Abstract
Histopathology image processing, analysis and computer-aided diagnosis have been shown as effective assisting tools towards reliable and intra-/inter-observer invariant decisions in traditional pathology. Especially for cancer patients, decisions need to be as accurate as possible in order to increase the probability of optimal treatment planning. In this study, we propose a new image collection library (HICL–Histology Image Collection Library) comprising 3831 histological images of three different diseases, for fostering research in histopathology image processing, analysis and computer-aided diagnosis. Raw data comprised 93, 116 and 55 cases of brain, breast and laryngeal cancer respectively collected from the archives of the University Hospital of Patras, Greece. The 3831 images were generated from the most representative regions of the pathology, specified by an experienced histopathologist. The HICL Image Collection is free for access under an academic license at http://medisp.bme.teiath.gr/hicl/ . Potential exploitations of the proposed library may span over a board spectrum, such as in image processing to improve visualization, in segmentation for nuclei detection, in decision support systems for second opinion consultations, in statistical analysis for investigation of potential correlations between clinical annotations and imaging findings and, generally, in fostering research on histopathology image processing and analysis. To the best of our knowledge, the HICL constitutes the first attempt towards creation of a reference image collection library in the field of traditional histopathology, publicly and freely available to the scientific community.
Keywords: Histology image collection, Cancer, Computer-aided diagnosis, Microscopy, Brain cancer, Breast cancer, Laryngeal cancer
Introduction
Cancer is a major cause of morbidity and mortality worldwide: according to the World Health Organization (WHO), approximately 14 million new cases were diagnosed and 8.2 million deaths were registered in 2012. The overall cancer incidence rate is expected to increase from 14 million to 19.3 million in the next two decades, which implies an even deadliest impact of the disease [1]. Although numerous initiatives have been taken to promote better outcomes involving state-of-art diagnostic technologies for early detection [2, 3] and innovative treatments for improving survival [4, 5], death rates have not been yet reduced and the quality of life of affected patients has not been significantly enhanced.
It is well known that diagnosis, prognosis and treatment planning relies on traditional pathology practices. Although complex technologies, such as positron emission tomography (PET), magnetic resonance imaging (MRI) and x-ray computed tomography (CT) may provide indications regarding the presence of the disease, such technologies cannot be used for predicting the disease’s course and designing the patient’s treatment plan [6], even in cases for which these indications might be strong. Findings are always verified at the subsequent step of the microscopy examination using traditional pathology practices.
In traditional pathology practice, diagnostic decisions are made following visual inspection of the biological material under the microscope. Reviewing biological material with the microscope is a very complex process, time consuming and, most importantly, may result to diagnostic misinterpretations, which may lead to serious complications in patient management [7]. The effect of diagnostic errors has been recognized as a serious social and economic health care problem, which only in the USA costs dozens of billions of dollars and affect more than 1 million patients per year [8]. The risk of diagnostic errors is higher in more than 200 identified rare cancer types [9]. However, even in common cancer types, it is possible that optimal diagnostic decisions are not met since (a) in many cases the most representative part of the tumor is either not presented to the observing physician (poor sampling) or not visually identified [10]; (b) the criteria for diagnostic conclusions are vague (i.e., “numerous” multinucleated, “mild” cellularity); thus, decisions are objective and may differ from one physician to another (inter-observe variability); (c) since cancer evolves in a biological continuum, it is very difficult to establish the exact boundaries between the different grades or stages of the disease, although different grades and stages may affect decisive treatment planning [11]; (d) the experience of the observing physician is of paramount importance, especially for the rare cancer types for which many physicians may have limited training [12, 13].
Computer-aided diagnosis (CAD) systems have been shown as potential second opinion tools that may (a) guide physicians towards more accurate decisions, (b) reduce inter-and intra-observer variability, (c) efficiently manage and integrate the vast amount of information related to each patient (i.e., multiple images, electronic registries, laboratory tests) [14–16]. A successful paradigm of CAD systems may be found in radiology, especially in mammography [15, 17, 18], with commercial FDA-approved software solutions. One of the main reasons for boosting up the research in CAD systems in radiology was the publicly available image databases, such as the DDSM project [19], the MIAS mammogram database [20], the mammographic images database from LAPIMO EESC/USP [21], the Optimam Mammography Image Database [22] and the Image Database Resource Initiative [23]. Hundreds of computer aided-diagnosis and image processing research studies have utilized these databases as reference; thus, the impact of these databases may be considered as most important. Such reference image collections, as those described above, are lacking from the field of traditional histology and histopathology.
The purpose of this project is to create a publicly available resource of static histopathology images for medical image processing, analysis and computer-aided diagnosis research applications. The proposed image collection library, the Histology Image Collection Library (HICL), comprises 3831 distinct images from three different diseases (brain, breast and laryngeal cancer) with associated clinical annotations (grade, stage, survival, molecular factors, morphometrics, radiological findings, demographics etc). Interested investigators could exploit the library for research in image processing and analysis, in decision support systems, in statistical and correlative analysis of annotations with imaging findings, in comparing different image processing and analysis methodologies on the same data (as a reference dataset) and, generally, in fostering overall research on histopathology image processing and analysis. To the best of our knowledge, the HICL constitutes the first attempt towards creation of an image collection library in the field of traditional histopathology for image processing, analysis and decision support system research purposes, publicly and freely available to the scientific community.
Methods and Materials
Raw Data Collection and Processing
Raw data comprised 93 cases of brain cancers (astrocytomas, oligodendrogliomas, meningiomas), 116 cases of breast cancer and 55 cases of laryngeal cancer collected from the archives of the University Hospital of Patras, Greece. The study follows the guidelines of the ethics committee of the University of Patras. Each case corresponds to a different patient. For each case, on average, five stained sections were generated from the same material. Sections were placed on slides for microscopic examination.
Data Annotation/Diagnosis
Each case was diagnosed and annotated with associated clinical information by an experienced histopathologist (P.R.). All cases were checked for intra-observer concordance (blind readings of the same data by the same histopathologist following a time period greater than one (1) month from first reading). In cases where intra-observer variation was observed, the physician reviewed the slides on a multiheaded microscope with another histopathologist in order to accomplish unanimous decision concerning annotations. About 12% of brain cancer cases, 2% of laryngeal cases and 5% of breast cancer cases needed a second reader.
Image Digitization
The experienced histopathologist marked the most representative areas of the tumor. From these regions, images were digitized using two light microscopy imaging systems. The first one comprised a Zeiss Axiostar-Plus (Zeiss, Göttingen, Germany) microscope connected to a Leica DC 300F (Leica Microsystems GmbH) camera, and the second one consisted of a Leica DM 2500 microscope and a Leica DFC 420C camera (Leica Microsystems GmbH) (Fig. 1). Most of the images were generated using the second microscopy imaging system with specifications the tiff format, 1728 × 1296 pixels, pixel size 2.78 μm × 2.78 μm, horizontal and vertical resolution 96 dpi, 24-bit depth and file size around 6.40 MB. A part of breast cancer images were generated using the first microscopy imaging system with specifications the tiff format, 1300 × 1030 pixels, 6.7 μm × 6.7 μm, 150 dpi, 48-bit depth and file size around 7.66 MB.
Fig. 1.
Microscopy imaging system (Leica DM 2500 microscope and a Leica DFC 420C camera)
Image Collection Organization
Each image was anonymized and organized in the collection to associate with the following information:
ID: a unique identification number is assigned to each image
Staining: images have been processed with different stains (i.e., hematoxylin and eosin (Η&Ε), immunohistochemistry (IHC) for p63 and estrogen receptors (ER) expression)
Magnification factor: images have been digitized under different magnification factors (i.e., ×20, ×40)
Microscope equipment used for digitization and viewing: (i.e., Leica)
Diagnosis: Images have been related to case diagnosis (i.e., grade, stage, low-high risk)
Hospital information: (i.e., University Hospital of Patras)
Disease type: (i.e., brain cancer, breast cancer, laryngeal cancer)
Image number information: (i.e., from each ID we have extracted more than one images)
HICL Image Collection Access
The HICL is free for access under an academic license. The interested user may access the image collection at http://medisp.bme.teiath.gr/hicl/ (Fig. 2). The webpage contains information regarding the image collection contents, presents sample images, lists relevant publications and has an application form, in which the interested researcher fills up in order to get access to the collection.
Fig. 2.
HICL image collection webpage (http://medisp.bme.teiath.gr/hicl/)
Results
Brain Cancer Cases
All 93 brain cancer cases had undergone surgery at the University Hospital of Patras between 1993 and 2002. Patients’ ages ranged from 10 to 76 years. All patients were treated with partial or total tumor resection. Most patients with high-grade tumors were post-operatively treated with radiation and/or chemotherapy. Tumor grade was defined as I, II III or IV according to the WHO grading system [24]. Of the 93 cases, 36 were classified as low grade (grades I–II), 65 as high grade (grades III–IV) and 3 as between low and high grade (grades II–III). The most common neoplasm was glioblastoma multiforme (grade IV), which was dominant for patients exceeding 60 years old. Low-grade tumors appeared in a stable rate for patients younger than 40 years, with tendency to decrease at higher ages, since low-grade tumors usually recur and progress after initial tumor resection and/or treatment. The highest risk group constituted patients over 60 years old. It is worth mentioning the relatively high incidence rates for young people between 10 and 20 years old. Ninety-one (91) brain cancer cases were also reviewed for the estimation of eight (8) histological features on a Likert scale basis (Table 1).
Table 1.
Associated information for brain cancer cases
| Age | Gender | H&Ea (number of images) | Histological tumor grade (number of images) | ||||||||
| Male | Female | ×20 | ×40 | I | I–II | II | II–III | III | III–IV | IV | |
| 50 ± 16 | 58 | 33 | 1257 | 1291 | 255 | 167 | 418 | 100 | 801 | 77 | 730 |
| Cellularity (no. of cases) | Mitoses (no. of cases) | Apoptosis (no. of cases) | Multinucleated (no. of cases) | ||||||||
| Mild | Medium | Marked | Absent | Present | Absent | Present | Absent | Present | Numerous | ||
| 14 | 47 | 29 | 38 | 50 | 34 | 56 | 58 | 27 | 5 | ||
| Giant (no. of cases) | Vascular proliferation (no. of cases) | Necrosis (no. of cases) | Pleomorphism (no. of cases) | ||||||||
| Absent | Present | Numerous | Absent | Present | Marked | Absent | Present | Marked | Absent | Present | Marked |
| 47 | 34 | 9 | 1 | 69 | 21 | 34 | 36 | 21 | 51 | 27 | 12 |
a H&E hematoxylin and eosin
Breast Cancer Cases
All 116 breast cancer cases were infiltrative (invasive) ductal carcinomas. Tumor grade was carried out on H&E-stained sections following the WHO recommendations and employing the Elston and Ellis grading scheme [22]. Additionally, information regarding mammographic features and status of other molecular indices such as ER, PR, cerbB-2, p53, Ki-67, and cath-D was also retrieved for each case. ER expression was assessed on IHC-stained specimens, following the clinical routine protocol [25] that takes into consideration the percentage ratio of ER-expressed nuclei (brown colored) to the total number of expressed and non-expressed (blue) nuclei. Five percent was used as the cut-off value of ER expression for characterizing the case as having positive ER status (ER+). IHC evaluation was performed without taking into consideration the corresponding histological grade. Of the 116 cases, 31 were classified as grade I, 35 as grade II and 50 as grade III (Table 2).
Table 2.
Associated information for breast cancer cases
| H&Ea (number of images) | IHCb-ERc (number of images) | Histological tumor grade (number of cases) | Mammography findings | ||||||
| ×20 | ×40 | ×40 | I | II | III | Shading | Shading+ | Vagueness | Vagueness+ |
| 231 | 227 | 414 | 31 | 35 | 50 | 35 | 13 | 24 | 9 |
| Lymph size (cm) | ERc (%) | PRd (%) | Her-2e (%) | p53 (%) | Ki67 (%) | Cath Df (%) | |||
| 3.08 ± 2.04 | 52.1 ± 35.0 | 30.5 ± 29.4 | 40.3 ± 37.4 | 20.5 ± 29.8 | 26.4 ± 24.1 | 34.4 ± 32.1 | |||
a H&E hematoxylin and eosin
b IHC immunohistochemical staining
c ER estrogen receptors
d PR progesterone receptors
e Her-2 human epidermal growth factor receptor 2
f Cath D cathepsin D
Laryngeal Cancer Cases
All 55 laryngeal cancer patients had undergone biopsy examination at the University Hospital of Patras between 2008 and 2012. Patients’ ages ranged from 44 to 89 years. Clinical and pathological staging was defined according to the American Joint Committee on Cancer (AJCC) guidelines [26]. All lesions were diagnosed as laryngeal squamous cell carcinomas. P63 expression was assessed by visual inspection on the IHC-stained specimens. Cases with more than 50% positively expressed nuclei were considered as having positive P63 expression. During the IHC evaluation, histological grade was not taken under consideration. Finally, information regarding lesion site, staging, smoking habits, alcohol habits, profession and survival was also retrieved for each case (Table 3).
Table 3.
Associated information for laryngeal cancer cases
| Age (years) | IHCa-P63 (number of images) | Histological tumor grade (number of cases) | Lesion site (number of cases) | ||||||
| ×20 | ×40 | I | II | III | Glottic | Supraglottic | Spread to subsites | N/A | |
| 63.4 ± 11.0 | 224 | 226 | 21 | 18 | 16 | 35 | 11 | 3 | 6 |
| Stage (number of cases) | |||||||||
| T | N | ||||||||
| 2 | 3 | 4 | 0 | 1 | 2 | N/A | II | III | IV |
| 8 | 29 | 13 | 5 | 43 | 2 | 2 | 7 | 27 | 17 |
| Smoking habit (number of cases) | Alcohol habit (number of cases) | Survival (number of cases) | |||||||
| Cigarettes/day | Moderate | Heavy | years | N/A | Moderate | Present | Numerous | >5 years | <5 years |
| 46.6 ± 19.9 | 10 | 15 | 38.1 ± 12.6 | 14 | 58 | 27 | 5 | 26 | 16 |
a IHC immunohistochemical staining
Image Samples
Figures 3, 4, and 5 illustrate examples of images of different disease types, different magnification, and different diagnoses respectively. In total, 2548 H&E brain cancer images, 872 breast cancer images and 411 laryngeal cancer images were created.
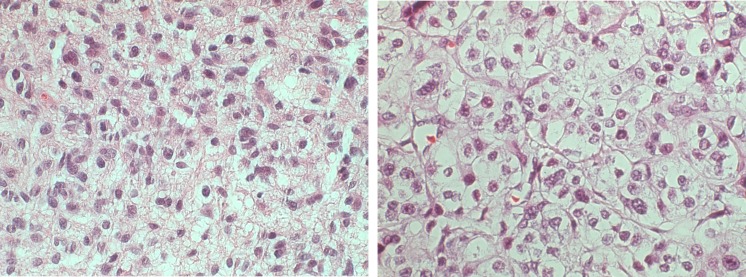
Fig. 3.
Left H&E brain cancer image from a high-grade case, ×400. Right H&E breast cancer image from a high-grade case, ×400
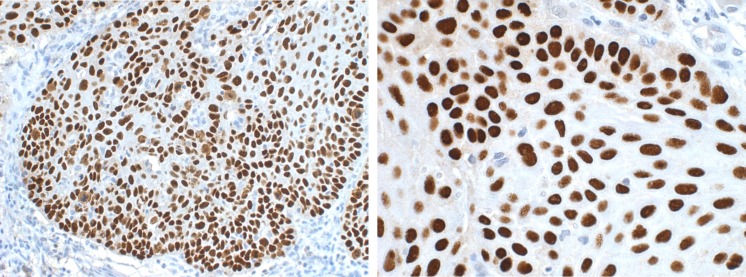
Fig. 4.
Left IHC p63-stained image, laryngeal cancer, ×200. Right IHC p63-stained image, laryngeal cancer, ×400
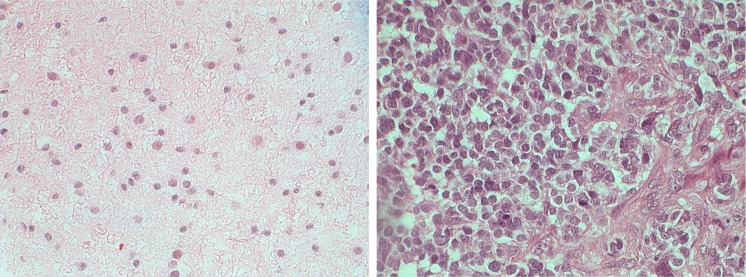
Fig. 5.
Left H&E low-grade brain cancer image (astrocytoma), ×400. Right H&E high-grade brain cancer image (astrocytoma), ×400
From the 93 brain cancer cases, 2548 H&E-stained images were generated (1257 at ×20 and 1291 at ×40), among which 827 were created from low-grade cases, 1612 images from high-grade cases and 100 images from ambiguous (low- to high-grade) cases.
From the 116 breast cancer cases, 872 images were generated, among which 414 were IHC stained at ×40 magnification and 458 were H&E stained (227 at ×20 and 231 ×40). From the 414 IHC-stained images, 112 were created from grade I cases, 148 from grade II cases and 121 from grade III cases. From the 458 H&E-stained images, 97 were created from grade I cases, 99 from grade II cases and 134 images from grade III cases.
From the 55 laryngeal cancer cases, 411 P63-stained images were generated, among which 168 were created from grade I cases, 131 from grade II cases and 112 from grade III cases.
Image Annotation
The ID, staining and magnification factor were organized in the title name of each image. An example of decoding the title of the image “67_HE_40X_LEICA_GIII_PATRA_BRAIN_CANCER_1.tif” is the following:
67: patient ID
HE: staining protocol
40X: magnification factor
Leica: microscope used
GIII: histological diagnosis–grade III
PATRA: hospital-city
BRAIN_CANCER: disease
1: the number of image at the specific ID
An explanatory excel file provides detailed information regarding each case, such as age, gender, molecular factors, habits, survival and any other existing additional clinical information (see summary in Tables 2 and 3).
Discussion
Reviewing of slides under the microscope is a complicated process, which in some cases may lead to diagnostic misinterpretations. Especially for cancer patients, decisions need to be as accurate as possible in order to increase the probability of a better and successful treatment planning. Image processing, analysis and decision support systems in histopathology have been shown as valuable assisting tools towards more accurate decisions, with numerous promising applications in brain cancer, breast cancer, leukemia, thyroid cancer, laryngeal cancer and other diseases [27–35].
The HICL attempts to create the first, to the best of our knowledge, reference image collection library in histopathology, freely available to the scientific community under an academic license. The applications of such a library may span into numerous research fields of medical image processing and analysis. It could be used to (a) design processing algorithms for contrast equalization of under or over stained images, correction of non-uniform illumination effects, removal of blurring, improvement of visualization of out-of-focus regions and enhancement of textures. Although effort was given to ensure uniformity of conditions for the preparation of the raw clinical material and for image digitization, the interested researcher will identify in the collection images which pose image processing challenges. Such images may originate from old cases (more than 10 years old data), for which the stain intensity has been sensibly reduced, over- or under-stained specimens, out-of-focus regions in parts of the image due to difference in tissue thickness and similar challenges that could be resolved following the application of the proper image processing algorithms; (b) to design and test segmentation algorithms for region of interest delineation (such as nuclei) and compare different segmentation approaches on the same data. Although nuclei appear darker than surrounding background, segmentation is not straightforward especially for cases with increased cellularity, multinucleated cells and irregular shape of heterogeneous texture nuclei (i.e., for higher-grade cases); (c) to design and test computer-aided diagnosis and decision support systems. In our image collection, we provide several clinical annotations for designing two-class or multi-class decision support systems, such as grading, staging and survival data; (d) to investigate potential meaningful correlations between histological annotations and other clinical annotations. From example, breast cancer cases are associated with grade, mammographic findings, size of nodules/masses, ER, PR, cerB, p53, ki67, cathD and demographics. Laryngeal cancer cases are accompanied with grading, staging, lesion site, smoking habits, alcohol habits, profession, demographics and survival. Finally, brain cancer cases are associated with grading, demographics and eight histological features. Besides these applications, depending on the imagination and resourcefulness of interested researchers, many other applications may emerge.
Effort has been given to ensure that all cases included the study have been reliably annotated. Towards this direction three measures were taken. The first measure comprised the collaboration with a highly experienced histopathologist (more than 30 years of clinical experience). The second measure consisted of securing high intra-observer rates. Data were re-evaluated by the same histopathologist following 1 month from initial reading. The third measure constituted the review of difficult cases under a multiheaded microscope with other histopathologist until a consensus decision was taken. Although the abovementioned steps may secure, to a certain extent, the reliability of clinical annotations, it is possible (and reasonable) that some images may divert from given annotations since it is well known that tumors are heterogeneous and develop along a biological continuum. The latter means that even in a high-grade tumor sample, one may find low-grade tumor regions. Thus, when generating images from a high-grade tumor-annotated case, it is possible to find low-grade alike associated image samples, a situation that resembles real-world conditions (i.e., tumors’ heterogeneous evolution).
It worth noticing that the HICL is a cumulative effort that lasted more than 10 years. The reasons are numerous: First, brain cancers that were included in the study are rare with incidence rates 4–5 on 100,000 persons (astrocytomas, oligodendrogliomas and meningiomas). Second, the laryngeal cases are presented with 5-year survival annotations. Third, although it may not be obvious, the effort required by the collaborating histopathologist to give all related clinical annotations was immense, considering that the completion of a typical case review required on average 30–40 min. In our study we have included 93 brain cancer cases, 116 breast cancer cases and 55 laryngeal cancer cases, thus, in total 264 cases, which correspond to about 130–175 h dedicated work for clinically assessing all cases. The above complexity may explain the fact that although numerous image collections/databases may be found available in various medical fields such as in radiology [20–22], dermatology [36, 37], such collections are difficult to find in histopathology. Most histology-related databases serve mainly educational purposes [38–40]. The HICL may be considered as a first attempt towards rendering histopathology data, which are so time consuming to assess and collect, publicly and freely available to the scientific community to benefit all interested researchers in the field of histopathology image processing, analysis and decision support system design.
Although the HICL has not been organized in a database form, it is very easy for the user to retrieve all necessary information for each image sample by performing a simple search based on the name of each sample, which includes information regarding the unique identification number of the originating case, the magnification, the diseases type, the staining procedure, the diagnostic annotation and the microscope imaging system type. Moreover, all associated clinical annotations are organized in excel files, in order to facilitate the user to filter, organize and present data based on preferred clinical characteristics (see Tables 2 and 3).
The HICL Histology Image Collection project is ongoing and will grow in the future by (a) organizing the images in a relational database in order to enable the user to search under specific criteria the content of the database (i.e., search by disease, by grade, by stage, by gender), (b) adding images from new diseases (currently, we are collecting HPV and colorectal cancer images), (c) creating template segmentation image masks, which will contain the exact coordinates of nuclei within each image.
Acknowledgements
This work was supported by the Research Committee of the Technological Educational Institution (TEI) of Athens, under the framework of the “Special Program for Research Grants” for the Support of TEI of Athens Researchers for 2015.
References
- 1.D. Price ID, C. Renaud and R. Dickinson. MRI scanners: a buyer’s guide. http://dev.ersnet.org/uploads/Document/8d/WEB_CHEMIN_2563_1194523150.pdf
- 2.Conway JR, Carragher NO, Timpson P. Developments in preclinical cancer imaging: innovating the discovery of therapeutics. Nat Rev Cancer. 2014;14:314–28. doi: 10.1038/nrc3724. [DOI] [PubMed] [Google Scholar]
- 3.Pandey AP, Girase NM, Patil MD, Patil PO, Patil DA, Deshmukh PK. Nanoarchitectonics in cancer therapy and imaging diagnosis. J Nanosci Nanotechnol. 2014;14:828–40. doi: 10.1166/jnn.2014.9014. [DOI] [PubMed] [Google Scholar]
- 4.Masoudi-Nejad A and Asgari Y. Metabolic cancer biology: structural-based analysis of cancer as a metabolic disease, new sights and opportunities for disease treatment. Semin Cancer Biol 2014 [DOI] [PubMed]
- 5.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124:30–9. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richard R. Carlton AMA. Principles of Radiographic Imaging: An Art and a Science, 5th edition. Cengage Learning; 2012
- 7.Bellocq JP, Anger E, Camparo P, Capron F, Chenard MP, Chetritt J, Chigot JP, Cochand-Priollet B, Coindre JM, Copin MC, Flejou JF, Galateau F, Gaulard P, Guiu M, Michiels JF, Saint-Andre JP, Scoazec JY, Vacher-Lavenu MC. Securising diagnosis in pathology in 2011. The diagnostic error: between rhetoric and reality. Ann Pathol. 2011;31:S92–4. doi: 10.1016/j.annpat.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 8.David E Newman-Toker KMM, David O Meltzer. How much diagnostic safety can we afford, and how should we decide? A health economics perspective. BMJ Qual Saf 22:2013 [DOI] [PMC free article] [PubMed]
- 9.Gemma G, MvdZ J, Paolo GC, Sabine S, Angelo Paolo DT, Ian K, Renner O, Lisa L, Sandra M, Andrea T, Annalisa T, Riccardo C. Rare cancers are not so rare: the rare cancer burden in Europe. European Journal of Cancer. 2011;47:2493–2411. doi: 10.1016/j.ejca.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Herfarth K, Gutwein S, Debus J. Postoperative radiotherapy of astrocytomas. Seminars in Surgical Oncology. 2001;20:13–23. doi: 10.1002/ssu.1012. [DOI] [PubMed] [Google Scholar]
- 11.Paulus W, Peiffer J. Intratumoral histologic heterogeneity of gliomas. A quantitative study. Cancer. 1989;64:442–7. doi: 10.1002/1097-0142(19890715)64:2<442::AID-CNCR2820640217>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 12.Coons W, Jhonson P, Sceithauer B, Yates A, Pearl D. Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer. 1997;79:1381–93. doi: 10.1002/(SICI)1097-0142(19970401)79:7<1381::AID-CNCR16>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 13.Prayson R, Agamanolis D, Cohen M, Estes M, Kleinschmidt-DeMasters B, Abdul-Karim F, McClure S, Sebek B, Vinay R. Interobserver reproducibility among neuropathologists and surgical pathologists in fibrillary astrocytoma grading. Journal of the Neurological Sciences. 2000;175:33–39. doi: 10.1016/S0022-510X(00)00274-4. [DOI] [PubMed] [Google Scholar]
- 14.Doi K. Computer-aided diagnosis in medical imaging: historical review, current status and future potential. Comput Med Imaging Graph. 2007;31:198–211. doi: 10.1016/j.compmedimag.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jalalian A, Mashohor SB, Mahmud HR, Saripan MI, Ramli AR, Karasfi B. Computer-aided detection/diagnosis of breast cancer in mammography and ultrasound: a review. Clin Imaging. 2013;37:420–6. doi: 10.1016/j.clinimag.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki K. A review of computer-aided diagnosis in thoracic and colonic imaging. Quant Imaging Med Surg. 2012;2:163–76. doi: 10.3978/j.issn.2223-4292.2012.09.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishikawa RM. Current status and future directions of computer-aided diagnosis in mammography. Comput Med Imaging Graph. 2007;31:224–35. doi: 10.1016/j.compmedimag.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Zhou C. Computer-aided diagnosis: radiologists' "second opinion" in breast cancer diagnosis on mammography. Indian J Med Res. 2006;124:123–4. [PubMed] [Google Scholar]
- 19.Heath M, Bowyer K, Kopans D, Moore R and Kegelmeyer P. The Digital Database for Screening Mammography. Proceedings of the Fifth International Workshop on Digital Mammography, 2001
- 20.Suckling J, Parker, J, Dance, DR, Astley, S, Hutt, I, Boggis, CRM, Ricketts, I, Stamatakis, E, Cerneaz, N, Kok, SL, Taylor, P, Betal, D, and Savage, J. The Mammographic Image Analysis Society Digital Mammogram Database. Exerpta Medica. International Congress Series−1994, 1994
- 21.Matheus BR, Schiabel H. Online mammographic images database for development and comparison of CAD schemes. J Digit Imaging. 2011;24:500–6. doi: 10.1007/s10278-010-9297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Looney PT, Young KC and Halling-Brown MD. Medxviewer: providing a web-enabled workstation environment for collaborative and remote medical imaging viewing, perception studies and reader training. Radiat Prot Dosimetry 169(1–4):32–7, 2015. [DOI] [PubMed]
- 23.Armato SG, 3rd, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, MacMahon H, Van Beeke EJ, Yankelevitz D, Biancardi AM, Bland PH, Brown MS, Engelmann RM, Laderach GE, Max D, Pais RC, Qing DP, Roberts RY, Smith AR, Starkey A, Batrah P, Caligiuri P, Farooqi A, Gladish GW, Jude CM, Munden RF, Petkovska I, Quint LE, Schwartz LH, Sundaram B, Dodd LE, Fenimore C, Gur D, Petrick N, Freymann J, Kirby J, Hughes B, Casteele AV, Gupte S, Sallamm M, Heath MD, Kuhn MH, Dharaiya E, Burns R, Fryd DS, Salganicoff M, Anand V, Shreter U, Vastagh S, Croft BY. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys. 2011;38:915–31. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleihues P, Burger PC, and Scheithauer BW. Histological Typing of Tumors of the Central Nervous System. Berlin: Springer-Verlag, 1993
- 25.Diaz LK, Sahin A, Sneige N. Interobserver agreement for estrogen receptor immunohistochemical analysis in breast cancer: a comparison of manual and computer-assisted scoring methods. Ann Diagn Pathol. 2004;8:23–7. doi: 10.1016/j.anndiagpath.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Diaz LK, Sneige N. Estrogen receptor analysis for breast cancer: current issues and keys to increasing testing accuracy. Adv Anat Pathol. 2005;12:10–9. doi: 10.1097/00125480-200501000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Ali S, Veltri R, Epstein JI, Christudass C, Madabhushi A. Selective invocation of shape priors for deformable segmentation and morphologic classification of prostate cancer tissue microarrays. Comput Med Imaging Graph. 2015;41:3–13. doi: 10.1016/j.compmedimag.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kostopoulos S, Glotsos D, Cavouras D, Daskalakis A, Kalatzis I, Georgiadis P, Bougioukos P, Ravazoula P, Nikiforidis G. Computer-based association of the texture of expressed estrogen receptor nuclei with histologic grade using immunohistochemically-stained breast carcinomas. Anal Quant Cytol Histol. 2009;31:187–96. [PubMed] [Google Scholar]
- 29.Loukas C, Kostopoulos S, Tanoglidi A, Glotsos D, Sfikas C, Cavouras D. Breast cancer characterization based on image classification of tissue sections visualized under low magnification. Comput Math Methods Med. 2013;2013:829461. doi: 10.1155/2013/829461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosquera-Lopez C, Agaian S, Velez-Hoyos A, Thompson I. Computer-aided prostate cancer diagnosis from digitized histopathology: a review on texture-based systems. IEEE Rev Biomed Eng. 2015;8:98–113. doi: 10.1109/RBME.2014.2340401. [DOI] [PubMed] [Google Scholar]
- 31.Rathore S, Hussain M, Aksam Iftikhar M, Jalil A. Novel structural descriptors for automated colon cancer detection and grading. Comput Methods Programs Biomed. 2015;121:92–108. doi: 10.1016/j.cmpb.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Singh A, Au J, Saeedi P, Havelock J. Automatic segmentation of trophectoderm in microscopic images of human blastocysts. IEEE Trans Biomed Eng. 2015;62:382–93. doi: 10.1109/TBME.2014.2356415. [DOI] [PubMed] [Google Scholar]
- 33.Wolberg WH, Street WN, Mangasarian OL. Computer-derived nuclear features compared with axillary lymph node status for breast carcinoma prognosis. Cancer. 1997;81:172–9. doi: 10.1002/(SICI)1097-0142(19970625)81:3<172::AID-CNCR7>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 34.Decaestecker C, Lopes BS, Gordower L, Camby I, Cras P, Martin JJ, Kiss R, VandenBerg SR, Salmon I. Quantitative chromatin pattern description in Feulgen-stained nuclei as a diagnostic tool to characterize the oligodendroglial and astroglial components in mixed oligo-astrocytomas. J Neuropathol Exp Neurol. 1997;56:391–402. doi: 10.1097/00005072-199704000-00008. [DOI] [PubMed] [Google Scholar]
- 35.Kostopoulos S, Konstandinou C, Sidiropoulos K, Ravazoula P, Kalatzis I, Asvestas P, Cavouras D, Glotsos D. Assessing the performance of four different categories of histological criteria in brain tumours grading by means of a computer-aided diagnosis image analysis system. J Microsc. 2015;260:37–46. doi: 10.1111/jmi.12264. [DOI] [PubMed] [Google Scholar]
- 36.Kassianos AP, Emery JD, Murchie P, Walter FM. Smartphone applications for melanoma detection by community, patient and generalist clinician users: a review. Br J Dermatol. 2015;172:1507–18. doi: 10.1111/bjd.13665. [DOI] [PubMed] [Google Scholar]
- 37.Dermofit. Nurs Stand 31:32, 2016 [DOI] [PubMed]
- 38.Hemminki K, Ji J, Brandt A, Mousavi SM, Sundquist J. The Swedish Family-Cancer Database 2009: prospects for histology-specific and immigrant studies. Int J Cancer. 2010;126:2259–67. doi: 10.1002/ijc.24795. [DOI] [PubMed] [Google Scholar]
- 39.Patel SG, Rosenbaum BP, Chark DW, Lambert HW. Design and implementation of a web-based, database-driven histology atlas: technology at work. Anat Rec B New Anat. 2006;289:176–83. doi: 10.1002/ar.b.20112. [DOI] [PubMed] [Google Scholar]
- 40.Foord K, Guy R, Apthorp L, How K, Trevethick P, Ziemann M. Updated audit database for breast imaging/histopathology correlation. Clin Radiol. 2001;56:755–62. doi: 10.1053/crad.2001.0769. [DOI] [PubMed] [Google Scholar]