Significance
The plant-signaling molecule ethylene is biosynthesized from 1-aminocyclopropane-1-carboxylic acid (ACC), as catalyzed by ACC oxidase, which is homologous to the 2-oxoglutarate (2OG) oxygenases, but which does not use a 2OG cosubstrate. Bacteria produce ethylene in a highly unusual reaction that involves oxidative 2OG fragmentation. Biophysical studies on a Pseudomonas ethylene-forming enzyme (EFE) reveal how structural and stereoelectronic factors enable the EFE to bias reaction away from normal 2OG oxygenase catalysis involving two-electron substrate oxidation concomitant with succinate formation, toward the arginine-dependent four-electron oxidation of 2OG to give ethylene. The results imply that negative catalysis, with respect to ethylene formation, has operated during the evolution of 2OG oxygenases and will be useful in protein engineering aimed at optimizing ethylene production.
Keywords: ethylene-forming enzyme, 2-oxoglutarate-dependent oxygenases, hydroxylase, plant development, oxidoreductase
Abstract
Ethylene is important in industry and biological signaling. In plants, ethylene is produced by oxidation of 1-aminocyclopropane-1-carboxylic acid, as catalyzed by 1-aminocyclopropane-1-carboxylic acid oxidase. Bacteria catalyze ethylene production, but via the four-electron oxidation of 2-oxoglutarate to give ethylene in an arginine-dependent reaction. Crystallographic and biochemical studies on the Pseudomonas syringae ethylene-forming enzyme reveal a branched mechanism. In one branch, an apparently typical 2-oxoglutarate oxygenase reaction to give succinate, carbon dioxide, and sometimes pyrroline-5-carboxylate occurs. Alternatively, Grob-type oxidative fragmentation of a 2-oxoglutarate–derived intermediate occurs to give ethylene and carbon dioxide. Crystallographic and quantum chemical studies reveal that fragmentation to give ethylene is promoted by binding of l-arginine in a nonoxidized conformation and of 2-oxoglutarate in an unprecedented high-energy conformation that favors ethylene, relative to succinate formation.
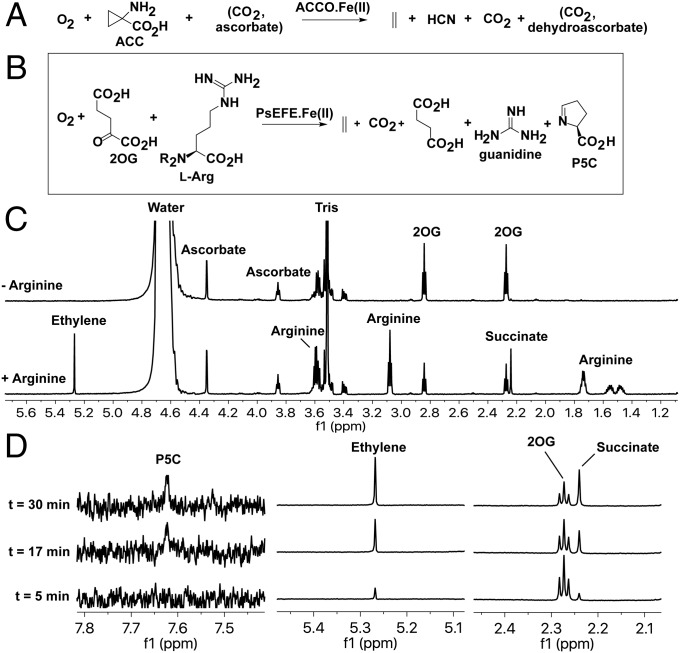
Ethylene is of industrial importance and is a vital signaling molecule in plants, where it has roles in germination, senescence, and stress responses (1). Commercial manipulation of the natural ethylene response is agriculturally important in controlling fruit ripening (2). In higher plants, ethylene is produced from methionine, via oxidation of 1-aminocyclopropane-1-carboxylic acid (ACC) in an unusual reaction catalyzed by the Fe(II)-dependent ACC oxidase (ACCO) (3, 4), which is part of the 2-oxoglutarate (2OG)-dependent oxygenase superfamily, although it does not use a 2OG cosubstrate (Fig. 1A) (5–7). Ethylene is also produced in some microorganisms by oxidation of 2-oxo-4-methylthiobutyric acid in a reaction not directly enzyme catalyzed (8, 9).
Fig. 1.
Ferrous iron-dependent oxygenase catalyzed ethylene formation. (A) ACCO, which catalyzes ethylene formation in plants and fungi, does not use 2OG, but belongs to the 2OG oxygenase superfamily. ACCO catalysis is stimulated by CO2, which is a coproduct; ascorbate is a proposed cosubstrate. (B) PsEFE catalyzes ethylene production in a manner stimulated by l-Arg; P5C is a coproduct. (C) 1H NMR showing the dependence of PsEFE on l-Arg for ethylene and succinate formation from 2OG (30 min). (D) Partial 1H NMR spectra (700 MHz) showing time-dependent formation of P5C, ethylene, and succinate by PsEFE. Note the relatively low P5C levels compared with succinate/ethylene.
In work aimed at producing industrial ethylene by biocatalysis, Pseudomonas strains, including plant pathogens, were shown to produce large amounts of ethylene (10–14). Bacteria engineered to produce ethylene using the Pseudomonas syringae pv. phaseolicola ethylene-forming enzyme (PsEFE) have been developed to ripen fruit as an alternative to the use of synthetic ethylene (15, 16). Ethylene-forming enzymes are being explored for biocatalysis in cyanobacteria (17–19). PsEFE-catalyzed ethylene production is 2OG-dependent and is stimulated by the addition of l-arginine (l-Arg), which is also converted by PsEFE into pyrroline-5-carboxylate (P5C; Fig. 1B) (13, 20). In contrast to the consensus 2OG oxygenase mechanism, which involves sequential binding of 2OG, substrate, and then oxygen, an unprecedented “dual circuit” mechanism is proposed for PsEFE (13).
We describe biochemical, structural, and modeling studies supporting a branched mechanistic pathway for PsEFE that can lead either to ethylene via oxidative fragmentation of 2OG or to succinate via a more typical 2OG oxygenase reaction, which sometimes results in P5C formation (Fig. 1B). The unusual Grob-type fragmentation of 2OG to give ethylene is reliant on l-Arg binding in a conformation unsuited for oxidation and on the 2OG C-5 carboxylate binding in an unprecedented high-energy conformation that promotes ethylene formation. The results will enable protein engineering efforts to optimize biocatalytic ethylene production by optimizing the branch of the bifurcating mechanism for ethylene and have implications for the evolution of ethylene-forming enzymes.
Results
Biochemical Studies.
A recombinant form of the ethylene-forming enzyme from PsEFE was produced in Escherichia coli and purified to near homogeneity. Using NMR and GC/MS-based assays, we found that, as reported (14, 21), PsEFE-catalyzed ethylene production is 2OG-dependent, stimulated by the addition of Fe(II) [some Fe(II) likely copurifies with PsEFE], and is increased by the addition of ascorbate or DTT (Fig. 1 C and D and SI Appendix, Fig. S1 A–E). Use of 13C- and 2H-labeled 2OG demonstrates that ethylene is derived from 2OG and that CO2 is coproduced with ethylene (SI Appendix, Fig. S1 F and G). Using a synthetic standard, we validated PsEFE-catalyzed P5C production from l-Arg (SI Appendix, Fig. S2 A–F). However, by NMR, we only observed relatively low levels of P5C formation compared with succinate and ethylene (SI Appendix, Fig. S3A); guanidine formation was observed in the presence of l-Arg (SI Appendix, Fig. S2 G and H) (22). Consistent with the literature (21), ethylene and succinate production depends on l-Arg, with no activity being observed with d-Arg (SI Appendix, Fig. S4 E and F). Of the other amino acids tested for their ability to promote ethylene formation, only close l-Arg derivatives, including homoarginine and N-methylated/hydroxylated guanidino group derivatives, manifested substantial ethylene production (SI Appendix, Fig. S4 A–D). In all cases, lower activities were observed compared with l-Arg. Kinetic analyses imply apparent substrate inhibition for l-Arg and 2OG, with Km values of 41.31 ± 5.39 and 22.22 ± 3.45 μM, respectively (SI Appendix, Fig. S1 C and D). Similar conclusions have recently been made by Martinez and Hausinger in work on the PsEFE from P. syringae pv. phaseolicola PK2 (21). These results imply a mechanism for ethylene production in which binding of arginine plays a key role. To investigate this, we performed structural studies on PsEFE.
Crystallographic Analyses of PsEFE.
We obtained three PsEFE crystal structures: in complex with manganese and 2OG (to 1.45 Å resolution, PsEFE:Mn:2OG), in complex with manganese and the buffer bis-Tris-propane (BTP) (to 1.55 Å resolution, PsEFE:Mn:BTP), and in complex with iron, l-Arg, and the unreactive 2OG analog, N-oxalylglycine (NOG; to 1.08 Å resolution, PsEFE:Fe:NOG:l-Arg; SI Appendix, Table S1). A structure was solved and a preliminary model of the PsEFE:Mn:BTP structure was built using single-wavelength anomalous diffraction data obtained from selenomethionine-derivatized PsEFE:Mn:BTP crystals. The preliminary model was used to obtain initial phases for isomorphous higher-resolution native data for PsEFE:Mn:BTP crystals, and the model was fully refined against these data. This structure was used in molecular replacement to determine the PsEFE:Fe:NOG:l-Arg structure. The PsEFE:Mn:BTP and PsEFE:Mn:2OG crystals have the same I222 space group, with one molecule in the asymmetric unit; eight residues at the termini were not modeled because of disorder (amino acids 1–2 and 345–350). The PsEFE:Fe:NOG:l-Arg crystals have a different morphology and crystal form (space group P1), with four molecules in the asymmetric unit; disordered regions not modeled (chain A, amino acids 1, 298, and 341–350; chain B, amino acids 299 and 341–350; chain C, amino acids 1–2 and 343–350; and chain D, amino acids 342–350). There are small differences in the overall folds between the two crystal forms, with Cα rmsds ranging from 0.13 to 0.63 Å (SI Appendix, Table S2). PsEFE is predominantly a monomer in solution, as shown by gel filtration.
Overall Structure.
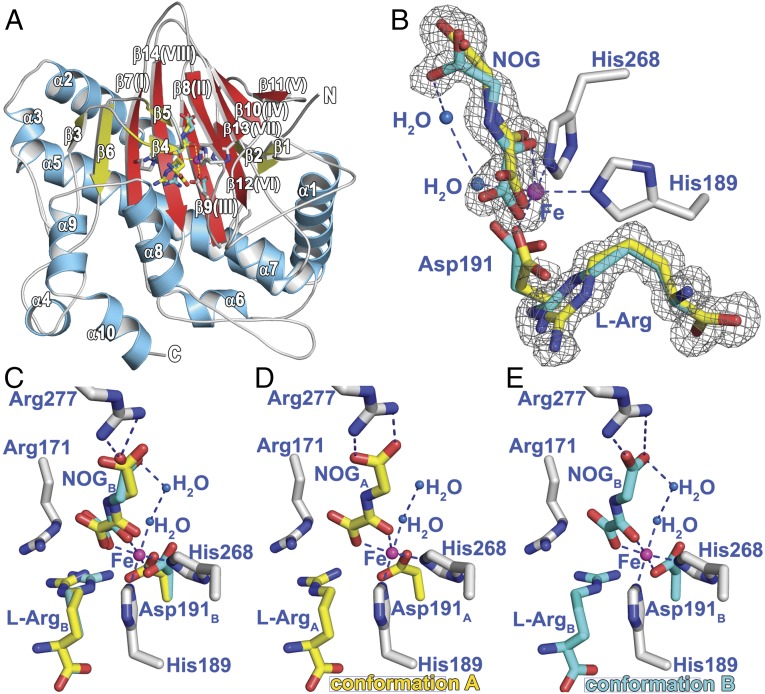
The overall PsEFE fold comprises 10 α-helices and 14 β-strands, of which eight β-strands (I–VIII) form the “major” and “minor” β-sheets of the conserved distorted double-stranded beta helix (DSBH): the 2OG oxygenase characteristic fold (Fig. 2A and SI Appendix, Figs. S5 and S6) (23–25). β-Strands β1 and β2 at the N-terminus extend the major β-sheet at the end of the DSBH away from the active site; β-strands β3 and β6 extend the other end of the major β-sheet close to the active site. α-Helices α2 and α5 bind across the surface of the major β-sheet and likely stabilize it. A loop region (residues 80–93), located between β3 and β6, which harbors β4 and β5, acts as a lid partially covering the active site and provides residues that bind the l-Arg cofactor/substrate. Three α-helices (α8, α9, α10) at the C terminus also contribute to the active site.
Fig. 2.
The overall fold of PsEFE and the two conformations observed for l-Arg and NOG. (A) The eight β-strands of the core DSBH are in red and labeled I–VIII. Non-DSBH β-strands are yellow, helices in marine blue, and loops in gray. The iron-ligating residues and substrates are sticks. (B) 2mFo-DFc electron density map contoured to 1σ, showing the two refined conformations of l-Arg and NOG. (C) Residues ligating to the iron (magenta) and involved in substrate binding. Two waters are present in the 2OG binding pocket with one coordinating to the metal. Conformations A (yellow) and B (cyan) are shown. (D and E) Conformations A and B of NOG and l-Arg. Conformation A, in which an l-Arg C-5 methylene hydrogen projects toward the metal, enables hydroxylation (leading to P5C); conformation B leads to ethylene. Note that Asp191 conformations A and B also interact differently with the metal.
The C-terminal α-helices α8, α9, α10; the β4–β5 loop; and the (β10–β11) DSBH IV–V “insert” loop directly interact with l-Arg (e.g., Arg316 in α8 with the l-Arg carboxylate; SI Appendix, Fig. S7 I–K). The role of these regions in PsEFE substrate binding is notable, because substrate binding by similar structural elements is a 2OG oxygenase subfamily characteristic (24, 26). The involvement of all these substrate recognition elements is very rare in other 2OG oxygenases. Thus, PsEFE should be regarded as a hybrid of subgroups I and II, in terms of its classification (25).
Active Site Geometry.
The overall metal-binding mode of PsEFE is relatively typical for 2OG oxygenases (Fig. 2), with the metal coordinated by His189 (C terminus of DSBH II), Asp191 (loop linking DSBH II and III), and His268 (N-terminus of DSBH VII), consistent with mutagenesis studies (27). However, in the high-resolution PsEFE:Fe:NOG:l-Arg structure, Asp191 adopts two conformations (see following). In the PsEFE:Mn:2OG and PsEFE:Fe:NOG:l-Arg structures, the metal is coordinated by 2OG/NOG in a bidentate manner: a single 2OG/NOG C-1 carboxylate oxygen ligates trans to His268, and the 2OG/NOG C-2 carbonyl ligates trans to Asp191 (SI Appendix, Fig. S8). The implied typical mode of Fe-chelation by 2OG is supported by spectroscopic studies (21). A water occupies the site trans to His189, likely where dioxygen binds. This arrangement suggests rearrangement to position the Fe(IV)=O intermediate, likely responsible for l-Arg oxidation (giving P5C), proximal to C-5 of the l-Arg; such rearrangement may be unnecessary for ethylene formation (see following) (28–30). In the PsEFE:Mn:BTP structure, two BTP hydroxyls and an amino group form tridentate metal coordination: the two hydroxyl groups ligate trans to His268 and Asp191, and the BTP amino group trans to His189.
Evidence for Induced Fit.
Despite the low overall rmsd for Cα atoms, comparison of structures with and without l-Arg reveals striking local differences in the backbone conformations (SI Appendix, Fig. S7 I and J), with regions surrounding the active site appearing to move to clamp l-Arg. In the PsEFE:Fe:NOG:l-Arg structure, the sidechain of Arg171 (located on DSBH I) is positioned to “π-π stack” with the l-Arg guanidine and form hydrogen bond and electrostatic interactions with the NOG C-1 carboxylate and Glu84 sidechain (an arginine at a similar position is involved in substrate/cofactor binding in some other 2OG oxygenases; Fig. 2 and SI Appendix, Fig. S9 C–E) (25). Glu84 (on the β4–β5 loop) rotates ∼90° about its Cα–Cβ bond (relative to the structure without l-Arg) into the active site to interact with the Arg171 guanidino group and both α-amino and guanidino groups of Conformation A (see following) of l-Arg (SI Appendix, Fig. S7K). Consistent with the proposed catalytic importance of these residues, the R171K, R171A, E84Q, and E84D variants do not produce ethylene (SI Appendix, Table S3). NMR assays indicate that neither succinate nor P5C is produced by the R171K and E84Q variants. Compared with its position in the absence of l-Arg, the sidechain of Tyr192 (on the DSBH II–III loop immediately after the HXD motif) rotates ∼90° about its Cα–Cβ bond into the active site to form a hydrogen bonding interaction with the l-Arg carboxylate and offset π-π stack with the l-Arg guanidino group (SI Appendix, Fig. S7J); consistent with a role for Tyr192 in catalysis, the Y192F variant manifests substantially reduced ethylene production (∼5% of wild type) (SI Appendix, Table S3).
The α8, α9, and α10 C-terminal helical region shows the largest local movement toward the active site (up to 3.0 Å), as observed in all molecules in the asymmetric unit (SI Appendix, Fig. S7 H and I). The observed movements involve Arg316 (on α8), the sidechain of which rotates ∼180° about its Cα–Cβ bond to form a salt bridge with the l-Arg carboxylate and hydrogen bond to the sidechain of Asn220 located on the DSBH IV–V loop (SI Appendix, Fig. S7K). The R316K and R316A variants are less active (∼13 and 4% of wild-type ethylene-forming activity) (SI Appendix, Table S3). The sidechain of Phe283 (on DSBH VIII) rotates ∼17° about its Cα–Cβ bond and ∼45° about its Cβ–Cγ bond to form part of the binding pocket for the l-Arg guanidino group (SI Appendix, Fig. S7J). Collectively, these results imply substantial induced fit on l-Arg binding and, as l-Arg is required for ethylene production, suggest that evolution of loop residues was important in the development of the ethylene-forming activity.
Alternative Substrate Conformations.
In all four molecules in the asymmetric unit of the high-resolution PsEFE:Fe:NOG:l-Arg structure, l-Arg and NOG are observed in two conformations (l-ArgA and l-ArgB, NOGA and NOGB), which correlate with two sidechain conformations of the metal-ligating Asp191 (each conformation was assigned an occupancy of 50%; Fig. 2 B–E and SI Appendix, Fig. S8). Analysis of the structures reveals l-ArgA/NOGA/Asp191A and l-ArgB/NOGB/Asp191B correlate, as the alternative combinations are not possible because of steric clashes. In both cases, monodentate metal coordination by Asp191 is observed and the shift in its carboxylate oxygen determines its interaction with the l-Arg guanidino group. Metal coordination of Asp191 in the two structures without l-Arg bound is most like conformation B in the l-Arg bound structure.
Both the α-amino acid and guanidino groups of l-Arg appear precisely bound in l-ArgA and l-ArgB. Key residues involved in l-Arg binding include Tyr192 and Arg316 (to its carboxylate), as well as Cys317 and Thr86 (to its α-amino group). Glu84 (l-ArgA only), Tyr192, Tyr318, Arg171, and Asp191 are important for interactions with the l-Arg guanidine (SI Appendix, Fig. S7K). Substitutions of these residues manifest substantially decreased ethylene production activity, except for the semiconserved Y318F variant (∼65% of wild type) (SI Appendix, Table S3).
The positions of the l-Arg α-amino acid group (up to the C-3/C-4 methylenes) are very similar in l-ArgA and l-ArgB, but the positions of their C-5 methylene and guanidino groups clearly differ (Fig. 2 B–E). In Conformation B, the C-5 methylene of l-ArgB is directed away from the metal; that is, it is not positioned for hydroxylation (Fig. 2E and SI Appendix, Figs. S8 A–D and 9B). Thus, l-ArgB likely correlates with ethylene formation (Fig. 3). In contrast, l-ArgA projects its C-5 hydrogens toward the metal (Fe to l-Arg C5 distance 4.5 Å; Fig. 2D and SI Appendix, Fig. S9A). l-ArgA is thus predicted to be the conformation yielding C-5 hydroxylation of l-Arg leading to P5C (Fig. 3).
Fig. 3.
Proposed outline mechanism. Ethylene and succinate formation occur in the presence of l-Arg (Fig. 1). Variations are possible [e.g., fragmentation of the cyclic peroxide intermediate II to give ethylene (21)], but are less likely, given the prevalence of intermediates of type III in 2OG oxygenase catalysis. NOG (and by implication 2OG) and l-Arg were observed in two conformations correlating with different Asp191 binding modes (Fig. 2). Formation of the bidentate 2OG intermediate complex I is supported by spectroscopic work (21). Turnover of 2OG to succinate also occurs (Fig. 1); succinate can be released from III, possibly with subsequent formation of H2O2. Formation of P5C occurs at a lower level than ethylene/succinate formation, via reaction of an Fe(IV)=O species (III), with l-Arg C-5. The hydroxylated product (IV) fragments and condenses to form P5C (21).
The distance and geometry between the l-ArgA and metal (4.3–4.5 Å) are similar to that in substrate structures of other 2OG oxygenases catalyzing hydroxylation, including l-Arg (derivative) hydroxylases VioC [PDB code 2WBO (31)] and clavaminic acid synthase [CAS; PDB code 1DRY (28); SI Appendix, Fig. S9 D and E]. However, VioC and CAS both catalyze hydroxylation at C-3, and their substrates are bound flipped ∼180° relative to the PsEFE binding mode. Most residues involved in l-Arg binding in PsEFE are not conserved in VioC/CAS, with the exception of the Arg171 equivalents, which bind the α-amino acid group as opposed to the guanidine group in PsEFE.
PsEFE-catalyzed C-5 l-Arg hydroxylation gives an unstable C-5 hydroxylated product, equivalent to the hemiaminal intermediate in histone demethylase catalysis (32). This PsEFE intermediate, as yet not observed by NMR, can fragment to give guanidine and glutamate semialdehyde, which is in equilibrium with 5-hydroxyproline and P5C, with the latter being preferred at neutral pH (33). Fragmentation of the nascent C-5 hydroxylated product could occur in solution or at the active site; in the latter regard, the apparently constrained bent conformation adopted by the arginine sidechain (in both l-ArgA and l-ArgB; Fig. 2B) is notable, suggesting aldehyde formation and cyclization at the active site (22, 33).
2-Oxoglutarate Binding.
There are striking differences between the PsEFE 2OG pocket and those of other 2OG oxygenases (SI Appendix, Fig. S11) (25). The PsEFE 2OG binding pocket is highly and unusually hydrophobic, being lined by the sidechains of Leu173, Phe175, Ile186, Ala199, Leu206, Val270, Ala279, Ala281, and Phe283 (SI Appendix, Fig. S10). A single, polar residue, Arg277, is positioned at the base of the pocket, the guanidino group that interacts with the 2OG/NOG C-5 carboxylate. For NOGA this interaction is bidentate, whereas for 2OG and NOGB, it is monodentate. Other 2OG oxygenases have a similarly positioned Arg or Lys, but have additional polar residues (Tyr/Ser/Thr) involved in 2OG binding, via hydrogen bonds, as exemplified in the 2OG C-5 carboxylate binding “RXS motif” in the CAS subfamily (SI Appendix, Fig. S11) (23).
There are several waters in the 2OG pocket (Fig. 2 B–E and SI Appendix, Fig. S11). In the NOG structure, a water hydrogen bonds with the NOG C-5 carboxylate; in the PsEFE:Mn:2OG structure, two waters hydrogen bond with the C-5 carboxylate (SI Appendix, Fig. S7D). A water-filled channel leads from the exterior to the 2OG C-5 carboxylate, possibly reflecting an oxygen entry/CO2 exit route (SI Appendix, Fig. S8E). Another water in the NOG/2OG pocket bridges between the metal ligated water and the C-5 carboxylate in 2OG/NOGB (2.8 Å) (Fig. 2 B–D); the combined binding of the C-5 bound water and that ligating the metal may in part reflect a dioxygen binding pocket. During 2OG oxygenase catalysis, after ligating to Fe(II), the dioxygen reacts with the 2OG ketone to give a cyclic peroxide, which then collapses to give the well-characterized Fe(IV)=O intermediate effecting substrate hydroxylation (Fig. 3). With PsEFE, the results imply that the Fe(IV)=O intermediate (or possibly, but less likely, a cyclic peroxide) undergoes Grob-type fragmentation to give ethylene and three CO2 molecules (Fig. 3). The question arises as to why ethylene formation occurs in PsEFE and not other 2OG oxygenases, where it has not been reported.
With PsEFE the methylenes of 2OG and NOG adopt an extended conformation, as observed in other 2OG oxygenases (SI Appendix, Fig. S11) (25). Strikingly, however, in contrast to other 2OG oxygenase structures, in which the C-5 carboxylate plane (C-5/O-3/O-4) of 2OG/NOG is coplanar with the C-5/C-4/C-3 carbon plane (SI Appendix, Fig. S11), with both 2OG and NOG, the C-5 carboxylate is not coplanar with the 2OG/NOG C-5/C-4/C-3 carbons. In the refined conformations of NOG, in the four chains (A–D) in the asymmetric unit of PsEFE:Fe:NOG:l-Arg structure, the torsion angles between the carboxylate O/C-5 and C-4/C-3 bonds are consistently close to 45°; in chain A: NOGA: 39.7°, 38.5° and NOGB: 45.3°, 43.7°; chain B: NOGA: 40.8°, 41.2° and NOGB: 46.0°, 44.5°; chain C: NOGA: 44.7°, 43.8° and NOGB: 46.0°, 45.0°; and chain D, NOGA: 47.6°, 46.8° and NOGB: 40.5°, 42.0°. In the PsEFE:Mn:2OG complex, the corresponding torsion angles for 2OG are 71.0°.
Molecular Dynamics Simulations and QM.
We investigated the stability of the unusual “strained” 2OG conformation observed in the PsEFE structures by molecular dynamics simulations using a PsEFE:Fe:2OG:l-Arg model based on the high-resolution PsEFE:Fe:NOG:l-Arg structure (SI Appendix, Fig. S12). 2OG maintained the strained conformation and its interactions with Arg277 for 94% of the simulation time (assuming an O-H hydrogen bond distance limit of 2.2 Å; SI Appendix, Table S4 and Fig. S12), suggesting the crystallographically observed conformation is catalytically relevant.
Surprisingly, the conformational preference for Grob-type fragmentation (34), involving entropically favored decarboxylation to give alkenes, has not been investigated in detail, even for small molecules. To compare the reactivity of the coplanar and strained conformations, we carried out quantum calculations of propionate derivatives with chloro, bromo, and trimethylamino groups with appropriate torsion angles 0°, 45°, and 90° (SI Appendix, Table S4 and Fig. S13). We also studied analogous reactions with (tert-butyl)-cyclohexanecarboxylate derivatives with the carboxylate and leaving group locked in a trans diaxial arrangement. The differences in the ΔG‡ (transition state free energy difference) values for fragmentation were consistently in favor of a torsion angle of 90°, with differences in ΔG‡ between the two conformations ranging from −2.7 to −4 kcal·mol−1 (mean, −3.6) (SI Appendix, Table S4). A similar, although smaller, trend was observed for the 45° torsion angle (SI Appendix, Table S4). These results support the potential for fragmentation of the crystallographically implied strained 2OG cosubstrate conformation to give ethylene.
Discussion
The overall fold of PsEFE is relatively typical of 2OG oxygenases (Fig. 2A); However, it contains highly distinctive features, including elements from group I and II 2OG oxygenases (25), which reveal PsEFE to have a hybrid fold (SI Appendix, Fig. S6). Notably, the conformation of PsEFE and the inclusion of a β(IV)–β(V) insert loop involved in substrate recognition is clearly distinct from that of ACCO, the only other enzyme known to directly catalyze ethylene production (35). Interestingly, sequence alignments of predicted EFEs (65–50% sequence identity [ID] to PsEFE), a related EFE-like subfamily (22–20% ID), and ACCOs (16–17% ID) show conservation in some of the key l-Arg interacting motifs and differences in the RXS motif suggesting they may not share a PsEFE 2OG fragmenting mechanism, but do share a common ancestor (SI Appendix, Figs. S14 and S15). The difference in the overall folds of PsEFE and ACCO (as well as more typical 2OG oxygenases) is mirrored in specific differences at their active sites, which reflect the different reactions they catalyze. The active site differences leading to oxidative fragmentation of 2OG to give ethylene rather than the normal succinate formation relate to the enzyme-bound conformation of 2OG and subsequently derived intermediates. The overall shape of the PsEFE active site is similar to that of normal 2OG oxygenases; indeed, PsEFE catalyzes a normal reaction (i.e., l-Arg hydroxylation). Thus, it appears likely that most 2OG oxygenases have actively evolved not to catalyze fragmentation of 2OG to give ethylene. The evolution of such “negative catalysis” (36) may have occurred because succinate is a useful byproduct, which can be recycled (e.g., by the TCA cycle), whereas CO2 and gaseous ethylene are not primary metabolites (as far as is known). In support of this, recent work on cancer metabolism suggests that, at least in animals, 2OG oxygenase catalysis may be metabolically linked to the TCA cycle (37).
A standout feature at the PsEFE active site concerns its 2OG/NOG-binding mode (Fig. 2 B–E and SI Appendix, Figs. S7, S10, and S11). 2OG/NOG binds in an unusual highly hydrophobic pocket and adopts a strained conformation that promotes Grob-type fragmentation of the iron-ligated succinate at the ferryl intermediate stage in the reaction (Fig. 3) to give two molecules of CO2 and ethylene. Ethylene formation is promoted by the presence of l-Arg, which is not oxidized during ethylene formation. Thus, in the ethylene-forming pathway, 2OG undergoes a four-electron oxidation. This differs from the ACCO reaction, in which ACC undergoes a two-electron oxidation, with reaction cycles for ACCO being proposed to be completed by ascorbate oxidation (38). Four-electron substrate oxidations are rare in 2OG oxygenase superfamily catalysis, but occur in isopenicillin N synthase-catalyzed oxidation of a dipeptide to isopenicillin-N; ironically, the isopenicillin N synthase structure is more closely related to ACCO than PsEFE (39).
Quantum calculations reveal that the crystallographically implied conformation of the 2OG-derived intermediate in PsEFE is crucial in enabling fragmentation to give ethylene, rather than substrate hydroxylation and succinate, as in normal 2OG oxygenase catalysis (SI Appendix, Fig. S13). There may also be other features at the iron center that are important in promoting ethylene formation, as evidenced by the different ligation predicted for the iron-ligating Asp191 in the ethylene-forming pathway (Fig. 2 B–E).
In addition to ethylene production, PsEFE also catalyzes a typical 2OG oxygenase type hydroxylation, albeit one leading to an interesting cyclization/fragmentation to give P5C and guanidine (Fig. 3). However, as shown by NMR analyses (Fig. 1D), the extent of P5C formation is lower than that of ethylene and succinate in our current assays. Thus, P5C formation appears substantially uncoupled from succinate formation.
Most crystallographic analyses of 2OG oxygenases imply that the 2OG C-5 carboxylate is near coplanar with the C-5/C-4/C-3 plane; that is, there is a near 0° syn-torsion angle (SI Appendix, Fig. S11). In PsEFE, the corresponding torsion angle has a mean of 44° in the NOG structure and is 71° in the 2OG structure, which, although is not the predicted optimum of 90° for ethylene formation, will promote fragmentation to ethylene (the torsion angle during solution catalysis by PsEFE is unknown and may be closer to 90°). The observation of a torsion angle less than the optimal of 90° for ethylene production is consistent with the observation of succinate formation and some hydroxylation to give P5C (Fig. 1). The balance between fragmentation to give either ethylene or succinate formation thus appears poised.
The structural insights into PsEFE will help in optimizing biocatalytic ethylene production by minimizing succinate formation to promote ethylene formation and by developing PsEFE variants not requiring arginine for ethylene-forming activity. Such work is of interest both with respect to industrial biocatalytic ethylene production (17) and to produce biological ethylene for agricultural applications (15).
PsEFE is proposed to produce ethylene to regulate plant physiology (e.g., ripening) beneficial to the bacteria (16). Some 2OG oxygenases have metabolic (e.g., in fatty acid metabolism) or sensing (e.g., in hypoxia sensing) roles in eukaryotes (40, 41). PsEFE has considerable potential for a sensing role, as in addition to using 2OG and oxygen and producing ethylene, succinate, and CO2, it requires a redox sensitive metal cofactor and converts arginine to P5C, a proline/glutamate/glutamine precursor.
The results raise the possibility that other 2OG oxygenases make ethylene, perhaps in addition to catalyzing more typical reactions. It is possible that the unusual conformations of 2OG observed with PsEFE are diagnostic of ethylene formation. Interestingly, the PsEFE active-site architecture has similarities with those of the ten-eleven translocation (TET) 2OG oxygenases (for PsEFE and the TET from Neurospora crassa Cα rmsd = 1.2 Å over 185 residues) (42). Arg171 of PsEFE is conserved in the two enzymes; in the NcTET structure, the analogous Arg190 interacts with its nucleosome substrate and with the 2OG C-1 carboxylate (SI Appendix, Fig. S9C). Substrate binding by NcTET involves Tyr217/Phe292 in a manner similar to that in which Tyr192/Phe283 of PsEFE interact with l-Arg (SI Appendix, Fig. S9C). However, the RXS motif that binds the 2OG C-5 carboxylate in many 2OG oxygenases, including the TETs, is not present in PsEFE, where the serine is replaced by an alanine (SI Appendix, Figs. S5 and S14). Thus, although it is unlikely that TETs catalyze ethylene production, other 2OG oxygenases may do so. The possibility of additional ethylene-forming enzymes is of interest in plant biology/agriculture, and it cannot be ruled out that some animals make ethylene. An early report describes ethylene formation in animal tissues, although the mechanism has yet to be validated (microbial/nonenzymatic processes are possible) (43). We are currently exploring the possibility of other 2OG oxygenase-related ethylene-forming enzymes.
Experimental Section
A synthetic PsEFE DNA sequence optimized for E. coli expression based on the gene sequence from P. syringae pv. phaseolicola (UNIPROT ID P32021; GeneArt Thermo Fisher) was subcloned into pCold-1 (Clontech) or pETite (Lucigen) vectors. Variants were made by mutagenesis using PCR. Vectors were transformed into E. coli strain BL21(DE3) for protein production. Proteins were purified by standard nickel affinity and gel filtration chromatography. Crystals were grown by the sitting drop vapor diffusion method (200–300-nL drops), and data were collected at the Diamond Light Source (Didcot U.K.). The initial PsEFE:Mn:BTP structure was determined by single-wavelength anomalous diffraction and refined in Phenix. Initially, PsEFE activity was assayed using an optimized version of the reported GC method (20). Purified PsEFE (between 10 and 15 μM) was added to 150 μL of 50 mM Hepes buffer at pH 7.2, containing 5 mM ascorbate or DTT, 5 mM 2-oxoglutarate, 2 mM l-Arg, and 200 μM (NH4)2Fe(SO4)2. 1H-NMR assays (at 298 K) were performed using a Bruker Avance III 700-MHz machine with a TCI inverse cryoprobe; water suppression used presaturation. Molecular dynamics simulations used the AMBER program (version 12) and force field parameters of Case et al. (44). The PsEFE:Fe:NOG:l-Arg crystal structure was used for QM studies; all structures corresponding to energy minima and transition states for 3-chloropropanoate fragmentation were optimized at the B3LYP/6–31+G* level using Gaussian 09 (45). For full experimental details, see SI Appendix.
Supplementary Material
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council Networks in Industrial Biotechnology and Bioenergy Proof of Concept Award BB/L013711/1 and the Wellcome Trust for funding, and the staff at the Diamond Light Source for access and technical assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates have been deposited with the Protein Data Bank under the accession codes 5LSQ, 5MOF, and 5LUN.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617760114/-/DCSupplemental.
References
- 1.Abeles FB, Morgan PW, Saltveit ME. Ethylene in Plant Biology. 2nd Ed Academic Press; San Diego: 1992. [Google Scholar]
- 2.Bleecker AB, Kende H. Ethylene: A gaseous signal molecule in plants. Annu Rev Cell Dev Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher-plants. Annu Rev Plant Physiol. 1984;35:155–189. [Google Scholar]
- 4.Kende H. Ethylene biosynthesis. Annu Rev Plant Physiol. 1993;44:283–307. [Google Scholar]
- 5.McRae DG, Coker JA, Legge RL, Thompson JE. Bicarbonate/CO2-facilitated conversion of 1-amino-cyclopropane-1-carboxylic acid to ethylene in model systems and intact tissues. Plant Physiol. 1983;73:784–790. doi: 10.1104/pp.73.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong JG, Fernández-Maculet JC, Yang SF. Purification and characterization of 1-aminocyclopropane-1-carboxylate oxidase from apple fruit. Proc Natl Acad Sci USA. 1992;89:9789–9793. doi: 10.1073/pnas.89.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, Ren JS, Clifton IJ, Schofield CJ. Crystal structure and mechanistic implications of 1-aminocyclopropane-1-carboxylic acid oxidase–the ethylene-forming enzyme. Chem Biol. 2004;11:1383–1394. doi: 10.1016/j.chembiol.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Primrose SB. Evaluation of the role of methional, 2-keto-4-methylthiobutyric acid and peroxidase in ethylene formation by Escherichia coli. J Gen Microbiol. 1977;98:519–528. doi: 10.1099/00221287-98-2-519. [DOI] [PubMed] [Google Scholar]
- 9.Ogawa T, Takahashi M, Fujii T, Tazaki M, Fukuda H. The role of NADH-Fe(iii)EDTA oxidoreductase in ethylene formation from 2-keto-4-methylthiobutyrate. J Ferment Bioeng. 1990;69:287–291. [Google Scholar]
- 10.Goto M, Hyodo H. Ethylene production by cell-free-extract of the Kudzu strain of Pseudomonas-syringae pv. Phaseolicola. Plant Cell Physiol. 1987;28:405–414. [Google Scholar]
- 11.Goto M, Ishida Y, Takikawa Y, Hyodo H. Ethylene production by the Kudzu strains of Pseudomonas syringae pv. Phaseolicola causing halo blight in Pueraria-lobata (Willd) Ohwi. Plant Cell Physiol. 1985;26:141–150. [Google Scholar]
- 12.Fukuda H, Fujii T, Ogawa T. Preparation of a cell-free ethylene-forming system from Penicillium digitatum. Agr Biol Chem Tokyo. 1986;50:977–981. [Google Scholar]
- 13.Fukuda H, et al. Two reactions are simultaneously catalyzed by a single enzyme: The arginine-dependent simultaneous formation of two products, ethylene and succinate, from 2-oxoglutarate by an enzyme from Pseudomonas syringae. Biochem Biophys Res Commun. 1992;188:483–489. doi: 10.1016/0006-291x(92)91081-z. [DOI] [PubMed] [Google Scholar]
- 14.Nagahama K, et al. Purification and properties of an ethylene-forming enzyme from Pseudomonas syringae pv. phaseolicola PK2. J Gen Microbiol. 1991;137:2281–2286. doi: 10.1099/00221287-137-10-2281. [DOI] [PubMed] [Google Scholar]
- 15.Digiacomo F, et al. Ethylene-producing bacteria that ripen fruit. ACS Synth Biol. 2014;3:935–938. doi: 10.1021/sb5000077. [DOI] [PubMed] [Google Scholar]
- 16.Weingart H, Ullrich H, Geider K, Völksch B. The role of ethylene production in virulence of Pseudomonas syringae pvs. glycinea and phaseolicola. Phytopathology. 2001;91:511–518. doi: 10.1094/PHYTO.2001.91.5.511. [DOI] [PubMed] [Google Scholar]
- 17.Eckert C, et al. Ethylene-forming enzyme and bioethylene production. Biotechnol Biofuels. 2014;7:33. doi: 10.1186/1754-6834-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong W, et al. The plasticity of cyanobacterial metabolism supports direct CO2 conversion to ethylene. Nat Plants. 2015;1:5053. [Google Scholar]
- 19.Ungerer J, et al. Sustained photosynthetic conversion of CO2 to ethylene in recombinant cyanobacterium Synechocystis 6803. Energy Environ Sci. 2012;5:8998–9006. [Google Scholar]
- 20.Nagahama K, et al. L-Arginine is essential for the formation in vitro of ethylene by an extract of Pseudomonas syringae. J Gen Microbiol. 1991;137:1641–1646. [Google Scholar]
- 21.Martinez S, Hausinger RP. Biochemical and spectroscopic characterization of the non-heme Fe(II) and 2-oxoglutarate dependent ethylene-forming enzyme from Pseudomonas syringae pv. Phaseolicola PK2. Biochemistry. 2016;55:5989–5999. doi: 10.1021/acs.biochem.6b00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batchelar ET, et al. Thioester hydrolysis and C-C bond formation by carboxymethylproline synthase from the crotonase superfamily. Angew Chem Int Ed Engl. 2008;47:9322–9325. doi: 10.1002/anie.200803906. [DOI] [PubMed] [Google Scholar]
- 23.Clifton IJ, et al. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J Inorg Biochem. 2006;100:644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 24.Aik W, McDonough MA, Thalhammer A, Chowdhury R, Schofield CJ. Role of the jelly-roll fold in substrate binding by 2-oxoglutarate oxygenases. Curr Opin Struct Biol. 2012;22:691–700. doi: 10.1016/j.sbi.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Aik W-S, et al. Introduction to structural studies on 2-oxoglutarate-dependent oxygenases and related enzymes. In: Hausinger RP, Schofield CJ, editors. 2-Oxoglutarate-Dependent Oxygenases. The Royal Society of Chemistry; London: 2015. pp. 59–94. [Google Scholar]
- 26.McDonough MA, et al. Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2) Proc Natl Acad Sci USA. 2006;103:9814–9819. doi: 10.1073/pnas.0601283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Que L., Jr One motif–many different reactions. Nat Struct Biol. 2000;7:182–184. doi: 10.1038/73270. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, et al. Crystal structure of a clavaminate synthase-Fe(II)-2-oxoglutarate-substrate-NO complex: Evidence for metal centered rearrangements. FEBS Lett. 2002;517:7–12. doi: 10.1016/s0014-5793(02)02520-6. [DOI] [PubMed] [Google Scholar]
- 29.Bollinger JMJ, Chang W-C, Matthews RJ, Boal AK, Krebs C. Mechanisms of 2-oxoglutarate-dependent oxygenases: The hydroxylation paradigm and beyond. In: Hausinger RP, Schofield CJ, editors. 2-Oxoglutarate-Dependent Oxygenases. The Royal Society of Chemistry; London: 2015. pp. 95–122. [Google Scholar]
- 30.Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 31.Helmetag V, Samel SA, Thomas MG, Marahiel MA, Essen LO. Structural basis for the erythro-stereospecificity of the L-arginine oxygenase VioC in viomycin biosynthesis. FEBS J. 2009;276:3669–3682. doi: 10.1111/j.1742-4658.2009.07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopkinson RJ, Hamed RB, Rose NR, Claridge TD, Schofield CJ. Monitoring the activity of 2-oxoglutarate dependent histone demethylases by NMR spectroscopy: Direct observation of formaldehyde. ChemBioChem. 2010;11:506–510. doi: 10.1002/cbic.200900713. [DOI] [PubMed] [Google Scholar]
- 33.Sleeman MC, Schofield CJ. Carboxymethylproline synthase (CarB), an unusual carbon-carbon bond-forming enzyme of the crotonase superfamily involved in carbapenem biosynthesis. J Biol Chem. 2004;279:6730–6736. doi: 10.1074/jbc.M311824200. [DOI] [PubMed] [Google Scholar]
- 34.Grob CA, Baumann W. Die 1,4-eliminierung unter fragmentierung. Helv Chim Acta. 1955;38:594–610. [Google Scholar]
- 35.Simaan AJ, Réglier M. In: 1-aminocyclopropane-1-carboxilic acid oxidase. 2-Oxoglutarate-Dependent Oxygenases. Hausinger RP, Schofield CJ, editors. The Royal Society of Chemistry; London: 2015. pp. 425–437. [Google Scholar]
- 36.Retey J. Enzymatic-reaction selectivity by negative catalysis or how do enzymes deal with highly reactive intermediates. Angew Chem Int Ed Engl. 1990;29:355–361. [Google Scholar]
- 37.Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: Mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rocklin AM, Kato K, Liu HW, Que L, Jr, Lipscomb JD. Mechanistic studies of 1-aminocyclopropane-1-carboxylic acid oxidase: Single turnover reaction. J Biol Inorg Chem. 2004;9:171–182. doi: 10.1007/s00775-003-0510-3. [DOI] [PubMed] [Google Scholar]
- 39.Rutledge P. Isopenicillin N synthase. In: Hausinger RP, Schofield CJ, editors. 2-Oxoglutarate-Dependent Oxygenases. The Royal Society of Chemistry; London: 2015. pp. 414–424. [Google Scholar]
- 40.Wanders RJA, Ferdinandusse S, Ebberink MS, Waterham HR. Phytanoyl-CoA hydroxylase: A 2-oxoglutarate-dependent dioxygenase crucial for fatty acid alpha-oxidation in humans. In: Hausinger RP, Schofield CJ, editors. 2-Oxoglutarate-Dependent Oxygenases. The Royal Society of Chemistry; London: 2015. pp. 338–349. [Google Scholar]
- 41.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 42.Li W, Zhang T, Ding J. Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi. Nucleic Acids Res. 2015;43:10026–10038. doi: 10.1093/nar/gkv979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ram Chandra G, Spencer M. Ethylene production by subcellular particles from rat liver, rat intestinal mucosa and Penicillium digitatum. Nature. 1963;197:366–367. doi: 10.1038/197366a0. [DOI] [PubMed] [Google Scholar]
- 44.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frisch MJ, et al. Gaussian 09. Gaussian, Inc.; Wallingford, CT: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.