Significance
Cell separation is a fundamental process in biomedicine, but is presently complicated, cumbersome, and expensive. We present a technique that can sort cells at a rate equivalent to or faster than gold-standard techniques such as fluorescence- and magnetic-activated cell sorting, but can do it label-free and with very low cell loss. The system uses dielectrophoresis to sort cells electrostatically, using an electrode chip that eschews microfabrication in favor of a laminate drilled with 397 electrode-bearing wells. This high level of parallelization makes the system immune to the bubbles that limit labs-on-chip, while also increasing capacity and throughput to levels comparable with high-throughput FACS and MACS, whereas the chip is cheap enough to be disposable, preventing interseparation contamination.
Keywords: dielectrophoresis, DEP, lab on a chip, FACS, MACS
Abstract
Currently, cell separation occurs almost exclusively by density gradient methods and by fluorescence- and magnetic-activated cell sorting (FACS/MACS). These variously suffer from lack of specificity, high cell loss, use of labels, and high capital/operating cost. We present a dielectrophoresis (DEP)-based cell-separation method, using 3D electrodes on a low-cost disposable chip; one cell type is allowed to pass through the chip whereas the other is retained and subsequently recovered. The method advances usability and throughput of DEP separation by orders of magnitude in throughput, efficiency, purity, recovery (cells arriving in the correct output fraction), cell losses (those which are unaccounted for at the end of the separation), and cost. The system was evaluated using three example separations: live and dead yeast; human cancer cells/red blood cells; and rodent fibroblasts/red blood cells. A single-pass protocol can enrich cells with cell recovery of up to 91.3% at over 300,000 cells per second with >3% cell loss. A two-pass protocol can process 300,000,000 cells in under 30 min, with cell recovery of up to 96.4% and cell losses below 5%, an effective processing rate >160,000 cells per second. A three-step protocol is shown to be effective for removal of 99.1% of RBCs spiked with 1% cancer cells while maintaining a processing rate of ∼170,000 cells per second. Furthermore, the self-contained and low-cost nature of the separator device means that it has potential application in low-contamination applications such as cell therapies, where good manufacturing practice compatibility is of paramount importance.
Since the separation of red and white blood cells was first reported in 1974 (1), the technology has become fundamental in the biomedical sciences. From isolation of diseased tissue to the identification of cells for therapeutic uses and the potential for regenerative cell-based therapies, cell-separation techniques are increasingly integrated into other fields such as biochemistry, electrical engineering, physics, and materials science (2). Three methods dominate cell separation. Density gradient methods exploit differences in density between populations such as red/white blood cells. Fluorescently activated cell sorting (FACS) uses fluorophore-conjugated antibodies as a discriminator; cells are launched in droplets, each containing one cell, through a fluorescence detection system to determine the cell type and are then electrostatically diverted into different output receptacles (3). FACS can sort up to 50,000 cells per second, with higher rates achievable at the cost of purity. Finally, magnetically activated cell sorting (MACS) uses magnetic microbeads conjugated with antibodies. These bind to targets on cell surfaces, which then can be extracted by applying a magnetic field (4).
However, the three methods have drawbacks. Only density gradient does not require the use of chemical labels; the others use fluorescent chemicals or antibodies to indicate the target population. These are expensive and may have limited specificity; in the case of MACS, the target protein must be present on the surface of the cells. Following separation, the labels may also persist in the cells, limiting their usefulness. Cell losses in FACS and MACS can exceed half the population, particularly at high sorting rates (5, 6). FACS machines (particularly high-throughput devices) are very expensive and are not easily cleaned, rendering cell populations separated by this method noncompliant with good manufacturing practice (GMP). Finally, as the cell preparation includes a period during which labels are added and then given sufficient time to conjugate, the total time to perform the sort can extend over several hours.
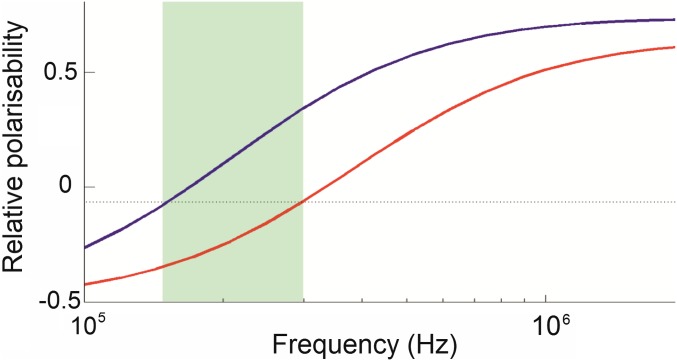
There have been many attempts to develop cell-separation techniques that are affordable to buy and use, do not require labels, and are able to retain significant numbers of cells. An alternative separation technique is dielectrophoresis (DEP). A polarizable particle suspended in a nonuniform electric field (7) interacts with the field gradient, inducing a dipole. The interaction of dipole and field gradient produces different Coulomb forces on either pole, inducing motion up or down the high field gradient according to the polarity of the dipole. The magnitude and polarity of the dipole itself are governed by the electrical properties of the cell (resistance and capacitance of membrane and cytoplasm) and medium, and the frequency of the electric field. DEP can be used both to characterize and separate cells according to the passive electrical properties, where two cell types experience a field gradient at a frequency such that one cell type is attracted to the electrodes, one repelled. The repelled cells pass through the device unaffected, but the others are attracted to, and retained by, the electrodes acting as an “electrostatic filter.” When the field is removed, these cells are released and can be collected separately. For example, Fig. 1 shows spectra of two identical cell types, save that one has double the membrane capacitance of the other. The highlighted frequency range indicates where one cell type experiences positive DEP, the other negative. DEP spectra can be determined by commercial devices such as the DEPtech 3DEP, and exploitable differences can be readily identified by visual inspection.
Fig. 1.
DEP spectra of two arbitrary cell types, whose properties are identical save for one (blue line) having a membrane capacitance twice that of the other (red line). In the frequency band (highlighted), the polarizabilities (and hence direction of force) of the two cells is different, enabling the cells to be separated.
DEP separation of live and dead yeast cells was first demonstrated in 1966 (8). Subsequent research demonstrated separation of cancerous and healthy cells, Gram-negative and Gram-positive bacteria, stem-cell subpopulations, and different types of viruses for sample preparation, whereas differences between cells indicated the potential for DEP to fractionate leukocyte subpopulations (9–20). However, few approaches to DEP have been able to compete with MACS or FACS: (i) implementations often suffer from cells being trapped in interconnecting tubing, devices requiring small chamber heights to ensure cells pass close to the electrodes, restricting throughput and ultimately limiting total cell capacity; and (ii) the high cost of manufacturing, making it difficult to mass-produce the separator as a reliable, disposable component. There are exceptions; the ApoStream method (21) can isolate significant numbers of circulating tumor cells from 12 million nucleated peripheral blood cells in 60 min (5,000 cells per second). Hu et al. (22) reported separating 10,000 cells per second, but required the use of chemical labeling. Lee et al. (23) reported a rate of 616 cells per second. Markx et al. (24) processed live and dead yeast cells at 107 cells per milliliter with purity in excess of 93%; however, the total capacity of the device was only 50 µl, limiting overall throughput. An alternative approach to DEP separation used 3D chips constructed from interleaved conducting and insulating sheets with holes or “wells” drilled through (25). The first prototype sorted 50:50 live and dead yeast to 86:14 at 0.4 mL min−1. A subsequent design (26) included remixing by passing through multiple wells, producing high cell recovery (>90) but low throughput (25 µL min−1).
To be comparable to FACS or MACS requires cell sorting at rates in excess of 105 s−1 with minimal cell loss, high purity, low cost, and highly robust. Here we describe an electrophysiology-activated cell enrichment (EPACE) method using a chip with 397 holes of 400-µm-diameter drilled through a laminate of 12 conducting layers separated by insulators, through which 397 wells of 400-µm-diameter were drilled. As shown in Fig. 2, this creates 397 parallel paths through the chip, each with 12 electrodes along the bore. This high degree of parallelization, and the fact that all cells are no more than 200 µm from the electrodes, allows very high cell-processing rates while minimizing cell loss, and creates immunity from bubbles (which will, at worst, block only one channel). Consequently, the system can separate cells at rates substantially in excess of 320,000 cells s−1 measured across all cell types studied, separating populations in excess of 108 cells into output receptacles in under 30 min with cell losses as low as 0.3%, and separation efficiencies as high as 96.4%, achieving more than 30× higher throughput of previous DEP devices.
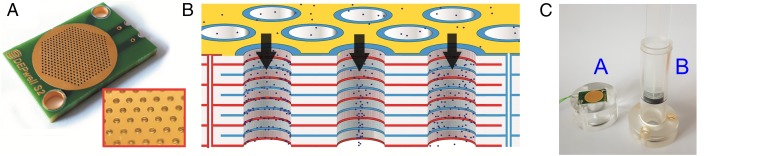
Fig. 2.
(A). Photograph of the DEP separation chip. The chip size is 30 mm × 20 mm. (Inset) Red section shows a close-up of the chip, showing the electrodes along the inside of the wells. The section of the chip in the image is ∼5 mm square. (B) Schematic of the chip, showing the three modes of DEP behavior. Cells flow from top to bottom through the wells; in the left well, cells can be seen experiencing positive DEP, are attracted to the electrodes, and held; in the center well, cells experience negative DEP, are repelled into the center of the well, and pass through. In the third well, cells experience no DEP force. In reality, cells of the same type will experience the same mode of behavior in all wells on the chip, but two different cell populations can exhibit behaviors different from each other. If one subpopulation experiences positive DEP and the others exhibit negative or neutral behavior, they can be separated. (C) The chip is loaded into a fluidic cartridge comprising two parts; an upper part B contains both housing and plunger, whereas a lower part A collects the cell solution. The chip fits between the two, sealed on both sides by O rings, and is clamped together by three Allen bolts.
Results
Cell Enrichment Using a Single-Pass Protocol.
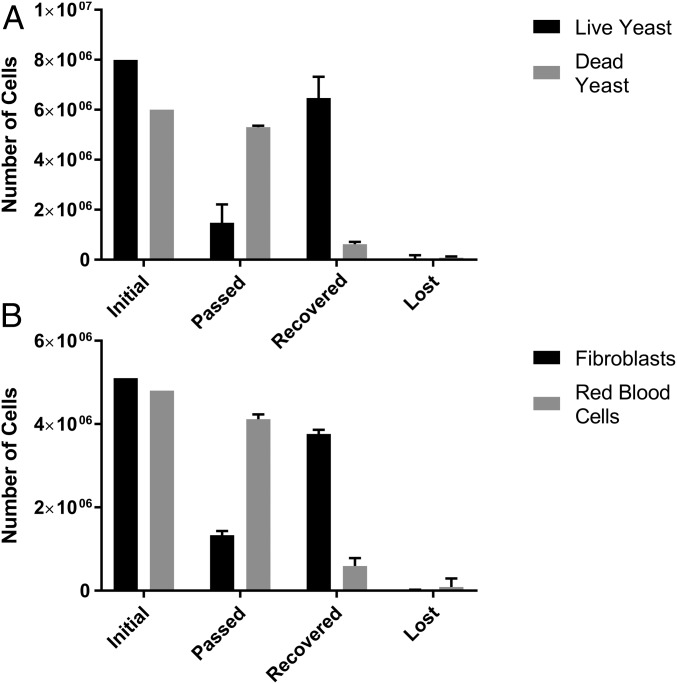
To assess separation efficacy, binary mixtures of cells were processed through the device in 5–15-mL units, processed at between 15,000 and 33,000 cells per second, the results of which are summarized in Fig. 3. Live and dead yeast showed mean recovery (the proportion of cells output in the correct outlet) of 81.3% and 89.5%, respectively; vole blood and fibroblasts showed separation efficiencies of 87.3% and 73.9%, respectively. Purities (the proportion of required cells in the output population) at the output for the above populations were 91.3% and 78.2% for live and dead yeast, and 75.6% and 86.4% for vole RBCs and fibroblasts. Whereas these values are low when considering the technique as a separation per se, they represent sufficiently high values to constitute effective cell enrichment. It is also significant that for the above population sorts, the mean cell losses (cells which were input but are unaccounted for at the output, due to damage, adherence to the electrodes or device casing, or being left in residual liquid in the device at the end of the experiment) were 1.4%, 2.7%, 4%, and 0.3%, respectively. These low cell losses are important for two reasons; first, multiple passes can be used to improve performance without significantly degrading cell numbers; second, unlike methods such as FACS, where a significant number of cells being separated are destroyed by the separation process (6), this protocol does not reduce cell number––an important issue where initial cell numbers are low. Experiments at 0.5, 0.8, and 1.0 mL min−1 yielded similar results, suggesting these metrics represented a plateau of trapping efficiency, and that the device was working well within its performance limit.
Fig. 3.
Average (n = 3) results of the enrichment of mixtures of (A) live and dead yeast and (B) RBCs and fibroblasts, showing the total cell number in each case. “Initial” refers to the numbers at the start of separation, “passed” represents the cells collected during the separation, “recovered” represents those cells collected by positive DEP and subsequently removed, and “lost” is those unaccounted for. Cells were processed through the device at ∼1 million cells in 1 mL/min.
To assess the viability of cells postsort, collected fibroblasts were suspended in fibroblast-growing medium, divided into two T75 flasks, and placed in the incubator to assess growth potential. The cells were observed 1 and 3 d after incubation and were found to have adhered to the flask and reached confluency, respectively. A DEP spectrum of the cells was obtained and a further passage performed to check for abnormalities; none was observed.
To identify the upper-throughput limit, live yeast cells were captured in the device in increasing concentrations, as it is the fraction of cells experiencing positive DEP that is limited by the capacity and efficacy of the DEP electrodes (the other, repelled, population simply passing through the device unaffected). Retaining the 1-mL min−1 flow rate, the concentration of cells per milliliter was increased from 106 mL−1 to ∼2 × 106, 5 × 106, 107, 2 × 107, and 108 cells per milliliter. Chip saturation was monitored in two ways: first, we measured the total number of cells retained in the chip and subsequently recovered. Second, cells were aliquoted in 1-mL fractions and analyzed to determine the cell ratio to observe trap saturation. Saturation can then be detected by the appearance of a higher proportion of live cells in the output stream. Any saturation effect would only affect cells collected by positive DEP on the electrodes; cells experiencing zero or negative DEP pass through the chip unimpeded and are collected in the receptacle below, and as a consequence there is no saturation limit on such cells.
At concentrations up to 2.7 × 107 mL−1 the device worked as for 1 × 106 mL−1. When concentrations of 2.7 × 107 mL−1 were used, the aliquots were normal and the mean cell recovery was 279 ± 12.2 million cells. When the input concentration was 5.9 × 107 mL−1, the number of cells increased dramatically (by over 300%) at the sixth aliquot, and the mean total of recovered cells was 321 ± 9.0 million cells; when 1.07 × 108 mL−1 were used, the device became saturated after the third aliquot (rising by 38% in aliquot 4, rising to >100% in aliquot 7 and later), and recovery indicated that the trap contained 359 ± 13.3 × 106 cells. After the separation run was completed, the cells collected in the chip were flushed and counted. In the three instances where the chip was saturated, the number of cells retained before the chip was saturated, with upper limits of ∼350 × 106 cells representing the capacity of the chip.
The difference in capacity between 59 million and 109 million cells per milliliter is interesting, as it suggests the trap capacity may be increased at high concentrations due to cell–cell interactions; however, it appears clear that a 300 × 106 cell sort (20 million cells per milliliter, 15 mL total volume) represents the maximum effective cell separation where the cell content is entirely unknown, although this represents the limit only on the positive DEP fraction; a sort of 300 million cells experiencing positive DEP from a further 300 × 106 (or more) experiencing negative DEP would in principle be entirely workable, raising the throughput past 0.6 × 109 cells per milliliter.
High-Volume Separation Performance Using Multipass Protocols.
Whereas enrichment on this scale is of interest, a second or third pass through the device enables the user to collect cells that were not retained in previous passes, significantly improving performance in a very short time. There are two strategies for multiple sorting passes: re-sorting the effluxed and retained cells, respectively. Here we examine these two protocols separately: a two-pass strategy reprocessing the effluxed cells to minimize cell loss while maximizing enrichment in the shortest possible time, and a three-pass strategy where the collected cells are reprocessed.
Enhancing the negative DEP population.
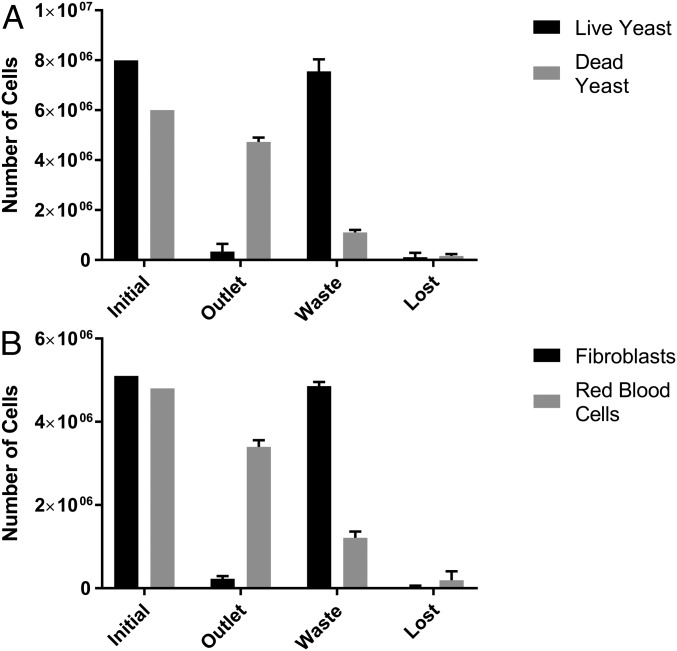
We sorted mixtures of 9.6–14 × 106 cells (similar to the cell number in a confluent T175 flask) at concentrations of 1–2 × 106 cells per milliliter and passed through the EPACE twice. After each run, the chip was flushed with fresh medium to recover cells retained by positive DEP before the output was reprocessed. Negative DEP was used to select the dead cells from mixtures of live and dead yeast cells, or RBCs from mixtures of RBCs and fibroblasts, the results of which can be seen in Fig. 4. After the second pass the purity of dead yeast was increased to 93.4% while the RBC fraction increased to 93.8%. However, recovery was reduced, falling to 81% and 73.7%, respectively, although these are still substantially above recovery rates for FACS and MACS. Overall cell losses (cells not appearing in either outlet) were 2.7% and 4%, with the remaining cells appearing in the other (waste) output. We postulated that the values of separation efficiency for the populations experiencing negative DEP were adversely affected by cells being trapped in the dead volume between the chip and outlet, which were then recovered with the retained cells rather than passing to the effluxed portion. To verify this, we performed a separation of 20 mL of RBC/fibroblast cells at the same concentration. After the second pass, the recovery of RBCs was broadly similar at 87.8%, but the purity rose to 96.6%.
Fig. 4.
Average (n = 3) results of a second stage of separation of mixtures of (A) live and dead yeast and (B) RBCs and fibroblasts, showing the total cell number in each case. For the second step, the cell population experiencing negative DEP was reprocessed. “Initial” refers to the numbers at the start of separation, “outlet” represents the cells collected during the separation, “waste” represents those cells collected by positive DEP and subsequently removed, and “lost” is those unaccounted for. Cells were processed through the device at 1 mL/ min−1; with an initial cell concentration of ∼1 million cells per milliliter.
Enhancing the positive DEP population.
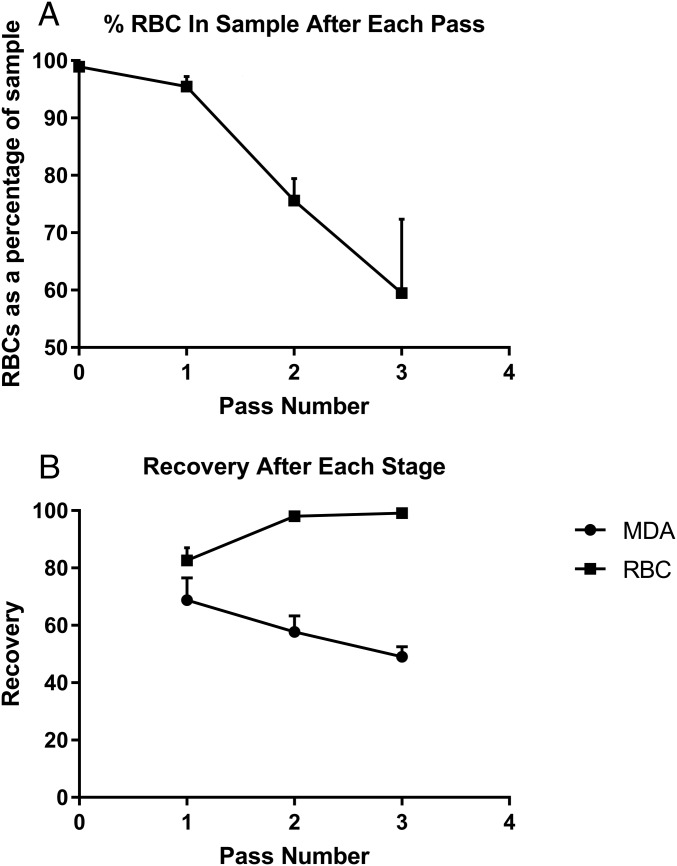
An important application of cell separation is the enrichment of relatively rare subpopulations. To use the EPACE platform to enrich highly asymmetrical cell concentrations a three-pass strategy was used, with the cell fraction retained in the electrodes being retrieved and then subject to two further iterations (total time taken ∼30 min). The cells collected were counted after each pass, and the ratio of cell types was determined. These results are summarized in Fig. 5. Human RBCs were spiked with MDA-MB-231 breast cancer cells, to a final concentration of 1.1% cancer to 98.9% RBCs, at a total cell concentration of ∼2 × 107 mL−1, to a total sample volume of 4 mL. Cells were passed through the device at 0.5 mL min−1. After the cell mixture had completed its first pass through the device, the fraction retained in the device was released in 1 mL of fresh medium, which was topped up to 3 mL with fresh medium after cell counting. Sampling indicated that the concentration of RBCs had dropped to 95.5% of the output. The 3 mL of solution was then subject to a second pass and resuspension, at which point RBCs had dropped to 75.6%, and after the third pass, RBCs made up 59.5% of the cell sample, with cancer cells forming the remaining 40.5% of the cells, an effective removal rate of the RBCs of 99.1%. At the end of the process, on average 47.7% of the initial population of cancer cells were recovered. However, those that had not been collected in the initial sort would be available for resorting in the waste from the previous passes, which could be used to significantly increase cell recovery if required, by adding an additional pass on the eluted cells.
Fig. 5.
Enrichment of MDA cancer cells cancer cells from RBCs, at an initial ratio of 1:99, average (n = 3) results using a three-pass protocol. (A) The percentage of RBC cells in the sample, per run. (B) The mean (n = 3) overall recovery of the two cell types after each stage; the recovery of RBCs reaches 99.1%, suggesting that the cancer cells have been significantly enriched.
Discussion
Analysis of Performance.
Whereas purity of sample at output is the final arbiter of the quality of a separation/enrichment method, it is difficult to use as a guide as it depends on the concentration of the two cell types at the input. Instead, we can look at the concentration of both the desired and undesired cells. Analyzing the two-pass protocol to enrich live and dead yeast and RBC/fibroblast cells by negative DEP, the population experiencing negative DEP has recovery of ∼90% for the desired cells and 20% for the undesired fraction for each round of enrichment. After two passes, these are approximately squared, such that ∼0.81× the desired and 0.04 undesired cells are present at the output, leading to a purity of 0.81/(0.81 + 0.04) = 95.3%, in line with experimental results. Similarly, for enrichment of cells experiencing positive DEP, across the three rounds of separation the same process was observed, with an average of ∼80% of MDA cells and 20% of RBCs appearing at the output for each round of enrichment. Over successive passes, this predicts RBC proportions of 96%, 86%, and 60%, in line with observations. The cell losses are sufficiently low for them to be disregarded in the calculation.
Effect of Design on Optimum Performance.
From our results, it is possible to identify ways to optimize the cartridge design and increase separator performance by improving the values of cell recovery for the wanted and unwanted cells. Notably, whereas the peak values of recovery and purity are high, it is the passed cells that have the highest purity, whereas retained cells always have the highest recovery. As configured, the device contains two “dead volumes” of sample that cannot be recovered at the end of the experiment. One occurs due to the use of a conical syringe plunger; at maximum insertion the tip of the cone touches the chip and 0.56 mL of unseparated cells remain surrounding the plunger tip. Similarly, ∼0.54 mL of postseparation cells remain in the space between the chip and outlet. Consider a mixture of two populations A and B, where population A experiences negative DEP and passes through the chip while population B is retained by positive DEP; the mixture is contained in a 10-mL sample. After the solution is passed through, 1.1 mL (11%) of cells remain in the dead volume, limiting maximum recovery of A to 89%. However, purity is unaffected because only the cells which have been passed through by the chip are collected. When the solution has been processed, we then draw 1 mL into the device and release population B. However, the device also contains 1.2 mL of the original cell mixture, containing cells from population A. This means that the recovery rate for mixture B is as high as can be obtained (because all of the cells are in the extracted volume), but the purity is downgraded by the presence of population A cells in the dead volumes. In effect, the dead volumes act to take cell mixtures intended for the output for population A, and deposit them in the output to population B. Whereas a future embodiment of the device could reduce the dead volumes by redesigning the plunger and outlet path, we can calculate the effective peak separation by mathematically removing the cells in the dead volumes. We estimate that using this approach, the recovery of RBCs and purity of fibroblasts in the separation described above would equal or exceed the same parameters for the two parameters unaffected by the dead volumes, which is to say that all purities and recoveries would exceed 95% for a two-pass strategy.
The design outlined in this paper is a proof of concept, but a number of design modifications suggest themselves to improve simplicity, throughput, and capacity. For example, the device presented here is loaded manually by drawing the syringe plunger upward. This means there is little control of the flow rate during loading; hence, during this time the chip is not energized. However, with the application of a bidirectional syringe pump that can both push and pull, it would be possible to separate on both draw and expel cycles, making separation simpler and more efficient. The single inlet of the cartridge also lends itself to combining with simple liquid handing /fraction collection systems, allowing automation of complex multipass protocols without user intervention. It is also evident that the design presented here could be modified to increase throughput. For example, a second chip, independently energized, could be mounted below the first to permit a second stage of separation; this could either be at the same frequency as the first, doubling the separation rate by removing the necessity for two separation passes, or at a different frequency to allow two-parameter sorting, while requiring no further antibodies (as would be required for FACS and MACS), making for a negligible cost increase over a single-chip version. Similarly, the throughput and total cell capacity could be increased by enlarging the chip and increasing the number of wells; this could be achieved by making a single electrode disk larger, or otherwise increasing the number of wells For example, a chip with ∼3,500 wells could potentially separate up to 1010 cells in a similar 30-min period, potentially allowing separation on industrial scales, or clinical applications such as sorting of stem cells from bone marrow.
Comparison with Other Separation Methods.
As described earlier, cell-separation methods requiring more sensitivity than density gradients can be addressed by two methods: FACS and MACS. A high-throughput FACS system is expensive; both FACS and MACS require expensive labels, and can only separate cells on the basis of this labeling. The EPACE system presented here separates on the basis of physical parameters without the need for labels, and both the instrumentation (pump and generator) and consumables required (media, chips) are an order of magnitude less expensive than either method. Furthermore, the system presented here could be manufactured as a fully assembled, sterile, integrated cartridge; with no cross-over possible with other reparation runs and no residual chemical labels to adversely affect cells, this allows the cells produced to be fully compliant with GMP.
Cell-throughput rates also compare well with other methods. High-throughput FACS cell sorters can process up to 100,000 events per second, although a relatively small proportion of these may actually contain cells. MACS is a bulk method rather than processing cells serially, but the time taken to prepare and perform separation is comparable to the method presented in this paper. Similarly, the number of cells to be separated by high-volume MACS systems is 109 cells; although no upper limit for FACS has been identified, a wide review of the literature reports few studies exceeding 2 × 107 cells. Baseline EPACE performance is comparable to MACS, and better than FACS. For example, the capacity of the device is ∼4 × 108 yeast cells; balancing the relatively small size (and hence higher packing density) of such cells against the fact that DEP trapping force scales with cell volume, a capacity of >108 mammalian cells is certainly not unreasonable. In terms of throughput, if we divide the number of cells processed by the time required for a two-pass sort, we have an effective sorting rate of 167,000 cells per second; adding in a third sort, as in the case of the rare cell study, still gives a net throughput comparable to high-throughput FACS. Considering cell loss, EPACE is shown to be comparable to, or better than, either existing method. In no case were more than 7% of cells lost, and in many cases fewer than 3% of cells were lost. By comparison, FACS typically loses >50% of cells through cell damage during droplet formation or rejection due to incorrect scanning, particularly at higher flow rates (5). When used in three-pass mode, cell recovery was lower, but the cells not initially captured by the device will be available for resorting in the outlet stream by using a more complex multiple-run separation strategy.
Comparing our method to published DEP methods, the EPACE process is 30× faster than the Hu et al. (22) system, 6× faster than the Gupta et al. (21) system, and 600× faster than the system presented by Lee (23), with comparable or better cell recovery and cell losses. The cell concentration and recovery were comparable to the Markx et al. system (24); without additional data we cannot compare throughput or cell loss. As these devices were microfluidic, we suggest that the system presented here should be significantly more robust; whereas the function of planar microfluidic devices can be compromised by the presence of a single bubble, the highly parallel design of our chip means that if a bubble appears it has no more effect than potentially blocking just one of the 397 holes.
Unlike FACS and MACS, which sort cells on the basis of specific recognition of membrane proteins, DEP separates on the basis of differences in the electrophysiology of the two cell types. This offers advantages over these existing methods, as many differences in cell types (and changes in cells in response to external stimuli) exhibit corresponding differences in the cell’s electrophysiome. Examples have included the differentiation of neural stem cells, where electrical changes in the membrane allow sorting of cells according to differentiation fate far in advance of conventional marker-based methods, e.g., refs. 27–29; indeed, stem-cell sorting may be an area where DEP-based separation may dominate over existing methods, whereas DEP separations in cancer cells have been reported for some decades (13), both for diagnosis and development of interventions. Many other cases of differential DEP response have been identified in the past 50 y, and all past demonstrations can be performed using this platform (30). Once differential electrophysiology is established by DEP profiling, it can be exploited through the use of ion channel blockers to emphasize difference and enhance DEP separation (31).
In conclusion, we have presented a DEP-based cell-separation technique that has a capacity and throughput comparable to the fastest MACS and FACS machines, requires no chemical labels, offers GMP compatibility, significantly lower cell loss, and significantly lower capital and running costs. Given the opportunity to exploit differences in cell electrophysiology in fields such as stem-cell therapy and cancer, we believe this offers significant promise as a new standard benchtop laboratory technique.
Materials and Methods
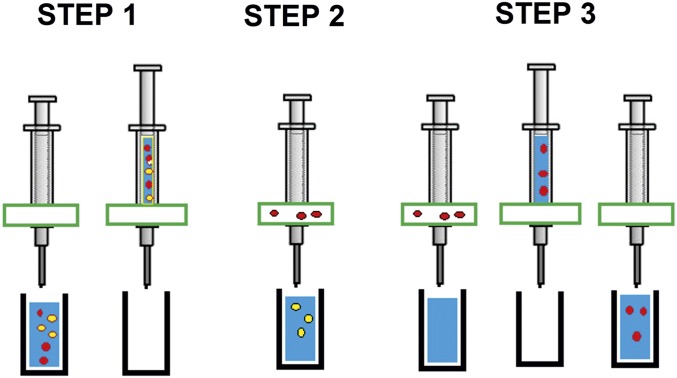
Experiments followed the steps outlined in Fig. 6. The cartridge and chip were cleaned with ethanol before assembly. The chip manufacture and cell mixtures are described in Supporting Information. A sample containing a mixture of the two cell types A and B was loaded into the fluidic cartridge by inserting the tube into the solution, then manually withdrawing the plunger to load the reservoir. This was then placed in a vertically mounted syringe pump (Razel, maximum output of 1 mL min−1), and the solution was pumped through the chip while voltage was applied to the electrodes using a signal generator (Jupiter 2000, Blackstar), connected to the chip via a custom-made amplifier board to supply up to 18 peak-to-peak voltage at frequencies up to 1 MHz. The output (enriched for population A) was collected in a second receptacle. Then fresh solution was loaded into the cartridge, the field was deactivated, and the chip was flushed manually with fresh medium to dislodge and recover the cells from population B that had been collected by positive DEP. When using a second or third pass, it was possible to enrich either cell-A or -B samples by repeating the procedure with the enriched samples. The total time for the two-pass procedure was under 30 min for yeast, and under 15 min for RBC/fibroblasts. Cells were exposed to the electric field for an average of ∼5 min for the positive DEP fraction, or under 10 s for the negative DEP fraction. All experiments were repeated three times. For the three-pass experiments to enrich rare cells, at the end of the second and third passes the cells were recovered by switching the field off, manually pulling the plunger up slightly, aspirating air into the fluidic cartridge, and expelling the retained cells in the small sample volume that is still within the chip. Separations were evaluated for cell recovery (the proportion of desired cells which arrive at the correct outlet), purity (the proportion of desired cells in a given outlet), and cell loss (the number of cells which are unaccounted for at the end of the separation). The number of missing cells was obtained by subtracting the “passed” and “recovered” populations from the initial number of cells.
Fig. 6.
Schematic showing the separation procedure, which is divided into three steps. In step 1, a mixture of cells is drawn into the syringe through the chip. In step 2, the chip is activated and cells are expelled at typically 1 mL min−1. One cell type is retained in the electrodes while the other is eluted. In step 3, after the solution is fully expelled, fresh medium is drawn into the device and expelled manually at a higher rate while the retaining field is deactivated, allowing the retained cells to be collected.
DEP spectra of individual cell types were measured using a DEPtech 3DEP and a separation frequency midway between the cross-over frequencies (the frequency on the DEP spectrum where the response crosses zero) for the two cell types to be separated; for the live/dead yeast mixture, 1 MHz was selected; for the fibroblast/RBC mixture, 22 kHz was selected; and for the MDA/RBC mixture, 76 kHz was used. Cells in output mixtures were either identified by the use of Trypan blue (live and dead yeast) or by visual inspection of the morphologically quite different RBCs and fibroblasts/MDAs. Work on human and mammalian cells was approved by the University of Surrey Ethics Committee; further details are available in the Supporting Information. Human participants in the study were screened for relevant self-reported health issues. All participants provided written, informed consent after having received explanation of the various study procedures.
Cells
Live and Dead Yeast.
Yeast was grown using Tesco’s freeze-dried baker’s yeast grown in a yeast extract peptone dextrose (YPD) medium (Sigma-Aldrich) set overnight in an incubator at 37 °C. Yeast cells were killed by transferring live yeast in YPD medium to an incubator set to 70 °C for 3 h. Yeast was washed and suspended in deionized (DI) water before use in experiments; conductivity was not fixed, but was at or below 1 mS⋅m−1 (typically 0.2–0.5 mS⋅m−1).
Rodent Fibroblasts and RBCs.
Mouse fibroblasts (3T3), kindly donated by Dr. Andrew Liu, University of Memphis, Memphis, TN, were cultured in an incubator set at 37 °C and 5% CO2 in growth medium. Cells were cultured in DMEM (Sigma-Aldrich) with the addition of 10% FBS (Fisher Scientific), 1 mM sodium pyruvate (Sigma-Aldrich), and 1% penicillin-streptomycin (Sigma-Aldrich). Adult common voles were sourced from the University of Surrey’s breeding colony in the United Kingdom, which was based on animals kindly donated by Prof. M. P. Gerkema, University of Groningen, Groningen, The Netherlands (32–34).Voles were bred in a licensed establishment, and were housed according to the Home Office Code of Practice for the care and accommodation of animals. The animals did not undergo any regulated procedure, and blood was collected after killing using a Schedule 1 method, which was performed by a competent person. Whole vole blood (1 mL) was collected following published protocols (35) using cardiac puncture, and collected in heparin-coated tubes. Collection was followed by RBC isolation through three identical centrifugation steps (10 min, 1,620 × g, 4 °C), each followed by resuspension of the RBC-containing pellet in PBS. Finally, the two cell types were mixed and resuspended in DEP medium comprising DI water supplemented with 8.5/0.3% sucrose/dextrose (Sigma-Aldrich), then washed and resuspended in DEP medium. The medium had a similar conductivity to the DI water sample described above.
Human Cancer and RBCs.
Human blood was collected from donors at the University of Surrey and RBCs were isolated following published protocols (36). The RBCs, which were then washed twice in DEP medium consisting of 8.5% (wt/vol) sucrose, 0.5% (wt/vol) dextrose, 100 µM CaCl2, and 250 µM MgCl2. MDA-MB-231 cells were cultured in MEM medium (Sigma-Aldrich) supplemented with 10% heat-inactivated FBS (Invitrogen), 2 mM l-Glutamine, and 1% penicillin-streptomycin (Sigma-Aldrich). The cells were grown in T75 flasks in a standard cell culture incubator at 5% CO2, 95% humidified air at 37 °C and subcultured every 48 h when 80% confluent. To prepare for experiment, MDA-MB-231 cells were washed twice in a medium consisting of 8.5% (wt/vol) sucrose, 0.5% (wt/vol) dextrose, 100 µM CaCl2, and 250 µM MgCl2 and supplemented with NaOH to pH of 7.4. The medium conductivity was adjusted using PBS to 10 mS⋅m−1.
Preparation of Mixtures.
Cell samples were prepared by counting using a NanoEnTek C-chip disposable hemocytometer (Labtech), mixing to obtain an ∼50:50 ratio, then remeasured. The stock of live and dead yeast was found to contain 8.0 million live cells and 6.0 million dead cells and was processed as 15-mL samples (a concentration of 0.93 million cells per milliliter). Vole fibroblasts and RBCs were mixed, and the resultant mixture was measured to contain 5.1 million fibroblasts and 4.8 million RBCs in each of three 5-mL aliquots, a total concentration of 1.98 million cells per milliliter. A 20 mL sample of 11.1 million fibroblasts and 13.4 million RBCs was also prepared. Human RBCs were spiked with MDA-MB-231 breast cancer cells, to a final concentration of 1.1% cancer to 98.9% RBCs, at a total cell concentration of ∼2 × 107 mL−1, to a total sample volume of 4 mL.
Device Fabrication
The device comprised three principal parts: a fluidic chip, a casing, and the support instrument (including principally a signal generator, amplifier, and syringe driver). The chip comprised 10 layers of 70-µm-thick copper separated by 150-µm-thick glass fiber reinforced epoxy (FR4) layers, with two further 35-µm-thick copper layers at the top and bottom. All copper layers were patterned in a circle slightly larger than the area through which the separation wells were drilled, to minimize overlap and associated capacitive losses. Three hundred ninety seven wells with a 400-µm diameter were drilled in a hexagonal pattern at the center of the chip; larger holes for assembly and contacts were placed at the perimeter of the chip. Exposed copper areas were gold-plated to maintain stability and biocompatibility. The chip was housed in a cartridge machined from poly(methyl methacrylate), which comprised a reservoir containing a syringe plunger from a 20-mL syringe, a chip holder, and an outlet. The chip formed the base of the reservoir, such that when the plunger was pushed, cells were able to flow through all 397 holes. Below the chip, a collection cone funnelled the suspension into a 1.46-mm-diameter outlet tube to a collection receptacle. The chip was mounted between the top cylinder and bottom cone of the assembly and secured using Allen bolts, with a watertight seal provided by two 19.13-mm-diameter O rings.
Acknowledgments
We thank Mr. David Gould and Mr. Rowan Lonsdale for their assistance in constructing the system, and DEPtech for funding the manufacture of the chips.
Footnotes
Conflict of interest statement: The authors declare a conflict of interest. K.F.H. and M.P.H. are named on a patent (US Patent 8864973, granted 2014) describing the use of 3D multilayer laminate electrodes to construct DEP electrodes. This technology was used to construct the electrode chips used in this project.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700773114/-/DCSupplemental.
References
- 1.English D, Andersen BR. Single-step separation of red blood cells. Granulocytes and mononuclear leukocytes on discontinuous density gradients of Ficoll-Hypaque. J Immunol Methods. 1974;5:249–252. doi: 10.1016/0022-1759(74)90109-4. [DOI] [PubMed] [Google Scholar]
- 2.Tomlinson MJ, Tomlinson S, Yang XB, Kirkham J. Cell separation: Terminology and practical considerations. J Tissue Eng. 2013;4:2041731412472690. doi: 10.1177/2041731412472690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonner WA, Hulett HR, Sweet RG, Herzenberg LA. Fluorescence activated cell sorting. Rev Sci Instrum. 1972;43:404–409. doi: 10.1063/1.1685647. [DOI] [PubMed] [Google Scholar]
- 4.Miltenyi S, Müller W, Weichel W, Radbruch A. High gradient magnetic cell separation with MACS. Cytometry. 1990;11:231–238. doi: 10.1002/cyto.990110203. [DOI] [PubMed] [Google Scholar]
- 5.Emad A, Drouin R. Evaluation of the impact of density gradient centrifugation on fetal cell loss during enrichment from maternal peripheral blood. Prenat Diagn. 2014;34:878–885. doi: 10.1002/pd.4387. [DOI] [PubMed] [Google Scholar]
- 6.Brooks KH, Fernandez-Boltran R. Tissue and cell culture. In: Delves PJ, editor. Cellular Immunology Labfax. Academic Press; Oxford: 1994. [Google Scholar]
- 7.Pohl HA. The motion and precipitation of suspensoids in divergent electric fields. J Appl Phys. 1951;22:869–871. [Google Scholar]
- 8.Pohl HA, Hawk I. Separation of living and dead cells by dielectrophoresis. Science. 1966;152:647–649. doi: 10.1126/science.152.3722.647-a. [DOI] [PubMed] [Google Scholar]
- 9.Markx GH, Huang Y, Zhou XF, Pethig R. Dielectrophoretic characterization and separation of micro-organisms. Microbiol. 1994;140:585–591. [Google Scholar]
- 10.Markx GH, Pethig R. Dielectrophoretic separation of cells: Continuous separation. Biotechnol Bioeng. 1995;45:337–343. doi: 10.1002/bit.260450408. [DOI] [PubMed] [Google Scholar]
- 11.Stephens M, Talary MS, Pethig R, Burnett AK, Mills KI. The dielectrophoresis enrichment of CD34+ cells from peripheral blood stem cell harvests. Bone Marrow Transplant. 1996;18:777–782. [PubMed] [Google Scholar]
- 12.Talary MS, Mills KI, Hoy T, Burnett AK, Pethig R. Dielectrophoretic separation and enrichment of CD34+ cell subpopulation from bone marrow and peripheral blood stem cells. Med Biol Eng Comput. 1995;33:235–237. doi: 10.1007/BF02523050. [DOI] [PubMed] [Google Scholar]
- 13.Becker FF, et al. Separation of human breast cancer cells from blood by differential dielectric affinity. Proc Natl Acad Sci USA. 1995;92:860–864. doi: 10.1073/pnas.92.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiedler S, Shirley SG, Schnelle T, Fuhr G. Dielectrophoretic sorting of particles and cells in a microsystem. Anal Chem. 1998;70:1909–1915. doi: 10.1021/ac971063b. [DOI] [PubMed] [Google Scholar]
- 15.Muller T, Schnelle T, Gradl G, Shirley SG, Guhr G. Microdevice for cell and particle separation using dielectrophoretic field-flow fractionation. J Liq Chrom Relat Tech. 2000;23:47–59. [Google Scholar]
- 16.Cheng J, et al. Preparation and hybridization analysis of DNA/RNA from E. coli on microfabricated bioelectronic chips. Nat Biotechnol. 1998;16:541–546. doi: 10.1038/nbt0698-541. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, et al. Dielectric properties of human leukocyte subpopulations determined by electrorotation as a cell separation criterion. Biophys J. 1999;76:3307–3314. doi: 10.1016/S0006-3495(99)77483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan H, Hughes MP, Green NG. Separation of submicron bioparticles by dielectrophoresis. Biophys J. 1999;77:516–525. doi: 10.1016/S0006-3495(99)76908-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fatoyinbo HO, Hughes MP, Martin SP, Pashby P, Labeed FH. Dielectrophoretic separation of Bacillus subtilis spores from environmental diesel particles. J Environ Monit. 2007;9:87–90. doi: 10.1039/b614556f. [DOI] [PubMed] [Google Scholar]
- 20.Pethig R. Review—Where is dielectrophoresis (DEP) going? J Electrochem Soc. 2017;164:B3049–B3055. [Google Scholar]
- 21.Gupta V, et al. ApoStream(™), a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics. 2012;6:24133. doi: 10.1063/1.4731647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu X, et al. Marker-specific sorting of rare cells using dielectrophoresis. Proc Natl Acad Sci USA. 2005;102:15757–15761. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee D, Hwang B, Choi Y, Kim B. A novel dielectrophoresis activated cell sorter (DACS) to evaluate the apoptotic rate of K562 cells treated with arsenic trioxide (As2O3) Sens. Actuat. A. 2016;242:1–8. [Google Scholar]
- 24.Markx GH, Talary MS, Pethig R. Separation of viable and non-viable yeast using dielectrophoresis. J Biotechnol. 1994;32:29–37. doi: 10.1016/0168-1656(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 25.Fatoyinbo HO, Kamchis D, Whattingham R, Ogin SL, Hughes MP. A high-throughput 3D composite dielectrophoretic separator. IEEE Trans Biomed Eng. 2005;52:1347–1349. doi: 10.1109/TBME.2005.847553. [DOI] [PubMed] [Google Scholar]
- 26.Abdul Razak MA, Hoettges KF, Fatoyinbo HO, Labeed FH, Hughes MP. Efficient dielectrophoretic cell enrichment using a DEP-well based system. Biomicrofluidics. 2013;7:064110. doi: 10.1063/1.4842395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon MG, et al. Increasing label-free stem cell sorting capacity to reach transplantation-scale throughput. Biomicrofluidics. 2014;8:064106. doi: 10.1063/1.4902371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labeed FH, et al. Biophysical characteristics reveal neural stem cell differentiation potential. PLoS One. 2011;6:e25458. doi: 10.1371/journal.pone.0025458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vykoukal J, Vykoukal DM, Freyberg S, Alt EU, Gascoyne PR. Enrichment of putative stem cells from adipose tissue using dielectrophoretic field-flow fractionation. Lab Chip. 2008;8:1386–1393. doi: 10.1039/b717043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes MP. Fifty years of dielectrophoretic cell separation technology. Biomicrofluidics. 2016;10:032801. doi: 10.1063/1.4954841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duncan L, et al. Dielectrophoretic analysis of changes in cytoplasmic ion levels due to ion channel blocker action reveals underlying differences between drug-sensitive and multidrug-resistant leukaemic cells. Phys Med Biol. 2008;53:N1–N7. doi: 10.1088/0031-9155/53/2/N01. [DOI] [PubMed] [Google Scholar]
- 32.van der Veen DR, Saaltink DJ, Gerkema MP. Behavioral responses to combinations of timed light, food availability, and ultradian rhythms in the common vole (Microtus arvalis) Chronobiol Int. 2011;28:563–571. doi: 10.3109/07420528.2011.591953. [DOI] [PubMed] [Google Scholar]
- 33.van der Veen DR, et al. Impact of behavior on central and peripheral circadian clocks in the common vole Microtus arvalis, a mammal with ultradian rhythms. Proc Natl Acad Sci USA. 2006;103:3393–3398. doi: 10.1073/pnas.0507825103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Veen DR, et al. Circadian rhythms of C-FOS expression in the suprachiasmatic nuclei of the common vole (Microtus arvalis) Chronobiol Int. 2008;25:481–499. doi: 10.1080/07420520802254403. [DOI] [PubMed] [Google Scholar]
- 35.Hanson MS, et al. Phosphodiesterase 3 is present in rabbit and human erythrocytes and its inhibition potentiates iloprost-induced increases in cAMP. Am J Physiol Heart Circ Physiol. 2008;295:H786–H793. doi: 10.1152/ajpheart.00349.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]