Significance
Eukaryotes normally receive their genetic material from their parents but may occasionally, like prokaryotes do, acquire DNA from unrelated organisms through horizontal transfer (HT). In animals and plants, HT mostly concerns transposable elements (TEs), probably because these pieces of DNA can move within genomes. Assessing the impact of HTs on eukaryote evolution and the factors shaping the dynamics of these HTs requires large-scale systematic studies. We have analyzed the genomes from 195 insect species and found that no fewer than 2,248 events of HT of TEs occurred during the last 10 My, particularly between insects that were closely related and geographically close. These results suggest that HT of TEs plays a major role in insect genome evolution.
Keywords: horizontal transfer, transposable elements, insects, genome evolution, biogeography
Abstract
Horizontal transfer (HT) of genetic material is central to the architecture and evolution of prokaryote genomes. Within eukaryotes, the majority of HTs reported so far are transfers of transposable elements (TEs). These reports essentially come from studies focusing on specific lineages or types of TEs. Because of the lack of large-scale survey, the amount and impact of HT of TEs (HTT) in eukaryote evolution, as well as the trends and factors shaping these transfers, are poorly known. Here, we report a comprehensive analysis of HTT in 195 insect genomes, representing 123 genera and 13 of the 28 insect orders. We found that these insects were involved in at least 2,248 HTT events that essentially occurred during the last 10 My. We show that DNA transposons transfer horizontally more often than retrotransposons, and unveil phylogenetic relatedness and geographical proximity as major factors facilitating HTT in insects. Even though our study is restricted to a small fraction of insect biodiversity and to a recent evolutionary timeframe, the TEs we found to be horizontally transferred generated up to 24% (2.08% on average) of all nucleotides of insect genomes. Together, our results establish HTT as a major force shaping insect genome evolution.
Horizontal transfer (HT) is the transmission of genetic material between organisms through a mechanism other than reproduction. In prokaryotes, HT is pervasive, its mechanisms are well understood, and it is now viewed as one of the main forces shaping genome architecture and evolution (1, 2). In contrast, the study of HT in eukaryotes is less documented, but has been increasingly investigated. The majority of genes horizontally acquired by eukaryotes come from bacteria, but the extent to which these transfers have contributed to eukaryote evolution is still unclear (3, 4). Gene transfers from eukaryote to eukaryote appear to be largely limited to filamentous organisms, such as oomycetes and fungi (5, 6).
In animals and plants, very few cases of such horizontal gene transfers (HGTs) have been reported so far (7, 8). In fact, most of the genetic material that is horizontally transferred in animals and plants consists of transposable elements (TEs) (9–11), which are pieces of DNA able to move from a chromosomal locus to another (12). The greater ability of TEs to move between organisms certainly relates to their intrinsic ability to transpose within genomes, which genes cannot do. HT of TEs (HTT) may allow these elements to enter naive genomes, which they invade by making copies of themselves, and then escape before they become fully silenced by anti-TE defenses (13). A growing number of studies have identified such HTT (11, 14–16). However, a common drawback of these studies has been the inclusion of a limited set of TEs (11) or organisms (16), which hampers our understanding of the breadth of HTT, its contribution to genome evolution, and of the factors and barriers shaping these transfers in eukaryotes (13). In this study, we overcame these limitations by performing a large-scale, comprehensive analysis of HTT in insects. We focused on insects because a large number of whole-genome sequences are publicly available for this group and because insect genomes are known to harbor diverse and highly dynamic TE landscapes (17).
Results and Discussion
To detect HTT in insects, we de novo characterized TEs in all reference genome assemblies available in GenBank as of May 2016 (n = 195 species; Dataset S1 and Fig. S1) which represent 123 genera and 13 of the 28 insect orders. To minimize detection biases, we did not rely on established genome annotations that are available for only a subset of the species included in our study, and instead treated every species’ genome equally. This automatic characterization was performed with the RepeatModeler pipeline (18) and led to the identification of 53,452 TE families assigned to 98 superfamilies (Dataset S2). These exclude 3,417 families whose consensus sequences were found to include genes that may not belong to TEs (SI Materials and Methods), as well as all short interspersed element (SINE) consensus sequences, which might correspond to RNA pseudogenes (19). For each species, the consensus sequences of TE families were used to locate TE copies in genomic contigs. TE copies >100 base pairs were compared by pairwise reciprocal homology searches between every two species. After filtering out short and low-score alignments, and alignments between TEs from different superfamilies, we retained a total of ∼5.9 million hits, each of which indicated TE homology between two genomes.
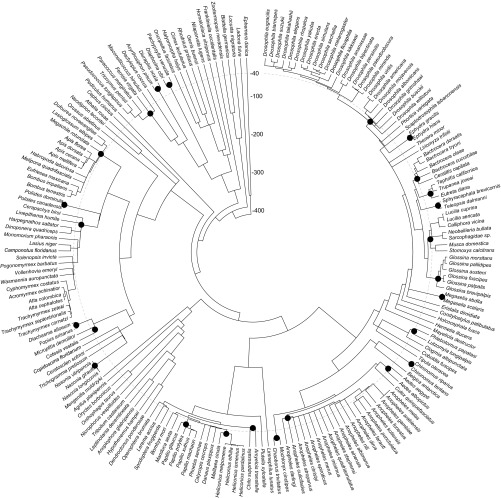
Fig. S1.
Time tree of the 195 insect species used in this study. Divergence times are taken from Misof et al. (23) and other references collected by timetree.org/. Times are shown in My from present. Black dots represent the most recent common ancestors of species that were collapsed into lineages within which homology at TEs was ignored, due to insufficient synonymous genetic divergence (dS) at core genes or because these species diverged in the last 40 My (dashed line).
TEs inherited from a recent common ancestor by descendent species, rather than horizontally transferred between these species, may present homology passing our filters. We identified clades of related insect species for which this situation may happen, by relying on the common assumption that inherited TEs evolve neutrally and similarly to synonymous sites of protein coding genes (20). This assumption implies that TEs showing higher interspecific homology than the synonymous sites of orthologous genes should share a more recent ancestor than the host species, and hence be the result of HT (16, 21). Conversely, TEs whose divergence is similar to the synonymous divergence of orthologous genes may be vertically inherited. We thus collapsed a clade of insect species into a lineage if (i) a fraction (>0.3%) of its core orthologous genes showed lower synonymous divergence than the highest nucleotide divergence of TEs or (ii) these species diverged in the last 40 My (Materials and Methods and Fig. S2). This collapsing resulted in the delineation of 81 lineages. We ignored all homologies within these lineages and considered the ∼1.46 million hits between species from distinct lineages to essentially result from HT.
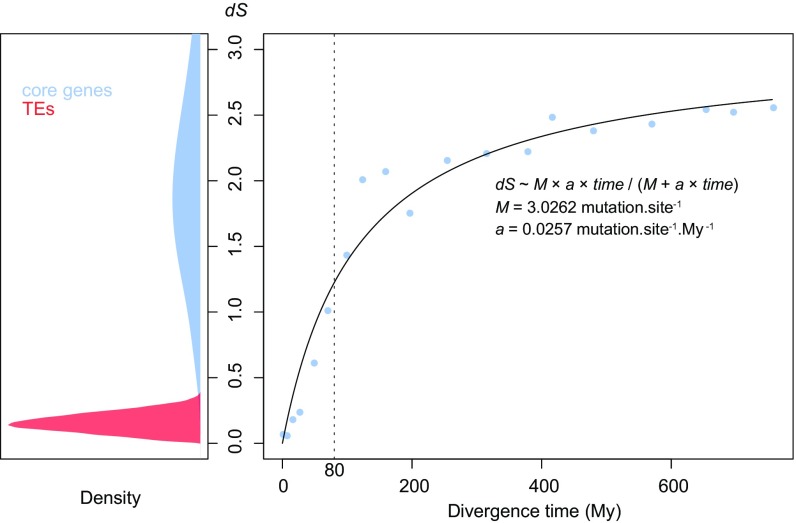
Fig. S2.
Divergence of horizontally transferred TEs and of orthologous cores genes in insects. (Left) Between-lineage synonymous nucleotide divergence (dS) of core insect genes (blue) according to Li (45) and between-lineage divergence of horizontally transferred TEs (red) according to Kimura 2-parameter distance (47). This distance is the model of substitution used by Li (45). (Right) dS of orthologous cores genes (all species considered) according to divergence times obtained from refs. 23 and 36, discarding dS values >9. dS was averaged over classes of divergence time that were defined to contain similar numbers of values. The fitted curve assumes initial linear relationship between dS and time (coefficient a) and a maximal value of dS (M) due to mutational saturation. According to fitted parameters, the maximal divergence of TEs (0.408) corresponds to a divergence time of 18.3 My, hence to transfers that have occurred <10 My ago. The vertical dashed line at 80 My represents the divergence time below which species were collapsed into lineages.
To translate these hits into a minimum number of HTT events, it must be considered that many hits may result from a single TE that transferred between two lineages and transposed into multiple similar copies. The resulting hits have a distinct feature: The copies they involve diverged within the recipient lineage after the transfer; hence, they should be more similar to each other than to copies from the other lineage. We applied a heuristic approach based on this principle (Materials and Methods) to cluster hits within each pair of lineages and TE superfamily into candidate HTT events. We also considered that a transferred TE might have degraded into nonoverlapping parts that share no sequence homology. To avoid counting these parts as several independently transferred elements, we further clustered hits on the basis of insufficient nucleotide or protein homology between copies. These two rounds of clustering yielded 4,499 HTT events for all pairs of insect lineages (Fig. 1), after discarding transfers that involved fewer than five TE copies per lineage. Taking pairs of lineages separately, each of these transfers represents an independent HTT event.
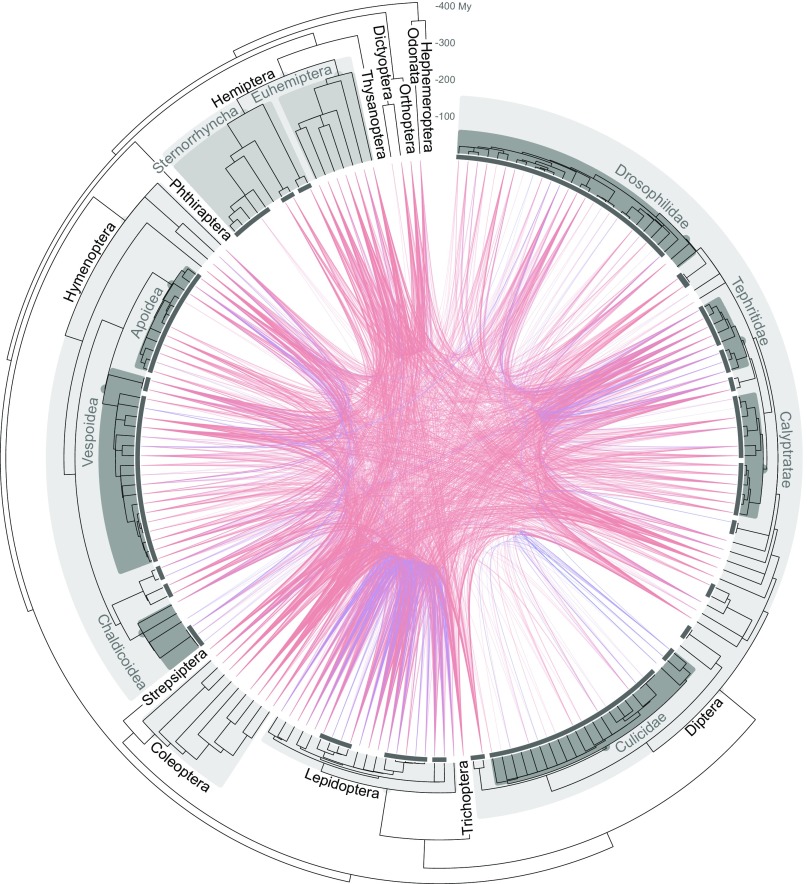
Fig. 1.
HTTs among insects. The concave tree, based on ref. 23, shows the phylogeny of the 195 insect species used in this study. Each dark segment next to tips encompasses related species between which HTT was not inferred, due the risk of confusion with vertical inheritance, and which were collapsed into a lineage. Each inferred HTT between two lineages, either direct or indirect (4,499 HTTs total), is shown by a curve. If several species of a lineage were found to share TEs due to the same HTT, we connected the two species (one per lineage) that presented the highest level of sequence identity for the transferred element. Blue curves represent transfers of retrotransposons (class I TEs), and red curves transfers of DNA transposons (class II TEs).
To infer the minimum number of HTT events across all 81 insect lineages, we considered that any HTT inferred in a pair of lineages might not reflect a direct transfer from one to the other, but separate acquisitions of TEs from other sources, which may already be counted as HTTs in other lineage pairs. There is no method that can discriminate between direct transfer and such “indirect” transfers. However, it is possible to avoid counting indirect HTTs, by constructing networks of insect species sharing similar TEs, in which the minimum number of HTT events is the number of lineages minus one. This parsimonious count resulted in 2,248 HTT events among the 81 insect lineages considered in our study (Fig. 2 and Dataset S2). This unprecedented figure is more than four times higher than the total number of HTT events reported so far in metazoans, plants, and fungi combined (10). Such a high number is still not unexpected, given that studies focusing on one or a few TEs often found one or more HTT events between multiple, distantly related taxa (11, 14–16, 22).
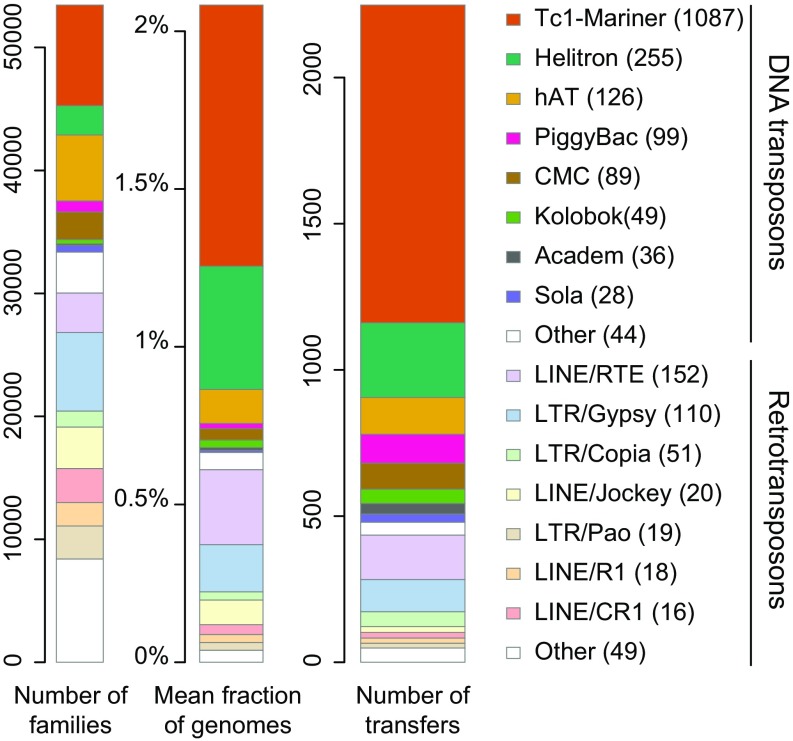
Fig. 2.
Characteristics of the TEs horizontally transferred between insects. Minimum numbers of transfer events are given in parentheses. The “Other” categories comprise all superfamilies of a given class of TEs [DNA transposons (class II) or retrotransposons (class I)] involved in <15 HTTs each. TE families (Left) correspond to those retrieved using RepeatModeler (Dataset S2). Center represents the mean fraction of an insect genome that is constituted by TEs from these families, which belong to a given super family.
Nonetheless, the actual number of HTT events that occurred in insects must largely exceed our inference, for at least four reasons. First, the inherent difficulty in resolving highly similar TE copies as distinct loci during genome assembly may have made some TE families undetectable or unable to pass our selection filters. Second, homology can only be found between recently diverged TEs, putting a maximum date on the HTT events that initiated the divergences. We estimate that the HTT events we uncovered mainly occurred in the last 10 My (assuming that TEs evolve similarly to synonymous sites for which we inferred an overall mutation rate; Fig. S2). Third, we conservatively collapsed many species into lineages within which HTT events have been characterized in previous studies (9). Fourth, the available insect genome sequences represent a very small fraction of the known diversity of insects. However, we found that 72 of the 81 lineages and 176 of the 195 species were involved in at least one HTT event. Therefore, HTT is not only widespread in insects, but the true number of HTT events is likely to be several orders of magnitude larger than the number we report.
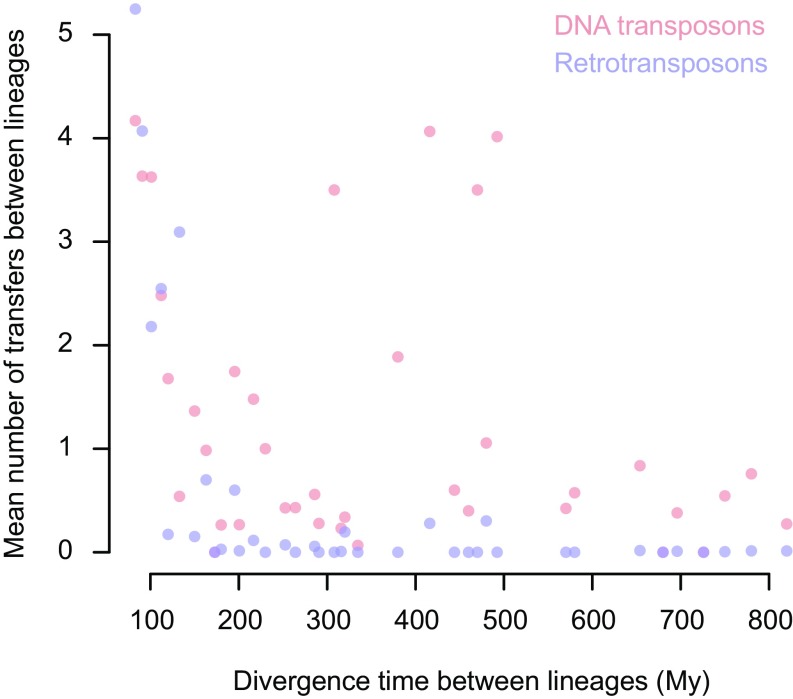
The tremendous amount of HTT events we uncovered in insects enabled us to statistically assess the impact of two main factors on this process: phylogenetic and geographic distance. For the former, we relied on recently estimated divergence times (23), and the latter involved assigning as many insect species as possible (n = 179) to the six main biogeographic realms. The effects of the two factors may not be confounded, because there is no positive correlation (R = −0.01) between species relatedness and geographic cooccurrence (originating from the same realm) among the 81 lineages we defined. We first tested whether HTT events were more likely to occur between less divergent lineages, as observed in bacteria (24, 25). Our analysis revealed a significant negative correlation between the number of HTT events and the divergence times of the lineages (Fig. 3). This pattern could, in theory, reflect vertical inheritance mistakenly inferred as HTT. Indeed, the number of vertically inherited TEs for which we can detect sequence similarities between lineages decreases with their divergence time. If that was the cause for the pattern shown in Fig. 3, the fraction of species per lineage that are found to share TEs with another lineage, due to an apparent HTT, should also decrease with the divergence of these lineages (SI Discussion). We did not observe such a decrease (Table S1), and therefore saw no evidence that homology between vertically inherited TEs was frequently counted as HTT. The phylogenetic effect we observed is also unlikely to be explained by reduced opportunities of HTT between phylogenetically distant species, because these are not more geographically distant on average. Instead, as proposed for HGT and HTT in bacteria (1, 25, 26), compatibility between TEs and recipient host cells may decrease as genetic distance from source lineages increases. Transposition is known to involve a number of interactions between TEs and host cellular factors (e.g., transcription factors and chromatin), which, depending on the type of TE, may be limited or very intricate (27–30). For example, some DNA transposons from the Tc1-Mariner superfamily only require their transposase to transpose in vitro (27). Weak dependence on host factors may explain why Tc1-Mariner TEs were found to be transferred between more or less distantly related taxa (31). Our systematic search supports this hypothesis as the highest number of HTT in insects was found for members of the Tc1-Mariner superfamily (Fig. 2). Interestingly, retrotransposons show lower transfer rates between distantly related lineages than DNA transposons (Fig. 3). Compared with DNA transposons, retrotransposons may require host factors that: (i) are more numerous and diverse, and/or (ii) tend to be less conserved between taxa. Such differences may explain the overall higher numbers of HTT events of DNA transposons than retrotransposons reported in our study (1,813 vs. 435 transfers; Fig. 2) and others (13, 32).
Fig. 3.
The number of HTTs between insects negatively correlates with insect divergence time. This trend is significant for both DNA transposons (red points) and retrotransposons (blue points) (P ∼ 4 × 10−3 and P < 10−4, respectively; two-sided Mantel tests). Number of transfers were averaged over pairs of lineages assigned to each 10 My-wide class of divergence time. To avoid bias due to differences in the number of species per lineage, we randomly sampled one species per lineage and computed averages over 104 samplings.
Table S1.
Correlation between the fraction of species per lineage possessing TEs resulting from a transfer and the divergence time with the other lineage involved in the transfer
| Lineage* | No. of transfers† | Mean fraction of species involved‡ | Coefficient§ | P value§ |
| Drosophila | 513 | 3.7/25 | 0.039 | 0.810 |
| Bactrocera | 736 | 2.3/5 | 0.126 | 0.999 |
| Lasius | 716 | 3.8/18 | 0.111 | 0.999 |
| Heliconius | 544 | 1.3/5 | −0.080 | 0.052 |
| Apis | 86 | 3.6/20 | −0.011 | 0.460 |
| Anopheles | 492 | 1.7/12 | 0.044 | 0.836 |
| Musca | 210 | 1.4/7 | 0.016 | 0.590 |
| Glossina | 28 | 2.2/6 | 0.137 | 0.756 |
The correlation was tested separately for the eight lineages comprising at least five species and involved in at least 10 inferred HTT events. Lineages are named after a representative genus (Fig. S1).
Transfers may represent either direct or indirect HTT (main text), and a given transfer may be used in more than one correlation test (row).
The denominator is the number of species in each lineage.
Correlations were assessed by Spearman rank tests, and a P value corresponds to the risk of inferring a negative correlation while there is none.
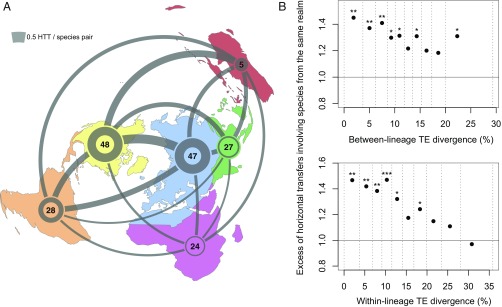
Finally, we assessed how geographic distance affects the likelihood of HTT at the global scale, by estimating the average number of transfers per pair of species assigned to biogeographic realms. The resulting map (Fig. 4A) shows that all pairs of realms comprise species involved in HTT at varying intensities. To test whether HTT was constrained by geographical distance, we estimated the extent to which two species that share TEs due to a HTT event preferentially originate from the same native realm. We also accounted for the fact that insect lineages may have moved to different geographic realms after exchanging TEs. This analysis required estimating the time since a transfer event. As proxies for this time, we took two independent measures of nucleotide divergence between TE copies originating from a HTT event: between- and within-lineage divergence. If insect lineages tend to move after exchanging TEs, the geographic cooccurrence of species that share TEs due to HTT should decrease with the estimated age of the transfer. Observations followed these predictions (Fig. 4B). Not only did HTT preferentially involve species belonging to the same realms overall, this tendency was more pronounced for the most recent transfers. These results confirm that geographic proximity generally favors HTT in insects. To some extent, this observation is reminiscent of that found in bacteria, where HGT occurs more frequently between lineages sharing the same habitat (25). Furthermore, our analysis likely underestimates the extent to which transfer events preferentially occur within geographic realms because sampled species may be more or less distantly related to, and geographically distant from, those that directly exchanged TEs.
Fig. 4.
Geographic proximity facilitates HTTs among insects. (A) Biogeographic realms to which 179 of the 195 studied insect species could be assigned, excluding Antarctica. The area of a disk is proportional to the number of species assigned to the corresponding realm, which is indicated inside the disk. Transfers involving species from different realms are represented by curves, and transfers involving species from the same realms are presented by circles, the thickness of which is proportional to the mean number of transfer per species pair. (B) Transfers more frequently involve species from the same realm, and significantly so for more recent transfers. Time since transfers is approximated by two measures of nucleotide divergence of TEs. For 10 defined classes of TE divergence (x axis), we plotted the fraction of transfers involving species from the same realms, divided by the mean of its null distribution obtained by permuting species across realms 104 times. Asterisks represent the portion (P) of permutations that yielded higher values than observations. *P < 0.05; **P < 0.01; ***P < 0.001. Class boundaries are shown by dashed lines and were defined so that classes encompass similar numbers of species pairs, to provide more comparable statistics.
Strikingly, the geographic cooccurrence of species sharing horizontally transferred TEs negatively correlated with the estimated time since the transfers (Fig. 4B). The correlation with the time inferred from within-lineage divergence of TEs is more significant (P < 0.004, permutation test) than with the time inferred from between-lineage divergence (p ∼ 0.06), possibly due to better accuracy of the former estimate (Materials and Methods). This correlation strongly suggests that older HTT events involve lineages whose descendants are more likely to be found in different realms, as expected if insect lineages had more time to migrate after exchanging TEs. Hence, historical and geographical patterns of HTs offer a rare illustration of the global movement of lineages across biogeographic realms over the past few million years.
Despite the conservative constraints imposed by our approach to detect HTT, we found that 2.08% of the nucleotides contained in insect genomes on average, and up to 24.6% in some species (Dataset S1), derived from the activity of TEs that were horizontally transferred within just the last ∼10 My. Extrapolating our estimates over the ∼480 My of insect evolution and the whole insect biodiversity points toward millions of HTT events generating substantial fractions of insect genomes. These inferences, combined with the pronounced impact TEs have on genome structure and dynamics (33, 34), establish HTT as a major factor driving insect molecular evolution. Our results call for further assessments of the influence of HTT on other taxonomic groups and of the ecological factors and relationships affecting HTT dynamics (35).
SI Text
SI Materials and Methods
Filtering Out TE Family Consensus Sequences Containing Non-TE Genes.
We aimed at identifying TE family consensus sequences containing non-TE genes, because the conservation of these genes may be interpreted as HTT. For this task, we performed homology searches of all TE consensus sequences generated by RepeatModeler against the nonredundant “nr” protein database of NCBI using Diamond blastx, retaining the five best hits per query sequence. We did a similar search against the RepBase database of known TE proteins (44). We discarded a TE consensus (hence family) if (i) it had homology to an nr protein over at least 90 amino acids in a region that did not show homology with a RepBase protein and if (ii) this nr protein did not show a homology of at least 40% over at least 200 amino acids with a RepBase protein. Homology between proteins was determined by a Diamond blastp search of nr proteins fulfilling criterion (i) against RepBase proteins. For all searches, we ignored alignments of e-value < 10−3.
Delineation of Protein Regions in TE Copies.
Most TEs include proteins that are needed for transposition. To delineate protein parts that remain in TE copies, we performed blastx homology searches of all TE copies, from the ∼1.46 million retained megablast hits, against the RepeatMasker protein database that is used by RepeatModeler for TE classification. For each resulting hit, we aggregated the consecutive HSPs that tend to be generated at each frameshift. We discarded aggregated HPSs <30 amino acids and any hit involving a protein that was not assigned to the TE superfamily of the copy it aligned to. To detect other proteins in copies (some types of TEs can contain several proteins), we extracted copy regions of at least 100 bp that were not involved in HSPs, and submitted them to the steps described above. In total, three iterative blastx searches were performed.
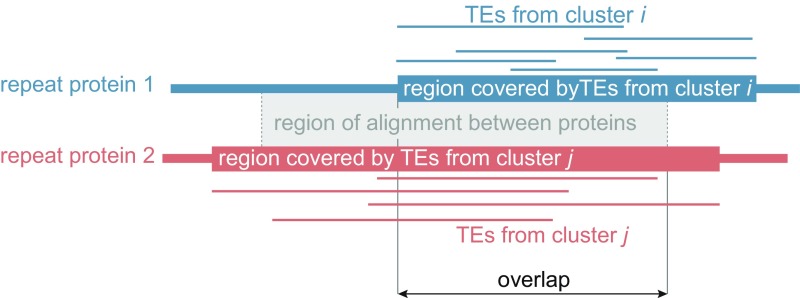
For all TE copies composing hits from each cluster, we combined the aligned regions of each protein into larger ones if these regions were separated by 10 amino acids or fewer. This aggregation resulted in 10,027 protein regions, representing 1,227 different proteins across all 8,713 clusters. We then assessed whether two protein regions from clusters i and j “overlapped.” This overlap was measured as the intersection between the protein regions and the coordinates of the HSP between the two corresponding proteins (Fig. S6). The latter was obtained by a blastp search comparing all 1,227 proteins against themselves with an e-value threshold of 10−4.
Fig. S6.
Determination of protein homology length (overlap) between TEs from two candidate transfers (clusters of hits). Blastx alignments (HSPs) of TE copies involved in each cluster on RepeatMasker proteins are represented by thin lines, and the blastp HSP between the full-length proteins is represented by the gray box with dashed contour. Protein regions covered by TE copies are the unions of the blastx HSPs that are distant from 10 amino acids or fewer. The overlap was taken as the length of the intersection between both covered regions and the blastp HSP.
Initial Clustering of Hits to Reduce Workload.
There could be >100,000 megablast hits involving a given pair of lineages for a given TE superfamily, requiring our clustering approach to make ∼5 billion pairwise comparisons (100,0002/2) between hits. Working around software and computer memory limitations (a list in R cannot contain >231-1 elements) to perform so many comparisons was deemed unproductive, considering that many of these comparisons are, in fact, unnecessary. Indeed, many hits share copies (several query copies aligned to the same target copy) and can already be attributed to the same HTT according to our criteria, because the highest within-lineage identity of two hits sharing a copy is 100%. To alleviate the workload, we discarded redundant hits by first clustering them via a single-linkage algorithm implemented in an R function (here, two hits are linked if they share a TE copy) that did not require generating pairs of hits. Then, within each formed cluster, we retained at least 100 hits (if as many were present) or enough hits to have 30 different TE copies per lineage. We retained hits that constituted reciprocal best hits between copies or that involved larger protein regions (as determined by our blastx procedure above). This selection reduced the number of hits from ∼1.46 million to 383,652.
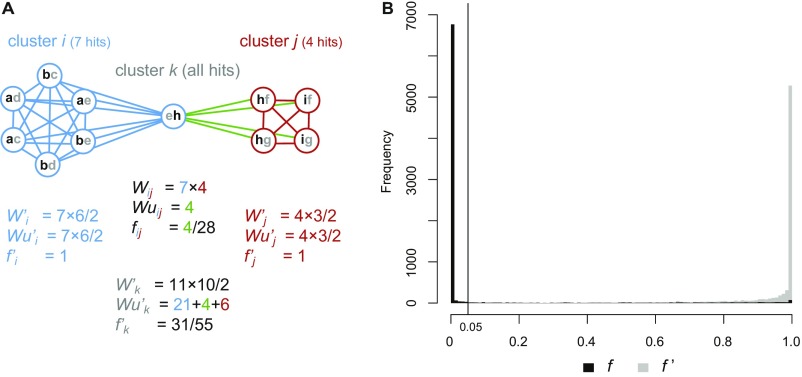
Connectivity Between and Within Clusters of Hits.
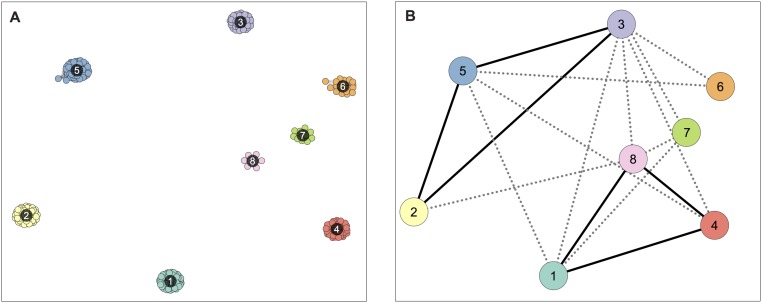
To quantify the connectivity between two clusters i and j in biological terms, we defined index fij = Wuij/Wij. Wij is the number of pairs of hits (each pair grouping one hit from each cluster) where copies within at least one lineage have nonnull identity (i.e., a blastn hit of at least 100 bp), and Wuij is the number of such hit pairs that are directly connected (i.e., which have higher within-lineage than between-lineage identity). To quantity the connectivity within clusters, we defined an equivalent index f′i = Wu′i/W′i, based on similar counts, but using pairs of hits assigned to a single cluster i. Both f and f′ vary from zero to unity. See Fig. S5A for an example. If clusters (hence transfers) are clearly distinguished, f should be low and f’ should be close to one, which was verified (Fig. S5B).
Fig. S5.
Distinctiveness between clusters of blast hits involving TE copies. (A) Indices of connectivity between hits assigned to different clusters i and j (fij) and between hits assigned to the same clusters (f′i, f′j), based on the hypothetical example of clustering shown in Fig. S3. The f′ index is also calculated as if all hits were assigned to a single cluster k. Indices are defined in SI Materials and Methods. It is assumed that all TE copies, labeled with lowercase letters (Fig. S3), show some homology detectable by blastn (hence W and W′ indices equal their highest possible values) and that the hit between copies e and h was assigned to cluster i. Connections between hits from different clusters are shown in green. (B) Distribution of the indices of connectivity between hits from different clusters (f) and from the same cluster (f′), based on real data. The vertical line represents the value of f above which two clusters are considered to possibly represent the same transfer. This threshold would group clusters i and j in same candidate transfer, because fij > 0.05.
Clusters i and j were considered to represent two separate HTT events if they had a protein overlap (Fig. S6) of at least 100 amino acids and if Wij = 0, meaning than no nucleotide homology was found between TE copies associated to the different clusters; or if fij ≤ 5% (Fig. S5).
Otherwise, we conservatively considered that these two clusters may represent the same HTT event.
Clique Formation to Delineate and Count Transfer Events.
Connecting any two clusters that may represent the same transfer event yields a graph of clusters (Fig. S4B). Logically, each transfer event is represented by an unconnected cluster or adjacent (directly connected) clusters. This is because nonadjacent clusters represent separate transfer events. In graph theory, a group of all-adjacent elements is a clique.
Fig. S4.
Example of delineation of HTT events from blast hits. (A) Clustering of hits between TE copies from the LINE/RTE-BOVB superfamily. These hits, each represented by a point, involve Bombyx mori and Chilo suppressalis. Height clusters, constituting candidate HTT events were delineated according to the method of Clauset et al. (48), based on higher within-lineage than between-lineage identity of the TEs they involve, and are represented as numbered groups of hits of different colors. The location of hits on the plot was defined by the layout_nicely() function of the igraph package (49) in R. (B) Connections of these clusters of hits into cliques, based on insufficient protein homology between TE copies from the clusters. Insufficient protein homology between two clusters is represented by connection lines. Plain lines connect clusters of hits included in the same clique. As a clique cannot contain nonadjacent clusters, the connection between clusters 2 and 3 has excluded cluster 6 from the clique, because it is not adjacent to cluster 2.
Cliques were established by associating elements with the algorithm described in Dataset S3. The composition of the resulting cliques depends on the elements we associate first in a clique. This is because the elements that can be added to a clique must be adjacent to all elements that are already present in the clique. Associations in cliques were set in priority for pairs of clusters whose copies aligned to very similar or identical proteins (based on the identity reported by our blastp search described above). These clusters were considered more likely to represent the retention of nonoverlapping parts of an ancestrally transferred TE.
We also relied on cliques to count the overall number of transfers among all insect lineages. As explained in Materials and Methods, this count required establishing networks of insect species involved in HTTs of similar TEs (Fig. S7). To avoid considering independent transfer events in the same network, we split a network into cliques. We achieved this by generating connections between every two transfers of the network, except for those involving the same pair of lineages (hence identified as independent). Here, priority to clique formation was given to nonindependent transfers that had a higher proportion of copies from shared TE families (Fig. S7). The minimum number of transfers events inferred from our data was the sum, across all 607 formed cliques, of the number of lineages in each clique minus one. This number was computed for each TE superfamily.
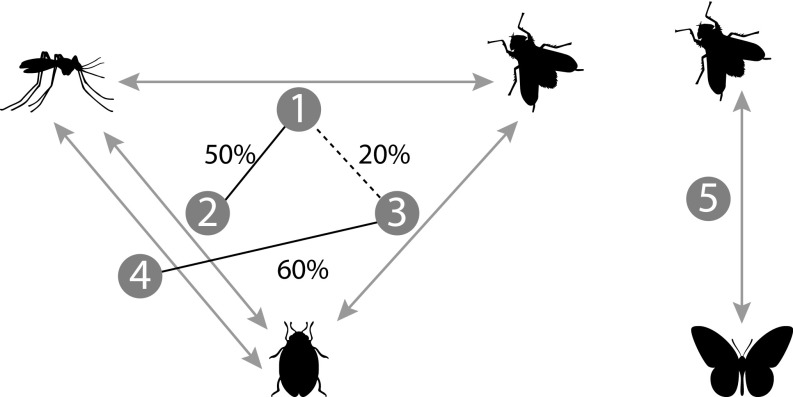
Fig. S7.
Hypothetical networks of insect species connected by HTs of similar TEs. Each transfer inferred between two species is represented by a gray arrow and a number. Transfer 5 forms a two-species network on its own. Lines connect two transfers that have at least one TE family in common. Each percentage next to a connection indicates the proportion of TE copies from one transfer (the one that involves fewer copies among the two) that is assigned to a TE family involved in the other transfer. Transfers 2 and 4 involve the same pair of lineages and cannot be connected, because they were previously determined as independent. Priority to connection (clique formation) is given to pairs of transfers that have a higher percentage of TE copies from shared families. Plain lines represent the connections that would be made, producing two cliques. These cliques cannot be merged because the two independent clusters 2 and 4 cannot be in the same clique. The two resulting cliques involve the three lineages each, yielding a minimum number of five HTT events to explain the data (3 − 1 + 3 − 1 + 2 − 1, counting the unconnected transfer on the right). Should transfers 1 and 3 be connected in priority, a minimum number of four HTT events would have been inferred (because a single clique of three lineages would include transfers 1, 3, and 4).
The algorithm in Dataset S3 processes pairs of adjacent elements one after the other to form cliques. Pairs are assumed to be sorted in the order of priority given to associations in a clique: elements from the pairs that are processed first are associated in cliques first.
SI Discussion
Heuristic Approach Used to Identify Transfers.
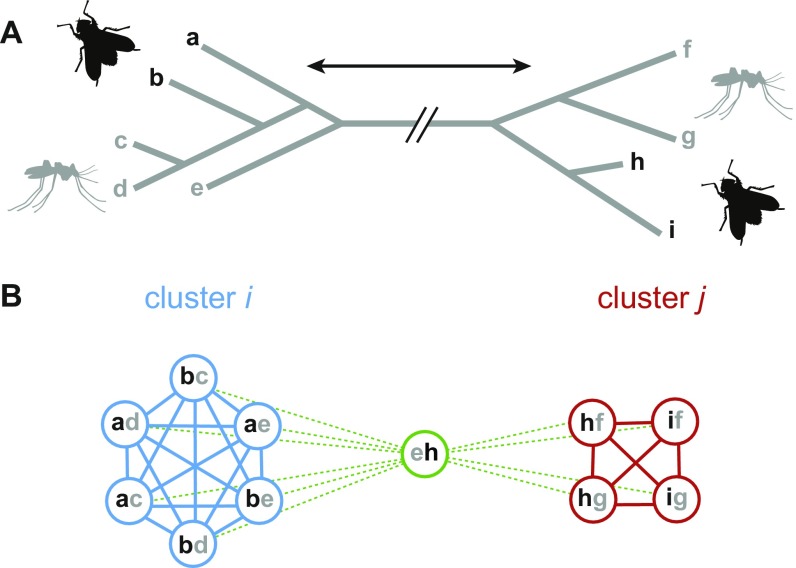
A single HTT event involving two lineages typically yields many homologous TE copies, hence many hits, due to transposition and speciation within lineages before and/or after the event. These events can be counted by clustering TE copies from several species and identifying clusters grouping TEs from sufficiently divergent lineages (11). We did not apply this approach to our large dataset because some TEs would bridge, via single-linkage clustering, other TEs that have no homology to each other (e.g., belonging to different classes). In addition, a single cluster may arise from several independent HTTs between the same lineages.
Our approach to infer HTT events by comparing within-lineage identity to between-lineage identity of TE copies should be more sensitive, because it explicitly considers that TE copies resulting from the same transfer should present lower divergence within a least one of the lineages involved than between the two lineages. This is because TE copies can diverge within a lineage only after the TE entered it. As shown on Fig. S3, our method may underestimate the number of HTT events if similar TEs were transferred separately between the same lineages. Although sensitivity could be increased by discarding all megablast hits below a certain identity threshold (eliminating the hit shown in green on Fig. S3), we did not apply such a filter to favor reliability of inferences.
Fig. S3.
Illustration of the approach used to cluster blast hits between TE copies into candidate HTT events. (A) Hypothetical distance relationships between TE copies from two lineages. Copies are identified with lowercase letters. (B) Megablast hits between TE copies from different lineages are shown as colored circles containing the identifiers of the two copies they involve. Connecting any two hits whenever they involve TE copies showing higher identity within one lineage than between lineages yields two clusters of hits (candidate HTT events) labeled i and j. Connections are shown as plain lines. If the overall distance between groups of copies corresponding to these clusters (represented by the double arrow in A) is not long enough, copies from the two clusters may be involved in megablast hits such as the one shown in green. These hits will connect to many others (dashed lines) because the TE copies they involve have lower identity than copies within lineages. These connections will lead to the inference of a single HTT (Fig. S5A). Hence, two HTT events can only be inferred if they involve two groups of TE copies that are quite distinct. Although this limits the sensitivity of the method (separate transfers of similar elements may be inferred as one), this also greatly reduces erroneous inference of separate HTT events from confused signals (e.g., homoplasy).
The risk of connecting hits resulting from distinct HTT events restricts the clustering to hits involving just two lineages. This, however, is not an issue because two HTTs identified in distinct pairs of insect lineages cannot represent the same HTT event, as long as no hit should reflect the vertical inheritance of TEs from a common ancestor to these lineages.
Vertical Inheritance Possibly Mistaken as HTT.
The apparent higher frequency of HTT events between more closely related lineages (Fig. 3) could be due, in principle, to vertical inheritance mistaken as HTT. This is because homology between sequences inherited from a common ancestor is mostly detectable (by megablast) between closely related lineages. To produce the observed effect (Fig. 3), such a mistake should be quite pervasive.
If this effect actually resulted from the decrease in similarity between vertically inherited TEs with divergence time (hence the ability to detect shared TEs), the fraction of species of a given lineage that share TEs with another lineage apparently involved in an HTT should negatively correlate with the divergence of both lineages, as we explain in the main text. Indeed, as similarity between TEs of two lineages decreases (due to sequence divergence or deletions), certain pairs of species lose all similar TEs before others do. The lack of negative correlation (Table S1) argues against this scenario, and hence against vertical inheritance of TEs frequently mistaken as HTT events.
To be compatible with our observations, a scenario of vertical inheritance would require the loss of similar TEs in all species of two lineages at approximately the same divergence time. This scenario does not involve a negative correlation between the fraction of species sharing similar TEs and the divergence time of lineages. However, most of the species in a lineage should appear involved in an inferred HTT event, or none of them (in the latter case, no HTT is inferred). This scenario is contradicted by the data: The fraction of species of a given lineage sharing TEs from a transfer is moderate to low (Table S1).
The negative correlation that we tested (Table S1) is also expected if the erroneous inference of HTT only involves the most closely related taxa, inferred HTT between more divergent taxa being real (distant lineages would share no homologous vertically inherited TEs). This is because TEs are horizontally acquired by the ancestor of only a fraction of the species belonging to a recipient lineage. This fraction is expected to be small if the transfer is recent. Indeed, we estimate that HTT have occurred in the past few My (Fig. S2), which is more recent than most divergence events within lineages (Fig. S1).
In principle, mistaking vertical inheritance as HTT could yield another correlation: DNA sequence divergence between TEs should correlate with that of vertically inherited genes in the same lineages. Although this correlation was not observed (we found it to be slightly negative and not significant), this is not a powerful test. Indeed, detected homology, be it between horizontally transferred or vertically inherited TEs, is constrained by the sensitivity of our megablast search, whereas homology between core genes (detected by blastp) is not.
Materials and Methods
Source Data and Time-Tree Construction.
We used the latest genome assemblies of 195 insect species (Dataset S1) at the contig level. These assemblies constitute all of the publicly available reference genome sequences of insects (Insecta) as of May 2016, excluding species for which the assembly size appeared too short. A time tree of these species (Fig. S1) was manually constructed by setting node ages to match divergence times obtained from timetree.org/ (36), using dates established by Misof et al. (23) when available.
The following steps were implemented in R scripts (37) calling other programs. Unless specified otherwise, program and function arguments were left at their default values, and homology searches used blast+ (38) algorithms, retaining only the best alignment per query.
Extraction of TEs from Genomes and Homology Search.
TE family consensus sequences were generated by RepeatModeler (18), setting “ncbi” as the search engine, and were provided as a custom library to RepeatMasker (39) to locate associated TE copies in each species’ genome, ignoring low complexity regions (option “-nolow”). Copies >100 bp were extracted from genomic contigs by using the Biostrings R package (40).
Each homology search was performed with the megablast algorithm. It used a given species’ TE copies as query and another species’ copies as target. This represented 37,830 searches (1952 − 195; that is, avoiding self-comparisons). In the following, a “hit” refers to an alignment (or high-scoring segment pair; HSP) resulting from this initial megablast search.
Defining Insect Lineages Among Which Hits Should Not Result from Vertical Inheritance of TEs.
To compare interspecific divergence at TEs to synonymous divergence of genes, we located core genes in each genome using the BUSCO pipeline (41) and its database of ancestral arthropod proteins. We concatenated exons into coding sequences (CDSs) based on coordinates reported for each complete gene. We used Megan 6 (42) to select translated CDSs that had the best homology to known arthropod proteins, and among this selection, we excluded proteins that had homologies to TEs (with an e-value of at most 10−3). These homologies were established by Diamond blastp searches (43) against the nonredundant protein database of National Center for Biotechnology Information (NCBI) and the TE database RepBase (44), respectively.
Protein sequences were compared between every two species by using reciprocal blastp searches, with an e-value threshold of 10−4. Proteins involved in reciprocal best hits and corresponding to the same ancestral protein (same BUSCO identifier) were considered orthologous. Alignments of <100 amino acids and between nonorthologs were discarded. We realigned the pair of protein regions covered by each hit with the pairwiseAlignment() function of Biostrings (40) and translated the resulting alignment into a nucleotide alignment with a custom R function. Rates of synonymous substitution (dS) between orthologous CDS were computed with Li’s method (45), as implemented by the kaks() function of package seqinr (46).
The distribution of dS for each insect clade was established on values obtained from all pairs of species spanning its two immediate subclades. To avoid pseudoreplication in dS values between a given CDS and all orthologs from the other subclade, we only used the dS value corresponding to the longest alignment of each CDS. A clade was collapsed (all TE homologies within it were ignored) if >0.3% of the dS values of orthologous core genes were lower than the highest divergence between TEs that we computed as described below.
Nucleotide divergence at horizontally transferred TEs was established on a random sample of 400,000 megablast hits obtained from pairs of species that diverged in the last 40 My (hence likely representing nonvertical transfers). We realigned TE regions based on the HSP coordinates using Biostrings and computed the distance between copies according to Kimura’s two-parameter model (47), which is the model of substitution used by Li’s method (45).
Identification of Independent HTT Events.
Candidate transfer events were identified by clustering hits involving a given pair of insect lineages and TEs from to the same superfamily, because hits between TEs from different superfamilies were discarded. See the SI Discussion and Fig. S3 for more detail on the clustering approach we used.
We first reduced the number of hits to obtain a manageable number of pairwise comparisons (SI Materials and Methods). Every two hits were “connected” if identity of TE copies within one lineage was equal to or higher than at least one of the two between-lineage identities associated with the hits. Within-lineage identity was assessed by a blastn homology search of all TE copies from the same lineage against themselves (i.e., set as both query and target) authorizing all hits for a given query. Alignments <100 bp were not recorded, and identity was considered as zero in that case. The resulting connections produced an undirected graph of hits, in which clusters were delineated by the algorithm (48) implemented in the cluster_fast_greedy() function of the igraph package (49), which maximizes within-cluster connectivity and minimizes between-cluster connectivity (SI Materials and Methods and Fig. S4 and S5). Across all TE superfamilies and lineage pairs, this yielded 8,713 clusters of hits.
To test whether any two clusters i and j represented the retention of nonoverlapping parts of an ancestral TE instead of separate transfer events, we compared protein regions identified in the TEs they involved (SI Materials and Methods). Clusters i and j were considered to represent separate HTTs if they had low connectivity (SI Materials and Methods and Fig. S5) and if protein regions overlapped by at least 100 amino acids (Fig. S6). Otherwise, these clusters were “connected”. Applying connections to every pair of clusters yielded an undirected graph of clusters where every HTT event would constitute either an unconnected cluster or a “clique” of clusters (Fig. S4B). A clique is a network whose elements (here, clusters of hits) are all directly connected (adjacent) to each other. Cliques were delineated by an algorithm (SI Materials and Methods and Dataset S3) implemented in an R function. This clustering resulted in 1,535 cliques and 5,340 unconnected clusters. We collectively refer to those as transfers or HTTs below.
To reduce the risk of cross-contamination of DNA between species seen as HTT, we imposed that the TE families involved in a transfer be represented, in each lineage, by at least five TE copies measuring at least half the length of their respective consensus. We further imposed that at least two of these copies, for each lineage, be present in the retained megablast hits.
Minimum Number of HTT Events.
The minimum number of HTT events, considering all insect lineages, was counted by establishing networks of lineages connected by transfers of similar TEs (Fig. S7). In such a network, every apparent transfer between two lineages may result from two acquisitions of TEs from (an)other lineage(s), which, according to parsimony, are already represented by transfers in the network. However, two transfers that were previously identified as independent, and involving the same pair of lineages, cannot both result from the same two acquisitions, and should be in different networks. To establish networks, every two transfers were connected if at least one given TE family was involved in both transfers and if these were not previously characterized as independent (SI Materials and Methods and Fig. S7). From the resulting graph, networks were delineated by single-linkage clustering. To avoid considering independent transfers in the same network, we split the network into cliques that cannot contain independent transfers (SI Materials and Methods).
Dating HTT Events.
We approximated the time since a transfer by the minimum between-lineage nucleotide divergence of copies resulting from a HTT. This proxy may overestimate the age of the transfer under a scenario where the two lineages considered have not directly exchanged TEs (and acquired these from a third party) or where the sampled species diverged from the donor of TEs before the transfer.
We thus used another proxy, based on the divergence of TEs within the supposed recipient lineage. This measure may underestimate the age of TE acquisition, but is less influenced by the species pair used to date the transfer. Within-lineage divergence associated to a transfer was taken, for each of the two lineages involved, as the ninth decile of the raw nucleotide divergence between TE copies included in the corresponding cluster of hits, which we previously estimated by blastn searches. We used the ninth decile rather than the maximum, because the latter would have put high weight on the two copies that diverged the most. To consider that TEs may have diverged within one of the two lineages (the donor) before the transfer, we used the lower decile value among the two. If the transfer was a clique of several cluster of hits, we used the value obtained from the cluster that comprised the hit having the highest identity, for consistency with the estimate based on between-lineage divergence.
Analysis of Biogeographic Data.
Native biogeographic realms of 179 insect species (Dataset S1) were obtained from several Internet sources. Within a lineage, closely related species occupying distinct realms may appear involved in the same HTT, due to speciation before or after the transfer. The two realms we associated to each HTT were those of the two species (one per lineage) that yielded the hit of highest identity, which we used to date the transfer (as described above). To avoid counting the same HTT several times, all other species were considered not involved in the transfer. This selection is equivalent to a random draw among the species descending from an ancestor that acquired or emitted TEs, should speciation have occurred after the transfer.
Correlation between time since a transfer and cooccurrence of the species involved (defined as originating from the same realm and encoded as a binary value) was estimated by Pearson’s R (n = 3,863 transfers between located species). To test its significance, we computed R 104 times after randomly permuting realms across species and compared it to the value obtained from real data.
Supplementary Material
Acknowledgments
We thank Bouziane Moumen and Mohamed Amine Chebbi for their help with bioinformatic procedures and Nicolas Bech for helping to create a world map. We thank the genotoul bioinformatics platform Toulouse Midi-Pyrénées (bioinfo.genotoul.fr/) and genouest (https://bipaa.genouest.org/) for providing computing and storage resources. This work was supported by Agence Nationale de la Recherche Grant ANR-15-CE32-0011-01 TransVir (to C.G.); the 2015–2020 State-Region Planning Contract and European Regional Development Fund; and intramural funds from the Centre National de la Recherche Scientifique and the University of Poitiers.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621178114/-/DCSupplemental.
References
- 1.Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: Building the web of life. Nat Rev Genet. 2015;16:472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 2.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: The agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 3.Ku C, Martin WF. A natural barrier to lateral gene transfer from prokaryotes to eukaryotes revealed from genomes: The 70 % rule. BMC Biol. 2016;14:89–100. doi: 10.1186/s12915-016-0315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keeling PJ. Functional and ecological impacts of horizontal gene transfer in eukaryotes. Curr Opin Genet Dev. 2009;19:613–619. doi: 10.1016/j.gde.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Richards TA, et al. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci USA. 2011;108:15258–15263. doi: 10.1073/pnas.1105100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szöllősi GJ, Davín AA, Tannier E, Daubin V, Boussau B. Genome-scale phylogenetic analysis finds extensive gene transfer among fungi. Philos Trans R Soc Lond Ser B Biol Sci. 2015;370:20140335. doi: 10.1098/rstb.2014.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham LA, Li J, Davidson WS, Davies PL. Smelt was the likely beneficiary of an antifreeze gene laterally transferred between fishes. BMC Evol Biol. 2012;12:190–202. doi: 10.1186/1471-2148-12-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christin PA, et al. Adaptive evolution of C(4) photosynthesis through recurrent lateral gene transfer. Curr Biol. 2012;22:445–449. doi: 10.1016/j.cub.2012.01.054. [DOI] [PubMed] [Google Scholar]
- 9.Wallau GL, Ortiz MF, Loreto EL. Horizontal transposon transfer in eukarya: detection, bias, and perspectives. Genome Biol Evol. 2012;4:689–699. doi: 10.1093/gbe/evs055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dotto BR, et al. HTT-DB: Horizontally transferred transposable elements database. Bioinformatics. 2015;31:2915–2917. doi: 10.1093/bioinformatics/btv281. [DOI] [PubMed] [Google Scholar]
- 11.El Baidouri M, et al. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 2014;24:831–838. doi: 10.1101/gr.164400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. AMS Press; Washington, DC: 2002. p. 1204. [Google Scholar]
- 13.Schaack S, Gilbert C, Feschotte C. Promiscuous DNA: Horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol. 2010;25:537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivancevic AM, Walsh AM, Kortschak RD, Adelson DL. Jumping the fine LINE between species: Horizontal transfer of transposable elements in animals catalyses genome evolution. BioEssays. 2013;35:1071–1082. doi: 10.1002/bies.201300072. [DOI] [PubMed] [Google Scholar]
- 15.Dupeyron M, Leclercq S, Cerveau N, Bouchon D, Gilbert C. Horizontal transfer of transposons between and within crustaceans and insects. Mob DNA. 2014;5:4–13. doi: 10.1186/1759-8753-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallau GL, Capy P, Loreto E, Le Rouzic A, Hua-Van A. VHICA, a new method to discriminate between vertical and horizontal transposon transfer: Application to the mariner family within Drosophila. Mol Biol Evol. 2016;33:1094–1109. doi: 10.1093/molbev/msv341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maumus F, Fiston-Lavier AS, Quesneville H. Impact of transposable elements on insect genomes and biology. Curr Opin Insect Sci. 2015;7:30–36. doi: 10.1016/j.cois.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Smit AFA, Hubley R. 2015 RepeatModeler Open-1.0. Available at www.repeatmasker.org/
- 19.Vassetzky NS, Kramerov DA. SINEBase: A database and tool for SINE analysis. Nucleic Acids Res. 2013;41:D83–D89. doi: 10.1093/nar/gks1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lampe DJ, Witherspoon DJ, Soto-Adames FN, Robertson HM. Recent horizontal transfer of mellifera subfamily mariner transposons into insect lineages representing four different orders shows that selection acts only during horizontal transfer. Mol Biol Evol. 2003;20:554–562. doi: 10.1093/molbev/msg069. [DOI] [PubMed] [Google Scholar]
- 21.Bartolomé C, Bello X, Maside X. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009;10:R22. doi: 10.1186/gb-2009-10-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robertson HM. Evolution of DNA transposons in eukaryotes. In: Craig NLea., editor. Mobile DNA II. ASM; Washington, DC: 2002. pp. 1093–1110. [Google Scholar]
- 23.Misof B, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346:763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 24.Hooper SD, Mavromatis K, Kyrpides NC. Microbial co-habitation and lateral gene transfer: What transposases can tell us. Genome Biol. 2009;10:R45–54. doi: 10.1186/gb-2009-10-4-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popa O, Dagan T. Trends and barriers to lateral gene transfer in prokaryotes. Curr Opin Microbiol. 2011;14:615–623. doi: 10.1016/j.mib.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 26.Wagner A, de la Chaux N. Distant horizontal gene transfer is rare for multiple families of prokaryotic insertion sequences. Mol Genet Genomics. 2008;280:397–408. doi: 10.1007/s00438-008-0373-y. [DOI] [PubMed] [Google Scholar]
- 27.Lampe DJ, Churchill ME, Robertson HM. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 28.Ivics Z, Izsvak Z. Sleeping beauty transposition. Microbiol Spectrum. 2015;3:MDNA3-0042-2014. doi: 10.1128/microbiolspec.MDNA3-0042-2014. [DOI] [PubMed] [Google Scholar]
- 29.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–627. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peddigari S, Li PW, Rabe JL, Martin SL. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res. 2013;41:575–585. doi: 10.1093/nar/gks1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartl DL, Lohe AR, Lozovskaya ER. Modern thoughts on an ancyent marinere: Function, evolution, regulation. Annu Rev Genet. 1997;31:337–358. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- 32.Silva JC, Loreto EL, Clark JB. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 2004;6:57–71. [PubMed] [Google Scholar]
- 33.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venner S, et al. Ecological networks to unravel the routes to horizontal transposon transfers. PLoS Biol. 2017;15:e2001536. doi: 10.1371/journal.pbio.2001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, VA: 2016. [Google Scholar]
- 38.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421–429. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smit AFA, Hubley R, Green P. 2015 RepeatMasker Open-4.0. Version 4.0. Available at www.repeatmasker.org/
- 40.Pagès H, Aboyoun P, Gentleman R, DebRoy S. 2016. Biostrings: String objects representing biological sequences, and matching algorithms. R package version 2.40.0.
- 41.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 42.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 44.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11–16. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li WH. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 46.Charif D, Lobry JR. 2007. Seqin{R} 1.0-2: A contributed package to the {R} project for statistical computing devoted to biological sequences retrieval and analysis. Structural Approaches to Sequence Evolution: Molecules, Networks, Populations, eds Bastolla U, Porto M, Roman HE, Vendruscolo M (Springer, New York), pp 207–232.
- 47.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 48.Clauset A, Newman MEJ, Moore C. Finding community structure in very large networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70:066111. doi: 10.1103/PhysRevE.70.066111. [DOI] [PubMed] [Google Scholar]
- 49.Csardi G, Nepusz T. 2006. The igraph software package for complex network research. InterJournal Compex Systems:1695.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.