Abstract
The development of accurate urinary biomarkers for the non-invasive detection of urothelial bladder cancer (UBC) could transform patient pathways by reducing reliance on cystoscopy, and the identification of highly prognostic (or even predictive) biomarkers could better guide patient management. A number of approaches are being utilised to address these challenges in both urinary- and plasma-borne tumour DNA (tDNA), so-called “liquid biopsies”. Next generation sequencing (NGS) and droplet digital PCR (ddPCR) allow detection of very low levels of such tDNA amongst a large excess of non-tumour DNA, the former permitting large mutation panels to be assessed and the latter potentially identifying ultrarare mutant alleles yet restricted for multiplexing. Christensen et al. recently published their data regarding a ddPCR approach for the detection of common FGFR3 and PIK3CA mutations in urinary cell-free DNA (cfDNA) and circulating tumour DNA (ctDNA). In this proof-of-principle study, levels of mutant cfDNA in the urine of non-muscle-invasive bladder cancer (NMIBC) patients were shown to be positively correlated with tumour stage, grade and size, and a high initial level of mutant urinary cfDNA indicated future disease progression. In a cystectomy patient group, high mutant urinary cfDNA predicted future disease recurrence, the association being more pronounced with ctDNA. In this Perspective, we discuss these data in more detail and in parallel with the study’s limitations. We set these findings within the context of the field as a whole, highlighting important data from other groups, the strengths and weaknesses of alternative approaches, and the exciting and potentially significant future utilities of these techniques.
Keywords: Liquid biopsy, urinary biomarker, bladder cancer
The development of accurate biomarkers for the non-invasive detection of urothelial bladder cancer (UBC) could transform patient pathways by reducing reliance on cystoscopy which is burdensome for patients and expensive for healthcare providers (1,2). In addition, the identification of highly prognostic (or even predictive) biomarkers could better guide patient management, and especially for those patients with high-risk non-muscle-invasive bladder cancer (HR-NMIBC) who represent a treatment challenge (3,4). However, despite decades of intensive research, the need for effective biomarkers for bladder cancer detection and prognostication remains unmet (2,5). A small number of soluble urinary protein biomarkers (NMP22, BTA) or exfoliated cell tests based on proteins (ImmunoCyt) or aneuploidy (UroVysion) have obtained Food and Drug Administration (FDA) approval but have not been widely adopted due to limited sensitivity and/or specificity (5). More recently, research studies analysing the DNA in urinary cell pellets for changes in copy number, methylation status and somatic mutations have all shown potential for accurate non-invasive detection of bladder cancer (6-8). These tests still require thorough validation both in the incident and recurrent disease settings. Pitfalls include obtaining sufficient high-quality DNA from all urine samples, dilution of tumour DNA (tDNA) with non-tumour DNA, the heterogeneity of bladder cancer and, for surveillance, verifying biomarkers that are absolutely cancer specific, i.e., are not seen in any “field effect” (9). The advantages and disadvantages of these approaches are summarised in Table 1.
Table 1. The advantages and disadvantages of various approaches to the analysis of urinary tDNA.
| Method | Disadvantages | Advantages |
|---|---|---|
| DNA methylation | Relatively large amount of DNA required | Very high specificity and sensitivity recently reported (7) |
| Influenced by other factors (age, smoking) | ||
| Copy number changes | Unable to detect low levels of tumour DNA in a high background of non-tumour DNA | High specificity and most UBCs have copy number changes (10) |
| Microsatellite analysis | Unable to detect low levels of tumour DNA in a high background of non-tumour DNA | Good sensitivity and specificity reported (11) |
| Multiple individual tests needed | ||
| Mutations | Multiple mutations must be analysed to achieve high sensitivity | High specificity and can detect low levels of tumour DNA in a high background of non-tumour DNA (8,12,13) |
tDNA, tumour DNA; UBCs, urothelial bladder cancers.
Disease heterogeneity requires that either multiple markers need to be measured for a broadly applicable test which detects as many tumours as possible (high sensitivity), or the biomarker test needs to be individualised to the characteristics of a patient’s disease. The latter approach relies on the hypothesis that recurrent disease will retain the same genomic aberrations as the primary tumour (9,14). The former approach may require measuring a combination of alterations in copy number, methylation, and mutations at very low mutant allele frequencies; further technological innovation is needed to allow these diverse changes to be detected using a single analytical platform.
Two recent technologies that allow detection of very low levels of tDNA amongst a large excess of non-tumour DNA are droplet digital PCR (ddPCR) and next generation sequencing (NGS). ddPCR is ideal for detecting ultra-rare alleles and gives absolute quantitation of the number of mutant alleles, but is limited in its ability to multiplex, i.e., large numbers of biomarkers cannot be assayed simultaneously. ddPCR is therefore ideal for developing individualized biomarker assays. Conversely, NGS is able to measure a large number of biomarkers (up to whole genome level), enabling the development of broad applicability tests, but is limited in its ability to detect very low mutant allele frequencies. With current technology and using high read depths, the lower of limit of detection of NGS is determined by the background noise and is of the order of 1% mutant allele frequency. NGS workflows that incorporate unique molecular identifiers (so that reads can be traced to original DNA molecules in the specimen) enable detection of one tDNA molecule amongst >1,000 wtDNA molecules (15); however, these approaches may be difficult to apply for multiple markers, especially when low nanogram quantities of DNA are available (as often occurs with urine), and require ultra-high read depths, thus increasing expense. Targeted NGS has been successfully used for the detection of mutations in FGFR3 and TERT individually, and as part of a 6-gene panel in urinary cell-pellet DNA (8,13,16). It should be noted that low DNA yields from some urine specimens will provide only a few hundred/low thousand genome equivalents.
Whatever analytical approach is used, it is essential that the biospecimen contains sufficient tDNA to be detected, and urine DNA can be considered as two types which are easily separated by centrifugation or filtration: genomic DNA in exfoliated cells (the cell pellet) and cell-free DNA (cfDNA) in the supernatant/filtrate. We have observed that urinary cfDNA consists of slightly larger fragments than plasma cfDNA with a broad size distribution peaking between 200 and 300 bp, and a median concentration of 4 ng/µL in UBC patients (unpublished data). Two studies have directly compared the utility of urinary cell pellet DNA and urinary cfDNA for detecting bladder cancer using microsatellite analysis or copy number changes (6,17). Both found a higher proportion of tDNA relative to non-tumour DNA in the supernatant than in the cell pellet. This raises the fascinating proposition that the various biomarker assays that have previously worked well on cell pellet DNA might work even better on supernatant cfDNA. Furthermore, the size-based capture and enrichment of tumour cells may also improve the identification and analysis of urinary tDNA (18), and the presence of circulating tumour cells have prognostic value (19).
In 2016, Birkenkamp-Demtröder et al. used NGS to detect chromosome breakpoints in the bladder tumours of 12 patients (20). They subsequently designed ddPCR assays targeting these variants and assayed cfDNA isolated from longitudinally collected urine and plasma samples. The somatic variants were detectable in both the plasma and urine of patients, even in non-muscle-invasive bladder cancer (NMIBC) patients. The levels of tDNA dropped in patients post-treatment and high levels were predictive of progression.
The same group have now expanded on the urine and plasma cfDNA work with a ddPCR study of common FGFR3 and PIK3CA mutations (21). Theoretically, these markers should be applicable for a large proportion of bladder cancer patients (especially those with low-grade NMIBC), although additional “personalised” markers based on tumour sequencing would have to be used for the remainder of patients [or, alternatively, a broader panel (8)]. Initially, Christensen et al. tested over 800 formalin-fixed paraffin-embedded (FFPE) tumour tissues for the S249C and Y373C FGFR3 and E545K PIK3CA mutations, with 172 tumours testing positive (36% of NMIBCs and 11% of patients undergoing cystectomy); 54 of these patients were selected for subsequent urine and plasma cfDNA analysis, with selection based upon the availability of “liquid biopsies” and disease course characteristics.
ddPCR data provide an absolute determination of the number of copies of mutant DNA per volume of body fluid. The absolute levels of mutant cfDNA in the urine of NMIBC patients were shown to be positively correlated with tumour stage, grade and size, although the association with European Organisation for Research and Treatment of Cancer (EORTC) risk category did not reach statistical significance (22). In the NMIBC patient group (n=25), tDNA was not detected in plasma but a high initial level of urinary supernatant tDNA indicated future disease progression (P=0.036). In the cystectomy patient group (n=27), high urinary supernatant tDNA also appears to be a predictor of future recurrence (P=0.031); this effect is more pronounced with plasma tDNA/ctDNA (P<0.001).
This is interesting research, although a number of aspects require clarification and further study. The small number of patients whose liquid biopsies were ultimately analysed were enrolled from 1992 to 2012, and so there is understandably a spectrum of treatment approaches over time. Intravesical Bacillus Calmette-Guérin (BCG) appears to have been utilised sparingly in those patients with T1 NMIBC, and cystectomy appears to have been applied inconsistently to those NMIBC patients who progressed to MIBC (3); furthermore, no patient undergoing cystectomy received neoadjuvant chemotherapy (23). All of these treatment factors would have a significant influence on outcomes and, potentially, the persistence or re-emergence of tDNA, or the ability for mutations to be detected by the assay. And despite the urgent need for better prognostic biomarkers for HR-NMIBC patients, very few initial stage T1 patients were analysed (n=5). Furthermore, as the authors highlight, urinary supernatant tDNA may originate from renal clearance of tDNA in the circulation, and so the presence of the E545K PIK3CA mutation may not be bladder cancer specific (24).
In addition, the concept of “anticipatory” or “pre-emptive” diagnosis is a well-known pitfall in the field of non-invasive UBC diagnosis, whereby test specificity can be adversely affected by either the “field effect” and persistence of tumour-related abnormalities in the urine after clearance of disease (9), or by recurrent or residual disease that is not visible by white light cystoscopy (25-29). These phenomena also have implications for prognostication. In this study the majority of NMIBC patients whose samples were selected for plasma and urinary tDNA analysis experienced recurrence or progression [Figure 3 in the paper (21)]: only 2 of the 25 (8%) patients experienced sustained event-free survival at a median follow-up of 59 months, and neither had persistence of plasma or urinary tDNA after the tumour episode. By not studying more patients with such a prolonged event-free survival, the authors have perhaps missed an opportunity to demonstrate the frequency of urinary tDNA persistence in patients whose bladders have seemingly been successfully treated. The selected NMIBC patients also experienced a very high rate of residual disease: almost two-thirds of T1 episodes were seemingly followed by recurrence or progression at the next cystoscopy episode after treatment. Therefore, although the data show that the presence of urinary tDNA in NMIBC patients is significantly associated with progression of disease, it is equally likely to be indicative of inadequate treatment by resection and/or intravesical therapy. A contemporary cohort where patients receive current guideline-directed treatment may reveal a lower frequency of urinary tDNA post-treatment and during surveillance, but the relationship with subsequent progression may be more robust. And a similar phenomenon might be observed in patients treated with neoadjuvant chemotherapy prior to cystectomy.
In the future, utilising genome-wide or panel-based approaches for the analysis of plasma or urine tDNA may permit the near real-time monitoring of tumour evolution during intravesical therapy, neoadjuvant chemotherapy or chemoradiotherapy (6,30), possibly permitting adjustments to therapeutic approaches (Figure 1).
Figure 1.
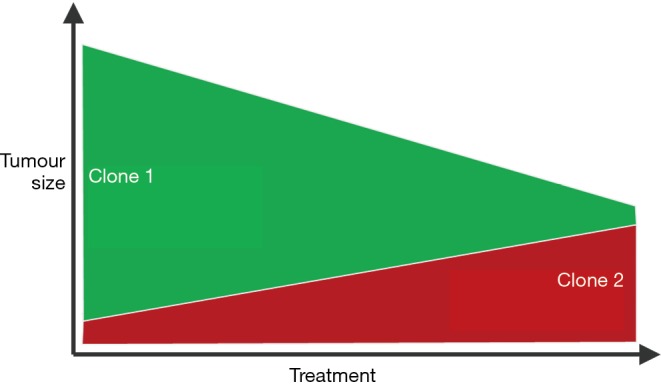
Illustration of how a therapy may eradicate the predominant clone within a tumour (clone 1, green), accompanied by tumour shrinkage, yet in parallel a potentially more aggressive treatment-resistant clone thrives (clone 2, red). It is feasible that genome-wide or panel-based approaches to the analysis of urinary or plasma tDNA during treatment may demonstrate such phenomena and could be performed regularly, whereas one would not invasively biopsy a bladder tumour on a regular basis to identify the same phenomena. A liquid biopsy may also capture disease heterogeneity better than a solid biopsy which only samples a single part of a single tumour. Clearly, in vivo, the scenario is far more complex with multiple clones within the same tumour vying for survival, and so this illustration is a gross oversimplification.
However, despite its limitations, the authors of this study are to be commended on advancing the non-invasive characterisation of bladder cancer. Whilst it is a proof-of-principle study analysing a small number of patients and their liquid biopsies, it clearly points to a bright future for cfDNA-based tests in improving the management of bladder cancer patients (6). The bladder cancer community should now focus on selecting the best biomarkers, optimising assay sensitivity, and large-scale validation of these novel tests.
Acknowledgements
The authors’ work cited in the manuscript is supported by a philanthropic donation to the University of Birmingham in support of bladder cancer research.
Provenance: This is a Guest Editorial commissioned by Editorial Board Member Dr. Xiongbing Zu, MD, PhD (Department of Urology, Xiangya Hospital, Central South University, Changsha, China).
Conflicts of Interest: RT Bryan has contributed to advisory boards and has received honoraria for teaching from Olympus Medical Systems on the topic of narrow band imaging (NBI) cystoscopy; the other author has no conflicts of interest to declare.
References
- 1.Svatek RS, Hollenbeck BK, Holmäng S, et al. The economics of bladder cancer: costs and considerations of caring for this disease. Eur Urol 2014;66:253-62. 10.1016/j.eururo.2014.01.006 [DOI] [PubMed] [Google Scholar]
- 2.D'Costa JJ, Goldsmith JC, Wilson JS, et al. A Systematic Review of the Diagnostic and Prognostic Value of Urinary Protein Biomarkers in Urothelial Bladder Cancer. Bladder Cancer 2016;2:301-17. 10.3233/BLC-160054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babjuk M, Böhle A, Burger M, et al. EAU Guidelines on Non-Muscle-invasive Urothelial Carcinoma of the Bladder: Update 2016. Eur Urol 2017;71:447-61. 10.1016/j.eururo.2016.05.041 [DOI] [PubMed] [Google Scholar]
- 4.Thomas F, Noon AP, Rubin N, et al. Comparative outcomes of primary, recurrent, and progressive high-risk non-muscle-invasive bladder cancer. Eur Urol 2013;63:145-54. 10.1016/j.eururo.2012.08.064 [DOI] [PubMed] [Google Scholar]
- 5.Schmitz-Dräger BJ, Droller M, Lokeshwar VB, et al. Molecular markers for bladder cancer screening, early diagnosis, and surveillance: the WHO/ICUD consensus. Urol Int 2015;94:1-24. 10.1159/000369357 [DOI] [PubMed] [Google Scholar]
- 6.Togneri FS, Ward DG, Foster JM, et al. Genomic complexity of urothelial bladder cancer revealed in urinary cfDNA. Eur J Hum Genet. 2016;24:1167-74. 10.1038/ejhg.2015.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feber A, Dhami P, Dong L, et al. UroMark-a urinary biomarker assay for the detection of bladder cancer. Clin Epigenetics 2017;9:8. 10.1186/s13148-016-0303-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ward DG, Baxter L, Gordon NS, et al. Multiplex PCR and Next Generation Sequencing for the Non-Invasive Detection of Bladder Cancer. PLoS One 2016;11:e0149756. 10.1371/journal.pone.0149756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer 2015;15:25-41. 10.1038/nrc3817 [DOI] [PubMed] [Google Scholar]
- 10.Larré S, Camparo P, Comperat E, et al. Diagnostic, staging, and grading of urothelial carcinomas from urine: performance of BCA-1, a mini-array comparative genomic hybridisation-based test. Eur Urol 2011;59:250-7. 10.1016/j.eururo.2010.10.007 [DOI] [PubMed] [Google Scholar]
- 11.van Tilborg AA, Kompier LC, Lurkin I, et al. Selection of microsatellite markers for bladder cancer diagnosis without the need for corresponding blood. PLoS One 2012;7:e43345. 10.1371/journal.pone.0043345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurst CD, Platt FM, Knowles MA. Comprehensive mutation analysis of the TERT promoter in bladder cancer and detection of mutations in voided urine. Eur Urol 2014;65:367-9. 10.1016/j.eururo.2013.08.057 [DOI] [PubMed] [Google Scholar]
- 13.Millholland JM, Li S, Fernandez CA, et al. Detection of low frequency FGFR3 mutations in the urine of bladder cancer patients using next-generation deep sequencing. Res Rep Urol 2012;4:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czerniak B, Dinney C, McConkey D. Origins of Bladder Cancer. Annu Rev Pathol 2016;11:149-74. 10.1146/annurev-pathol-012513-104703 [DOI] [PubMed] [Google Scholar]
- 15.Ståhlberg A, Krzyzanowski PM, Jackson JB, et al. Simple, multiplexed, PCR-based barcoding of DNA enables sensitive mutation detection in liquid biopsies using sequencing. Nucleic Acids Res 2016;44:e105. 10.1093/nar/gkw224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinde I, Munari E, Faraj SF, et al. TERT promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in urine. Cancer Res 2013;73:7162-7. 10.1158/0008-5472.CAN-13-2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szarvas T, Kovalszky I, Bedi K, et al. Deletion analysis of tumor and urinary DNA to detect bladder cancer: urine supernatant versus urine sediment. Oncol Rep 2007;18:405-9. [PubMed] [Google Scholar]
- 18.Andersson E, Steven K, Guldberg P. Size-based enrichment of exfoliated tumor cells in urine increases the sensitivity for DNA-based detection of bladder cancer. PLoS One 2014;9:e94023. 10.1371/journal.pone.0094023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gazzaniga P, Gradilone A, de Berardinis E, et al. Prognostic value of circulating tumor cells in nonmuscle invasive bladder cancer: a CellSearch analysis. Ann Oncol 2012;23:2352-6. 10.1093/annonc/mdr619 [DOI] [PubMed] [Google Scholar]
- 20.Birkenkamp-Demtröder K, Nordentoft I, Christensen E, et al. Genomic Alterations in Liquid Biopsies from Patients with Bladder Cancer. Eur Urol 2016;70:75-82. 10.1016/j.eururo.2016.01.007 [DOI] [PubMed] [Google Scholar]
- 21.Christensen E, Birkenkamp-Demtröder K, Nordentoft I, et al. Liquid Biopsy Analysis of FGFR3 and PIK3CA Hotspot Mutations for Disease Surveillance in Bladder Cancer. Eur Urol 2017;71:961-9. 10.1016/j.eururo.2016.12.016 [DOI] [PubMed] [Google Scholar]
- 22.Sylvester RJ, van der Meijden AP, Oosterlinck W, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol 2006;49:466-5; discussion 475-7. [DOI] [PubMed]
- 23.Witjes JA, Compérat E, Cowan NC, et al. EAU guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2013 guidelines. Eur Urol 2014;65:778-92. 10.1016/j.eururo.2013.11.046 [DOI] [PubMed] [Google Scholar]
- 24.McGranahan N, Favero F, de Bruin EC, et al. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med 2015;7:283ra54. 10.1126/scitranslmed.aaa1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuiverloon TC, Beukers W, van der Keur KA, et al. Combinations of urinary biomarkers for surveillance of patients with incident nonmuscle invasive bladder cancer: the European FP7 UROMOL project. J Urol 2013;189:1945-51. 10.1016/j.juro.2012.11.115 [DOI] [PubMed] [Google Scholar]
- 26.Allory Y, Beukers W, Sagrera A, et al. Telomerase reverse transcriptase promoter mutations in bladder cancer: high frequency across stages, detection in urine, and lack of association with outcome. Eur Urol 2014;65:360-6. 10.1016/j.eururo.2013.08.052 [DOI] [PubMed] [Google Scholar]
- 27.Yoder BJ, Skacel M, Hedgepeth R, et al. Reflex UroVysion testing of bladder cancer surveillance patients with equivocal or negative urine cytology: a prospective study with focus on the natural history of anticipatory positive findings. Am J Clin Pathol 2007;127:295-301. 10.1309/ADJL7E810U1H42BJ [DOI] [PubMed] [Google Scholar]
- 28.Bryan RT, Collins SI, Daykin MC, et al. Mechanisms of recurrence of Ta/T1 bladder cancer. Ann R Coll Surg Engl 2010;92:519-24. 10.1308/003588410X12664192076935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bryan RT, Shah ZH, Collins SI, et al. Narrow-band imaging flexible cystoscopy: a new user's experience. J Endourol 2010;24:1339-43. 10.1089/end.2009.0598 [DOI] [PubMed] [Google Scholar]
- 30.Murtaza M, Dawson SJ, Pogrebniak K, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 2015;6:8760. 10.1038/ncomms9760 [DOI] [PMC free article] [PubMed] [Google Scholar]


