic sensitivity of the newNAT PCR assay was reported in our previous study [Zeng et al., 2015]. By multiple testing with newNAT on serial dilutions of the same quantitative standard used in this study from 10 PFU/ml to 0.1 PFU/ml, the 95% limit of detection for the newNAT was 5.4 PFU/ml and the 50% limit of detection was 0.8 PFU/ml [Zeng P, 2015]. It could be speculated that higher sensitivity for the newNAT assay relative to comNAT may result from the difference in PCR procedures between the two assays. The comNAT assay is based on a one-step RT-PCR with 20μL reaction volume, whereas the new NAT assay employed a two-step RT-PCR with larger RT (120μL) and PCR (75μL) reaction volumes. In addition, the high specificity of the newNAT (previously reported as 99.98%) permits the test to use a higher Ct value as a cutoff, thus contributing to its greater sensitivity compared to comNAT.
The newNAT assay for SFTSV has been applied in large-scale screening among blood donors from endemic region (Xinyang, China) using four-sample mini-pools. After testing of 2490 pools (9960 samples from blood donors) and further resolution testing on reactive pools, two suspected SFTSV RNA positive donors with extremely low viral load (mean Cts of 39.3 and 39.7, respectively, viral loads<20PFU/ml) were identified. The study of SFTSV NAT performance on pools from blood donors documented the sensitivity and specificity of the newNAT assay and suggested that the newNAT assay was applicable to SFTSV surveillance and potentially blood screening in endemic regions after further optimization.
One limitation of this study is that we could not rule out whether the 5 newNAT positive/comNAT negative samples in ELISA negative patients were false positive by newNAT, since there was no follow-up sample to test for seroconversion. However, based on the application of newNAT to SFTSV screening among blood donors (see above), the specificity of the new NAT assay is greater than 99.9%, which reduces the likelihood of false positives in this study. A second limitation is that, compared with the comNAT assay, the testing procedure of the newNAT is more cumbersome with two-step real time RT-PCR; In general, the assay may need to be modified for automation to reach the demand of potential high-throughput screening at blood centers should donor screening be warranted. Furthermore, the low LOQD may require higher facility standards and stricter experimental procedures in the laboratory to avoid PCR contamination. For clinical diagnosis, however, the newNAT assay demonstrated sensitivity in detection of samples with viral loads lower than the LOQD of the comNAT assay.
In conclusion, we have developed and validated a sensitive and specific real-time RT-PCR assay for SFTSV RNA quantitative detection. The assay should be an important tool to help with clinical diagnosis of SFTS patients as well as evaluations of the need for blood screening to prevent potential transfusion transmitted SFTS infections.
Acknowledgments
The authors acknowledge the Centers for Disease Control (Fort Collins, CO, USA) for providing the cultured SFTSV panel and Guangdong blood center for providing the clinical samples of Dengue and Chikungunya fever virus. The authors also acknowledge Dr. Steve Kleinman and Dr. Roger Dodd for their careful review of the paper. The study was supported by the National Natural Science Foundation of China (Grant No. 81400096); PUMC Youth Fund and the Fundamental Research Funds for the Central Universities (Grant No.33320140190); Funding of Scientific support, Science and Technology Department of Sichuan province (Grant No. 2014SZ0025) and the NHLBI Recipient Epidemiology and Donor Evaluation Study-III (HHSN268201100008I and HHSN268201100001I).
References
- Bloch EM, Lee TH, Krause PJ, Telford SR, 3rd, Montalvo L, Chafets D, Usmani-Brown S, Lepore TJ, Busch MP. Development of a real-time polymerase chain reaction assay for sensitive detection and quantitation of Babesia microti infection. Transfusion. 2013;53(10):2299–2306. doi: 10.1111/trf.12098. [DOI] [PubMed] [Google Scholar]
- Chang MS, Woo JH. Severe fever with thrombocytopenia syndrome: tick-mediated viral disease. Journal of Korean medical science. 2013;28(6):795–796. doi: 10.3346/jkms.2013.28.6.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui N, Bao XL, Yang ZD, Lu QB, Hu CY, Wang LY, Wang BJ, Wang HY, Liu K, Yuan C, Fan XJ, Wang Z, Zhang L, Zhang XA, Hu LP, Liu W, Cao WC. Clinical progression and predictors of death in patients with severe fever with thrombocytopenia syndrome in China. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2014;59(1):12–17. doi: 10.1016/j.jcv.2013.10.024. [DOI] [PubMed] [Google Scholar]
- He JGCZ, Wu JB, Yang XX, Li Q, et al. The Epidemiological Investigation on a human to human anaplasmosis infection in South Anhui Province in 2006. World J Infect. 2006;(5):343–347. [Google Scholar]
- Jiao Y, Zeng X, Guo X, Qi X, Zhang X, Shi Z, Zhou M, Bao C, Zhang W, Xu Y, Wang H. Preparation and evaluation of recombinant severe fever with thrombocytopenia syndrome virus nucleocapsid protein for detection of total antibodies in human and animal sera by double-antigen sandwich enzyme-linked immunosorbent assay. Journal of clinical microbiology. 2012;50(2):372–377. doi: 10.1128/JCM.01319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, He B, Huang SY, Wei F, Zhu XQ. Severe fever with thrombocytopenia syndrome, an emerging tick-borne zoonosis. The Lancet Infectious diseases. 2014;14(8):763–772. doi: 10.1016/S1473-3099(14)70718-2. [DOI] [PubMed] [Google Scholar]
- Niu G, Li J, Liang M, Jiang X, Jiang M, Yin H, Wang Z, Li C, Zhang Q, Jin C, Wang X, Ding S, Xing Z, Wang S, Bi Z, Li D. Severe fever with thrombocytopenia syndrome virus among domesticated animals, China. Emerging infectious diseases. 2013;19(5):756–763. doi: 10.3201/eid1905.120245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Han MG, Yun SM, Park C, Lee WJ, Ryou J. Severe fever with thrombocytopenia syndrome virus, South Korea, 2013. Emerging infectious diseases. 2014;20(11):1880–1882. doi: 10.3201/eid2011.140888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage HM, Godsey MS, Jr, Lambert A, Panella NA, Burkhalter KL, Harmon JR, Lash RR, Ashley DC, Nicholson WL. First detection of heartland virus (Bunyaviridae: Phlebovirus) from field collected arthropods. The American journal of tropical medicine and hygiene. 2013;89(3):445–452. doi: 10.4269/ajtmh.13-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Chai C, Lv H, Lin J, Wang C, Chen E, Zhang Y, Chen Z, Liu S, Gong Z, Jiang J. Epidemiological characteristics of severe fever with thrombocytopenia syndrome in Zhejiang Province, China. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases. 2014;25:180–185. doi: 10.1016/j.ijid.2014.02.022. [DOI] [PubMed] [Google Scholar]
- Sun Y, Liang M, Qu J, Jin C, Zhang Q, Li J, Jiang X, Wang Q, Lu J, Gu W, Zhang S, Li C, Wang X, Zhan F, Yao W, Bi Z, Wang S, Li D. Early diagnosis of novel SFTS bunyavirus infection by quantitative real-time RT-PCR assay. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2012;53(1):48–53. doi: 10.1016/j.jcv.2011.09.031. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, Kamei T, Honda M, Ninomiya D, Sakai T, Senba T, Kaneyuki S, Sakaguchi S, Satoh A, Hosokawa T, Kawabe Y, Kurihara S, Izumikawa K, Kohno S, Azuma T, Suemori K, Yasukawa M, Mizutani T, Omatsu T, Katayama Y, Miyahara M, Ijuin M, Doi K, Okuda M, Umeki K, Saito T, Fukushima K, Nakajima K, Yoshikawa T, Tani H, Fukushi S, Fukuma A, Ogata M, Shimojima M, Nakajima N, Nagata N, Katano H, Fukumoto H, Sato Y, Hasegawa H, Yamagishi T, Oishi K, Kurane I, Morikawa S, Saijo M. The first identification and retrospective study of Severe Fever with Thrombocytopenia Syndrome in Japan. The Journal of infectious diseases. 2014;209(6):816–827. doi: 10.1093/infdis/jit603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen HL, Zhao L, Zhai S, Chi Y, Cui F, Wang D, Wang L, Wang Z, Wang Q, Zhang S, Liu Y, Yu H, Yu XJ. Severe fever with thrombocytopenia syndrome, Shandong Province, China, 2011. Emerging infectious diseases. 2014;20(1):1–5. doi: 10.3201/eid2001.120532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Liu L, Huang X, Ma H, Zhang Y, Du Y, Wang P, Tang X, Wang H, Kang K, Zhang S, Zhao G, Wu W, Yang Y, Chen H, Mu F, Chen W. Metagenomic analysis of fever, thrombocytopenia and leukopenia syndrome (FTLS) in Henan Province, China: discovery of a new bunyavirus. PLoS pathogens. 2011;7(11):e1002369. doi: 10.1371/journal.ppat.1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu BLCH, Zhu Q, Zhang J, Xia SL. The Epidemiological Investigation on the First Outbreak of Tsutsugamushi Disease in Henan Province. Henan J Prev Med. 2005;17:3. [Google Scholar]
- Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, Sun YL, Zhang L, Zhang QF, Popov VL, Li C, Qu J, Li Q, Zhang YP, Hai R, Wu W, Wang Q, Zhan FX, Wang XJ, Kan B, Wang SW, Wan KL, Jing HQ, Lu JX, Yin WW, Zhou H, Guan XH, Liu JF, Bi ZQ, Liu GH, Ren J, Wang H, Zhao Z, Song JD, He JR, Wan T, Zhang JS, Fu XP, Sun LN, Dong XP, Feng ZJ, Yang WZ, Hong T, Zhang Y, Walker DH, Wang Y, Li DX. Fever with thrombocytopenia associated with a novel bunyavirus in China. The New England journal of medicine. 2011;364(16):1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng PCL, Ma L, Zeng Z, Wang J. Evaluation of lower limit of detection by nucleic acid testing on severe fever with thrombocytopenia syndrome virus. Chin J Blood Transfusion. 2015;28(4):377–379. [Google Scholar]
- Zeng P, Ma L, Gao Z, Wang J, Liu J, Huang X, Yang Q, Cao R, Wen X, Zhu L, Ma H, Yang Z, Lee TH, Brambilla D, Yuan M, Glynn S, Ness P, Kleinman S, Busch M, Shan H. A study of seroprevalence and rates of asymptomatic viremia of severe fever with thrombocytopenia syndrome virus among Chinese blood donors. Transfusion. 2015;55(5):965–971. doi: 10.1111/trf.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]