Abstract
We built a natural language processing (NLP) method to automatically extract clinical findings in radiology reports and characterize their level of change and significance according to a radiology-specific information model. We utilized a combination of machine learning and rule-based approaches for this purpose. Our method is unique in capturing different features and levels of abstractions at surface, entity, and discourse levels in text analysis. This combination has enabled us to recognize the underlying semantics of radiology report narratives for this task. We evaluated our method on radiology reports from four major healthcare organizations. Our evaluation showed the efficacy of our method in highlighting important changes (accuracy 99.2%, precision 96.3%, recall 93.5%, and F1 score 94.7%) and identifying significant observations (accuracy 75.8%, precision 75.2%, recall 75.7%, and F1 score 75.3%) to characterize radiology reports. This method can help clinicians quickly understand the key observations in radiology reports and facilitate clinical decision support, review prioritization, and disease surveillance.
Keywords: Natural language processing, Radiology reports, Imaging informatics
Introduction
Reviewing and making sense of a large volume of text is a time-consuming and laborious task in many clinical settings. In the clinical domain, physicians and healthcare providers face an information overload problem. Often, clinicians need to review multiple narrative documents describing patients’ medical history, clinical laboratory, surgical pathology, and radiology results. Studies have shown that the volume of narrative and structured clinical data has been growing exponentially [1, 2], while clinicians have only a limited time to review and interpret the data. Therefore, natural language processing (NLP) methods that provide summaries of major clinical findings and characterize their important aspects can be instrumental to direct the focus of healthcare providers to clinically significant observations and help clinicians to review and understand the information efficiently.
Radiology reports are a particularly common source of information in many medical conditions. However, the majority of radiology reports remain in unstructured text format. Even in the presence of recent attempts to introduce structured templates for radiology reports, most of the information in these templates is in free text. Given the large volume of these texts, which can vary in size, purpose, modality, and source, it is a cumbersome task to review, comprehend, and prioritize the information in radiology reports. This large volume may cause information errors and is a major obstacle for healthcare provider’s efficiency and productivity. To tackle this problem, we propose an NLP system that will distill the clinical findings in a report, including important new observations and significant changes in previous observations.
A related field to our work is computer-based summarization, which started in the 1950s [3, 4]. There are multiple survey papers that discuss the different summarization techniques from early times to the recent era [5–9]. Summarization techniques are also developed and applied in the biomedical domain [10–12]. These methods mostly customize general text summarization methods in the biomedical domain. For example, Sarkar [13] uses features such as term frequency, title, and position for summarization; Reeve et al. [14] uses graph-based lexical chaining for biomedical text; Chuang and Yang [15] uses an array of supervised classifiers such as naïve Bayes, decision tree, and neural networks for biomedical summarization. A large number of these biomedical summarization methods focus on biomedical articles and literature summarization as their input rather than radiology reports and clinical notes [11, 16]. Electronic medical record (EMR) data and notes were also used to find and retrieve relevant biomedical literature, which was subsequently used as input to summarization methods [15, 17, 18]. There have been other methods for summarization of radiology reports [19–21]. However, these methods are mostly focused on a specific medical condition and they do not provide a characterization of clinical findings in radiology reports.
Despite the large number of existing text summarization and characterization methods, generally, they can be divided into three categories based on the level of abstraction in text analysis and their utilized textual features: (1) surface level, (2) entity level, and (3) discourse level [22]. Surface-level features include simple textual features such as term frequencies, term positions, and cue words. The entity-based features represent entities in text and their relationships such as term similarities, term’s dictionary-based memberships, and syntactic relationships. The discourse-level features focus on overall text structure and its semantics such as thread of topics and covered subjects [9]. Our NLP approach presented in this paper is unique, because our method covers all three levels of abstractions in text analysis, features, and criteria used for extracting and characterizing summaries. For example, as described in the “Material and Methods” section, we use surface-level features such as n-grams and frequency-based weights, entity-level features such as named-entity recognition annotations, and discourse-level analysis such as negation detection to extract and determine significance and change in clinical findings. As a result, our method provides a comprehensive approach to analyze the radiology reports at different abstraction levels compared to the previous summarization and characterization methods. The details of this method are described in the following section.
Material and Methods
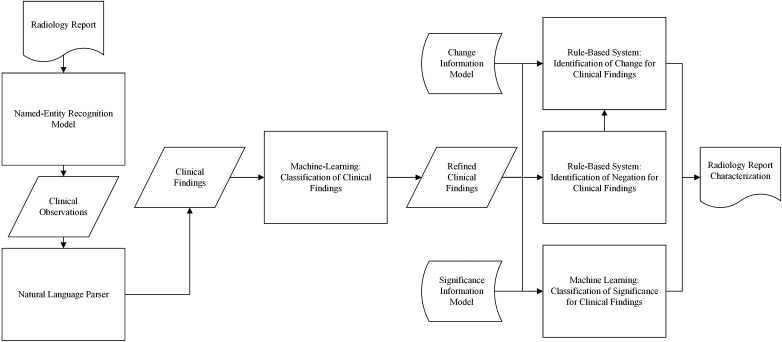
We use a combination of machine learning and rule-based methods to extract clinical findings in radiology reports and characterize their level of change and significance. Figure 1 shows the overview of our approach. The details of this approach are as follows.
Fig. 1.
Overview of our methodology for characterization of change and significance for clinical findings in radiology reports
Information Model
The information model in our method provides a coherent framework for radiology report information. The information model focuses on two major aspects of clinical findings in radiology reports: change and significance. This model is inspired by a previous work on radiology report assessment [23]. Change in our information model has four classes: new/worse, unchanged, improved, and indeterminate. These classes indicate clinically significant changes in the outputs of our characterization method compared to the most recent prior study. Significance in our information model has three classes: significant, normal/insignificant, and indeterminate. These finding classes indicate potential for harm and the need for follow-up, treatment, or change in management based on a provided summary elicited from the radiology report. Tables 1 and 2 describe each of these classes with examples from radiology reports for change and significance, respectively.
Table 1.
Change information model classes for radiology report summaries and their associated descriptions and examples
| Change class | Description | Example |
|---|---|---|
| New or worse | Finding was not present on the prior study or has progressed | “skeletal metastases a few of which are slightly more prominent” |
| Unchanged | Finding has not changed | “no significant change in the mild mediastinal and mild right hilar adenopathy” |
| Improved | Finding has partially resolved | “marked interval improvement in the bilateral pulmonary nodules” |
| Indeterminate | Change cannot be assessed | “calcified right hilar and mediastinal nodes with calcified granulomas in the liver and spleen” |
Table 2.
Significance information model classes for radiology report summaries and their corresponding descriptions and examples
| Significance class | Description | Example |
|---|---|---|
| Significant | Finding may or likely cause harm without initiation of treatment or change in management | “bilateral pulmonary and right pleural nodules suspicious for a metastases” |
| Normal or insignificant | Finding is normal or unlikely to cause harm without initiation of treatment or change in management | “minor airway secretions”, “no definite new nodules” |
| Indeterminate | Unable to draw conclusions from imaging | “small mediastinal lymph nodes that do not meet criteria for lymphadenopathy” |
Radiology Report Data Set
The source of the radiology reports in this work is the RadCore database and Stanford Translational Research Integrated Database Environment (STRIDE). RadCore and STRIDE were used jointly to build training and test data sets. RadCore is a multi-institutional database of radiology reports aggregated in 2007 from three major healthcare organizations: Mayo Clinic, MD Anderson Cancer Center, and Medical College of Wisconsin. RadCore radiology reports were collected under institutional review board approval from those three organizations. STRIDE database contains radiology reports from Stanford Health Care since 1998. The use of these data in our project was approved by our institutional review boards.
We use manually annotated radiology reports to build and evaluate our methodology. Given the large amounts of data in RadCore and STRIDE radiology report repositories and our limited resources, we restricted our focus to chest computed tomography (CT) radiology reports to keep the manual annotation requirements tractable. Therefore, we randomly selected ten radiology reports from each of the four organizations with chest CT study type. There were no major differences in the formatting of chest CT radiology reports in different organizations. In our manual annotations, a domain expert radiologist highlighted findings in selected 40 radiology reports’ text and rated their significance and change according to our information models.
To evaluate the quality of the manual annotations, we calculated interannotator agreement for a subset of our data set. We randomly selected 25% of our annotated radiology reports, and we asked an independent radiologist to annotate their findings according to the information models. We calculated the total percentage of agreements between two annotators. To remove the effect of agreements by chance, we also calculated Cohen’s kappa coefficient [24], a widely accepted agreement metric in NLP, for these two sets of annotations.
Clinical Observation Extraction
We use our previously developed NLP method to identify clinical observations in radiology reports [25]. In this method, a combination of semantic and syntactic features such as negations, word shapes, word stems, part of speech tags, n-grams, and RadLex [26] ontology memberships are used in a machine learning named-entity recognition model to identify terms and phrases that belong to radiological observations. This named-entity recognition model is based on a conditional random field (CRF) framework [27]. CRF is a discriminative sequence classifier, which is used in state-of-the-art part of speech tagging and named-entity recognition systems [28]. A CRF model includes an estimation of the conditional distribution of output labels given the input features with an associated graphical structure. This method uses a linear chain graphical structure to predict sequences of annotation labels for the sequences of input words from radiology reports. The CRF model considers previously assigned labels, surrounding terms, and their features as context for annotation of a single word. Our previous work showed strong performance of this method for annotating observations in chest CTs (precision 89.4%, recall 84.1%, and F1 score 86.7%) [25].
Inclusion of Grammatical Dependencies
To provide meaningful summaries, we need to include associated modifiers and dependents for extracted observations. To do that, we parse the radiology report sentences using Stanford Parser [29]. The Stanford Parser is an open-source and widely used probabilistic natural language parser that identifies grammatical roles of words in sentences. A probabilistic parser searches over the space of all possible candidate parses that represent grammatical roles of words of a sentence and derive the most probable parse by using dynamic programming [29]. After parsing the radiology report sentences, we expand extracted observations from the last step to their largest subsuming noun phrase in the parse trees. This expansion completes the extracted observations based on grammatical dependencies and complements the clinical findings.
Refinement of Clinical Findings
We process the outcomes of the grammatical expansions to filter out text snippets that are not representing clinical findings. For this purpose, we develop a text classifier that identifies extracted text snippets that refer to clinical findings. This text classifier is based on a support vector machine (SVM) framework [30]. SVM is one of the most effective classifiers in machine learning. SVM is a maximum-margin classifier, which finds the decision boundary with the largest separation between positive and negative training examples. To train this classifier, we partition our manually annotated radiology reports into separate training and test sets. Our training set contains 80% of radiology reports from each organization. The test contains the remaining 20% of the data.
To use radiology report content in an SVM classification framework, we need to model radiology report text quantitatively. We therefore modeled radiology reports as vectors in Euclidian space, where each vector dimension corresponds to an n-gram, which is a contiguous sequence of one, two, or three words in a report. If a report contains an n-gram, that n-gram has a non-zero weight in the report’s vector representation. The weight of each n-gram was computed using term frequency-inverse document frequency (tf-idf), a common weighting scheme in text mining [31]. A tf-idf weight increases proportionally by the n-gram frequency in the report and is scaled down by the commonality of the n-gram among all reports in the data set. In this work, we used LIBSVM, a widely used open-source machine learning library, with a linear kernel function to train our SVM classifier [32].
Determine Change in Clinical Findings for Summaries
To rate the degree of change in clinical findings of the extracted summaries, we utilized a simple key term matching approach accompanied with a negation detection tool. The rules and criteria used were decided in discussions with radiologist coauthors, based on the review of 20% of data set’s radiology reports as the training set, while the remaining 80% of the data set was held out as the test set for evaluation. To determine the negative context for our key terms, we used NegEx [32], a widely used clinical text-mining tool. NegEx first identifies negation triggers in text based on its dictionary, and then uses a set of rules to determine which terms fall within the scope of those triggering terms [33]. The list of change model classes and their deciding criteria are listed in Table 3.
Table 3.
Our rules and criteria to determine levels of change in radiology report summaries
| Change class | Rule |
|---|---|
| New or worse | Presence of “new”, “increase”, “develop”, “progress”, and “more” in positive context |
| Unchanged | Presence of “new”, “increase”, “develop”, “progress”, and “more” in negative context Presence of “change” in negative context Presence of “stable”, “remain”, and “persist” |
| Improved | Presence of “improve” and “decrease” |
| Indeterminate | Absence of other criteria |
Assess the Significance of Clinical Findings in Summaries
Significance for clinical findings is encoded in numerous ways in radiology. Because of this variety and the complexity of natural language associated with significance of clinical findings in extracted summaries, our radiologist collaborators could not identify a conclusive set of rules and key words to recognize their level of significance. Therefore, to address this problem, we developed an SVM text classifier to assess the significance of the clinical findings in extracted summaries. The previously constructed training and test sets for clinical finding refinement are also manually annotated by our collaborator radiologist according to our significance information model (Table 2). Similarly, we used tf-idf weights of n-grams in the annotated training set to train a linear kernel function SVM classifier to rate significance levels in extracted summaries.
Evaluation
We evaluated the developed classifiers for clinical finding refinement on the holdout test set. The developed methods for change and significance assessment were evaluated on both true clinical findings and clinical finding results from our refinement method on their associated test sets. Testing change and significance assessment on refinement results gives the end-to-end evaluation of our characterization method. In these evaluations, we measured standard machine learning evaluation metrics of accuracy, precision, recall, and F1 score [34]. 95% confidence intervals were calculated for these metrics using the asymptotic approach in R statistical toolbox [35].
Results
Table 4 shows the number of manual annotations that are used in our work to build and evaluate our method. Table 5 shows agreement percentage and kappa coefficient for the inter-annotator agreement evaluation on 25% of annotations.
Table 4.
The number of annotated classes in manual annotations in entire data set and the test set
| Annotation | Counts in the data set | Counts in the test set |
|---|---|---|
| Not a clinical finding | 161 | 35 |
| New or worse clinical finding | 23 | 21 |
| Unchanged clinical finding | 10 | 7 |
| Improved clinical finding | 65 | 50 |
| Indeterminate clinical finding | 593 | 488 |
| Significant clinical finding | 275 | 45 |
| Normal or insignificant clinical finding | 255 | 45 |
| Clinical finding with indeterminate significance | 0 | 0 |
| Total number of annotations | 1382 | 691 |
Table 5.
Inter-annotator agreement on manual annotations
| Annotation type | Agreement percentage (%) | Kappa coefficient (%) |
|---|---|---|
| Clinical finding | 96.8 | 83.9 |
| Change | 94.6 | 79.4 |
| Significance | 96.3 | 82.1 |
Table 6 shows accuracy, precision, recall, and the F1 score of our method for capturing clinical findings, characterizing their change and significance in radiology report summaries, and their corresponding 95% confidence intervals. This table shows the measurements for change and significance determination on true clinical findings, in addition to extracted clinical findings by our SVM classifier for an end-to-end evaluation.
Table 6.
Evaluation results of our method for extracting clinical findings for radiology reports and determining their characteristics with 95% confidence intervals (CIs)
| Results | Accuracy (CI) (%) | Precision (CI) (%) | Recall (CI) (%) | F1 score (CI) (%) |
|---|---|---|---|---|
| Extracting clinical findings | 78.4 (70.4–86.4) |
81.8 (74.3–89.3) |
90.0 (84.2–95.8) |
85.7 (78.9–92.5) |
| Determination of change on true clinical findings | 99.3 (98.2–100.0) |
97.3 (94.3–100) |
93.4 (88.6–98.2) |
95.2 (91.1–99.3) |
| Determination of change on extracted clinical findings (end-to-end) | 99.2 (97.9–100.0) |
96.3 (92.8–99.8) |
93.5 (88.8–98.2) |
94.7 (90.4–99.0) |
| Determination of significance on true clinical findings | 78.9 (71.0–86.8) |
79.3 (71.4–87.2) |
78.9 (71.0–86.8) |
78.8 (70.9–86.7) |
| Determination of significance on extracted clinical findings (end-to-end) | 75.8 (67.5–84.1) |
75.2 (66.8–83.6) |
75.7 (67.4–84.0) |
75.3 (66.9–83.7) |
We also measured the evaluation metrics and the 95% confidence intervals for all classes of change and significance for radiology report summaries in the end-to-end evaluation. As it was noted in the breakdown of the manual annotations (Table 4), because our data set does not contain clinical findings with indeterminate significance, we focused on normal/insignificant and significant classes in our analysis. Table 7 shows these results for change classes, and Table 8 shows the results for significance classes.
Table 7.
Results of end-to-end evaluation and the corresponding 95% CIs for each change class
| Results | Accuracy (CI) (%) | Precision (CI) (%) | Recall (CI) (%) | F1 score (CI) (%) |
|---|---|---|---|---|
| New or worse | 99.3 (98.2–100.0) |
92.6 (87.6–97.6) |
92.6 (87.6–97.6) |
92.6 (87.6–97.6) |
| Unchanged | 98.9 (97.3–100.0) |
93.1 (88.2–98.0) |
96.4 (92.9–99.9) |
94.7 (90.4–99.0) |
| Improved | 99.8 (99.4–100.0) |
100.0 (99.7–100.0) |
85.7 (78.9–92.5) |
92.3 (87.2–97.4) |
| Indeterminate | 98.2 (95.9–100.0) |
99.4 (98.4–100.0) |
98.5 (96.5–100.0) |
99.0 (97.5–100.0) |
Table 8.
Results of end-to-end evaluation and the corresponding 95% CIs for each significance class
| Results | Accuracy (CI) (%) | Precision (CI) (%) | Recall (CI) (%) | F1 score (CI) (%) |
|---|---|---|---|---|
| Normal/insignificant | 75.8 (67.5–84.1) |
81.5 (74.0–89.0) |
75.9 (67.6–84.2) |
78.6 (70.6–86.6) |
| Significant | 75.8 (67.5–84.1) |
68.9 (59.9–77.9) |
75.6 (67.2–84.0) |
72.1 (63.4–80.8) |
Discussion
The main contribution of this work is the use of NLP and machine learning frameworks to extract clinical findings and characterize their change and significance according to a radiology-specific information model. This method relies on our previously developed information extraction system, which annotates granular level concept classes such as observations in radiology reports and new NLP methods to extract and refine the clinical findings and rate their change and significance. Our results show that the presented approach can characterize key radiological synopses in radiology reports with high accuracy. We also demonstrated the generalizability of our radiology report characterization approach to different healthcare organizations by training and testing our method on data from different organizations. For an input radiology report, this automated pipeline generates an easy-to-read summarization text output, encompassing clinical findings and their level of change and significance, in a fraction of a second.
Our radiology report characterization method has many potential clinical applications. For example, our method can assist healthcare providers at the point of care as a part of an online clinical decision support system by providing the characterization for key clinical findings for decision-making based on radiology reports. The resulting characterizations can be combined with other information from electronic health records for review prioritization, disease surveillance, and content-based image retrieval. Given the performance of our method on multi-organizational radiology reports, our characterization method can provide an infrastructure to develop and improve various data-driven biomedical information systems that deal with information overload.
The reference standard annotations in this study were generated by one radiologist. This might have introduced bias into the annotation process. To explore these potential biases, we asked a second independent radiologist to annotate 25% of our data set. The inter-annotator agreement measures for this subset showed reasonably high agreements between the two annotators (agreement percentages were between 94.6 and 96.8% and kappa coefficients were between 79.4 and 83.9% for different types of annotations). This level of interannotator agreement shows the integrity of our reference standard annotations. The existing disagreements between the annotators demonstrate the challenges of the manual annotation process, caused by the complexity of radiology report language. We expect providing a comprehensive set of annotation guidelines with expressive examples will improve the quality of the reference standard annotations and the interannotator agreement.
As part of our error analysis, we reviewed the errors that occurred at each stage of our method. For capturing clinical findings (F1 score 85.7%), most of the errors were caused by invalid clinical observation annotations from the utilized CRF named-entity recognition system. We expect that expanding the training data for retraining both the CRF model and our finding refinement SVM model will improve the performance of the method. The errors for detecting changes (end-to-end F1 score 94.7%) are mostly due to the triggering of conflicting rules in summaries due to expressions such as “unchanged increased” (identified as new/worse by our method and unchanged by the radiologist) or “development of a small amount” (identified as unchanged by our method and new/worse by the radiologist). Adding new rules in our method to resolve the conflicts between different rules can resolve these errors. Other errors in change characterization were caused by underlying semantics of the summaries. For example, “as seen on the prior examination” indicates an unchanged status, or “more callus formation” is an indication of healing fracture and improving condition. Although new rules can address these errors case by case, we plan to develop a more sophisticated machine learning method for capturing text’s underlying semantics for this task through novel semantic text analysis frameworks such as deep neural networks [36] as future work. That said, considering the strong performance and relatively simple implementation of the rule-based approach, the current rule-based method is highly effective for the change characterization task. The errors in determining the significance of clinical findings (end-to-end F1 score 75.3%) are mostly due to new terms and phrases in the test set that were not observed in the training process. We expect that expanding the training set will significantly improve the performance of our SVM classifier for this task.
We also examined our method’s errors in our multi-organizational study through manual review. We observed the error types for identifying and characterizing radiology report summaries are similar in reports from different organizations. This is due to similarities in the patterns of our information model classes in radiology reports across different organizations. Of note, we did not observe any spelling errors in the review of the radiology reports. All reports in our data set are dictated by radiologists using speech recognition systems [37]. These speech recognition systems have built-in dictionaries, perform spell check, and therefore almost always include correctly spelled terms and phrases in radiology reports [30]. In addition, no errors caused by homophones were observed by our domain expert radiologists who reviewed and annotated the radiology reports in our data set.
Limitations and Avenues for Future Work
As future work, we plan to expand our annotated training and test data sets and enrich the NLP features to address the current errors of the NLP approach and improve the robustness of our method and its evaluation. The number of radiology reports for training and evaluating our method was small and limited. Expanding this data set will inform and refine the machine learning models for error cases in the training process and improve the performance of our method. Also, the increase of the test set will enhance the confidence intervals of the evaluation metrics. Adding new features that capture text semantics such as distributional semantics and term co-occurrence patterns [38, 39] will address the NLP errors even without observing similar cases in the training set. In addition, as mentioned in the “Material and Methods” section, the reference standard annotations in this work are generated by one domain expert. To address any potential biases in these annotations, we plan to leverage multiple annotators instructed with annotation guidelines and examples for manual annotation in the future extension of our work. We will use the majority vote among these overlapping annotations to remove potential disagreements, biases, and noise in annotations. We expect this will increase the reliability of our annotated training set and improve our machine learning models for radiology report information extraction and characterization.
As a limitation of this study, our training and test sets in this work are small and only focused on chest CT reports. This is due to the time-consuming nature of manual annotation and our limited resources for the manual annotation of radiology reports. Despite this restriction, chest CT report narratives cover clinical findings from many vital organs and conditions and are representative of the complexity of radiology report narratives for other imaging modalities and body regions. Even with a relatively small set of training data, our results showed the robustness and generalizability of our method on data from different organizations. In fact, none of the NLP techniques described in this work are specific to an information model, narrative, or organization. The developed techniques are applicable to other types of narratives with different information models and data sources as well. We plan to extend and apply this method beyond chest CT modality to other types of radiology reports and clinical notes in both structured and unstructured format.
As another future work, we plan to extend our characterization method to multiple radiology reports for each patient to capture the complete imaging history for patients. For this purpose, we will consider temporal patterns across various reports, non-monotonic reasoning, and measures such as pointwise mutual information [40] to remove the redundancies and contradictions in the summarization results from multiple radiology reports. We also plan to expand the richness of our information model. This includes adding more concept classes, such as classes to describe urgency of clinical findings, for more detailed characterization of radiology reports.
Conclusions
We developed an NLP method to extract clinical findings from the radiology report narrative and determine the level of change and significance of these clinical findings according to an information model. This radiology-specific information model covers different levels of change and significance required for an informed clinical decision-making process. Our method uses a combination of machine learning and rule-based approaches and leverages various features and abstractions at surface, entity, and discourse levels in text analysis. For evaluation, we applied our method on radiology reports from four major healthcare organizations. Our results showed the strength of our method in summarizing radiology reports and rating their level of change (accuracy 99.2%, precision 96.3%, recall 93.5%, and F1 score 94.7%) and significance (accuracy 75.8%, precision 75.2%, recall 75.7%, and F1 score 75.3%). Considering the evaluation results, the method provided a coherent characterization framework for radiology reports in the presence of various wording and stylistic variations in radiology reports in different organizations. The extracted clinical findings and their associated characterizations can enable clinicians to understand radiology reports better and prioritize the report review process to rapidly identify reports that need further follow-up. Our method can facilitate automated identification of patients for clinical trials, accelerate disease surveillance, and enable real-time clinical decision support and content-based image retrieval systems.
References
- 1.Smith R. Strategies for coping with information overload. Bmj. 2010;341:c7126. doi: 10.1136/bmj.c7126. [DOI] [PubMed] [Google Scholar]
- 2.Davidoff F, Miglus J. Delivering clinical evidence where it’s needed: building an information system worthy of the profession. JAMA. 2011;305(18):1906–7. doi: 10.1001/jama.2011.619. [DOI] [PubMed] [Google Scholar]
- 3.Luhn HP. The automatic creation of literature abstracts. IBM Journal of research and development. 1958;2(2):159–65. doi: 10.1147/rd.22.0159. [DOI] [Google Scholar]
- 4.Baxendale PB. Machine-made index for technical literature: an experiment. IBM Journal of Research and Development. 1958;2(4):354–61. doi: 10.1147/rd.24.0354. [DOI] [Google Scholar]
- 5.Das D, Martins AF. A survey on automatic text summarization. Literature Survey for the Language and Statistics II course at CMU. 2007;4:192–5. [Google Scholar]
- 6.Mitkov R.(2005) The Oxford handbook of computational linguistics. Chapter 32, Oxford University Press; Jan 13
- 7.Gupta V, Lehal GS. A survey of text summarization extractive techniques. Journal of Emerging Technologies in Web Intelligence. 2010;2(3):258–68. doi: 10.4304/jetwi.2.3.258-268. [DOI] [Google Scholar]
- 8.Elfayoumy S, Thoppil J. A survey of unstructured text summarization techniques. The International Journal of Advanced Computer Science and Applications. 2014;5(7):149–54. doi: 10.14569/IJACSA.2014.050716. [DOI] [Google Scholar]
- 9.Lloret E.(2008) Text summarization: an overview. Paper supported by the Spanish Government under the project TEXT-MESS (TIN2006-15265-C06-01).
- 10.Afantenos S, Karkaletsis V, Stamatopoulos P. Summarization from medical documents: a survey. Artificial intelligence in medicine. 2005;33(2):157–77. doi: 10.1016/j.artmed.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 11.Mishra R, Bian J, Fiszman M, Weir CR, Jonnalagadda S, Mostafa J, Del Fiol G. Text summarization in the biomedical domain: a systematic review of recent research. Journal of biomedical informatics. 2014;52:457–67. doi: 10.1016/j.jbi.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pivovarov R, Elhadad N. Automated methods for the summarization of electronic health records. Journal of the American Medical Informatics Association. 2015;22(5):938–47. doi: 10.1093/jamia/ocv032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarkar K. Using domain knowledge for text summarization in medical domain. International Journal of Recent Trends in Engineering. 2009;1(1):200–5. [Google Scholar]
- 14.Reeve L, Han H, Brooks AD (2006). BioChain: lexical chaining methods for biomedical text summarization. In Proceedings of the 2006 ACM Symposium on Applied Computing Apr 23 (pp. 180–184). ACM
- 15.Chuang WT, Yang J (2000). Extracting sentence segments for text summarization: a machine learning approach. In Proceedings of the 23rd Annual International ACM SIGIR Conference on Research and Development in Information Retrieval Jul 1 (pp. 152–159). ACM
- 16.Fiszman M, Rindflesch TC, Kilicoglu H (2004). Abstraction summarization for managing the biomedical research literature. In Proceedings of the HLT-NAACL Workshop on Computational Lexical Semantics May 6 (pp. 76–83). Association for Computational Linguistics.
- 17.Elhadad N, Kan MY, Klavans JL, McKeown KR. Customization in a unified framework for summarizing medical literature. Artificial intelligence in medicine. 2005;33(2):179–98. doi: 10.1016/j.artmed.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 18.McKeown KR, Elhadad N, Hatzivassiloglou V (2003). Leveraging a common representation for personalized search and summarization in a medical digital library. In Proceedings of the 3rd ACM/IEEE-CS Joint Conference on Digital Libraries May 27 (pp. 159–170). IEEE Computer Society
- 19.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. Journal of the American Medical Informatics Association. 1994;1(2):161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automatic detection of acute bacterial pneumonia from chest X-ray reports. Journal of the American Medical Informatics Association. 2000;7(6):593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendonça EA, Haas J, Shagina L, Larson E, Friedman C. Extracting information on pneumonia in infants using natural language processing of radiology reports. Journal of biomedical informatics. 2005;38(4):314–21. doi: 10.1016/j.jbi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Mani I. Maybury MT. Advances in automatic text summarization: MIT press; 1999. [Google Scholar]
- 23.Zafar HM, Chadalavada SC, Kahn CE, Jr, et al. Code abdomen: an assessment coding scheme for abdominal imaging findings possibly representing cancer. Journal of the American College of Radiology: JACR. 2015;12(9):947. doi: 10.1016/j.jacr.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carletta J. Assessing agreement on classification tasks: the kappa statistic. Computational linguistics. 1996;22(2):249–54. [Google Scholar]
- 25.Hassanpour S, Langlotz CP. Information extraction from multi-institutional radiology reports. Artificial intelligence in medicine. 2016;66(1):29–39. doi: 10.1016/j.artmed.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langlotz CP. RadLex: a new method for indexing online educational materials. Radiographics. 2006;26(6):1595–7. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 27.Lafferty J, McCallum A, Pereira FC (2001). Conditional random fields: probabilistic models for segmenting and labeling sequence data, Proceedings of the Eighteenth International Conference on Machine Learning, San Francisco. (pp. 282–289). Morgan Kaufmann Publishers Inc.
- 28.Sutton C, McCallum A. An introduction to conditional random fields for relational learning. Introduction to statistical relational learning. 2006:93-128.
- 29.Klein D, Manning CD (2003). Accurate unlexicalized parsing. In Proceedings of the 41st Annual Meeting on Association for Computational Linguistics-Volume 1 Jul 7 (pp. 423–430). Association for Computational Linguistics.
- 30.Cortes C, Vapnik V. Support-vector networks. Machine learning. 1995;20(3):273–97. [Google Scholar]
- 31.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST). 2011;2(3):27. [Google Scholar]
- 32.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of biomedical informatics. 2001;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 33.Powers DM (2011). Evaluation: from precision, recall and F-measure to ROC, informedness, markedness and correlation. (1): 37–63
- 34.R Core Team (2013). R: a language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria
- 35.Deng L, Yu D. Deep learning: methods and applications. Foundations and Trends in Signal Processing. 2014;7(3–4):197–387. doi: 10.1561/2000000039. [DOI] [Google Scholar]
- 36.Pezzullo JA, Tung GA, Rogg JM, Davis LM, Brody JM, Mayo-Smith WW. Voice recognition dictation: radiologist as transcriptionist. Journal of digital imaging. 2008;21(4):384–389. doi: 10.1007/s10278-007-9039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. In Advances in neural information processing systems 2013 (pp. 3111-3119).
- 38.Pennington J, Socher R, Manning CD. Glove: global vectors for word representation. In EMNLP 2014 (Vol. 14, pp. 1532-1543).
- 39.Church KW, Hanks P. Word association norms, mutual information, and lexicography. Computational linguistics. 1990;16(1):22–9. [Google Scholar]
- 40.Manning CD, Schütze H. Foundations of statistical natural language processing. Cambridge: MIT press; 1999. [Google Scholar]