Targeting bacterial cell wall carbohydrates
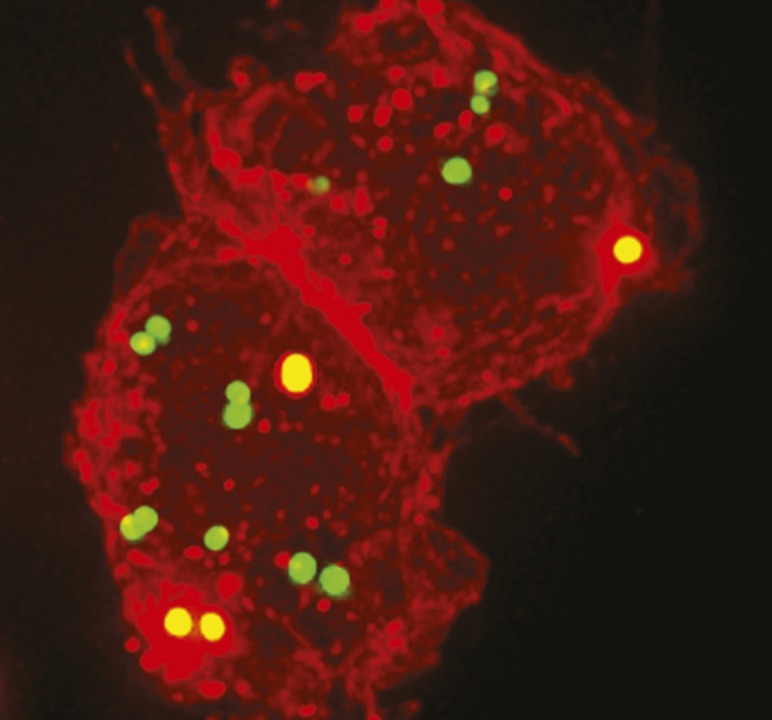
MRSA (green) engulfed by macrophages (red) following treatment with a lysibody.
The cell walls of Gram-positive bacteria contain numerous surface carbohydrates that are often highly conserved across species, providing a potential therapeutic target for the attachment of high-affinity antibodies. However, the poor immune response provoked by these bacterial targets has made obtaining such antibodies a challenge. Assaf Raz et al. (pp. 4781–4786) demonstrate the ability of binding domains from autolysins and lysins—enzymes that break down bacterial cell walls—to bind with the human antibody IgG fragment crystallizable region. The resulting molecule, termed a lysibody, is a functional homodimer that binds with high-affinity and specificity to carbohydrate determinants on the bacterial surface. Furthermore, the authors demonstrate the reproducibility of the approach with three separate binding domains for methicillin-resistant Staphylococcus aureus (MRSA). The lysibodies induced complement fixation on the surface of MRSA cells, and promoted phagocytosis and killing of MRSA by macrophages and human polymorphonuclear neutrophils. Lysibodies also protected mice from infection with MRSA in two models. According to the authors, lysibodies might aid the treatment of a range of bacterial infections. — C.S.
Tethered antibodies and virus resistance
Antibodies can be anchored to the plasma membrane of cells for specific functions. Jia Xie et al. (pp. 4655–4660) used such an approach to bind membrane-tethered antibodies (MTA) to cell surface receptors that doubled as virus receptors to render cells resistant to viral infection. The authors delivered MTAs that targeted the rhinovirus A and B receptor into HeLa cells before exposing the cells to rhinovirus. Of the surviving cells, cells expressing protective MTAs were more likely to survive rhinovirus exposure than cells with MTAs that had been selected only for binding. By adjusting the antibody’s antigen-binding site, the authors improved the likelihood of T-cell attachment, while maintaining virus-neutralizing capabilities. Further, the authors engineered HeLa-derived cells to express MTA-configured CD4 antibodies. The cells demonstrated almost complete inhibition of infection for four of six HIV isolates tested as well as significant protection for the remaining isolates. In both experiments, cells were more likely to survive if they contained MTAs than if they contained the same antibodies in soluble form. According to the authors, attaching MTAs to virus receptors may help generate virus-resistant cells, and resistant cells may have a selective advantage. — L.C.
Codon position and mRNA translation efficiency
The genetic code of life consists of RNA codons, which specify amino acids and sites of termination of protein synthesis. Several factors, including the relative positions of codons, can influence the efficiency of codon translation during protein synthesis. Fabienne Chevance and Kelly Hughes (pp. 4745–4750) report the effects of synonymous codon changes on the expression of the Salmonella regulatory protein FlgM, coded by the flgM gene. At a codon site in flgM, the authors found that substituting the Thr6 and Pro8 codons with synonymous alternates produced a 600-fold range in FlgM regulatory activity. Similar synonymous changes in the Thr6 and Leu9 codons resulted in a twofold range in FlgM regulatory activity. In a subsequent in vivo assay, the authors linked the level of FlgM activity produced by any codon arrangement to ribosome stalling at synonymous codons. Furthermore, the codon arrangements that affected FlgM protein levels were independent of flgM messenger RNA (mRNA) stability or expected mRNA secondary structures. The findings suggest that the efficiency of mRNA translation might depend on a triplet-of-triplet genetic code, in which translation of a given codon is influenced by the flanking codons. According to the authors, the effects of codon position on mRNA translation efficiency might be explained by codon recognition interactions. — C.S.


