Abstract
Due to their cell membrane-mimicking properties, liposomes have served as a versatile research tool in science, from membrane biophysics and drug delivery systems to bottom-up synthetic cells. We recently reported a novel microfluidic method, Octanol-assisted Liposome Assembly (OLA), to form cell-sized, monodisperse, unilamellar liposomes with excellent encapsulation efficiency. Although OLA provides crucial advantages over alternative methods, it suffers from the presence of 1-octanol droplets, an inevitable by-product of the production process. These droplets can adversely affect the system regarding liposome stability, channel clogging, and imaging quality. In this paper, we report a density-based technique to separate the liposomes from droplets, integrated on the same chip. We show that this method can yield highly pure (>95%) liposome samples. We also present data showing that a variety of other separation techniques (based on size or relative permittivity) were unsuccessful. Our density-based separation approach favourably decouples the production and separation module, thus allowing freshly prepared liposomes to be used for downstream on-chip experimentation. This simple separation technique will make OLA a more versatile and widely applicable tool.
INTRODUCTION
Liposomes are supramolecular assemblies where a microscopic aqueous volume is confined within a lipid bilayer that separates it from the surrounding aqueous environment. Since the lipid bilayer is the universal component of cell membranes in living systems, liposomes serve as an excellent model system for cells. They have been instrumental in a wide variety of basic sciences such as biomimetic membrane biochemistry (e.g., membrane protein reconstitution, microreactors for enzymatic reactions, and protein-lipid bilayer interactions), biomembrane physics (e.g., vesicle growth and shape changes, membrane fusion and fission, and membrane dynamics), and also as a bottom-up model system for synthetic cells (e.g., reconstituting the minimal bacterial divisome machinery, assembling eukaryotic cytoskeletal elements, and mRNA and protein synthesis).1–9 Liposomes have already found applications in the medical field, acting as carriers for drug and gene delivery,10,11 and in bioanalytical systems, especially in integrated microfluidic devices.12,13
We recently reported a new microfluidic method, Octanol-assisted Liposome Assembly (OLA), to produce liposomes in a controlled and efficient manner.14 There are a number of important advantages of OLA over existing bulk methods (extrusion,15 hydration,16 and electroformation17) and microfluidic methods:18–23 (1) The use of a biocompatible organic solvent that enables an efficient and quick de-wetting of the double-emulsion droplets within a few minutes. (2) The liposomes are unilamellar and monodisperse. (3) Their diameter is of a biologically relevant size (5–20 μm) and can be controlled by altering the flow velocities of the inner and outer aqueous phases. (4) The method has an excellent encapsulation efficiency.
OLA uses 1-octanol as the lipid-carrying organic solvent which eventually gets separated spontaneously from the double-emulsion droplet in the form a droplet containing excess lipids, leaving behind a fully matured liposome. Thus, for every liposome, a single 1-octanol droplet (henceforth referred to as droplet) forms as a by-product. There are several problems associated with such a mixed system of liposomes and droplets. (1) Stability: Owing to the small but finite water-solubility (0.46 g l−1) of 1-octanol24 and the constant interaction of droplets with channel walls, the droplets gradually make the channel walls hydrophobic, rendering the environment less stable for liposomes. (2) Clogging: At low flow rates, the droplets tend to stick to each other, resulting in aggregates that eventually can clog the channel. (3) Imaging: Due to the excess fluorescent lipids present in the droplets, they are very bright. The presence of these bright objects in close proximity to the liposomes adversely affects the imaging contrast. These problems severely hinder the on-chip manipulation of liposomes. Therefore, it is highly desirable to separate the droplets, in order to obtain a system containing only liposomes.
In order to separate two objects on a microfluidic chip, it is necessary to distinguish them based on certain physical attributes, and subsequently design a microfluidic method capable of separating objects based on that specific attribute. Comparing liposomes and droplets, we identified the following distinctive properties. (1) Size: In OLA, droplets are consistently smaller than the corresponding liposomes. (2) Density: The density of 1-octanol25 is 827 kg/m3, whereas a liposome has the same density as that of water (1000 kg/m3), which if needed can be increased by encapsulating high-density molecules such as sucrose. (3) Relative permittivity: The dielectric constant of 1-octanol24 is 10.3, while that of the liposome (without any added salts and ignoring the contribution of the lipid bilayer) can be assumed to be very close to water, i.e., about 80. (4) Deformability: Liposomes are more deformable owing to their low bending rigidity (∼10−19 N m),26,27 as compared to the surface tension-governed stiffness (∼10−2 N/m) of droplets.25 (5) Conductivity: For a liposome containing pure water, the conductivity will be close to 0.6 × 10−5 S/m, as compared to 1.4 × 10−5 S/m for 1-octanol.25 (6) Fluorescence: The droplets are brighter due to the excess fluorescent lipids they contain. We mainly focused on the size, density, and relative permittivity to tackle the problem of separating the liposomes from droplets.
Numerous active and passive microfluidic particle separation methods have been developed over the years.28 Active methods, such as Standing Surface Acoustic Waves (SSAW; separation by size and density)29,30 and dielectrophoresis (DEP; separation by relative polarizability)31,32 use an external force to achieve particle separation. These active methods can be modified to include a fluorescence-activated component, as in Surface Acoustic Wave Actuated Cell Sorting (SAWACS)33 or Fluorescence Activated Droplet Sorting (FADS).34
Passive methods, on the other hand, rely solely on aptly designed microfluidic circuits and control of fluid flows. Examples of passive methods and the attributes that underlie their separation mechanism include Deterministic Lateral Displacement (DLD; size, deformability and shape),35 Pinched Flow Fractionation (PFF; size),36,37 hydrodynamic particle filtration (size),38 continuous inertial focusing (size),39 Density Difference Amplification-based Cell Sorting (dDACS, density),40 and Microvortex Manipulation (MVM, density).41 We explored a variety of previously reported separation techniques with the OLA design, but the attempts were unsuccessful. The main problem we encountered was the incompatibility between the optimal flow rates required for liposome production and those for the separation techniques.
We finally came up with a very effective density-based technique to remove the droplets, which yielded a clean system (up to 96% pure samples) of liposomes. The technique is solely based on the density difference between the droplets and the liposomes, is integrated on the same chip, and is fully compatible with the flow rates required for optimal working of OLA. Furthermore, it greatly decouples the production module and the separation module, thus making the two modules work independently of each other and allowing a large variety of downstream on-chip experiments with liposomes.
In this paper, we first describe the density-based method which we developed and successfully integrated on chip. Subsequently, we also briefly describe our unsuccessful attempts to integrate several established active and passive techniques (DLD, PFF, and DEP) and report the reasons for their incompatibility with OLA.
RESULTS
Density-based separation works efficiently
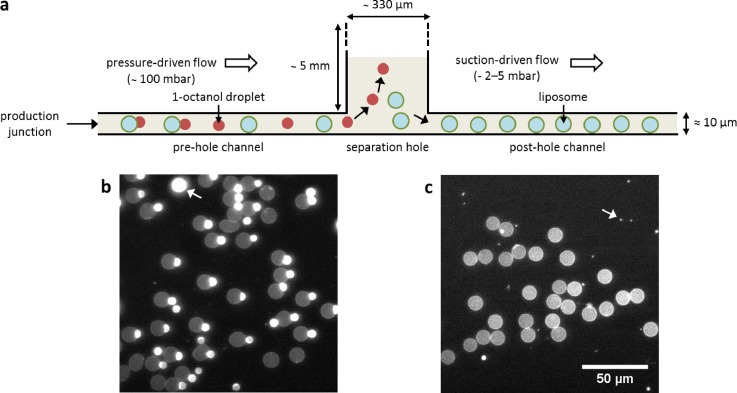
Since 1-octanol droplets are less dense than the aqueous phase in which they are suspended, they should float to the top of the channel, whereas liposomes will distribute homogenously throughout the channel due to having a similar density to their surroundings. The microfluidic channels used for OLA are typically about 8–10 μm in height, similar to or only slightly larger than the size of liposomes and droplets. As a result, we could not use the standard-height OLA-design for separation, even though there is a difference in the density between liposomes and droplets. Thus, we needed to locally increase the height of the device significantly to create enough space for the droplets to separate from the liposomes. To do so, we punched a hole, of a lateral size much larger (∼330 μm in diameter) than the channel width (50 μm), across the post-junction channel (Fig. 1(a)). As the typical height of the polydimethylsiloxane (PDMS) block used for making microfluidic devices is ∼5 mm, this hole created a large reservoir providing ample space for the droplets to float to the top of the reservoir and separate from the liposomes, which remained near the bottom. In order to help the liposomes to move from the hole into the post-hole channel, we applied a small suction (an underpressure of 2–4 mbar) at the exit using negative-pressure pumps.
FIG. 1.
On-chip density-based separation of 1-octanol droplets from OLA-based liposomes. (a) Side-view schematic of the device, showing the technique leading to the separation of droplets from liposomes. The double-emulsion droplets produced at the junction give rise to liposomes and 1-octanol droplets, which coexist in the pre-hole channel. As they enter the separation hole, the droplets drift upwards, while the liposomes are slowly sucked into the post-hole channel. (b) Fluorescence image of the pre-hole channel showing individual liposomes and droplets, and double-emulsion droplets on the verge of budding-off. The droplet marked by a white arrow is an example of a larger 1-octanol droplet produced by an instability at the junction. (c) Fluorescence image of the post-hole channel predominantly showing liposomes and some very small droplets (indicated by white arrow).
The result is shown in Fig. 1(c) (see also supplementary material, Movie S1), which shows a snapshot of the post-hole channel. Due to the excellent separation, it is largely filled with liposomes and only a tiny fraction of droplets. For comparison, Fig. 1(b) shows a snapshot of the pre-hole channel, where an equal amount of liposomes and droplets is observed. This density-based separation worked well for all conditions where the suction was strong enough to pull only the liposomes into the post-hole channel but not the droplets that were drifting upwards in the hole. If the suction pressure was too high, both liposomes and droplets got sucked into the post-hole channel and no separation occurred. Alternatively, we applied suction at both the separation hole and the exit, and achieved separation as well, as long as the effective pressure (peff = phole – pexit) was within the pressure of 2–4 mbar. Furthermore, it was possible to apply a small positive pressure at the separation hole and achieve separation. However, this was more complicated to implement, since the separation hole had to be first fully filled with water in order to prevent potential air bubbles from entering the channels.
The separation process depended on two parameters: the drift velocity of the droplets and the volumetric flow rate through the post-hole channel. The drift velocity, vdrift, of the droplets is given by the relation vdrift = f/ζ where f is the net force acting on the droplet and ζ is the viscous friction coefficient. In this case, f is the buoyancy force exerted on the droplet due to the density difference between the droplet and the surrounding medium (ρnet = ρmedium – ρ1–octanol) and equals f = ρnetVg, where V is the volume of the droplet and g is the gravitational acceleration constant. For a spherical droplet, the friction coefficient equals ζ = 6πηR, where η is the viscosity of the surrounding aqueous phase and R is the radius of the droplet. The relation for the drift velocity due to the buoyancy force thus becomes vdrift = 2ρnetR2g/9η. The diameter of a droplet varied between 4 and 10 μm (see Fig. 2(c)), resulting in the corresponding drift velocity of a droplet between 0.9 and 5.5 μm/s.
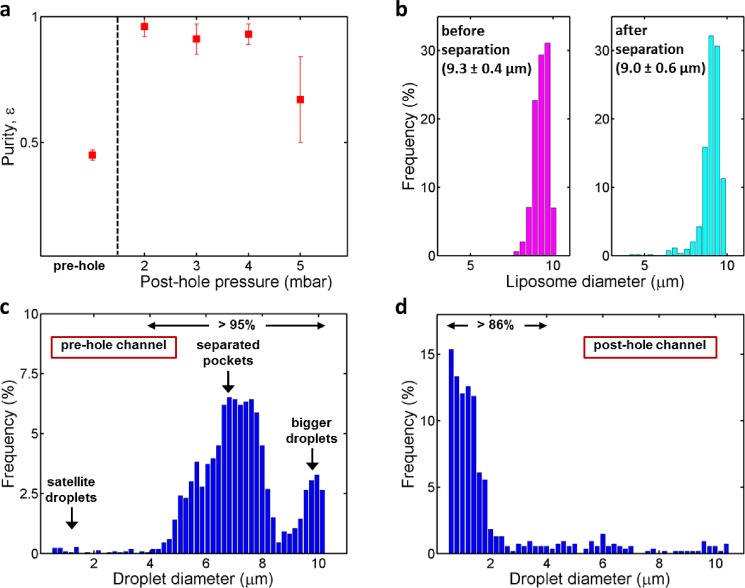
FIG. 2.
Efficiency of the density-based separation technique. (a) Purity of liposomes obtained at different post-hole pressures. The pre-hole purity is also shown for comparison. The purity obtained is an average of at least three separate runs (number of particles analysed for each data set is >1500). Error bars indicate standard deviations. (b) An example showing the frequency histograms of the liposomes in pre-hole (magenta, n = 1028) and post-hole (cyan, n = 541) channels. (c) Example of the size distribution of droplets in the pre-hole channel (n = 2195). One can clearly see three discrete populations, belonging to—from left to right—smaller satellite droplets, droplets that are a result of the budding-off process from the liposomes, and bigger droplets that are formed as a result of instability at the production junction. The latter two populations (liposomes >3.9 μm) make up more than 95% of the population. (d) Example of the size distribution of the contaminating droplets in the post-hole channel (n = 540), after the separation takes place. More than 86% of the droplets are below 3.9 μm, the cut-off value used for the purity analysis (see main text).
The volumetric flow rate Q is inversely proportional to the channel resistance R. Because the resistance of a microchannel decreases non-linearly with increasing diameter, the punched hole has a dramatically lower resistance than the post-hole channel. The 330 μm wide and 5 mm high hole has a ∼104 times lower resistance than the typical post-hole channel (h = 10 μm, w = 200 μm, and L = 2 mm; see Materials and Methods). As a result, the volumetric flow rate of the fluid into the post-hole channel is negligible and the fluid predominantly accumulated in the separation hole. Note also that the hydrostatic pressure generated as a result of fluid accumulating in the hole (∼0.5 mbar, for a fluid reservoir 5 mm high) was clearly not sufficient to make the liposomes move into the post-hole channel. We facilitated the flow of liposomes into the post-hole channel by applying a small suction pressure at the exit. Using the resistances of the post-hole channel and cross-sectional area of the hole, one can calculate the flow velocity at which the fluid will enter the post-hole channel. For a pressure difference of 1 mbar, this velocity is about 0.4 μm/s, which is smaller than the upward drift velocity of the droplets in the hole. This means that the droplets will float to the top of the hole and stay there, whereas the liposomes will travel to the post-hole channel.
We experimentally quantified the separation efficiency of the process by calculating the fraction of liposomes relative to the total number of particles present, i.e., ε = nliposomes/(nliposomes + ndroplets). Fig. 2(a) shows the purity ε obtained in the pre-hole channel as well as the post-hole channel at different suction pressures. The purity in the pre-hole channel was 0.45 ± 0.02 (mean ± standard deviation, ntotal = 6912), very close to the maximum of 0.5 (by default, each liposome is accompanied by one droplet in the formation process). The reason that the value is slightly less than 0.5 is (1) the occasional formation of small satellite droplets42,43 along with double-emulsion droplets, (2) occasional production of a droplet instead of a double-emulsion due to a temporary instability at the junction, and (3) occasional bursting of the liposomes. After letting the liposome-droplet mixture pass through the separation hole, the purity in the post-hole channel increased dramatically to ε = 0.93 ± 0.03, clearly indicating successful separation of the liposomes from the droplets. The purity values for specific underpressures were ε = 0.96 ± 0.04 at peff = 2 mbar (ntotal = 2810); ε = 0.91 ± 0.06 at peff = 3 mbar (ntotal = 2792); and ε = 0.93 ± 0.04 at peff = 4 mbar (ntotal = 1580). We could reach the purities as high as 0.98, in individual experiments (peff = 2 mbar, ntotal = 178). The separation efficiency was sensitive to the applied suction pressure. For higher pressures, the purity decreased considerably (ε = 0.67 ± 0.17 at peff = 5 mbar, ntotal = 3763) as the post-hole flow velocity (2 μm/s) became comparable to the upward drift velocity of the droplets in the separation hole.
We also investigated if the separation process affected the size distribution of liposomes. Fig. 2(b) shows an example of the size distributions of liposomes before and after the separation took place. The liposome before the separation had a very narrow distribution (9.3 ± 0.4 μm). After the separation, the liposomes were of the same size but the distribution became slightly broader (9.0 ± 0.6 μm). The mean diameter marginally decreased due to the appearance of a fraction of smaller liposomes. The minor increase in the dispersity might be due to the shearing experienced by the liposomes while re-entering the microfluidic channel from the separation hole, which could result in some loss of volume or unintentional liposome splitting, leading to smaller liposomes. We conclude that the separation method hardly affects the monodispersity of the liposomes, and the slight broadening of the size distribution is a minor effect in view of the excellent overall system improvement gained by the separation of liposomes from droplets.
The complex size distribution of the droplets in the pre-hole and post-hole channels was taken into consideration while evaluating the purity. From Fig. 2(c), it is clear that the majority of the droplets in the pre-hole channel were 4–10 μm in diameter, with 95% of the population larger than 3.9 μm. Smaller droplets (<3.9 μm) corresponded to the satellite droplets and the remnants of the occasional bursting of liposomes. The middle fraction (4–8 μm) corresponded to the normal 1-octanol pockets that budded off from the double-emulsion droplet, while the bigger droplets (9–10 μm) usually resulted from temporary instabilities at the junction. The latter two fractions are the main ones adversely affecting the OLA system, and which we aimed to separate out. The size distribution of the (unwanted) droplets in the post-hole channel, on the other hand, was very different, with more than 86% of the population smaller than 3.9 μm (Fig. 2(d)). Thus, most of the residual droplets that unintentionally ended up in the post-hole channel were small satellite droplets or remnants of burst liposomes. These small droplets (1–4 μm) have a relatively low upward drift velocity (<0.9 μm/s) and therefore the suction flow managed to extract them from the hole into the post-hole channel. Fortunately, as they were very small, they did not negatively affect the stability and visualization of the liposomes. Since the presence of these smaller droplets does not reflect the true measure that characterizes the efficiency of our separation technique, we ignored them for calculating the purity in the post-hole channel, and only considered the remaining part of the droplet population (>3.9 μm).
An important advantage of our separation method is the decoupling of the production module from the post-separation module, as the fluid velocity is virtually reduced to zero in the separation hole. Therefore, the small suction applied at the exit (and optionally at the separation hole) hardly affects the double-emulsion droplet production. This means that the production can work independently from all the processing steps on the liposomes after the separation. For example, a batch of liposomes can be sucked out of the separation hole on demand, while the liposome production remains continuous with newly formed liposomes accumulating in the separation hole. As will be clear from the next section (Separation based on size or relative permittivity was unsuccessful), this lack of decoupling between the production and the separation module was a major reason why several other separation methods were unsuccessful.
Separation based on size or relative permittivity was unsuccessful
Before we successfully achieved droplet liposome separation based on density, we unsuccessfully attempted a number of methods reported in the literature. Here, we report three of them, namely, DLD, PFF, and DEP. We started by integrating, on the OLA chip, the well-established technique of Deterministic Lateral Displacement (DLD), first developed by Huang et al.44 Here, size- and deformability-based separation of particles is achieved in an array of pillars, with a carefully chosen diameter, spacing, and phasing, introducing a migration angle in the array and a threshold radius. The presence of the pillars generates streamlines that follow a precisely defined path through the device. Particles with a radius below the threshold flow in “zig-zag-mode,” and thus more or less straight through the array, while particles with a radius above the threshold will be “bumped” or displaced to the next streamline, effectively travelling in “bump-mode” through the array along a specific migration angle. At the end of the array, small and large particles are therefore spatially separated.45
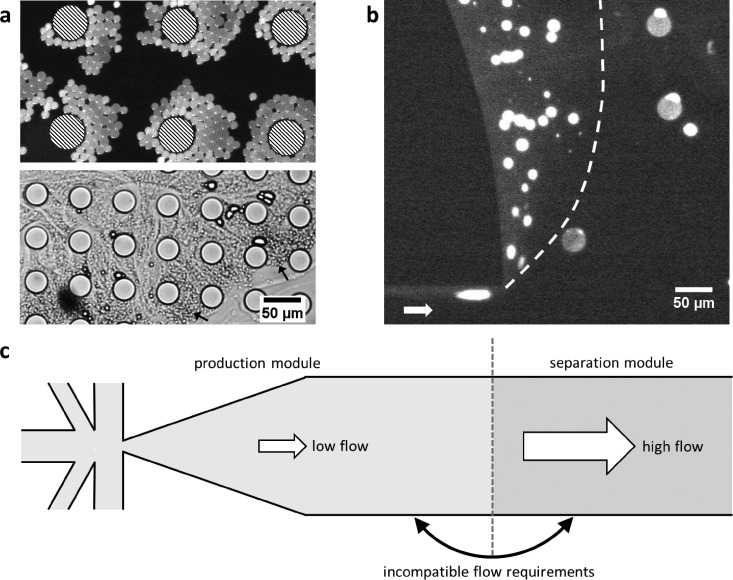
Although we tried several different DLD designs (arrays with a mirror design46 or with a sheath flow47), we were not able to achieve separation of liposomes from droplets. There were three main reasons why DLD was found to be incompatible with OLA: (1) Problems in coupling the liposome production with the DLD array: Since the OLA production junction was directly connected to the DLD array, the flow rates in these different modules needed to be compatible with each other. OLA production works optimally at relatively low pressures, while DLD needs high flow velocities to achieve successful separation. Due to the low flow rates necessary for OLA production, the droplets tended to aggregate at the pillars of the DLD array instead of flowing through in “zig-zag” mode, ultimately leading to a clogged array (Fig. 3(a)). Also, the production of liposomes often has a start-up phase during which the production is not yet stable and mostly 1-octanol droplets are produced, which worsened the aggregation and clogging of the DLD array. Using higher flow rates to prevent aggregations in the array was not an option, because then the production module would not work optimally. (2) Problems with the surface treatment of the array: The dense pattern of pillars in the array caused problems with the polyvinyl alcohol (PVA) treatment (see Materials and Methods). Often, small amounts of PVA adhered to the pillars and could not be removed with vacuum suction. These PVA remnants hardened upon baking and altered the precise fluid flows necessary for the DLD array to work, thereby destroying its functionality (Fig. S1, supplementary material). (3) Deformability of liposomes: DLD can be used to separate particles based on deformability.35 However, in the case of OLA, this would be an unwanted effect. Once a liposome encounters a pillar in the array, it becomes slightly deformed, decreasing its hydrodynamic radius and bringing its size closer to that of the droplet. Obviously, this would decrease any separation efficiency of liposomes and droplets.
FIG. 3.
DLD and PFF did not achieve efficient liposome-droplet separation. (a) 1-octanol droplets aggregated at the supporting pillars (striped circles) before the DLD array (top, fluorescence image) and even more in the DLD array, destroying its functionality (bottom, bright-field image). The arrows indicate prominent aggregates. Increasing the flow rates in the DLD array stalled the liposome production. (b) Very low number of liposomes (only those with the 1-octanol pocket still attached) could be separated from droplets using PFF (dashed line acts as a guideline and white arrow indicates the flow direction). Virtually, every liposome burst due to the high flow rates in the pinched section. Decreasing the flow rate in the pinched section, to prevent bursting, led to an effectively zero separation efficiency. (c) Top-view schematic showing the main problem of implementing DLD and PFF to separate droplets and liposomes, namely, the incompatible flow requirements. The liposome production using OLA needs low flow rates, but the DLD and PFF methods require higher flow rates and additional sheath flows. Due to the physical connection between the production module and the module with the separation method, it becomes impossible to satisfy both requirements, leading to the problems displayed in the other panels.
Next, we turned our attention to Pinched Flow Fractionation (PFF), a different method to separate particles by size, originally developed by Yamada et al.36 The basic design elements of the method are a narrow channel, called the pinched segment, that opens up into an expansion chamber. The particle mixture is pressed against the sidewall of the pinched segment by a sheath flow. Particles of different sizes will have their hydrodynamic centre located in streamlines nearer or further from the sidewall. At the entrance to the expansion chamber, the streamlines fan out, carrying the particles with them: small particles travel near the walls of the chamber, while large particles end up near the middle of the chamber. In short, small differences in particles' positions in the pinched segment are amplified upon entrance to the expansion chamber.36 The method can be modified to include a density-based separation mechanism, called sedimentation-based PFF.37 This works similar to normal PFF regarding size-based separation but further gains a density-based separation through the use of a curved expansion chamber and high fluid flows, which result in an outward pointing inertial force on particles in the expansion chamber. Particles with higher density then experience a larger inertial force and therefore “sediment” towards the outside of the expansion chamber.37
We implemented sedimentation-based PFF on our OLA chip but again failed to achieve a high-quality separation of the liposomes from droplets. The reasons that PFF was incompatible with OLA were similar to those for DLD: (1) Production-separation coupling: PFF required considerably higher flow rates than optimal for the liposome production, and most of the liposomes did not survive the shear forces present in the pinched segment (Fig. S2, supplementary material). Only a small fraction of liposomes, notably those with the 1-octanol pocket still attached, were observed to enter the expansion chamber and were successfully separated from the droplets (Fig. 3(b)). By lowering the flow rate of the sheath flow, most liposomes could survive the pinched segment, but then the separation mechanism did not work anymore. Previous studies37 of sedimentation-based PFF mention that in order to achieve density-based separation, flow rates of up to 2 ml/h are needed, which are several orders of magnitude higher than the typical flow rates at which OLA operates. (2) Problems with the surface treatment: PFF required the expansion chamber to have multiple outlets to drain the chamber homogeneously. However, multiple outlets complicated the surface treatment, in particular, the removal of the PVA with vacuum suction. This resulted in most microfluidic devices having one or more clogged outlets, rendering them non-functional. (3) Deformability of the liposomes: Liposomes were compressed in the pinched segment to a size comparable to the droplets, thereby rendering the size-based separation less effective (Fig. S2, supplementary material). This incompatibility in the flow requirements required for optimal liposome production and optimal separation process (for both DLD and PFF) is schematically shown in Fig. 3(c).
Due to the failure of two established passive methods, we decided to attempt an active method based on dielectrophoresis (DEP), an effect first studied in depth by Pohl.48 We chose to implement the DEP method to keep the experimental setup as simple as possible. In a non-uniform electric field, a neutral particle will experience a net force, the direction of which is determined by the relative polarizability of the medium and the particle. A particle in a more polarizable medium will move towards areas of high field gradient, while in the opposite case, when the particle is more polarizable than the surrounding medium, it will move to areas of the low field gradient. The relative polarizability of the medium and the particle is determined by the conductivity and dielectric constant of each and the frequency of the applied field.49 Since liposomes are mostly composed of water, they can be assumed to behave similarly to the surrounding medium. However, 1-octanol and water have sufficiently dissimilar dielectric constants and conductivities for the paths of 1-octanol droplets to be, in principle, diverted by DEP.
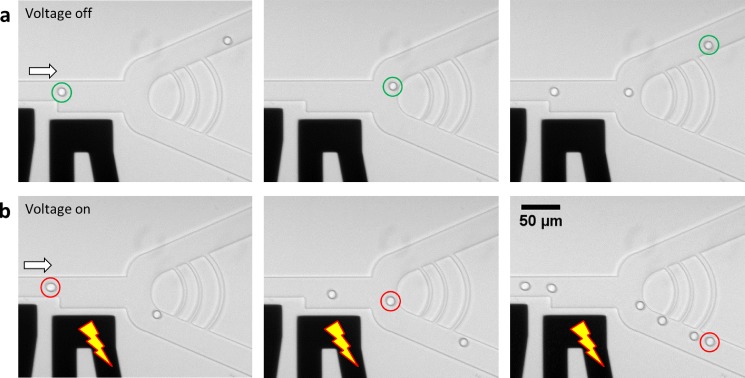
Most examples in the literature relate to the sorting of water-in-oil droplets.32,34 In the case of OLA, however, we wanted to sort octanol-in-water droplets. In order to ensure that the device design (inspired from Weitz lab designs34) was functional, initial experiments were performed using an inverted system, i.e., water-in-octanol droplets. It was verified that these droplets indeed could be sorted with DEP using AC fields (300 V, 5 kHz) using liquid metal electrodes composed of eutectic Galium-Indium50 (Sigma-Aldrich Co.) patterned in the same plane as the microfluidic channels (Fig. 4, Movie S2, supplementary material). However, attempts to similarly sort octanol-in-water droplets were all unsuccessful, even when varying the field frequencies between DC and 10 kHz (Movie S3, supplementary material). Voltages even up to 1.4 kV DC did not have any effect, with voltages higher than 1.4 kV leading to the breakdown of the polydimethylsiloxane (PDMS) barrier between the electrodes and the microfluidic channel. We suggest that the conductive medium surrounding the 1-octanol droplet screened the electric field to such an extent that the dielectrophoretic force became too small to divert the paths of these 1-octanol droplets.
FIG. 4.
Diverting the path of water-in-octanol droplets using DEP. (a) The DEP junction consists of a low-resistance (top) and a high-resistance (bottom) channel. If the voltage on the electrodes (the black channels) is off, the droplets follow the low-resistance path. (b) Once the voltage is turned on (300 V, 5 kHz), the dielectrophoretic force diverts the droplets into the high-resistance channel at the bottom. Two droplets are encircled (in green and in red) at consecutive time-frames, as a guide to the eye. The arrows indicate the flow direction. The time difference between consecutive frames is 100 ms. Note that this technique worked well for water droplets in 1-octanol but was unsuccessful for 1-octanol droplets in water.
DISCUSSION
Following up on our recently reported novel microfluidic method (OLA) to produce liposomes on chip,14 we have, in this paper, addressed how to tackle the removal of 1-octanol droplets that adversely affect the system, in terms of the stability of liposomes, clogging of the microfluidic channels, and imaging quality. We have presented a straightforward method to remove the 1-octanol droplets, yielding a system with only liposomes. Our separation technique is based on the density difference between the droplets and liposomes. Using a reservoir with a big hole in the middle of the microfluidic channel, the lighter 1-octanol droplets floated upwards, while the liposomes remained near the bottom. A small suction pressure (-2–4 mbar) ensured that the liposomes entered the post-hole channel, ready for visualization and further on-chip manipulation. A high separation efficiency (an average purity of 0.93 ± 0.03) was realized as compared to a purity of about 0.45 for non-separated samples. Our separation method is conveniently integrated on the same microfluidic chip where the production takes place and does not require any complicated fabrication techniques or equipment. It also decouples the production and the separation module to a great extent so that they can work independently of each other.
Due to their similarities with biological cell membranes, liposomes play an integral part in studying membrane biophysics, membrane-related chemistry, and bottom-up synthesis of artificial cells. In terms of direct visualization of chemical reactions within a single vesicle and real-time monitoring of the morphological changes, giant unilamellar vesicles (>1 μm) are particularly useful as compared to small unilamellar vesicles (SUVs, ≤100 nm), and large unilamellar vesicles (100–1000 nm). With OLA being readily able to produce ∼10 μm diameter liposomes, efficient removal of the 1-octanol droplets ensures the availability of a clean batch of liposomes that can be visualized and manipulated within just a few minutes after their preparation. When compared with another microfluidic technique, Microvortex Manipulation (MVM),41 also designed to sort particles based on their densities, our technique has a number of advantages. MVM focusing requires multi-height lithography and a stringent condition on the densities of the particles to be separated with respect to the medium (ρparticle1 > ρmedium, and ρparticle2 < ρmedium). The technique presented here does not require multi-height lithography and there is no need for the liposomes to be heavier than the surrounding medium. Also, the critical herringbone pattern required for MVM focusing can potentially pose a problem for the surface treatment that is essential for proper functioning of OLA.
Although separation is also possible off-chip,20 retaining the liposomes inside the microfluidic chip has a great advantage as it saves purification time, eliminates the possible loss of material during off-chip handling, and, most importantly, enables downstream on-chip experimentation. Also, given that OLA is a microfluidic technique, the liquid volume under consideration is very small (volumetric flow rate ∼5 μl/h), making off-chip separation highly impractical. A range of bulk techniques (molecular-sieve chromatography,51 sedimentation field flow fractionation,52 high-speed centrifugation,53 and density-gradient centrifugation54) have been developed to homogenize the liposome samples, i.e., to separate liposomes of similar size. Such size separation is not necessary in the case of the OLA technique since it generates monodisperse liposomes. Furthermore, all of these bulk methods are employed to purify SUVs, while we aimed at purifying micron-sized liposomes. Additionally, these processes tend to have low yields (10%−60%).53,55 Thus, our on-chip liposome separation technique has clear advantages over known off-chip bulk techniques.
It is certainly possible to add further complexity to our production-separation modules. For example, a “feeder channel” that flows trigger molecules (e.g., adenosine triphosphate as an energy source or melittin peptides to form a membrane pore) can be fused to the post-hole channel in order to trigger a specific response in the liposomes. Such a system would be ideal, for example, to study the dynamics of proteins crucially involved in the bacterial cell division (FtsZ,5 MinCDE56), antibiotic transport across the lipid bilayer,12 or attempting a controlled growth of liposomes via external supply of lipids.6 We believe that the density-based separation of 1-octanol droplets from liposomes, presented here, will make OLA more user-friendly and accessible for a broad range of experiments.
MATERIALS AND METHODS
LO phase preparation
Lipids, dissolved in chloroform, were purchased from Avanti Polar Lipids. Chloroform was evaporated by passing a gentle stream of nitrogen and the lipids were further dried by desiccating for at least two hours. A stock concentration (100 mg/ml) was prepared by dissolving the lipids in ethanol and stored at −20 °C under the nitrogen atmosphere. 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) was used as the lipid source. A fluorescent lipid, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rh-PE), was added for visualization (DOPC:Rh-PE = 99.9:0.1, molar ratio). During experiments, the stock solution was dissolved in 1-octanol (Sigma-Aldrich Co.) to a final concentration of 2 mg/ml.
Soft lithography
Patterns were fabricated in silicon using e-beam lithography and dry etching as follows. The surface of a clean, 4″ diameter silicon wafer was first primed with hexamethyldisilazane (BASF SE) by spin-coating (1000 rpm for 1 min) and baking at 200 °C for 2 min. This was done in order to enhance the subsequent resist adhesion. A negative resist NEB22A (Sumitomo Chemical Co., Ltd) was then spin-coated (1000 rpm for 1 min) and the wafer was pre-baked at 110 °C for 3 min. A Leica EBPG 5200 (acceleration voltage 100 kV, aperture 400 μm) was used to write the desired pattern on the coated wafer with a dose of 16–18 μC/cm2. The wafer was immediately post-baked at 105 °C for 3 min. MF322 (The Dow Chemical Company) was used for developing the patterns (40 s), followed by rinsing the wafer in diluted MF322 solution (10 v% MF322 + 90 v% water) for 20 s and finally in deionized water for 20 s.
Dry etching was done using Bosch deep reactive ion etching, with an inductive coupled plasma (ICP) reactive-ion etcher (Adixen AMS 100 I-speeder). The process consisted of alternate etching (sulphur hexafluoride, SF6) and passivation (octafluorocyclobutane, C4F8) cycles. The pressure was kept around 0.04 mbar. The temperature of the wafer was kept at 10 °C, while the temperature of the main chamber was maintained at 200 °C. The sample holder was located at 200 mm from the plasma source. The etching step involved 200 sccm SF6 for 7 s with the ICP power set to 2000 W, while the capacitive coupled plasma (CCP) power (biased power) was switched off. The passivation step was 80 sccm C4F8 for 3 s with the ICP power set to 2000 W and the CCP power in chopped low frequency bias mode: 80 W for 10 ms and 0 W for 90 ms. The wafer was then cleaned in the same machine at a pressure of 0.04 mbar, ICP power set at 2500 W with a biased power of 60 W, a source-target distance of 200 mm, temperature set at 10 °C, using O2 gas at 200 sccm for 3–5 min. The height of the etched-structures was measured using a stylus profiler, DektakXT (Bruker Corporation) and was between 8 and 10 μm, for different wafers. Finally, the wafer was cleaned with acetone, rinsed in deionized water, and spin-dried. The wafer was then rendered hydrophobic by exposing it to (tridecafluoro-1,1,2,2-tetrahydrooctyl) trichlorosilane (abcr GmbH & Co.) in partial vacuum for at least 12 h.
Microfluidic devices were made by pouring PDMS, mixed with a curing agent at a mass ratio 10:1 (Sylgard 184, Dow Corning GmbH), on the wafer and baking at 80 °C for at least 4 h. The PDMS block was then peeled off from the wafer and holes were punched into it using a biopsy punch (World Precision Instruments, inner diameter ∼750 μm). The separation hole was punched with a biopsy punch with a smaller diameter (World Precision Instruments, inner diameter ∼330 μm) and the process was facilitated by observing the channel through a dissecting microscope (Zeiss, Stemi DV4 SPOT) with 35× magnification. The PDMS block was then cleaned with isopropanol and dried with nitrogen. Glass slides were also coated with a thin PDMS layer as described previously.14 The PDMS block and the PDMS-coated glass slide were then exposed to oxygen plasma for ∼10 s (3–4 SCFH O2, 200 W) using a Plasma-Preen system (Plasmatic Systems, Inc.). Immediately after the plasma treatment, the glass slide was bonded to the PDMS block. The device was further baked at 80 °C for ∼20 min. A microfluidic flow control system (positive pressure: 0–1000 mbar, negative pressure: 0–800 mbar, Fluigent GmbH) along with the MAESFLO software (version 3.2.1) was used to flow the solutions into the microfluidic device using appropriate metal connectors (BD microlance needles, outer diameter 640 μm, cut into ∼1 cm pieces) and tubing (Tygon Microbore Tubing, inner diameter 510 μm).
Surface treatment and experimentation
Channels downstream of the liposome-producing junction were rendered hydrophilic by adsorbing PVA polymers (Sigma-Aldrich Co.) to the PDMS surface, as described elsewhere.14 It was observed that reducing the time between the bonding of the microfluidic device and the surface treatment, to 3–4 h, substantially increased the effectiveness of the treatment. At the start of the experiment, it was made sure that the outer aqueous phase reached the production junction first and already flowed past the separation hole before the liposome production started. Also, sufficient flow was maintained in the pre-hole channel so as to not form any possible aggregates of liposomes and/or droplets before they reached the separation hole.
Image acquisition and processing
An Olympus IX81 or IX71 inverted microscope equipped with epifluorescence illumination, appropriate filter sets, and either a 10× (UPlanFLN, numerical aperture 0.30) or a 20× (UPlanSApo, numerical aperture 0.75) objective (Olympus) was used to perform the experiments. The images were recorded using either a Neo sCMOS camera or a Zyla 4.2 PLUS CMOS camera (Andor Technology Ltd.) and a micromanager software (version 1.4.14).57 Image processing was performed using ImageJ and MATLAB using self-written scripts.
For determining the diameter of liposomes and droplets, corresponding fluorescence images were properly thresholded and binarized, and the cross-sectional area of each of the particles was calculated. The diameter, either for free particles or when the particle was squeezed inside the microfluidic channel, was calculated as described previously.14
The resistances of the separation hole and the post-hole channel were calculated as Rhole = 8·η·L/πr4 and Rchannel = 12·η·l/w·h3·(1 – 0.63 h/w), respectively, where η is the fluid viscosity, L and r are the separation hole length and radius, and l, w, and h, respectively, are the channel length, width, and height.58 Flow velocities were calculated using the relation v = Q/a, where v is the flow velocity, Q is the volumetric flow rate, and a is the cross-sectional area.
Solution compositions
Inner aqueous phase (prepared in Milli-Q water): 15 v% glycerol, 0.04 mg/ml Alexa Fluor 647-Dextran (molecular weight: 10 000 Da, Sigma-Aldrich Co.); Lipid-carrying organic phase: 2 mg/ml lipids (99.9 mol. % DOPC + 0.1 mol. % Rh-PE) in 1-octanol; Outer aqueous phase (prepared in Milli-Q water): 50 mg/ml poloxamer 188 (Sigma-Aldrich Co.), 15 v% glycerol. Alexa Fluor 647-Dextran was added to visualize the liposomal lumen and further assist the analysis.
V. SUPPLEMENTARY MATERIAL
See supplementary material for problem with the surface treatment of DLD design (Fig. S1); undesired deformation and bursting of liposomes during PFF (Fig. S2); on-chip, density-based separation of 1-octanol droplets from liposomes (Movie S1); DEP working for water-in-octanol droplet system (Movie S2); and DEP not working for octanol-in-water droplet system (Movie S3).
ACKNOWLEDGMENTS
We would like to thank Kasper Spoelstra, Federico Fanalista, and Yaron Caspi for fruitful discussions. This work was supported by the NWO TOP-PUNT grant (No. 718014001), the Netherlands Organisation for Scientific Research (NWO/OCW) and European Research Council Advanced Grant SynDiv (No. 669598).
References
- 1. Walde P., Cosentino K., Engel H., and Stano P., ChemBioChem 11, 848 (2010). 10.1002/cbic.201000010 [DOI] [PubMed] [Google Scholar]
- 2. van Nies P., Nourian Z., Kok M., van Wijk R., Moeskops J., Westerlaken I., Poolman J. M., Eelkema R., van Esch J. H., Kuruma Y., Ueda T., and Danelon C., ChemBioChem 14, 1963 (2013). 10.1002/cbic.201300449 [DOI] [PubMed] [Google Scholar]
- 3. Loiseau E., Schneider J. A. M., Keber F. C., Pelzl C., Massiera G., Salbreux G., and Bausch A. R., Sci. Adv. 2, e1500465 (2016). 10.1126/sciadv.1500465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Veshaguri S., Christensen S. M., Kemmer G. C., Ghale G., Moller M. P., Lohr C., Christensen A. L., Justesen B. H., Jorgensen I. L., Schiller J., Hatzakis N. S., Grabe M., Pomorski T. G., and Stamou D., Science 351, 1469 (2016). 10.1126/science.aad6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cabré E. J., Sánchez-Gorostiaga A., Carrara P., Ropero N., Casanova M., Palacios P., Stano P., Jiménez M., Rivas G., and Vicente M., J. Biol. Chem. 288, 26625 (2013). 10.1074/jbc.M113.491688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peterlin P., Arrigler V., Kogej K., Svetina S., and Walde P., Chem. Phys. Lipids 159, 67 (2009). 10.1016/j.chemphyslip.2009.03.005 [DOI] [PubMed] [Google Scholar]
- 7. Noireaux V., Bar-Ziv R., Godefroy J., Salman H., and Libchaber A., Phys. Biol. 2, P1 (2005). 10.1088/1478-3975/2/3/P01 [DOI] [PubMed] [Google Scholar]
- 8. Lemière J., Guevorkian K., Campillo C., Sykes C., and Betz T., Soft Matter 9, 3181 (2013). 10.1039/c3sm27812c [DOI] [Google Scholar]
- 9. Haluska C. K., Riske K. A., Marchi-Artzner V., Lehn J.-M., Lipowsky R., and Dimova R., Proc. Natl. Acad. Sci. U.S.A. 103, 15841 (2006). 10.1073/pnas.0602766103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allen T. M. and Cullis P. R., Adv. Drug Delivery Rev. 65, 36 (2013). 10.1016/j.addr.2012.09.037 [DOI] [PubMed] [Google Scholar]
- 11. Zylberberg C. and Matosevic S., Drug Delivery 23, 3319 (2016). 10.1080/10717544.2016.1177136 [DOI] [PubMed] [Google Scholar]
- 12. Cama J., Schaich M., Al Nahas K., Hernandez-Ainsa S., Pagliara S., and Keyser U. F., Sci. Rep. 6, 32824 (2016). 10.1038/srep32824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edwards K. A., Bolduc O. R., and Baeumner A. J., Curr. Opin. Chem. Biol. 16, 444 (2012). 10.1016/j.cbpa.2012.05.182 [DOI] [PubMed] [Google Scholar]
- 14. Deshpande S., Caspi Y., Meijering A. E., and Dekker C., Nat. Commun. 7, 10447 (2016). 10.1038/ncomms10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Olson F., Hunt C. A., Szoka F. C., Vail W. J., and Papahadjopoulos D., Biochim. Biophys. Acta 557, 9 (1979). 10.1016/0005-2736(79)90085-3 [DOI] [PubMed] [Google Scholar]
- 16. Reeves J. P. and Dowben R. M., J. Cell Physiol. 73, 49 (1969). 10.1002/jcp.1040730108 [DOI] [PubMed] [Google Scholar]
- 17. Angelova M. I. and Dimitrov D. S., Faraday Discuss. Chem. Soc. 81, 303 (1986). 10.1039/dc9868100303 [DOI] [Google Scholar]
- 18. Arriaga L. R., Datta S. S., Kim S. H., Amstad E., Kodger T. E., Monroy F., and Weitz D. A., Small 10, 950 (2014). 10.1002/smll.201301904 [DOI] [PubMed] [Google Scholar]
- 19. Stachowiak J. C., Richmond D. L., Li T. H., Liu A. P., Parekh S. H., and Fletcher D. A., Proc. Natl. Acad. Sci. U.S.A. 105, 4697 (2008). 10.1073/pnas.0710875105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deng N. N., Yelleswarapu M., and Huck W. T. S., J. Am. Chem. Soc. 138, 7584 (2016). 10.1021/jacs.6b02107 [DOI] [PubMed] [Google Scholar]
- 21. Karamdad K., Law R. V., Seddon J. M., Brooks N. J., and Ces O., Lab Chip 15, 557 (2015). 10.1039/C4LC01277A [DOI] [PubMed] [Google Scholar]
- 22. Teh S. Y., Khnouf R., Fan H., and Lee A. P., Biomicrofluidics 5, 044113 (2011). 10.1063/1.3665221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Swaay D. and deMello A., Lab Chip 13, 752 (2013). 10.1039/c2lc41121k [DOI] [PubMed] [Google Scholar]
- 24.Knovel Critical Tables, 2nd ed. http://app.knovel.com/hotlink/toc/id:kpKCTE000X/knovel-critical-tables/knovel-critical-tablesa (Knovel Corporation, 2008).
- 25. Smallwood I. M., Handbook of Organic Solvent Properties (Arnold, London, 1996). [Google Scholar]
- 26. Seifert U., Adv. Phys. 46, 13 (1997). 10.1080/00018739700101488 [DOI] [Google Scholar]
- 27. Oglęcka K., Rangamani P., Liedberg B., Kraut R. S., and Parikh A. N., Elife 3, e03695 (2014). 10.7554/eLife.03695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sajeesh P. and Sen A. K., Microfluid. Nanofluid. 17, 1 (2014). 10.1007/s10404-013-1291-9 [DOI] [Google Scholar]
- 29. Shi J., Huang H., Stratton Z., Huang Y., and Huang T. J., Lab Chip 9, 3354 (2009). 10.1039/b915113c [DOI] [PubMed] [Google Scholar]
- 30. Nam J., Lim H., Kim C., Kang J. Y., and Shin S., Biomicrofluidics 6, 024120 (2012). 10.1063/1.4718719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gascoyne P. R. C. and Vykoukal J., Electrophoresis 23, 1973 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ahn K., Kerbage C., Hunt T. P., Westervelt R. M., Link D. R., and Weitz D. A., Appl. Phys. Lett. 88, 024104 (2006). 10.1063/1.2164911 [DOI] [Google Scholar]
- 33. Franke T., Braunmüller S., Schmid L., Wixforth A., and Weitz D. A., Lab Chip 10, 789 (2010). 10.1039/b915522h [DOI] [PubMed] [Google Scholar]
- 34. Baret J.-C., Miller O. J., Taly V., Ryckelynck M., El-Harrak A., Frenz L., Rick C., Samuels M. L., Hutchison J. B., Agresti J. J., Link D. R., Weitz D. A., and Griffiths A. D., Lab Chip 9, 1850 (2009). 10.1039/b902504a [DOI] [PubMed] [Google Scholar]
- 35. Beech J. P., Holm S. H., Adolfsson K., and Tegenfeldt J. O., Lab Chip 12, 1048 (2012). 10.1039/c2lc21083e [DOI] [PubMed] [Google Scholar]
- 36. Yamada M., Nakashima M., and Seki M., Anal. Chem. 76, 5465 (2004). 10.1021/ac049863r [DOI] [PubMed] [Google Scholar]
- 37. Morijiri T., Sunahiro S., Senaha M., Yamada M., and Seki M., Microfluid. Nanofluid. 11, 105 (2011). 10.1007/s10404-011-0785-6 [DOI] [Google Scholar]
- 38. Yamada M. and Seki M., Lab Chip 5, 1233 (2005). 10.1039/b509386d [DOI] [PubMed] [Google Scholar]
- 39. Di Carlo D., Irimia D., Tompkins R. G., and Toner M., Proc. Natl. Acad. Sci. U.S.A. 104, 18892 (2007). 10.1073/pnas.0704958104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song J., Song M., Kang T., Kim D., and Lee L. P., Biomicrofluidics 8, 064108 (2014). 10.1063/1.4902906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hsu C.-H., Di Carlo D., Chen C., Irimia D., and Toner M., Lab Chip 8, 2128 (2008). 10.1039/b813434k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shi X. D., Brenner M. P., and Nagel S. R., Science 265, 219 (1994). 10.1126/science.265.5169.219 [DOI] [PubMed] [Google Scholar]
- 43. Tan Y.-C. and Lee A. P., Lab Chip 5, 1178 (2005). 10.1039/b504497a [DOI] [PubMed] [Google Scholar]
- 44. Huang L. R., Cox E. C., Austin R. H., and Sturm J. C., Science 304, 987 (2004). 10.1126/science.1094567 [DOI] [PubMed] [Google Scholar]
- 45. McGrath J., Jimenez M., and Bridle H., Lab Chip 14, 4139 (2014). 10.1039/C4LC00939H [DOI] [PubMed] [Google Scholar]
- 46. Loutherback K., D'Silva J., Liu L., Wu A., Austin R. H., and Sturm J. C., AIP Adv. 2, 042107 (2012). 10.1063/1.4758131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Davis J. A., Inglis D. W., Morton K. J., Lawrence D. A., Huang L. R., Chou S. Y., Sturm J. C., and Austin R. H., Proc. Natl. Acad. Sci. U.S.A. 103, 14779 (2006). 10.1073/pnas.0605967103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pohl H., Dielectrophoresis: The Behavior of Neutral Matter in Nonuniform Electric Fields ( Cambridge University Press, Cambridge, 1978). [Google Scholar]
- 49. Beech J. P., Microfluidics Separation and Analysis of Biological Particles (Lund University, 2011). [Google Scholar]
- 50. Dickey M. D., Chiechi R. C., Larsen R. J., Weiss E. A., Weitz D. A., and Whitesides G. M., Adv. Funct. Mater. 18, 1097 (2008). 10.1002/adfm.200701216 [DOI] [Google Scholar]
- 51. Huang C., Biochemistry 8, 344 (1969). 10.1021/bi00829a048 [DOI] [PubMed] [Google Scholar]
- 52. Kirkland J. J., Yau W. W., and Szoka F. C., Science 215, 296 (1982). 10.1126/science.7053578 [DOI] [PubMed] [Google Scholar]
- 53. Barenholz Y., Gibbes D., Litman B. J., Goll J., Thompson T. E., and Carlson F. D., Biochemistry 16, 2806 (1977). 10.1021/bi00631a035 [DOI] [PubMed] [Google Scholar]
- 54. Goormaghtigh E. and Scarborough G. A., Anal. Biochem. 159, 122 (1986). 10.1016/0003-2697(86)90316-7 [DOI] [PubMed] [Google Scholar]
- 55. Güven A., Ortiz M., Constanti M., and O'Sullivan C. K., J. Liposome Res. 19, 148 (2009). 10.1080/08982100802674419 [DOI] [PubMed] [Google Scholar]
- 56. Caspi Y. and Dekker C., Elife 5, e19271 (2016). 10.7554/eLife.19271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Edelstein A., Amodaj N., Hoover K., Vale R., and Stuurman N., Curr. Protoc. Mol. Biol. 92, 14.20.1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bruus H., Theoretical microfluidics ( Cambridge University Press, New York, 2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for problem with the surface treatment of DLD design (Fig. S1); undesired deformation and bursting of liposomes during PFF (Fig. S2); on-chip, density-based separation of 1-octanol droplets from liposomes (Movie S1); DEP working for water-in-octanol droplet system (Movie S2); and DEP not working for octanol-in-water droplet system (Movie S3).