Abstract
Cancer is one of the leading causes of deaths worldwide. While cancers may initially show good response to chemotherapy or radiotherapy, it is not uncommon for them to recur at a later date. This phenomenon may be explained by the existence of a small population of cancer stem cells, which are inherently resistant to anti-cancer treatment as well as being capable of self-renewal. Therefore, while most of the tumour bulk consisting of cells that are not cancer stem cells respond to treatment, the cancer stem cells remain, leading to disease recurrence. Following this logic, the effective targeting of cancer stem cells holds promise for providing long-term cure in individuals with cancer. Cancer stem cells, like normal stem cells are endowed with mechanisms to protect themselves against a wide range of insults including anti-cancer treatments, such as the enhancement of the DNA damage response and the ability to extrude drugs. It is therefore important to develop new strategies if cancer stem cells are to be eradicated. In this review, we describe the strategies that we have developed to target cancer stem cells. These strategies include the targeting of the histone demethylase jumonji, AT rich interactive domain 1B (JARID1B), which we found to be functionally significant in the maintenance of cancer stem cells. Other strategies being pursued include reprogramming of cancer stem cells and the targeting of a functional cell surface marker of liver cancer stem cells, the aminopeptidase CD13.
Keywords: cancer stem cell, cellular reprogramming, epigenomics, therapeutics, JARID1B/KDM5B, CD13
Introduction
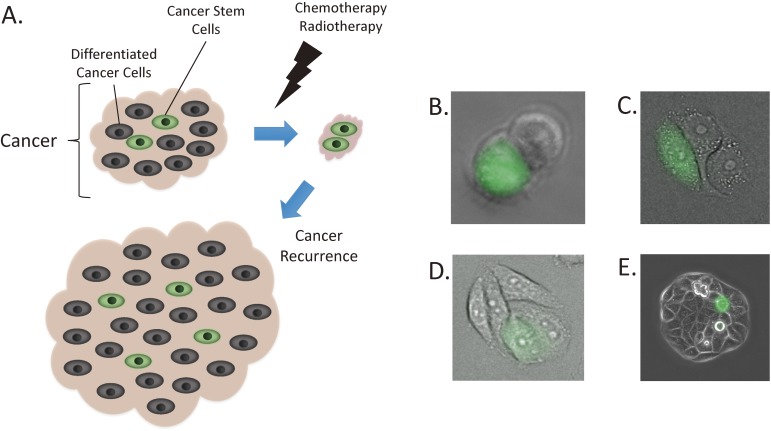
Cancer remains to be one of the leading causes of deaths worldwide.1) Although cancers may initially show good response to conventional treatments such as chemotherapy or radiotherapy, they quite commonly recur at a later date. One explanation for this phenomenon is the existence of a small population of cancer stem cells that are resistant to radiation and chemotherapy, allowing them to withstand treatment and cause recurrence, sometimes decades following the initial treatment2–4) (Fig. 1). An important notion is that similar to any proliferative tissues in adults, tumour growth is sustained only by a small number of stem cells that are responsible for self-renewal. While most of the tumour bulk consisting of cells which are not stem cells respond to treatment, the cancer stem cells remain. Following this logic, the eradication of cancer stem cells is absolutely critical for offering long-term cure to patients with cancer.2,5,6) Most encouragingly, the emerging evidence support the notion that targeting cancer stem cells could cure what would otherwise be a terminal disease.7)
Figure 1.
Cancers are made up of cancer stem cells (fluorescent green cells) and differentiated cancer cells (non-fluorescent cells). Conventional cancer treatment is able to eradicate the differentiated cancer cells but not the cancer stem cells. Cancer stem cells cause recurrence at a later date (A). Cancer stem cells demonstrate asymmetrical cell division, giving rise to one fluorescent cancer stem cell and non-fluorescent daughter cell (B). The daughter cell continues to divide multiple times (C and D), to make up the bulk of the cell population (E). While this results in there being only one cancer stem cell, this cell is capable of surviving chemotherapy and regenerating the entire cellular population.
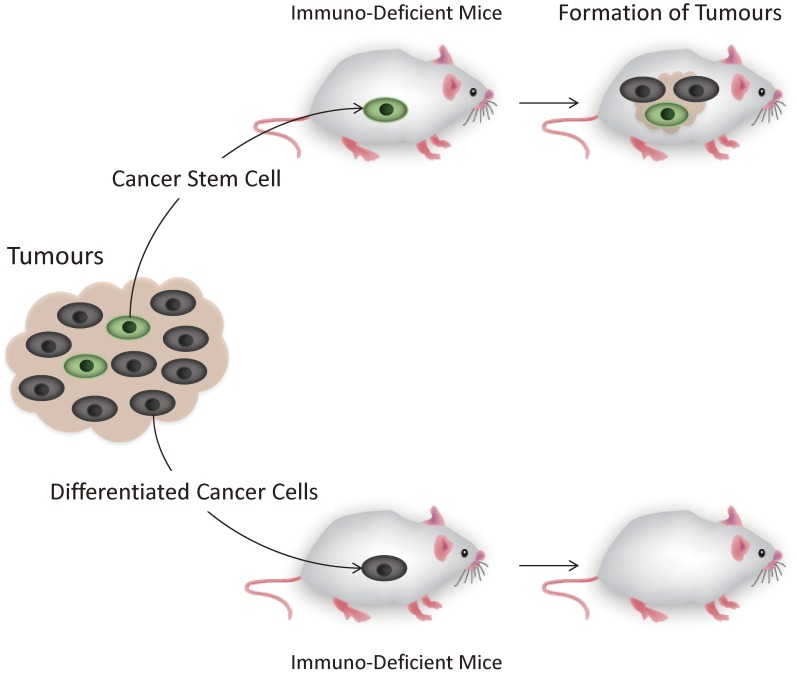
It was demonstrated as early as in the 1930s by Furth and Khan that single cancer cells could establish heterogeneous tumour mass resembling the cancers from which they were derived from.8) Subsequent experiments demonstrated that the frequency of the tumour initiating cells were low (one per 103 to 107 cells).9–11) However, it was not until around two decades ago that cancer stem cells could be isolated from cancers with flow cytometry using cell surface markers specific for cancer stem cells. This in turn allowed the biology of cancer stem cells to be studied in greater depth. In 1994, Dick and his colleagues were the first to isolate a small population of leukaemia cells that could grow tumours when xenotransplanted in immuno-deficient mice,12) and this was followed a few years later by the discovery of similar cells in solid cancers by Clarke and his colleagues.13,14) Most importantly for both these examples, it was only the cancer stem cells that could grow tumours in mice, and the transplantation of the differentiated cancer cells that made up the major proportion of the cancer mass did not result in tumour formation in mice (Fig. 2).
Figure 2.
The mouse xenograft model allows the presence of cancer stem cells to be discerned. This is because only the cancer stem cells are able to form tumours when transplanted into immuno-deficient mice.
It was the isolation of cancer stem cells that allowed scientists to determine that they are more resistant to radio- and chemotherapy.15–18) Cancer stem cells, like normal stem cells are endowed with mechanisms to protect themselves against a wide range of insults including anti-cancer treatments, such as the enhancement of both the DNA damage response and the ability to extrude drugs. On top of this, they are more resistant to the induction of apoptosis. Furthermore, it has been found that cancers with higher expression of stem cell markers are associated with increased rates of recurrence19,20) and poorer prognosis,21) supporting the notion that cancer stem cells are responsible for treatment resistance and recurrence.
In this review, we first describe a system that we developed to visualise cancer stem cells in situ, without disturbing their environment. We also describe the strategies being developed at Osaka University to target cancer stem cells. These strategies include the targeting of the histone demethylase jumonji, AT rich interactive domain 1B (JARID1B), which we found to be functionally significant in the maintenance of cancer stem cells. Other strategies being pursued include reprogramming of cancer stem cells and the targeting of a functional cell surface marker in liver cancer stem cells, the aminopeptidase CD13.
1. Visualisation of cancer stem cells
Much of the research in cancer stem cells have relied on their isolation using flow cytometry, which requires the cells to be taken away from their environment, potentially altering their biology through increased stress. We set out to develop a system for visualising cancer stem cells in situ, in vitro or in vivo, to aid research into their true behaviour.
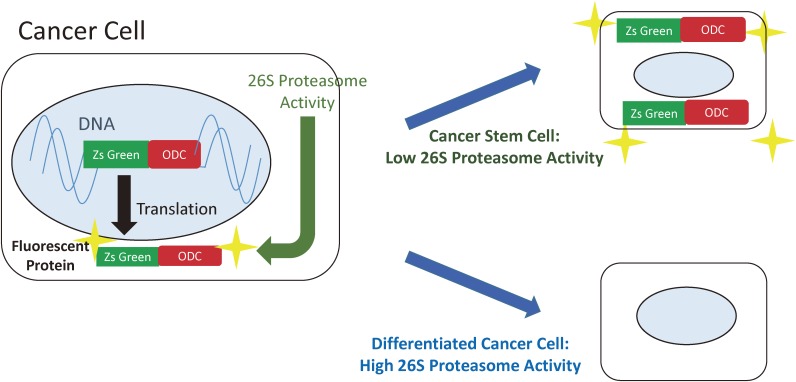
This was made possible by taking advantage of one of the characteristics of cancer stem cells, which is that they are quiescent with a low protein turnover and a downregulated 26S proteasome activity.22) Cell lines from colorectal cancer,16) cervical cancer17) and osteosarcoma18) were transfected with a vector coding for a fusion protein consisting of a green fluorescent protein, ZsGreen, and the C-terminal degron of the ornithine decarboxylase (ODC) that is normally destroyed by proteasomes. Cancer stem cells with low 26S proteasome activity were predicted to retain the green fluorescence due to decreased degradation of the fusion protein (Fig. 3). As expected, the fluorescent cells (ZsGreen-ODC positive) from the three cancers mentioned above demonstrated features of stemness, including the ability to form tumours as mice xenotransplants and to undergo asymmetric cell division. Furthermore, the fluorescent cells were more chemo- and radioresistant compared to the non-fluorescent cells,17,18) an important feature of cancer stem cells as mentioned previously. Others have also reported ZsGreen-ODC positive cells to harbour features of cancer stem cells including in pancreatic cancer,23) glioma and breast cancer,24) demonstrating the utility of this system across a wide range of cancers.
Figure 3.
Cancer stem cells can be visualised because they have downregulated 26S proteasome activity. Cells are transfected with a vector coding for a fusion protein consisting of ZsGreen, and the C-terminal degron of the ornithine decarboxylase. Degron directs the destruction of the fluorescent protein by proteasomes in differentiated cancer cells. In cancer stem cells, the fusion protein is not destroyed and the cells are fluorescently labelled.
Such visualisation of cancer stem cells using the ZsGreen-ODC system allows in addition for drug screening to search for novel agents that are able to eradicate cancer stem cells.
2. Drug development targeting the histone demethylase JARID1B
We and others have previously found the highly conserved histone demethylase, jumonji AT rich interactive domain 1B (JARID1B) to be a functional marker of cancer stem cells.25–27) Histone demethylases remove methyl groups from histone, and this post-transcriptional modification of the histones can affect gene expression. This is because DNA is wound around the histone protein, and the modification to the histone proteins can alter whether the DNA it is packaging can be made available for transcription. In this way, JARID1B is a powerful regulator of gene expression and is also involved in normal tissue development as well as the maintenance of cancer stem cells.25,28–33)
JARID1B belongs to a family of Jarid1 proteins that are highly homologous, and there is at least partial redundancy between Jarid1b and Jarid1a in demethylating H3K4.34,35) In melanoma, JARID1B was found to be a marker of a small proportion of cells with a slow cell turnover, but ones that gave rise to a progeny with high turnover.36) Knockdown of JARID1B lead to the exhaustion of tumour growth, suggesting that the histone demethylase was functionally important in maintaining continuous tumour growth.25) Furthermore, JARID1B was found to be critical in conferring the melanoma cells resistance against the chemotherapy cisplatin.37)
In colorectal cancer cell lines, we confirmed that knockdown of JARID1B compromised the growth of the tumours in the mouse xenograft model, as well as reducing the sphere forming capacity, suggesting the functional significance of JARID1B in cancer stem cells. JARID1B suppression was also found to increase trimethylation of histone 3 lysine 4 (H3K4) at the promoter region of the tumour suppressor p16/INK4A, resulting in the increased expression of the gene. p16/INK4A is also a key inducer of cellular senescence, and indeed promoted this cellular phenotype after knockdown of JARID1B expression.26) These results suggest JARID1B to be a good target against cancer stem cells. Moreover, the expression of JARID1B in the normal tissue is restricted to the testes and ovaries, which suggests that targeting JARID1B may be associated with minimal side effects.25)
We are therefore at present searching for small molecule inhibitors of JARID1B, with the aim of developing a drug that could be used clinically for the eradication of cancer stem cells. Such medications may require the co-administration with therapies aimed at inactivating the function of drug transporters that actively pump drugs out of cancer stem cells.38,39)
3. Treatment based on cell reprogramming
A. The concept.
The Nobel laureate Yamanaka and his colleagues made the landmark discovery that differentiated cells could be reprogrammed to become induced pluripotent stem (iPS) cells by the transfection of four transcription factors, Oct4, Sox2, Klf4 and c-Myc (hereon referred to as Yamanaka factors);40,41) this demonstrated a powerful method of transforming cellular phenotype through epigenetic manipulation. Cancers arise as a result of epigenetic dysregulation as well as genetic mutations,42) and stemness is controlled by epigenetics. We explored the effect of reprogramming cancer cells and cancer stem cells by the transfection of the Yamanaka factors, to see if their malignant potential could be diminished through the interference of their epigenetic signature.
Despite the abundance of genetic mutations harboured by cancer cell lines, we found they could be reprogrammed to become pluripotent cells, or induced pluripotent cancer (iPC, not to be confused with iPS) cells. Cancer cell lines transfected with the Yamanaka factors expressed markers of embryonic stem cells such as glycolipid carbohydrate epitope stage specific embryonic antigen 4 (Ssea-4), signifying the reprogramming from differentiated cells to pluripotent stem cells. What is more, the iPC cells could be manipulated to differentiate into cells of different phenotypes by controlling the environment in which they were grown in. The reprogrammed cancer cells could be guided to differentiate to cells of epithelial, mesenchymal, neural and adipose lineages by controlling the conditions of the culture mediums.43) In this way, we were able to demonstrate that even the iPC cells had the capacity for multipotent differentiation.
At first we thought that the iPC cells would revert to their original phenotypes. To our surprise however, we found that the iPC cells lost the ability to initiate tumours as subcutaneous xenotransplant in mice. Alongside this, iPC cells became more sensitive to chemotherapy.43) These findings suggest that reprogramming is a promising method of diminishing the function of cancer stem cells.
B. Reprograming of cancer cells using microRNAs.
In order to make use of the reprogramming strategy to eradicate cancer stem cells, we had to overcome some challenges. One problem associated with the administration of the Yamanaka factors to humans is the requirement for viral vectors for transfection, with the associated risk of random insertions of DNA sequences into the genome. The creation of both iPS and iPC cells are also highly inefficient.
In order to bypass the problems above associated with the use of the Yamanaka factors, we developed a method of reprogramming mice and human fibroblasts to pluripotent stem cells through the use of a specific combination of microRNAs.44) MicroRNAs (miRNAs) are small non-coding RNAs that silence gene expression, and regulate development, differentiation and cancer cell biology. Specific miRNAs have been characterized to control pluripotency-related genes,45–47) which prompted us to search for miRNAs that could reprogram differentiated cells to pluripotent stem cells.
To begin with, we used unbiased analysis to search for miRNA that are involved with the induction and maintenance of iPS cells. We first searched for miRNA that are highly expressed in mouse iPS cells and mouse ES cells, but with low or undetectable expression in differentiated mouse adipose stromal cells. These candidate miRNAs that were picked up from the first screen were then applied to cells derived from transgenic mice with green fluorescent protein (GFP) inserted downstream of the promoter for octamer-binding transcription factor 3/4 (Oct3/4), a transcription factor and master regulator of pluripotency.48) In this way, cells becoming pluripotent through the activation of Oct3/4 would become fluorescent, allowing the identification of miRNAs that could reprogram the cells to pluripotency. As a result, we found that a combination of three miRNAs (mir-200c, mir-302s and mir-369s) could reprogram both mouse and human cells to pluripotency.
The achievement of pluripotency was confirmed by demonstrating the ability of the miRNA induced pluripotent stems cells to differentiate to cells of different lineages, as well as being able to generate chimera mouse.
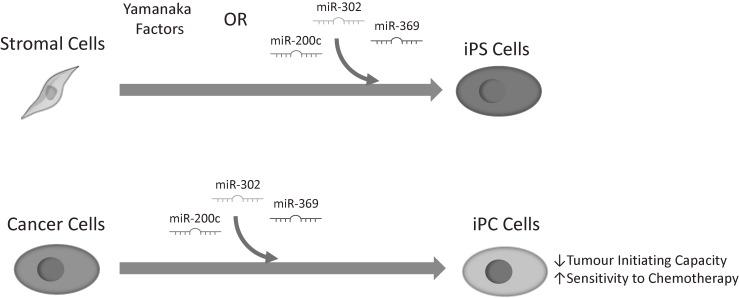
It was also possible to reprogram cancer cells through the administration of the three miRNAs, demonstrated by the expression of the pluripotency markers Oct3/4, Sox2 and Nanog. As was the case with the Yamanaka factors, cancer cell lines reprogrammed by miRNA also demonstrated decreased tumour initiating capacity and became sensitive to chemotherapy (Fig. 4).49) Most significantly, we have also been able to confirm the feasibility, safety and the effectiveness of administering the three miRNAs in vivo. In a mouse model of colorectal cancer, we were able to demonstrate that the three miRNAs are able to suppress tumourigenesis, suggesting that this therapy could be useful for both preventing and treating cancer.
Figure 4.
Stromal cells can be reprogrammed to inducible pluripotent stem (iPS) cells through the transfection of Yamanaka factors (Oct4, Sox2, Klf4 and c-Myc), or through three miRNAs (miR-200c, miR-302, miR-369). Cancer cells can also be reprogrammed to pluripotent cells, so called inducible pluripotent cancer (iPCs) cells, which have reduced tumour initiating capabilities and increased chemosensitivity.
One problem associated with miRNA based treatment is the presence of RNases in the bloodstream leading to the rapid degradation of RNA, making them unsuitable for systemic therapy when used in isolation. We therefore designed a drug delivery system, called the super carbonate apatite nanoparticles, which is capable of preventing the degradation of RNA. The super carbonate apatite nanoparticles are also able to selectively deliver RNAs to tumours because they preferentially release RNA in the low pH conditions found in tumours.50) In order to further increase the stability and the efficacy of the miRNA based therapy, we are also at present exploring the use of synthetic miRNA.
C. The functional mechanisms of the three miRNAs.
In order to understand the molecular mechanisms behind the effectiveness of the reprogramming therapy in suppressing cancer, we are examining the role of each of the three miRNAs.
mir-302s was found to repress the expression of multidrug resistance-associated proteins, which are the transmembrane protein transporters that expel a number of chemotherapeutic agents from the cytoplasm of the cancer cells.49) Such action of mir-302s may be the reason behind the increased sensitivity to chemotherapy in the reprogrammed cancer cells.
On the other hand, mir-369s was found to reprogram the cellular metabolism of cancer cells by promoting the Warburg effect, which is the preferential fermentation of sugars over oxidative phosphorylation, despite the presence of oxygen.51,52) The Warburg effect is a metabolic trait associated with stem cells, and mir-369s was found to promote this through the stabilisation of the expression of the isoform of an enzyme pyruvate kinase 2 (PKM2). PKM2 regulates the final rate-limiting step of glycolysis, allowing the diversion of glycolytic flux into the pentose phosphate pathway instead of the tricarboxylic acid cycle, thus driving the Warburg effect.53) Although it remains to be elucidated whether the altered metabolism directly affects carcinogenesis or chemoresistance, it seems likely that metabolism plays an important role in reprogramming cancer cells to the pluripotent state.
The mir-200 family are well known to block epithelial-mesenchymal transition (EMT), a cellular phenotype which is associated with the cancer cells acquiring the ability to invade the surrounding tissues and enter the bloodstream, leading to distant organ metastasis.54) Indeed, cancer cells transfected with mir-200c demonstrated decreased expression of the mesenchymal markers ZEB1, ZEB2, SNAIL, SLUG, increased expression of the epithelial marker E-cadherin, and decreased invasive capacity of the cells.49)
These results demonstrate that the three miRNAs target various aspects of cancer biology, and corroborates our hypothesis that these miRNAs are capable of changing the fundamental biology of the cancer cells.
4. Targeting CD13
Another approach to targeting cancer stem cells for treatment is to target the cell surface molecules that are specifically expressed in these cells. We were the first to isolate cancer stem cells from liver cancer, by identifying CD13 as a cell surface marker of cancer stem cells. This was achieved by performing a gene expression profile analysis of a side-population of liver cancer cells with a low cell turnover. The side-population of cells have enhanced ability to extrude dye, and this is a reliable method for isolating stem cells, including cancer stem cells.55,56) This revealed CD13 to be highly expressed in the population of cells with a low cellular turnover. Furthermore, CD13 positive cells were confirmed to have the characteristics of cancer stem cells, such as the ability to initiate tumours as xenografts in mice, form spheres in culture and chemoresistance.
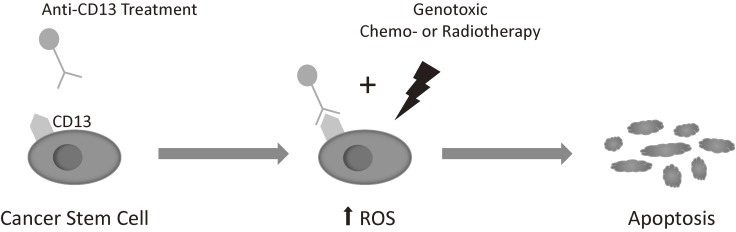
Furthermore, CD13 was proven to be functionally significant in the maintenance of cancer stem cells. CD13 was found to reduce reactive oxygen species (ROS)-induced DNA damage after genotoxic chemo- or radiotherapy, preventing apoptosis. The inhibition of CD13 by the commercially available neutralizing antibody ubenimex resulted in the increase of ROS in response to chemotherapy, leading to the significant reduction in the size of tumours in subcutaneous mouse xenograft models (Fig. 5). These results suggest that combining CD13 treatment with ROS inducing chemo- or radiotherapy, thereby targeting the cancer stem cells as well as the differentiated cancer cells, may lead to a better outcome in liver cancer.57,58)
Figure 5.
CD13 is a functional marker of cancer stem cells, reducing reactive oxygen species (ROS)-induced DNA damage after genotoxic chemo- or radiotherapy, preventing apoptosis. The inhibition of CD13 by the commercially available neutralizing antibody ubenimex results in the increase of ROS, leading to apoptosis.
Concluding remarks
Much has been achieved in the field of cancer stem cells in the last two decades. We and others have shown that treatment strategies targeting cancer stem cells can eradicate cancers. Through such work, we hope in the future that we will be able to overcome the problems of treatment resistance and recurrence in cancer, providing long-term cure from this prevalent disease.
Acknowledgments
We thank Dr. Naohiro Nishida, Dr. Naotsugu Haraguchi, Prof. Yuichiro Doki, and Prof. Hideshi Ishii from Osaka University for their assistance with the preparation of this manuscript. We thank them also for their fruitful discussions.
Biographies
Profile
Hugh Shunsuke Colvin was born in Tokyo in 1983 and graduated with a degree in Medicine from Cambridge University, UK in 2007. After internship, he majored in general surgery, especially colorectal surgery, and became a member of the Royal College of Surgeons of England in 2008. He came to the Gastroenterological Department in Osaka University in 2014 to conduct basic scientific research under the supervision of Prof. Masaki Mori. He is at present investigating the role of oncometabolites in colorectal cancer as part of his Ph.D. project. He is a Young Scientist Research Fellow of the Japan Society for the Promotion of Science and a fellow of the Interdisciplinary Program for Biomedical Sciences at Osaka University.

Masaki Mori was born in Kagoshima in 1956. He received his M.D. and Ph.D. degrees from Kyusyu University School of Medicine in 1980 and 1986, respectively. He is currently the Chairman of the Department of Gastroenterological Surgery at Osaka University Graduate School of Medicine, working as a surgeon and a cancer researcher. During his career, he has studied human surgical pathology for 4.5 years at Kyusyu University with his mentor, Prof. Munetomo Enjoji, and molecular biology for 1.5 years at New England Deaconess Hospital and Dana-Farber Cancer Institute in Boston with his mentors, Prof. Glenn D Steele and Prof. Lan Bo Chen. His major in surgery is the treatment of digestive organ cancers including of the large intestine, stomach, esophagus, liver and pancreas. With respect to cancer research, he has made efforts to bridge the gap between basic and clinical sciences for the advancement of treatment for cancer patients. For example, he has paid much attention to cancer stem cell research of gastrointestinal cancers. He became the chairman of the organizing committee of the 47th International Symposium of the Princess Takamatsu Cancer Research Fund, of which the title was “Current Status and Perspective of Cancer Stem Cell Research”, held in November 2016. For his achievements in cancer and surgical research, he has received the Medical Award of the Japan Medical Association, Princess Takamatsu Cancer Research Fund Prizes, Special Award from Kobayashi Foundation for Cancer Research, Special Award from SGH Foundation, and other awards. He was the President of the Japanese Society of Gastroenterological Surgery (2011–2015), and is the Vice-President of the Japan Surgical Society (2016–) and the Vice-President of the Japanese Cancer Association (2016–). He is also a member of the Science Council of Japan (2014–).

References
- 1).Torre L.A., et al. (2015) Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108. [DOI] [PubMed] [Google Scholar]
- 2).Clevers H. (2011) The cancer stem cell: premises, promises and challenges. Nat. Med. 17, 313–319. [DOI] [PubMed] [Google Scholar]
- 3).Wagle N., et al. (2011) Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 29, 3085–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Waclaw B., et al. (2015) A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature 525, 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Kreso A., et al. (2014) Self-renewal as a therapeutic target in human colorectal cancer. Nat. Med. 20, 29–36. [DOI] [PubMed] [Google Scholar]
- 6).Lobo N.A., Shimono Y., Qian D., Clarke M.F. (2007) The biology of cancer stem cells. Annu. Rev. Cell Dev. Biol. 23, 675–699. [DOI] [PubMed] [Google Scholar]
- 7).Nakanishi Y., et al. (2013) Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat. Genet. 45, 98–103. [DOI] [PubMed] [Google Scholar]
- 8).Furth J., Kahn M.C., Breedis C. (1937) The transmission of leukemia of mice with a single cell. Am. J. Cancer 31, 276–282. [Google Scholar]
- 9).Makino S. (1956) Further evidence favoring the concept of the stem cell in ascites tumors of rats. Ann. N. Y. Acad. Sci. 63, 818–830. [DOI] [PubMed] [Google Scholar]
- 10).Bruce W.R., van der Gaag H. (1963) A quantitative assay for the number of murine lymphoma cells capable of proliferation in vivo. Nature 199, 79–80. [DOI] [PubMed] [Google Scholar]
- 11).Hewitt H.B. (1958) Studies of the dissemination and quantitative transplantation of a lymphocytic leukaemia of CBA mice. Br. J. Cancer 12, 378–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Lapidot T., et al. (1994) A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 367, 645–648. [DOI] [PubMed] [Google Scholar]
- 13).Al-Hajj M., Wicha M.S., Benito-Hernandez A., Morrison S.J., Clarke M.F. (2003) Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Li C., et al. (2007) Identification of pancreatic cancer stem cells. Cancer Res. 67, 1030–1037. [DOI] [PubMed] [Google Scholar]
- 15).Fukusumi T., et al. (2014) CD10 as a novel marker of therapeutic resistance and cancer stem cells in head and neck squamous cell carcinoma. Br. J. Cancer 111, 506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Munakata K., et al. (2016) Cancer stem-like properties in colorectal cancer cells with low proteasome activity. Clin. Cancer Res. 22, 5277–5286. [DOI] [PubMed] [Google Scholar]
- 17).Hayashi K., et al. (2014) Visualization and characterization of cancer stem-like cells in cervical cancer. Int. J. Oncol. 45, 2468–2474. [DOI] [PubMed] [Google Scholar]
- 18).Tamari K., et al. (2014) Identification of chemoradiation-resistant osteosarcoma stem cells using an imaging system for proteasome activity. Int. J. Oncol. 45, 2349–2354. [DOI] [PubMed] [Google Scholar]
- 19).Merlos-Suárez A., et al. (2011) The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8, 511–524. [DOI] [PubMed] [Google Scholar]
- 20).de Sousa E., Melo F., et al. (2011) Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 9, 476–485. [DOI] [PubMed] [Google Scholar]
- 21).Takahashi H., et al. (2011) Significance of Lgr5(+ve) cancer stem cells in the colon and rectum. Ann. Surg. Oncol. 18, 1166–1174. [DOI] [PubMed] [Google Scholar]
- 22).Pan J., Zhang Q., Wang Y., You M. (2010) 26S proteasome activity is down-regulated in lung cancer stem-like cells propagated in vitro. PLoS One 5, e13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Adikrisna R., et al. (2012) Identification of pancreatic cancer stem cells and selective toxicity of chemotherapeutic agents. Gastroenterology 143, 234–45.e7. [DOI] [PubMed] [Google Scholar]
- 24).Vlashi E., et al. (2009) In vivo imaging, tracking, and targeting of cancer stem cells. J. Natl. Cancer Inst. 101, 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25).Roesch A., et al. (2010) A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 141, 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Ohta K., et al. (2013) Depletion of JARID1B induces cellular senescence in human colorectal cancer. Int. J. Oncol. 42, 1212–1218. [DOI] [PubMed] [Google Scholar]
- 27).Kano Y., et al. (2013) Jumonji/Arid1b (Jarid1b) protein modulates human esophageal cancer cell growth. Mol. Clin. Oncol. 1, 753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Lu P.J., et al. (1999) A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 274, 15633–15645. [DOI] [PubMed] [Google Scholar]
- 29).Vogt T., et al. (1999) Deficiency of a novel retinoblastoma binding protein 2-homolog is a consistent feature of sporadic human melanoma skin cancer. Lab. Invest. 79, 1615–1627. [PubMed] [Google Scholar]
- 30).Christensen J., et al. (2007) RBP2 belongs to a family of demethylases, specific for tri- and dimethylated lysine 4 on histone 3. Cell 128, 1063–1076. [DOI] [PubMed] [Google Scholar]
- 31).Iwase S., et al. (2007) The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 32).Klose R.J., et al. (2007) The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128, 889–900. [DOI] [PubMed] [Google Scholar]
- 33).Yamane K., et al. (2007) PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell 25, 801–812. [DOI] [PubMed] [Google Scholar]
- 34).Cloos P.A.C., Christensen J., Agger K., Helin K. (2008) Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22, 1115–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Chicas A., et al. (2012) H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 109, 8971–8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Roesch A., et al. (2005) Retinoblastoma-binding protein 2-homolog 1: a retinoblastoma-binding protein downregulated in malignant melanomas. Mod. Pathol. 18, 1249–1257. [DOI] [PubMed] [Google Scholar]
- 37).Roesch A., et al. (2013) Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 23, 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Dean M. (2009) ABC transporters, drug resistance, and cancer stem cells. J. Mammary Gland Biol. Neoplasia 14, 3–9. [DOI] [PubMed] [Google Scholar]
- 39).Moitra K. (2015) Overcoming multidrug resistance in cancer stem cells. BioMed Res. Int. 2015, 635745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- 41).Takahashi K., et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. [DOI] [PubMed] [Google Scholar]
- 42).Hahn W.C., Weinberg R.A. (2002) Rules for making human tumor cells. N. Engl. J. Med. 347, 1593–1603. [DOI] [PubMed] [Google Scholar]
- 43).Miyoshi N., et al. (2010) Defined factors induce reprogramming of gastrointestinal cancer cells. Proc. Natl. Acad. Sci. U.S.A. 107, 40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Miyoshi N., et al. (2011) Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8, 633–638. [DOI] [PubMed] [Google Scholar]
- 45).Houbaviy H.B., Murray M.F., Sharp P.A. (2003) Embryonic stem cell-specific MicroRNAs. Dev. Cell 5, 351–358. [DOI] [PubMed] [Google Scholar]
- 46).Judson R.L., Babiarz J.E., Venere M., Blelloch R. (2009) Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 27, 459–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Suh M.-R., et al. (2004) Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 270, 488–498. [DOI] [PubMed] [Google Scholar]
- 48).de Jong J., Looijenga L.H.J. (2006) Stem cell marker OCT3/4 in tumor biology and germ cell tumor diagnostics: history and future. Crit. Rev. Oncog. 12, 171–203. [DOI] [PubMed] [Google Scholar]
- 49).Ogawa H., et al. (2015) MicroRNAs induce epigenetic reprogramming and suppress malignant phenotypes of human colon cancer cells. PLoS One 10, e0127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50).Wu X., et al. (2015) Innovative delivery of siRNA to solid tumors by super carbonate apatite. PLoS One 10, e0116022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51).Warburg O. (1956) On the origin of cancer cells. Science 123, 309–314. [DOI] [PubMed] [Google Scholar]
- 52).Vander Heiden M.G., Cantley L.C., Thompson C.B. (2009) Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53).Konno M., et al. (2015) Embryonic microRNA-369 controls metabolic splicing factors and urges cellular reprograming. PLoS One 10, e0132789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54).Kalluri R., Weinberg R.A. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55).Goodell M.A., Brose K., Paradis G., Conner A.S., Mulligan R.C. (1996) Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 183, 1797–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56).Goodell M.A., et al. (1997) Dye efflux studies suggest that hematopoietic stem cells expressing low or undetectable levels of CD34 antigen exist in multiple species. Nat. Med. 3, 1337–1345. [DOI] [PubMed] [Google Scholar]
- 57).Haraguchi N., et al. (2010) CD13 is a therapeutic target in human liver cancer stem cells. J. Clin. Invest. 120, 3326–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58).Yamashita M., et al. (2016) A CD13 inhibitor, ubenimex, synergistically enhances the effects of anticancer drugs in hepatocellular carcinoma. Int. J. Oncol. 49, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]