Significance
The Hippo pathway restricts cell proliferation and plays key roles in organ size control and tissue homeostasis. The crucial step of this pathway is the regulation of the transcriptional coactivators YAP/TAZ by LATS1/2 kinases. We report in the present study that deubiquitinase YOD1 acts as a positive regulator of YAP/TAZ in controlling organ size. The significance of our study is the discovery of a regulatory mechanism for the Hippo pathway: high cell density miR-21−YOD1−ITCH−LATS signaling cascade, which is parallel to a previously known high cell density MST1/2−LATS signaling cascade. Data from harnessing a mouse model that allowed inducible human YOD1 expression in mouse liver and liver cancer patients suggest mechanistic insights to expand our understanding about regulation of the Hippo pathway.
Keywords: Hippo signaling, deubiquitinase, YOD1, ITCH, cell density
Abstract
Hippo signaling controls the expression of genes regulating cell proliferation and survival and organ size. The regulation of core components in the Hippo pathway by phosphorylation has been extensively investigated, but the roles of ubiquitination−deubiquitination processes are largely unknown. To identify deubiquitinase(s) that regulates Hippo signaling, we performed unbiased siRNA screening and found that YOD1 controls biological responses mediated by YAP/TAZ. Mechanistically, YOD1 deubiquitinates ITCH, an E3 ligase of LATS, and enhances the stability of ITCH, which leads to reduced levels of LATS and a subsequent increase in the YAP/TAZ level. Furthermore, we show that the miR-21-mediated regulation of YOD1 is responsible for the cell-density-dependent changes in YAP/TAZ levels. Using a transgenic mouse model, we demonstrate that the inducible expression of YOD1 enhances the proliferation of hepatocytes and leads to hepatomegaly in a YAP/TAZ-activity-dependent manner. Moreover, we find a strong correlation between YOD1 and YAP expression in liver cancer patients. Overall, our data strongly suggest that YOD1 is a regulator of the Hippo pathway and would be a therapeutic target to treat liver cancer.
The Hippo pathway plays an essential role in animal development and tissue homeostasis. The Hippo pathway can be initiated by changes in cell density, G protein-coupled receptor signaling, or mechanical stimuli, thereby activating a kinase cascade consisting of MST1/2 and LATS1/2 to maintain precise organ size (1–3). As a result, the dysregulation of the Hippo pathway may result in tumor formation and tissue degeneration. The key step of this pathway is the regulation of the transcriptional coactivators YAP/TAZ by LATS1/2 kinases. LATS1/2 phosphorylates YAP/TAZ to induce its cytoplasmic retention and subsequent β-TrCP-mediated proteasomal degradation. Inactivation of the Hippo pathway by genetic mutation or cytoskeletal rearrangement inhibits YAP/TAZ phosphorylation and allows YAP/TAZ to enter the nucleus, where it activates genes involved in cell proliferation. These findings show that regulation of the Hippo pathway is achieved at various levels via posttranslational modifications such as phosphorylation and ubiquitination.
The ubiquitin-mediated proteolytic pathway plays an essential role in controlling the abundance of several proteins and in maintaining normal cellular functions. ITCH has emerged as an E3 ubiquitin ligase that promotes the degradation of LATS kinases. Although the destabilization of LATS1 by ITCH leads to a significant increase in YAP/TAZ activation and is associated with tumorigenic traits (4, 5), the upstream signaling and biological process that regulates ITCH has not been investigated. The regulation of substrates by ubiquitination can be tightly reversed by deubiquitinases (DUBs), which can remove ubiquitin from the substrates (6). Notably, several studies have suggested that the DUB-E3 ligase complex modulates cellular protein dynamics by mediating E3 ligase stability and activity (7–9).
Here, we report that the DUB YOD1 is a regulator of the Hippo pathway. YOD1 removes ubiquitin from ITCH and promotes its stabilization, inducing LATS degradation and YAP/TAZ activation. The YOD1−ITCH−YAP/TAZ signaling axis is controlled by the differential expression of miR-21 in a cell-density-dependent manner. We show that the inducible expression of YOD1 in the liver enhances the proliferation of hepatocytes and leads to hepatomegaly in a YAP/TAZ-activity-dependent manner, suggesting that YOD1 is an intrinsic regulator of the Hippo pathway in vivo. Furthermore, our clinical findings that YOD1 expression is high and that it shows a strong positive correlation with YAP in liver cancer patients suggest that YOD1 would be a therapeutic target.
Results
DUB YOD1 Is Required for YAP/TAZ Transcriptional Activity.
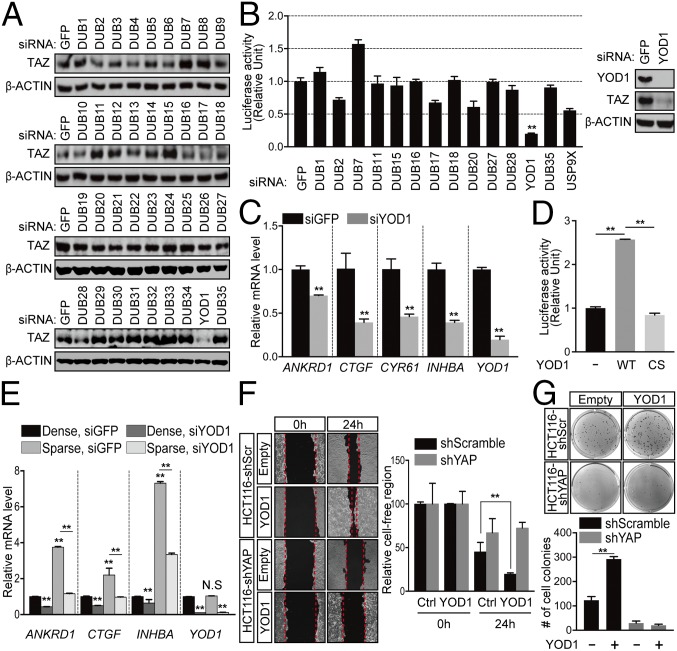
To investigate the involvement of DUBs in the Hippo pathway, we selected 36 representative DUBs and conducted unbiased siRNA screening by monitoring the levels of TAZ, a terminal modulator of the Hippo pathway. Among those tested, we focused on YOD1 (also known as OTUD2 or OTU1) because its siRNA-mediated depletion markedly reduced TAZ abundance (Fig. 1A) (10). Consistent with Fig. 1A, the knockdown of YOD1 reduced YAP/TAZ reporter activity and the expression of its target genes, including ANKRD1, CTGF, CYR61, and INHBA (Fig. 1 B and C). The knockdown of YOD1 reduced YAP/TAZ reporter activity to a greater degree than was caused by the knockdown of USP9X (Fig. 1B), which was used as a positive control in our screening (11). The overexpression of YOD1, but not the catalytic mutant version (YOD1-C160S), was sufficient to induce YAP/TAZ reporter activity, indicating that deubiquitinating enzyme activity is required for YAP/TAZ transcriptional activity (Fig. 1D). The subcellular localization and transcriptional activity of YAP/TAZ are regulated by cell density (2). In accordance with previous reports, the expression of YAP/TAZ target genes was found to be higher in sparsely cultured cells than the expression levels in densely cultured cells (Fig. 1E). However, the knockdown of YOD1 in sparsely cultured cells significantly reduced expression of YAP/TAZ target genes similar to the expression in densely cultured cells, which suggests that YOD1 controls YAP/TAZ transcriptional activity (Fig. 1E).
Fig. 1.
DUB siRNA screening identifies YOD1 as a potent activator of YAP/TAZ. (A) The level of TAZ in HEK293T cells transfected with siRNAs for different DUBs. (B) HEK293T cells were cotransfected with the siRNA indicated in the figure, pRL-TK, and 8xGTIIC luciferase reporter constructs. One day after transfection, luciferase activity was measured and normalized relative to Renilla activity. Endogenous YOD1 and TAZ protein levels were measured via Western blotting. (C) The levels of the mRNAs indicated in the figure were analyzed via reverse transcription real-time PCR (RT-qPCR), and the results were normalized relative to β-actin mRNA. (D) HEK293T cells were transfected with empty vectors, Flag-YOD1-WT, or Flag-YOD1-CS, and 8xGTIIC luciferase activity was measured. (E) HEK293 cells were transfected with siRNAs for 24 h and then replated in either low- or high-density conditions. At 48 h after siRNA transfection, the cells were harvested, and the levels of mRNAs indicated in the figure were analyzed via RT-qPCR. (F) (Left) HCT116 cells expressing shScramble or shYAP were transfected with the plasmids described in the figure, and the wounded areas were measured at 0 and 24 h using the TScratch program. (Magnification: 10×.) (Right) Quantification of unfilled areas. (G) (Top) Representative images from colony forming assays using cells described in F. Colony forming assay plates were taken without magnification using color camera. (Bottom) Quantification of the number of colonies. The statistical results represent average values from a representative experiment performed in triplicate. Error bars indicate SDs of triplicate assays. CS, C160S (YOD1 mutant); Ctrl, control; N.S, not significant; **P < 0.01.
Because our results suggest that YOD1 is required for YAP/TAZ activity, which is linked to the progression of tumors (12, 13), we examined whether YOD1 has oncogenic properties. The overexpression or depletion of YOD1 resulted in cells migrating faster or slower, respectively, than control cells, as measured by wound closure in wound-healing assays (Fig. 1F and Fig. S1A). The enhanced migration resulting from the overexpression of YOD1 depends on the presence of YAP, as shown by the fact that it was abolished by a concomitant loss of YAP (Fig. 1F). To further demonstrate the oncogenic function of YOD1, we performed an anchorage-independent growth assay. Here, YOD1 significantly enhanced colony formation in an enzymatic-activity-dependent manner (Fig. S1B). More importantly, colony formation induced by YOD1 expression was completely abolished when YAP was depleted, indicating that YOD1 exerts oncogenic activity through YAP (Fig. 1G). Taken together, these data suggest that YOD1 is a regulator of the Hippo pathway that has an oncogenic property through YAP/TAZ.
Fig. S1.
(Related to Fig. 1.) YOD1 has oncogenic properties. (A) (Left) HEK293 cells were transfected with siGFP or siYOD1, and the wounded areas were measured at 0 and 14 h using the TScratch program. (Magnification: 10×.) (Right) Values for unfilled areas. (B) (Left) Representative images of colony-forming assay results from HEK293 cells transfected with empty vectors (−), wild-type YOD1 (YOD1), and catalytic mutant YOD1 (YOD1-C160S). Colony forming assay plates were taken without magnification using color camera. (Right) The number of colonies. The statistical results represent average values from a representative experiment performed in triplicate. Error bars indicate SDs of triplicate assays. **P < 0.01. CS, C160S (YOD1 mutant); N.S, not significant.
YOD1 Promotes LATS Kinase Degradation Through ITCH.
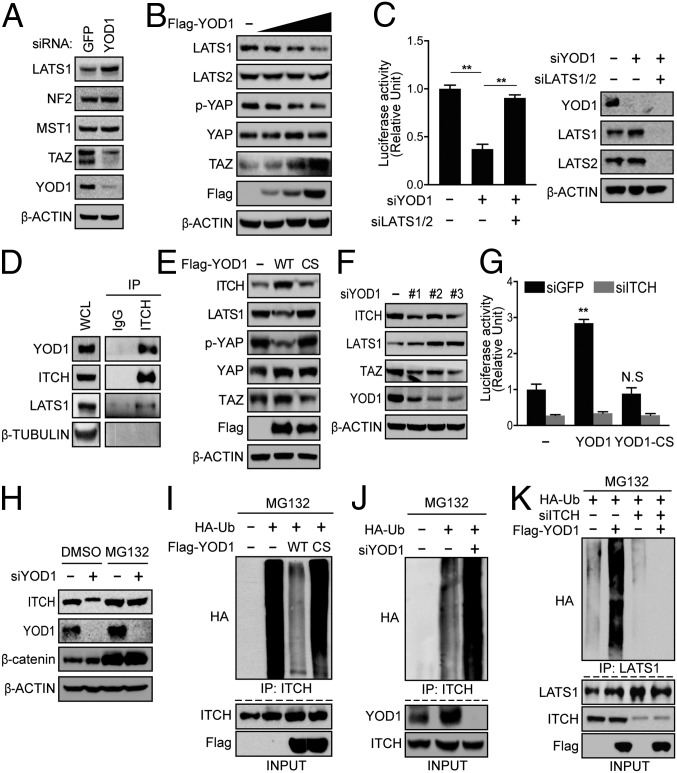
To identify the molecular target of YOD1, we first examined the levels of Hippo pathway components, such as NF2, MST1, and LATS1 in HEK293T cells and found that the loss or overexpression of YOD1 specifically increased or decreased LATS1 kinase, respectively, without altering the levels of the other components (Fig. 2 A and B). Knockdown of YOD1 in the human hepatocyte Huh7 cells resulted in a similar pattern, with a notable difference of significantly increased levels of LATS2 in addition to LATS 1 (Fig. S2A). Inducible expression of YOD1 in mice liver also showed down-regulation of both LATS1 and LATS2 (Fig. S4D), which suggests that the regulation of LATS2 by YOD1 is context-dependent. The finding that the reduction of reporter activity caused by YOD1 depletion was fully rescued by LATS1/2 knockdown further supports the possibility that YOD1 might potentiate YAP/TAZ transcriptional activity through LATS (Fig. 2C). Because LATS kinase is known to directly phosphorylate and induce the β-TrCP-mediated degradation of YAP/TAZ in a ubiquitin-dependent manner, we initially speculated that LATS would be a substrate of YOD1. However, contrary to our expectation, we did not observe an interaction between YOD1 and LATS1 (Fig. S2B). Because YOD1 is a DUB, we looked for ubiquitin E3 ligase(s) that is known to destabilize LATS kinase. Of the known E3 ligases, including ITCH, NEDD4, SIAH2, and SMURFs, we found that YOD1 interacted with only ITCH (Fig. 2D) and increased its abundance (Fig. S2C) (4, 5, 14–16). YOD1 overexpression was sufficient for the induction of increase in ITCH levels and subsequent decreases in LATS1 levels in an enzymatic-activity-dependent manner (Fig. 2E). Conversely, the knockdown of YOD1 by three siRNAs targeting different regions of the mRNA of YOD1 showed opposite effects on ITCH and LATS levels (Fig. 2F). Among the 36 DUBs tested, we observed that ITCH levels were strongly down-regulated when only YOD1 was depleted (Fig. S2D). More importantly, the up-regulation of YAP/TAZ transcriptional activity caused by YOD1 overexpression was completely abolished by the depletion of ITCH (Fig. 2G). The loss of YOD1 did not further reduce reporter activity when ITCH was codepleted in cells (Fig. S2E, Left). The finding that the increase in LATS1 level caused by ITCH loss was not further enhanced by the depletion of YOD1 indicates that ITCH mediates the effect of YOD1 in the regulation of Hippo signaling (Fig. S2E, Right).
Fig. 2.
YOD1 regulates LATS1 stability in an ITCH-dependent manner. (A) Assessment of levels of major Hippo components in HEK293T cells transfected with siRNAs for GFP or YOD1. (B) HEK293T cells were transfected with increasing amounts of YOD1 and analyzed via Western blotting for the indicated proteins. (C) (Left) HEK293T cells were cotransfected with the indicated siRNAs, pRL-TK, and 8xGTIIC luciferase. (Right) A representative Western blot indicating the successful knockdown of proteins by siRNAs. (D) Protein lysates from HEK293T cells were immunoprecipitated with an anti-ITCH antibody and analyzed via immunoblotting for the indicated proteins. WCL, whole-cell lysates. (E) Empty vectors, Flag-YOD1-WT, or Flag-YOD1-CS were expressed in HEK293T cells followed by Western blotting with the indicated antibodies. (F) HEK293T cells were transfected with a scrambled siRNA or three different YOD1 siRNAs targeting different regions. One day after siRNA transfection, the cells were harvested and analyzed via Western blotting for the indicated proteins. (G) HEK293T cells were transfected with either siGFP or siITCH. One day after siRNA transfection, the cells were cotransfected with the indicated plasmids and reporter constructs. Reporter activity was measured 24 h after the latter transfection. (H) HEK293T cells transfected with either siScrambled or siYOD1 were treated with either DMSO (control) or a proteasome inhibitor (MG132). (I) HA-ubiquitin was cotransfected with empty vectors, Flag-YOD1-WT, or Flag-YOD1-CS into HEK293T cells, which were treated with 25 µM MG132 for 4 h before being harvested. Total cell lysates were immunoprecipitated with an anti-ITCH antibody, followed by immunoblotting with an anti-HA antibody. (J) The experiments were performed as described in I except that siRNA targeting YOD1 was used. (K) HEK293T cells were transfected with the indicated plasmids and siRNAs and were treated with 25 µM MG132 for 4 h before being harvested. Total cell lysates were immunoprecipitated with an anti-LATS1 antibody, followed by immunoblotting with an anti-HA antibody. The statistical results represent average values from a representative experiment performed in triplicate. Error bars indicate SDs of triplicate assays. **P < 0.01. CS, C160S (YOD1 mutant); IP, immunoprecipitation.
Fig. S2.
(Related to Fig. 2.) YOD1 regulates LATS stability through ITCH. (A) Huh7 cells were transfected with a scrambled siRNA or three different YOD1 siRNAs targeting different regions. One day after siRNA transfection, the cells were harvested and analyzed via Western blotting for the indicated proteins. (B) HEK293T cells were cultured for 24 h after transfection with the plasmids indicated in the figure, and the cell lysates were immunoprecipitated with an anti-Flag antibody followed by immunoblotting with an anti-GFP antibody. Asterisk indicates IgG heavy chain. (C) HEK293T cells were transfected with the plasmids indicated in the figure, and lysates were analyzed via Western blotting for the indicated proteins. (D) The level of ITCH in HEK293T cells transfected with siRNAs for various DUBs. (E) (Left) HEK293T cells were cotransfected with the siRNAs indicated in the figure and with reporter constructs (pRL-TK and 8xGTIIC luciferase). (Right) A representative Western blot with antibodies for the proteins indicated in the figure. **P < 0.01; error bars indicate the mean ± SD. Student t test was used for statistical analysis. N.S, not significant.
Fig. S4.
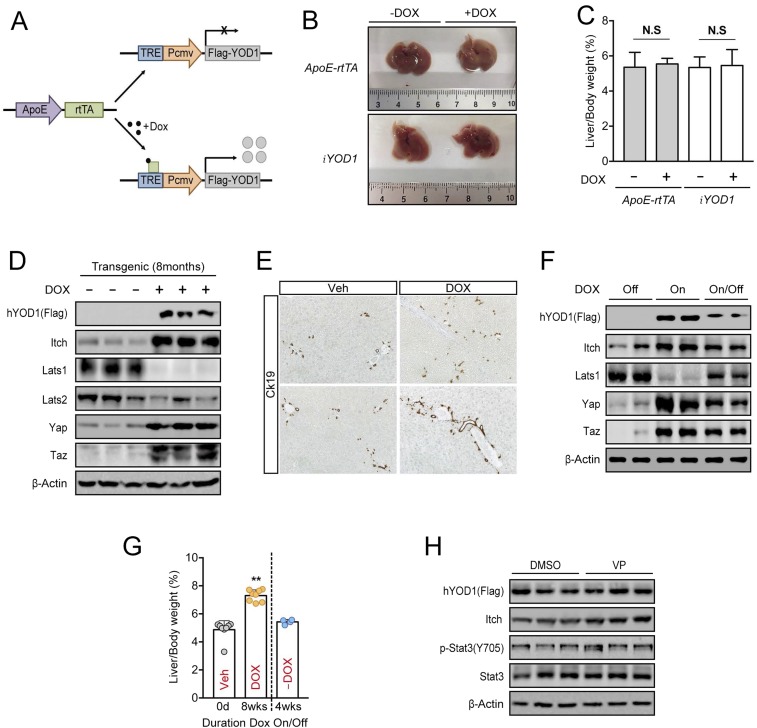
(Related to Fig. 4.) Reversible control of liver size by YOD1. (A) Schematic diagram for the inducible expression of YOD1 in ApoE-rtTA;iYOD1 transgenic mice. (B) Livers from ApoE-rtTA or iYOD1 mice fed without or with Dox for 8 wk starting at 4 wk of age. (C) Quantification of liver-to-body weight ratios for the animals analyzed in B. Values represent the mean ± SD (n ≥ 3). (D) Lysates from the livers of ApoE-rtTA;iYOD1 transgenic mice fed without or with Dox for 8 mo were immunoblotted with the antibodies indicated in the figure. (E) Expression of Ck19 by immunohistochemistry in liver section of ApoE-rtTA;iYOD1 transgenic mice fed without or with Dox for 8 mo at 4 wk of age. All images were taken at 200× magnification. (F) Four-week-old ApoE-rtTA;iYOD1 transgenic mice were fed Dox for 8 wk, followed by a withdrawal of Dox from their drinking water for 4 wk. Lysates from the livers of ApoE-rtTA;iYOD1 transgenic mice were immunoblotted with the antibodies indicated in the figure. (G) Liver and body weights were measured at the times indicated in the figure. **P < 0.01; error bars indicate the mean ± SD (n ≥ 4). (H) Western blot analysis of hYOD1 levels in livers from DMSO- or VP-injected ApoE-rtTA;iYOD1 transgenic mice that were used in Fig. 4F. The lysate for each lane was obtained from the liver of an individual mouse. DOX, doxycycline; N.S, not significant; Veh, vehicle.
The finding that YOD1 regulates ITCH levels prompted us to investigate the underlying molecular mechanism. ITCH is known to undergo autoubiquitination and self-degradation (7), and the down-regulation of ITCH by YOD1 depletion can be fully restored by treatment with the proteasome inhibitor MG132 (Fig. 2H), suggesting that YOD1 may enhance the stability of ITCH through a deubiquitination event. Therefore, we next investigated whether YOD1 promotes ITCH stabilization by deubiquitination of ITCH. To compare the status of ubiquitination on ITCH, we normalized total ITCH levels by blocking ubiquitin/proteasomal degradation of ITCH with MG132 treatment. ITCH was heavily ubiquitinated at the basal level, which was abolished by transfection with YOD1-WT but not YOD1-C160S (Fig. 2I). Conversely, ITCH ubiquitination was strongly increased in YOD1-depleted cells after treatment with MG132 (Fig. 2J). Interestingly, the YOD1-induced increase in LATS1 ubiquitination was impaired by the loss of ITCH, indicating that ITCH mediates YOD1-dependent LATS regulation (Fig. 2K). Overall, our data strongly suggest that YOD1 specifically regulates ITCH-dependent LATS degradation, thereby inhibiting the Hippo signaling pathway.
Cell-Density-Dependent Regulation of the miR-21−YOD1−ITCH Axis.
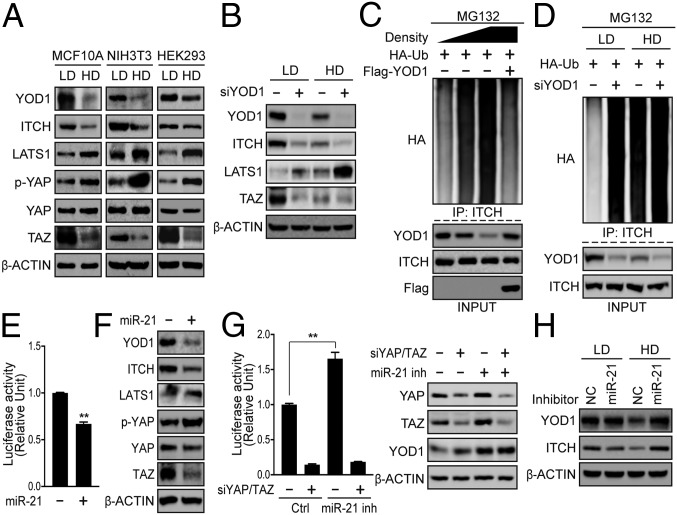
As cell density is a well-known intrinsic factor that regulates the Hippo pathway, and our findings show that YOD1 is required for YAP/TAZ activity, we examined the possibility that YOD1 can be controlled in a cell-density-dependent manner. Interestingly, the level of YOD1 in cells grown at a high density was much lower than in cells grown at a low density (Fig. 3A), whereas the level of YOD1 mRNA was not different between the conditions (Fig. S3A). Along with YOD1 levels, cells grown at a high cell density exhibited reduced levels of ITCH and TAZ (Fig. 3A). The reduced levels of ITCH and TAZ observed at a high cell density were restored by YOD1 overexpression, whereas the LATS1 level was decreased to the basal level (Fig. S3B). Inversely, the knockdown of YOD1 led to a marked decrease in the ITCH level at a low cell density and a further reduction at a high cell density (Fig. 3B). Next, we examined whether the decrease of YOD1 at a high cell density leads to an increase in ITCH ubiquitination. As shown in Fig. 3C, the ITCH ubiquitination status was increased along with the gradual reduction of YOD1 at a high cell density, and the ubiquitination of ITCH was fully restored following overexpression of YOD1 to endogenous levels. In addition, the depletion of YOD1 led to a dramatic increase in ITCH ubiquitination at a low cell density, whereas it did not further enhance ITCH ubiquitination at a high cell density (Fig. 3D). These results suggest that ITCH-mediated LATS1 ubiquitination and degradation is regulated by YOD1 in a cell-density-dependent manner.
Fig. 3.
Cell-density-dependent regulation of miR-21 is responsible for the down-regulation of YOD1. (A) MCF10A, NIH 3T3, and HEK293 cells were cultured at low and high cell densities, and cell extracts were analyzed via Western blotting for the indicated proteins. (B) HEK293 cells were transfected with siYOD1 for 24 h and then reseeded under either low or high cell-density conditions. At 48 h after siRNA transfection, the cells were harvested and analyzed via Western blotting for the proteins indicated in the figure. (C) HEK293 cells transfected with the indicated plasmids were cultured at low, middle, and high densities and then treated with MG132 for 4 h before being harvested, and the ubiquitination of ITCH was examined. (D) HEK293 cells were cotransfected with HA-Ubiquitin and siYOD1. One day after transfection, the cells were reseeded under either low or high cell-density conditions, and the ubiquitination of ITCH was examined after 1 d. (E) HEK293T cells were cotransfected with miR-21 mimics and reporter constructs. One day after transfection, luciferase activity was measured. (F) HEK293T cells were transfected with miR-21 mimics. After 24 h, the cell lysates were analyzed via Western blotting for the indicated proteins. (G) HEK293T cells were cotransfected with the indicated siRNAs, pRL-TK, and 8xGTIIC luciferase. (Left) One day after transfection, cells were treated with negative control locked nucleic acid (LNA) oligonucleotides or an miR-21 inhibitor for 48 h (50 nM). (Right) A representative Western blot indicating the efficiency of the siRNA and miR-21 inhibitor. (H) HEK293 cells cultured at low and high cell densities were treated with negative control LNA or a miR-21 inhibitor for 48 h (50 nM). Total cell lysates were analyzed via Western blotting for the indicated proteins. NC, negative control. The statistical results represent average values from a representative experiment performed in triplicate. **P < 0.01; error bars indicate the mean ± SD. Student t test was used for statistical analysis. Ctrl, control; HD, high density; IP, immunoprecipitation; LD, low density.
Fig. S3.
(Related to Fig. 3.) miR-21-YOD1-ITCH axis is regulated in a cell-density-dependent manner. (A) MCF10A, NIH 3T3, and HEK293 cells were cultured at low and high cell densities, and the levels of CTGF and YOD1 mRNAs were analyzed via RT-qPCR. (B) HEK293 cells were transfected with empty vectors or Flag-YOD1 for 24 h and then reseeded under either low or high cell-density conditions. At 48 h after plasmid transfection, the cells were harvested and analyzed via Western blotting for the proteins indicated in the figure. (C) HEK293T cells were transfected with the siRNAs indicated in the figure, and lysates were analyzed via Western blotting. (D) The location of the target sites of miR-21, 125a, and 30b in the YOD1 3′-UTR. The target sites were predicted using the TargetScan program. (E) The relative levels of mature miR-21 were quantitated in low versus high density of HEK293, HeLa, NIH 3T3, and MCF10A cells. The results were normalized relative to the small nuclear RNA U6. (F) The miR-21 levels in HEK293T cells were compared after transfection of the miR-21 mimic, inhibitor, and control and were normalized to U6 snRNA. (G) HEK293T cells were transfected with negative control scrambled siRNA (NC), miR-21 mimics, or YOD1 siRNA. The YOD1 mRNA expression levels were detected via RT-qPCR and were normalized to β-actin. (H) HEK293T cells were transfected with (Left) miR-21 mimics or (Right) an miR-21 inhibitor for 48 h (50 nM). The cell lysates were analyzed via Western blotting for the indicated proteins. (I) (Left) HEK293T cells were transfected with the miRNA mimics and analyzed via Western blotting for YOD1. (Right) RT-qPCR was used to analyze mature miRNA expression levels, and the results were normalized relative to the small nuclear RNA U6. (J) The levels of mature miRNAs indicated in the figure were analyzed via RT-qPCR, and the results were normalized relative to U6 snRNA. (K) The relative expression of mature miRNAs was quantitated at low and high densities. (Left) RT-qPCR data normalized to U6 snRNA. (L) HEK293T cells were transfected with the miRNA mimics and analyzed via Western blotting for YOD1. (Right) RT-qPCR was used to analyze mature miRNA expression levels, and the results were normalized relative to the small nuclear RNA U6. The statistical results represent average values from a representative experiment performed in triplicate. Error bars indicate SDs of triplicate assays. **P < 0.01. HD, high density; LD, low density.
To explore the molecular mechanism underlying density-dependent YOD1 regulation, we first examined whether YOD1 could be regulated by the components of the Hippo pathway that are known to sense and mediate cell−cell contact inhibition. However, the knockdown of Hippo core components had no influence on YOD1 levels (Fig. S3C). Previous work has demonstrated that miRNA biogenesis is globally activated by cell−cell contact (17). Here, we focused on microRNA-21 (miR-21) expression because it contains two regions targeting YOD1 and because miR-21 levels were increased in high cell density across various cell lines (Fig. S3 D and E). Furthermore, its role in the regulation of the Hippo pathway has not previously been examined (18, 19). The addition of miR-21 significantly reduced YAP/TAZ transcriptional activity (Fig. 3E and Fig. S3F). Consistent with this, miR-21 expression led to reductions in the levels of YOD1 without changing the level of its mRNA, indicating that miR-21 may not induce YOD1 mRNA degradation, but may inhibit its translation (Fig. 3F and Fig. S3G). Consistent with down-regulation of YOD1 by miR-21, ITCH and TAZ were markedly reduced, whereas LATS1 and YAP phosphorylation on Ser127 was increased (Fig. 3F). We further observed that the blockade of miR21 activity via the addition of a chemically modified miR-21 inhibitor led to an increase in the YAP/TAZ activities (Fig. 3G and Fig. S3F). In addition, treatment with the miR-21 inhibitor blocked the reduction of YOD1 levels at a high cell density (Fig. 3H). Upon examining the levels of upstream components of LATS, such as NF2 and MST1, we found that the addition or blockade of miR-21 did not alter the levels of these components (Fig. S3H). Taken together, these results suggest that the endogenous YOD1−ITCH−LATS−YAP/TAZ axis is regulated by the level of miR-21 in a cell-density-dependent manner.
In addition to miR-21, miR-125a and miR-30b contain predicted sequences targeting YOD1 and markedly reduced levels of YOD1 when overexpressed (Fig. S3 D and I). However, unlike miR-21, miR-125a and miR-30b expression was not increased in high cell density (Fig. S3J). Furthermore, we examined other miRNAs increased in high cell density, such as miR-15a and miR-29b (17). In accordance with previous reports, miR-15a and miR-29b were highly expressed in high cell density but did not reduce YOD1 levels (Fig. S3 K and L). Taken together, our data strongly suggest that miR-21 is the only miRNA that can respond to cell density and regulate the Hippo pathway by controlling the level of YOD1.
Inducible Expression of YOD1 in the Liver Leads to Hepatomegaly.
Because our findings show that YOD1 is required for YAP/TAZ activity (Fig. 1) and Hippo signaling is known to play essential roles in the control of mammalian organ size, we examined the physiological role of YOD1 in the regulation of organ size. To this end, we chose mouse liver as our in vivo model organ because our results suggested that YOD1 is a potential activator of YAP/TAZ and because it has been shown that inducible expression of YAP in liver leads to hepatomegaly (13). We generated transgenic mice carrying human YOD1 cDNA (hYOD1), the expression of which is controlled by a tetracycline (Tet)-responsive element fused with the minimal CMV promoter, and a reverse tetracycline transactivator (rtTA) under the control of the liver-specific ApoE promoter (Fig. S4A). These transgenic mice (hereafter referred to as ApoE-rtTA:iYOD1) allowed for the doxycycline (a tetracycline derivative)-dependent expression of hYOD1 in mouse livers (Fig. 4A). Consistent with results observed in cell culture experiments, we found that inducing the expression of hYOD1 via Dox intake for 8 wk resulted in marked increases in ITCH, YAP, and TAZ levels and a reduction in LATS1 levels (Fig. 4A), thereby up-regulating the expression of YAP/TAZ target genes, including Acta2, Ctgf, and Cyr61 (Fig. 4B). Oval cells are known as liver progenitor/stem cells, and they express EpCAM and Sox9 (20, 21). An increase in YAP in Mst1/Mst2-deleted mice caused a marked expansion of oval cells (22, 23). We also observed that the expression of the oval cell markers EpCAM and Sox9 was dramatically increased in the livers of Dox-treated mice (Fig. 4B). In agreement with these molecular changes, Dox-treated ApoE-rtTA:iYOD1 mice displayed hepatomegaly compared with the mice fed a normal diet (Fig. 4 C and D), whereas the Dox-treated ApoE-rtTA or iYOD1 mice had no changes in liver size (Fig. S4 B and C). The hepatomegaly of Dox-treated mice is in accordance with the observed up-regulation of hepatocyte proliferation, as determined by Ki-67 staining (Fig. 4E). Cytokeratin (CK) 19 is specifically expressed in proliferating oval cells. The liver of Dox-treated ApoE-rtTA:iYOD1 mice exhibited an increased number of cells positive for CK19 expression around the portal vein (Fig. S4E). These results indicate that hepatic overexpression of YOD1 results in proliferation and expansion of oval cells. We next questioned whether YOD1-induced hepatomegaly was dependent on continuous YOD1 expression. To this end, we stopped administering Dox in the drinking water after the initial 8 wk of Dox feeding. The expression levels of hYOD1, ITCH, YAP, and TAZ were reduced, and the enlarged livers returned to almost their normal size 4 wk after Dox withdrawal (Fig. S4 F and G). These data suggest that YOD1 is sufficient to induce liver overgrowth and it is reversible.
Fig. 4.
YOD1 expression leads to hepatomegaly in mice and is correlated with YAP level in the tumor tissues of liver cancer patients. (A) Lysates from the livers of nontransgenic and ApoE-rtTA;iYOD1 transgenic mice fed without or with Dox for 8 wk were immunoblotted with the antibodies indicated in the figure. (B) Total RNAs from the livers of ApoE-rtTA;iYOD1 transgenic mice fed without or with Dox for 8 wk were isolated, and RT-qPCR analyses for the indicated genes were performed. Data are presented as mean ± SD; **P < 0.01. Student t test was used for statistical analysis. (C) Livers from ApoE-rtTA;iYOD1 transgenic mice fed without or with Dox for 8 mo, starting at 4 wk of age. (D) Liver and body weights were measured at the times indicated in the figure. **P < 0.01, error bars indicate the mean ± SD (n ≥ 5). (E) Histogram shows the relative number of Ki67-positive hepatocytes in the liver sections from ApoE-rtTA;iYOD1 transgenic mice fed without or with Dox for 8 mo. Ten fields were chosen randomly for analysis in liver sections from each animal. Data are presented as mean ± SD (n = 8). **P < 0.01. Student t test was used for statistical analysis. (F) (Left) Representative pictures of livers from control (DMSO) and VP-injected ApoE-rtTA;iYOD1 transgenic mice fed Dox for 8 wk and (Right) values for liver-to-body weight ratios at the end of the 8-wk period. **P < 0.01; values represent the mean ± SD (n ≥ 7). (G) RT-qPCR analysis of the expression of the indicated genes in the livers described in F. **P < 0.01; error bars indicate the mean ± SD. (H) (Left) Histogram shows the relative number of Ki67-positive hepatocytes in the liver sections from DMSO- or VP-injected ApoE-rtTA;iYOD1 transgenic mice. Ten fields were chosen randomly for analysis in liver sections from each animal. (Right) Representative Ki67 immunohistochemistry staining of liver sections from each group. **P < 0.01; error bars indicate the mean ± SD (n = 5). (I) RT-qPCR analysis of the expression of EpCAM and Sox9 in livers from DMSO- or VP-injected ApoE-rtTA;iYOD1 transgenic mice. Data are presented as mean ± SD. **P < 0.01. Student t test was used for statistical analysis. (J) Immunohistochemical (IHC) staining of human liver cancer tissue samples. Images represent the IHC staining of three liver cancer patients stained with YOD1 and YAP. The staining data for (Left) normal tissues adjacent to tumors and (Right) tumor tissues are presented. All images were taken at 400×. Ctrl, control; DOX, doxycycline; Veh, vehicle.
To investigate whether the hepatomegaly caused by the inducible expression of YOD1 depends on YAP/TAZ transcriptional activity, we used the small molecule Verteporfin (VP), which is known to inhibit the YAP−TEAD interaction and YAP-induced liver overgrowth (24). When the ApoE-rtTA:iYOD1 mice were fed Dox for 8 wk, they were also given VP (50 mg/kg) via i.p. injections for the last 4 wk. The liver overgrowth and increased expression of YAP/TAZ target genes caused by the inducible expression of hYOD1 were significantly relieved in mice injected with VP (Fig. 4 F and G). In support of these results, hepatocyte proliferation and the expression of the oval cell markers EpCAM and Sox9 were significantly reduced in the livers of VP-injected mice, without changes in the levels of YOD1 (Fig. 4 H and I and Fig. S4H). VP is also known to inhibit cytokine induced STAT3 activity to reduce the viability of colorectal cancer cells (25). To exclude the possibility that blocking of hepatomegaly by the injection of VP was due to inhibition of STAT signaling rather than inhibition of YAP activity, we examined the level of STAT3 activation (pSTAT3Y705) in lysates of livers that were used in Fig. 4F. The levels of STAT3 and pSTAT3Y705 were not changed by the VP injection (Fig. S4H). Collectively, these results suggest that YOD1 leads to liver overgrowth by regulating YAP/TAZ transcriptional activity.
Our findings harnessing a mice model suggest that YOD1 may act as an oncogene in cancer patients. To examine the clinical relevance of our findings, we examined the expression of YOD1 and YAP in the tissues of 20 hepatocellular carcinoma patients. The levels of YOD1 were higher in tumors than in contiguous normal tissues in 12 out of 20 patients (Table S1, P value = 0.00008). Significantly, higher levels of YOD1 in tumor tissues exhibited a stronger correlation with the levels of YAP (Table S1, P value = 0.00049). Three cases of immunostaining data are presented in Fig. 4J. These results strongly suggest that YOD1 is an intrinsic positive regulator of YAP and works as an oncogene, at least in liver tissue.
Table S1.
Expression of YOD1 and YAP in hepatocellular carcinoma patients
| Patient case number | Normal tissue | Tumor | ||
| YOD1 | YAP | YOD1 | YAP | |
| I | 1 | 1 | 1 | 3 |
| II | 1 | 1 | 1 | 1 |
| III | 1 | 0 | 2 | 1 |
| IV | 1 | 1 | 2 | 2 |
| V | 1 | 0 | 1 | 0 |
| VI | 1 | 1 | 2 | 1 |
| VII | 1 | 0 | 2 | 0 |
| VIII | 1 | 0 | 2 | 3 |
| IX | 1 | 0 | 3 | 2 |
| X | 1 | 0 | 2 | 2 |
| XI | 1 | 0 | 1 | 3 |
| XII | 1 | 1 | 2 | 3 |
| XIII | 1 | 0 | 1 | 2 |
| XIV | 1 | 0 | 3 | 3 |
| XV | 2 | 2 | 2 | 2 |
| XVI | 1 | 0 | 3 | 3 |
| XVII | 2 | 1 | 3 | 2 |
| XVIII | 2 | 0 | 2 | 3 |
| XIX | 1 | 0 | 2 | 0 |
| XX | 1 | 0 | 1 | 2 |
The expression levels of YOD1 and YAP were examined in 20 hepatocellular carcinoma patients. Protein levels were examined in tumors and normal tissues contiguous to tumors. The number stands for the intensity of staining: 0, negative; 1, weak; 2, moderate; 3, strong.
Discussion
Although there have been recent advances clarifying how the Hippo pathway is regulated, our understanding is limited due to its complexity under physiologically relevant conditions. Identifying regulators and validating their modes of action are essential steps to fully understand Hippo signaling. In the present study, by using both biochemical, genetic approaches and clinical samples, we identified YOD1 as a mediator that links physiological signals with Hippo-YAP/TAZ signaling.
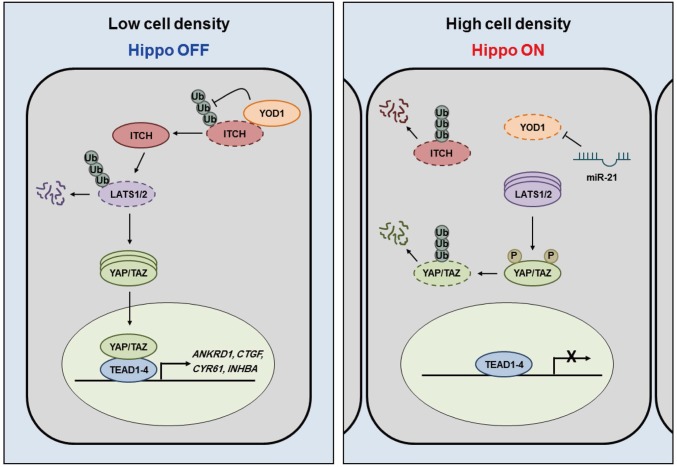
We found that YOD1 deubiquitinates and stabilizes ITCH, an E3 ligase of LATS1/2, and that the level of YOD1 is changed in a cell-density-dependent manner (Figs. 1−3). Cell density is a well-known intrinsic factor that activates the Hippo pathway via the enhancement of both the level and kinase activity of LATS (26, 27). Importantly, LATS kinase is the only tumor suppressor regulated by ITCH while controlling YAP. However, it is still unclear how cell density regulates the cellular abundance of LATS kinases. Here, we show that increases in miR-21 levels at a high cell density are crucial for the down-regulation of YOD1 and the accompanying destabilization of ITCH, which leads to increases in the level of LATS. Our findings suggest a clear mechanism for the regulation of the Hippo pathway: At low cell densities, high levels of YOD1 keep the level of LATS low, which prevents unnecessary activation of the Hippo pathway and allows for high expression levels of YAP/TAZ target genes (Fig. 5, Left). At high cell densities, however, low levels of YOD1 resulting from increases in the level of miR-21 lead to an increase in the level of LATS. Greater LATS activity, resulting from phosphorylation by activated MST1/2, increases the potential for the phosphorylation of YAP/TAZ, which may completely shut down the expression of YAP/TAZ-mediated target genes (Fig. 5, Right). In short, the activation of the Hippo pathway via cell-density-dependent stimuli is mediated in two ways: the activation of MST1/2 and the regulation of YOD1 by miR-21, which results in enhanced LATS activity. However, further studies are needed to understand the nature of the upstream signal and whether the processes mentioned above share the same signal.
Fig. 5.
Schematic model. At low cell densities, high levels of YOD1 keep the level of LATS1 low through deubiquitination of ITCH, which allows for high expression levels of YAP/TAZ target genes. At high cell densities, low levels of YOD1 resulting from increases in the level of miR-21 lead to an increase in the level of LATS1; this increases the phosphorylation of YAP/TAZ, which shuts down the expression of YAP/TAZ-mediated target genes.
ITCH has been found to be involved in multiple signaling pathways associated with cancer and immune responses (28–30), whereas YOD1 has been proposed to regulate endoplasmic reticulum-associated protein degradation (10). Although the phenotype of YOD1 knockout mice has not been reported, antigen-presenting cells of transgenic mice that express YOD1-C160S show enhanced antigen cross-presentation (31). The molecular target of YOD1 was not identified in this study, but it would be interesting to test whether ITCH is responsible for this phenotype (31). We focused on the role of YOD1 in the regulation of the Hippo pathway; however, because ITCH is involved in a variety of biological processes, diverse roles for YOD1 are expected under different cellular contexts.
Materials and Methods
The study was approved by the Medical Ethics Committee of Asan Medical Center (2016-0577). All human data and samples used in this study were anonymized. All identifiers of human data or samples were irreversibly stripped via an arbitrary alphanumeric code, making it impossible for anyone to link the samples to their sources. Therefore, no informed consent was obtained from the research participants. All experimental procedures were performed according to animal care and ethics legislation, and the study was approved by the Animal Care Committee of the University of Seoul. Details of materials and methods are described in SI Materials and Methods.
SI Materials and Methods
Study Approval.
The study was approved by the Medical Ethics Committee of Asan Medical Center (2016-0577). All human data and samples used in this study were anonymized. All identifiers of human data or samples were irreversibly stripped via an arbitrary alphanumeric code, making it impossible for anyone to link the samples to their sources. Therefore, no informed consent was obtained from the research participants. All experimental procedures were performed according to animal care and ethics legislation, and the study was approved by the Animal Care Committee of the University of Seoul.
Plasmids and Reagents.
Flag-YOD1 and Flag-YOD1C160S were obtained from Addgene. EGFP-YOD1 was created via PCR using Flag-YOD1 as a template. Flag-LATS1, EGFP-YAP, HA-SIAH2, HA-SMURF1/2, and HA-Ub were all generated using a standard PCR-based cloning method. The following plasmids were generous gifts: Myc-ITCH from Xiaolong Yang, Queen's University, Kingston, Canada; HA-NEDD4 from Fiona J. McDonald, Victoria University, Wellington, New Zealand; and 8xGTIIC-luciferase from Dae-Sik Lim, Korea Advanced Institute of Science and Technology, Daejeon, Republic of Korea. MG132 was purchased from Sigma. The LNA-modified miR-21 inhibitor was provided by Exiqon.
Cell Culture and Transfection.
HEK293T, HEK293, HeLa, NIH 3T3, HCT116-shScr, and HCT-116-shYAP cells were maintained in DMEM supplemented with 10% FBS (Gibco) and 1× antibiotic-antimycotic (Gibco) and were maintained at 37 °C in a humidified incubator with 5% CO2. HCT116-shScr and HCT-116-shYAP cells were kindly provided by Hei Cheul Jeung, Yonsei University College of Medicine, Seoul, Korea. MCF10A cells were maintained in DMEM/F-12 medium supplemented with 5% horse serum, 20 ng/mL of epidermal growth factor, 0.5 μg/mL of hydrocortisone, 10 μg/mL insulin, 100 ng/mL of cholera toxin (Sigma), and 1× penicillin/streptomycin (Gibco). Transfections were performed using Lipofectamine 2000 transfection reagent (Invitrogen), Turbofect (Fermentas) or calcium phosphate methods. In most cases, cells were collected and lysed 24 h after transfection. Information for the sequences of the siRNAs and miRNAs is available in Table S2.
Table S2.
List of siRNAs and miRNAs used in this study
| siRNA or miRNA | Sequence |
| GFP | GUUCAGCGUGUCCGGCGAG |
| YOD1#1 | GGGAGGAGCAAUAGAGAUA |
| YOD1#2 | GAAUGAGGGUUGAAGCCUA |
| YOD1#3 | GCAAUAGAGAUAUCGAUUU |
| ITCH | AAGUGCUUCUCAGAAUGAUGA |
| NF2 | UGGCCAACGAAGCACUGAU |
| MST1 | GGAUGUAAGUAAUCAAUUGAGGUCA |
| LATS1 | GTCTGCTTCATACATTCCTAA |
| LATS2 | AAAGGCGTATGGCGAGTAG |
| YAP | GACATCTTCTGGTCAGAGA |
| TAZ | ACGUUGACUUAGGAACUUU |
| hsa-miR-21 | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-125a | UCCCUGAGACCCUUUAACCUGUGA |
| hsa-miR-15a | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-29b | UAGCACCAUUUGAAAUCAGUGUU |
| hsa-miR-30b | UGUAAACAUCCUACACUCAGCU |
| Scramble | UCACAACCUCCUAGAAAGAGUAGA |
The siRNAs and miR mimics were synthesized by STpharm and Genolution, respectively. hsa, Homo sapiens.
Western Blot, Immunoprecipitation, and Ubiquitination Assays.
Cells were lysed in a lysis buffer [20 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and 1 μg/mL of leupeptin]. Lysates were cleared via centrifugation (14,200 × g, 10 min, 4 °C), and the supernatant was collected and used for Western blotting and immunoprecipitation. Total protein from livers was extracted with a NucleoSpin RNA/Protein Kit (Macherey-Nagel) following the manufacturer’s instructions. For immunoprecipitation, lysates were incubated with antibodies, followed by the addition Protein G agarose (Millipore) and a 2-h incubation. Agarose beads were washed with lysis buffer five times, after which the immunoprecipitated complexes were separated via the addition of a loading dye and boiling. Ubiquitination assays were performed using the same immunoprecipitation method described above. Equal amounts of protein were resolved via 6%, 8%, or 10% SDS/PAGE and then transferred to PVDF membranes (ATTO). The antibodies used in this study are described in Table S3.
Table S3.
List of antibodies used in this study
| Antigen | Company | Catalog no. |
| NF2 | Santa Cruz | sc-55575 |
| MST1 | Cell signaling | 3682 |
| ITCH | BD Bioscience | 611199 |
| LATS1 | Cell signaling | 9153 |
| mLATS1 (C66B5) | Cell signaling | 3477S |
| LATS2 | Cell signaling | 5888 |
| YAP | Santa Cruz | sc-101199 |
| p-YAP (S127) | Cell signaling | 4911 |
| TAZ | BD Bioscience | 560235 |
| YOD1 | Abgent | AP11304b |
| p-STAT3 (Y705) | Cell signaling | 9145 |
| STAT3 | Cell signaling | 4904 |
| β-catenin | BD Bioscience | 610154 |
| β-ACTIN | Sigma-Aldrich | A5441 |
| β-TUBULIN | Santa Cruz | sc-9104 |
| Flag | Sigma-Aldrich | F3165 |
| Myc | Applied Biological Materials | G019 |
| HA | Santa Cruz | Sc-732 |
RNA Extraction, cDNA Synthesis, and qPCR Protocol.
Total RNA was isolated from cultured mammalian cells and liver using TRIzol reagent (Invitrogen) and a NucleoSpin RNA/Protein Kit (Macherey-Nagel), respectively. To quantify the expression levels of genes, equal amounts of cDNA were synthesized following a standard RT-PCR method and were mixed with Power SYBR Green PCR master mix (Applied Biosystems) and 2 pmol each of both forward (for) and reverse (rev) primers. The threshold cycle (Ct) value for each gene was normalized to the Ct value for β-ACTIN, mGapdh, or U6 snRNA. Relative mRNA expression levels were calculated using the ΔΔCt method.
The following PCR primers were used for the mouse samples: Acta2, for: AGGGCTGTTTTCCCATCCATCG and rev: TCTCTTGCTCTGGGCTTCATCC; Amotl2, for: AGAGATTGGAATCGGCAAAC and rev: TTCTCCTGTTCCTGTTGCTG; Ctgf, for: CAAGGACCGCACAGCAGTT and rev: AGAACAGGCGCTCCACTCTG; Cyr61, for: ATGATGATCCAGTCCTGCAA and rev: TAGGCTGTACAGTCGGAACG; Gapdh, for: CGACTTCAACAGCAACTCCCACTCTTCC and rev: TGGGTGGTCCAGGGTTTCTTAC TCCTT; EpCAM, for: TTGCTCCAAACTGGCGTCTA and rev: ACGTGATCTCCGTGTC CTTGT; and Sox9, for: CAGCAAGAACAAGCCACACGTCAA and rev: TTGTCCGTTCTTCACCGACTTCCT. Primers for human samples were as follows: ANKRD1, for: AGTAGAGGAACTGGTCACTGG and rev: TGGGCTAGAAGTGTCTTCAGAT; CTGF, for: AGGAGTGGGTGTGTGACGA and rev: CCAGGCAGTTGGCTCTAATC; CYR61, for: CCTTGTGGACAGCCAGTGTA and rev: ACTTGGGCCGGTATTTCTTC; INHBA, for: TTGCCAACCTCAAATCGTGCT and rev: CCCACACTCCTCCACGATCAT; YOD1, for: CAGCGAATCCTCGTCGGATAC and rev: AAGCACAGGCAAAGTTTCCCT; and β-ACTIN, for: GCGGGAAATCGTGCGTGACATT and rev: GATGGAGTTGAAGGTAGTTT CGTG. The primers used for the miRNA assays are described in Table S4.
Table S4.
List of primers for miRNA assays
| miRNA | RT primer | PCR forward primer | PCR reverse primer |
| U6 snRNA | AAAATATGGAACGCTTCACGAATTTG | CTCGCTTCGGCAGCACATATACT | ACGCTTCACGAATTTGCGTGTC |
| has-miR-125a | CCTCACAACGATTCCACAAGCACAGGTTA | GTCCTCACAACGATTCCACAAG | GTTGATTCTCCCTGAGACCCTTTA |
| has-miR-15a | CCCTATCGGTTCGAGACACCACAAACCA | CTACCCTATCGGTTCGAGACAC | GGAATCCTGTAGCAGCACATAATG |
| has-miR-21 | CACCGTTCCCCGCCGTCGGTGTCAACA | CCCGCCTAGCTTATCAGACTG | GCCGTCGGTGTCAACATCA |
| has-miR-29b | CAAGCCCTAGTATTCCTCGACAACACTGATT | GCAAGCCCTAGTATTCCTCGAC | AGCTCCGTATAGCACCATTTGAAA |
| has-miR-30b | GTCGAGATTGTTGACGCCAGCTGAGTG | GTATGTCGAGATTGTTGACGCC | CGGTGCATTGTAAACATCCTACAC |
Luciferase Reporter Assay.
Cells seeded in 24-well plates were cotransfected with 100 ng of 8xGTIIC promoter luciferase, 10 ng of pTK-Renilla luciferase (Promega), and the indicated plasmids or siRNAs. Assays were performed in accordance with the manufacturer’s dual-luciferase assay protocols (Promega). Renilla luciferase activity was used to normalize firefly luciferase activity.
Wound-Healing Assay.
Cells transfected with the indicated DNAs or RNAs were seeded in 24-well plates and cultured until reaching 90% confluence. Cells were scratched with a 200-µL pipette tip and washed with media to remove debris, and fresh medium was added to each well. After the indicated times, cell-free regions were measured using TScratch software (32).
Colony-Forming Assay.
Cells were transfected with the indicated plasmids for 24 h before being split into 12-well plates. One day after transfection, 2,000 cells per well mixed in a 0.35% agarose/complete media suspension were seeded onto 0.7% agarose/complete media bottom layers; 7–10 d later, media were removed and stained with 0.005% crystal violet in 80% methanol. Colony formation was measured using ImageJ to quantify the mean pixel intensity within each well.
Human Liver Cancer Tissues.
For immunohistochemical analysis, 4-μm-thick sections on silanized charged slides were performed by heat-induced epitope retrieval method using Cell Conditioning 1 buffer for 32 min and incubated for 16 min with anti-YOD1 (ABGENT) and anti-YAP (NOVUS) in BenchMark ULTRA automatic immunostaining device (Ventana Medical Systems). Antigen−antibody reaction was visualized using Ventana OptiView 3,3′-Diaminobenzidine tetrahydrochloride (DAB) IHC Detection Kit (Ventana Medical Systems). Counterstaining was performed by using Ventana Hematoxylin II for 24 min and Ventana Bluing reagent for 4 min. All slides were removed from the stainer, dehydrated, and cover-slipped for microscopic examination.
Transgenic Mice and Drug Treatment.
To generate the Tet-On inducible YOD1 transgene, Flag-tagged human YOD1 cDNA (Addgene) was cloned into pBI-EGFP vectors (Clontech). Liver-specific ApoE-rtTAs-M2 expression plasmid was kindly provided by Sarah Comerford, University of Texas Southwestern Medical Center, Dallas, TX. Both transgenes were excised from their respective vectors and injected into fertilized (C57BL/6N) F2 hybrid mouse eggs. ApoE-rtTA mice were crossed with TRE-hYOD1 mice to generate double transgenic mice for the liver-specific inducible expression of hYOD1. Genotyping was performed on genomic DNA obtained from mouse tail-clip samples using a tissue lysis buffer [10 mM Tris (pH 8.8), 50 mM KCl, 1 mM MgCl2, 0.5% Nonidet P-40, 0.5% Tween 20]. The presence of ApoE-rtTA and TRE-hYOD1 alleles was determined via PCR using the following primers: YOD1, for: GTTTTGACCTCCATAGAAGAC and rev: CAGAACCCTTTTGGTATATCC; and rtTA-SM2, for: CCATGTCTAGACTGGACAAGA and rev: CTCCAGGCCACATATGATTAG. In this Tet-on system, 4-wk-old transgenic littermates were fed 200 µg/mL doxycycline (Sigma) in drinking water supplemented with 2.5% sucrose. Verteporfin (Sigma) was administered via i.p. injection three times a week at a dose of 50 mg/kg. Control animals were injected with 10% DMSO in PBS at the same volume and on the same schedule as verteporfin.
Mouse Liver Collection and Tissue Processing.
Mice were deeply anesthetized via i.p. injections of Zoletil (30 mg/kg) and xylazine (10 mg/kg). The liver was then removed, and pieces of extracted livers were immersed in 10% neutral-buffered formalin.
Mouse Immunohistochemistry.
Paraffin sections were deparaffinized in xylene and rehydrated in solutions of gradually decreasing ethanol concentrations. The antigen epitopes were then unmasked using a sodium citrate buffer (pH 6.0). Subsequently, the sections were incubated overnight at 4 °C with either anti-Ki-67 (Cell Signaling) or anti-Cytokeratin 19 (CK19; Abcam) primary antibodies. After the primary incubation, sections were incubated with the appropriate biotinylated secondary antibodies followed by treatment with freshly prepared DAB substrates (Vector Laboratories). Sections were lightly counterstained with hematoxylin and mounted. Ki-67-positive cells in the sections were counted using a microscope (Eclipse Ti; Nikon) at 200× magnification.
Acknowledgments
This work was supported by the National R&D Program for Cancer Control, Ministry of Health & Welfare Grant 1420060 (to E.-H.J.), and the National Research Foundation of Korea (Grant NRF-2016R1E1A1A01943544) (to E.-H.J.). S.-J.M. was supported by Grant HI15C3078 from Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health & Welfare, Republic of Korea. J.-R.K. was supported by the 2016 sabbatical year research grant of the University of Seoul. The biospecimen and data used in this study were provided by Asan Bio-Resource Center, Korea Biobank Network [2016-07(119)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620306114/-/DCSupplemental.
References
- 1.Yu FX, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 4.Ho KC, et al. Itch E3 ubiquitin ligase regulates large tumor suppressor 1 stability [corrected] Proc Natl Acad Sci USA. 2011;108:4870–4875. doi: 10.1073/pnas.1101273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salah Z, Melino G, Aqeilan RI. Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res. 2011;71:2010–2020. doi: 10.1158/0008-5472.CAN-10-3516. [DOI] [PubMed] [Google Scholar]
- 6.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Mouchantaf R, et al. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem. 2006;281:38738–38747. doi: 10.1074/jbc.M605959200. [DOI] [PubMed] [Google Scholar]
- 8.Xie Y, et al. Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem. 2013;288:2976–2985. doi: 10.1074/jbc.M112.430066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Yen L, Irwin L, Sweeney C, Carraway KL., 3rd Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol Cell Biol. 2004;24:7748–7757. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst R, Mueller B, Ploegh HL, Schlieker C. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol Cell. 2009;36:28–38. doi: 10.1016/j.molcel.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim M, Kim M, Park SJ, Lee C, Lim DS. Role of Angiomotin-like 2 mono-ubiquitination on YAP inhibition. EMBO Rep. 2016;17:64–78. doi: 10.15252/embr.201540809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamaratoglu F, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 13.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma B, et al. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat Cell Biol. 2015;17:95–103. doi: 10.1038/ncb3073. [DOI] [PubMed] [Google Scholar]
- 15.Cao L, et al. Ubiquitin E3 ligase dSmurf is essential for Wts protein turnover and Hippo signaling. Biochem Biophys Res Commun. 2014;454:167–171. doi: 10.1016/j.bbrc.2014.10.058. [DOI] [PubMed] [Google Scholar]
- 16.Salah Z, Cohen S, Itzhaki E, Aqeilan RI. NEDD4 E3 ligase inhibits the activity of the Hippo pathway by targeting LATS1 for degradation. Cell Cycle. 2013;12:3817–3823. doi: 10.4161/cc.26672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang HW, Wentzel EA, Mendell JT. Cell−cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci USA. 2009;106:7016–7021. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye X, et al. Coxsackievirus-induced miR-21 disrupts cardiomyocyte interactions via the downregulation of intercalated disk components. PLoS Pathog. 2014;10:e1004070. doi: 10.1371/journal.ppat.1004070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S, et al. miR-21 regulates chronic hypoxia-induced pulmonary vascular remodeling. Am J Physiol Lung Cell Mol Physiol. 2012;302:L521–L529. doi: 10.1152/ajplung.00316.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okabe M, et al. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development. 2009;136:1951–1960. doi: 10.1242/dev.031369. [DOI] [PubMed] [Google Scholar]
- 21.Furuyama K, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 22.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song H, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci USA. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, et al. Tumor-selective proteotoxicity of verteporfin inhibits colon cancer progression independently of YAP1. Sci Signal. 2015;8:ra98. doi: 10.1126/scisignal.aac5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das Thakur M, et al. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci USA. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oberst A, et al. The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase Itch. Proc Natl Acad Sci USA. 2007;104:11280–11285. doi: 10.1073/pnas.0701773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu L, et al. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J Biol Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- 30.Rossi M, et al. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci USA. 2006;103:12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sehrawat S, et al. A catalytically inactive mutant of the deubiquitylase YOD-1 enhances antigen cross-presentation. Blood. 2013;121:1145–1156. doi: 10.1182/blood-2012-08-447409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gebäck T, Schulz MM, Koumoutsakos P, Detmar M. TScratch: A novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques. 2009;46:265–274. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]