Significance
The spliceosome, which catalyzes pre-mRNA splicing via a two-step process, must balance the need for high-fidelity splice-site selection with the need for rapid, efficient splicing. We propose that the RNaseH domain (RH) of Prp8 contributes to this balance by toggling between two different conformations throughout the splicing cycle. Using a set of previously published prp8 alleles, we link alleles that stabilize one conformation of RH to high-fidelity, low-efficiency splicing and those that stabilize the other to low-fidelity, high-efficiency splicing. This model is consistent with recent data that indicate the conformation of the spliceosome is similar at both catalytic steps and provides an example of a structural basis for splicing fidelity.
Keywords: spliceosome, Prp8, splicing fidelity, splicing efficiency, lariat sequencing
Abstract
Pre-mRNA splicing is an essential step of eukaryotic gene expression that requires both high efficiency and high fidelity. Prp8 has long been considered the “master regulator” of the spliceosome, the molecular machine that executes pre-mRNA splicing. Cross-linking and structural studies place the RNaseH domain (RH) of Prp8 near the spliceosome’s catalytic core and demonstrate that prp8 alleles that map to a 17-aa extension in RH stabilize it in one of two mutually exclusive structures, the biological relevance of which are unknown. We performed an extensive characterization of prp8 alleles that map to this extension and, using in vitro and in vivo reporter assays, show they fall into two functional classes associated with the two structures: those that promote error-prone/efficient splicing and those that promote hyperaccurate/inefficient splicing. Identification of global locations of endogenous splice-site activation by lariat sequencing confirms the fidelity effects seen in our reporter assays. Furthermore, we show that error-prone/efficient RH alleles suppress a prp2 mutant deficient at promoting the first catalytic step of splicing, whereas hyperaccurate/inefficient RH alleles exhibit synthetic sickness. Together our data indicate that prp8 RH alleles link splicing fidelity with catalytic efficiency by biasing the relative stabilities of distinct spliceosome conformations. We hypothesize that the spliceosome “toggles” between such error-prone/efficient and hyperaccurate/inefficient conformations during the splicing cycle to regulate splicing fidelity.
Pre-mRNA splicing occurs via two transesterification reactions catalyzed by the spliceosome, a large, dynamic ribonucleoprotein complex. The catalytically active spliceosome is composed of three small nuclear ribonucleoprotein (snRNP) complexes (U2, U5, and U6), the Nineteen Complex and its related proteins, and a small number of accessory splicing proteins (1). In the first catalytic step of pre-mRNA splicing the 5′ splice site (5′SS) is cleaved, forming a lariat-3′ exon intermediate in which the 5′SS is covalently linked via a 2′,5′ phosphodiester bond to the branch site adenosine (BrA). In the second catalytic step the 3′ splice site (3′SS) is cleaved and the 5′ and 3′ exons are ligated. The excised lariat intron is then released from the spliceosome. Splicing is performed with high fidelity and high efficiency, and it is thought that core spliceosomal components contribute directly to maintenance of splicing fidelity and efficiency (1, 2).
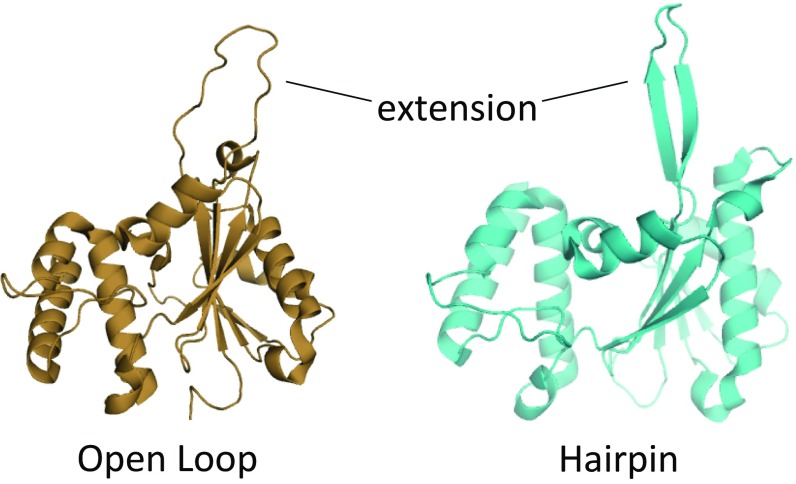
Prp8 is the largest and most highly conserved protein in the spliceosome and is a component of the U5 snRNP. Recent cryoelectron microscopy structures show that Prp8 acts as a platform at the heart of the spliceosome and undergoes considerable conformational rearrangements during the splicing cycle (3–11). Genetic screens have identified many alleles in Prp8 that exhibit compromised splicing fidelity or efficiency (summarized in ref. 12). A subset of these mutations maps to a unique and essential 17-aa extension within the RNaseH domain (RH) of Prp8 (13–20). Structural studies reveal that the RH extension can exist either in a β-hairpin form or as a disordered loop, and that it adopts these conformations at distinct steps of the splicing cycle (Fig. 1) (3, 4, 7–11, 19). Of the subset of prp8 alleles that map to the RH extension for which structural data are available, those that preferentially stabilize either the β-hairpin or disordered loop forms of RH correlate with distinct genetic phenotypes previously proposed to arise from unique first-step and second-step catalytic conformations of the spliceosome (Fig. 1 and Table S1) (17–20).
Fig. 1.
The 17-aa extension in the RNaseH subdomain of Prp8 adopts two different forms: an open loop (tan, Left) and a β-hairpin (cyan, Right). Structures are modified from Protein Data Bank ID code 4JK7 (19).
Table S1.
Summary of previous data and characterization of prp8 toggle alleles
| Yeast/(human) | Refs. | Original identification | Toggle classification | Information from human RH structure | Structure favored | Growth |
| V1860D (V1788) | 13, 14 | U4-cs1 suppressor | Transitional/first | Structure solved: H-bond with Y1786 stabilizes hairpin | Hairpin (19) | As WT |
| V1860N (V1788) | 14 | U4-cs1 suppressor | Transitional | Predicted H-bond with Y1786 | Hairpin (our prediction) | As WT |
| V1862D (I1790) | 13, 14 | U4-cs1 suppressor | Transitional | Predicted H-bond with T1800 | Hairpin (our prediction) | As WT |
| T1865K (T1793) | 13 | Designed | Transitional/first | Predicted H-bond with N1797 | Hairpin (our prediction) | As WT |
| A1871E (T1799) | 13 | Designed | Transitional/first | Predicted H-bond with H1791 | Hairpin (our prediction) | As WT |
| T1872E (T1800) | 13 | Designed | Transitional/first | Structure solved: H2O-mediated H-bond with Y1786 stabilizes hairpin | Hairpin (19) | As WT |
| T1861P (T1789) | 15, 19 | U4-cs1 suppressor | Catalytic/second | Structure solved: proline disrupts hairpin | Loop (19) | As WT |
| V1862Y (I1790) | 14, 19 | U4-cs1 suppressor | Catalytic/second | Structure solved: H-bond with N1797 stabilizes loop | Loop (19) | As WT |
| H1863E (H1791) | 13 | Designed | Catalytic/second | No prediction | Unknown | As WT |
| K1864E (K1792) | 16 | 5′SS suppressor | Catalytic/second | No prediction | Unknown | As WT |
| N1869D (N1797) | 16, 17, 20 | 5′SS suppressor | Catalytic/second | No prediction | Unknown | Ts (slightly) |
| V1870N (L1798) | 17, 18 | BrG suppressor | Catalytic/second | Predicted H-bond with G1796 | Loop (19) | As WT |
| I1875T (I1803) | 15 | U4-cs1 suppressor | Catalytic | Predicted H-bond with R1787 | Loop (our prediction) | As WT |
| Δ1860–1875 | This study | Designed | unclassified | Removes hairpin/loop | —– | Lethal |
The 13 prp8 toggle alleles used in this study and their human homologs are listed, along with references to data concerning each allele. The column headed “Original identification” indicates how the allele was initially discovered. The “Toggle classification” column indicates whether an allele was originally classified as either first or second step, along with classification of the allele as either transitional or catalytic. Structural information includes data from the crystal structures of the human Prp8 RH (19) showing how an allele affects the structure of the RH extension as well as our predictions. Finally, growth phenotypes in rich media are indicated. Ts indicates temperature sensitive growth at 37 °C.
However, recent structural and biochemical studies of the spliceosome and its evolutionary precursor, the group II intron, reveal that the spliceosome’s catalytic core is similar at both catalytic steps (21, 22). Structural data further suggest the group II intron passes through an obligatory intermediate transitional structure between two highly similar first- and second-step catalytic structures (22). Conservation argues that some components of the spliceosome might adopt a similar transitional intermediate, “toggling” between catalytic and transitional intermediate conformations during a typical splicing reaction, and that these conformations would be executed through specific structural toggles in core spliceosomal components (17).
Here we provide evidence for two distinct classes of prp8 RH alleles, which we refer to as “catalytic” and “transitional” in keeping with the nomenclature established for the group II intron. Specifically, catalytic prp8 alleles that stabilize the loop structure of the RH extension exhibit high-efficiency, low-fidelity splicing, whereas transitional prp8 alleles that stabilize the β-hairpin structure exhibit low-efficiency but high-fidelity splicing both on reporter constructs and at endogenous splice sites genomewide. We propose that the spliceosome cycles between catalytic and transitional conformations during each splicing cycle, and we implicate the RH extension of Prp8 as a structural toggle. This model is supported by recent high-resolution structural studies of the spliceosome that show that the RH extension adopts a loop form in the catalytically active Bact spliceosome (5) and a β-hairpin form in other noncatalytically active conformations (3, 4, 7–9). Our data provide a specific, mechanistic example of how distinct conformations of a core spliceosomal component can affect both splicing fidelity and efficiency.
Results
Creation of prp8 Toggle Allele Strains.
We chose 13 published prp8 RH extension alleles for characterization: V1860D, V1860N, T1861P, V1862D, V1862Y, H1863E, K1864E, T1865K, N1869D, V1870N, A1871E, T1872E, and I1875T. Together, we refer to these alleles as the “toggle” alleles. Whereas most of these alleles were discovered through genetic screens conducted to identify alleles that suppress growth defects caused by the mutation of the 5′SS, branch point (Br), or 3′SS motifs of a reporter gene (16–18, 20, 23), a subset was deliberately designed based on structural data (13) (Table S1). Structural data are available for a subset of these alleles (19); for the other alleles, we made predictions based on the available data (Table S1). Most alleles grew similarly to WT yeast in rich media at all temperatures tested, consistent with previous results. A strain in which the entire 17-aa RH extension was deleted failed to grow at any temperature (Table S1).
ACT1-CUP1 Reporter Assay Demonstrates That prp8 Toggle Alleles Sort into Two Distinct Classes.
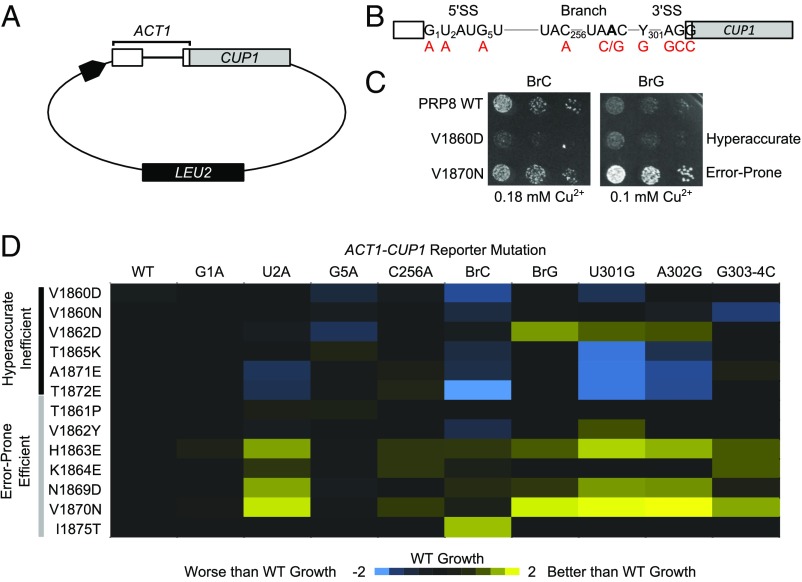
To determine the effects of the prp8 toggle alleles on splicing fidelity and efficiency we used the well-characterized ACT1-CUP1 splicing reporter system (Fig. 2A and Table S2) (24). In this system, the ability of yeast to grow on otherwise lethal concentrations of copper-containing media is directly proportional to splicing efficiency. ACT1-CUP1 reporters containing nonconsensus splicing sequences at the 5′SS (G1A, U2A, and G5A), Br (C256A, BrC, and BrG), or 3′SS (U301G, A302G, and G303/304C) were used to measure fidelity (Fig. 2 B and C). A false-color representation of the data is shown in Fig. 2D (25), and the results are consistent with reported data for the subset of toggle alleles previously examined (Fig. S1) (13, 16–20).
Fig. 2.
(A) Schematic of ACT1-CUP1 reporter. (B) Diagram of ACT1-CUP1 reporter intron. The 5′SS, BrA, and 3′SS are shown with mutations made to test fidelity in red. (C) Growth of prp8 toggle alleles in the presence of BrC (Left) or BrG (Right). (D) [Cu2+max] that supports growth was determined for each prp8 toggle allele and reporter. Values were transformed log2([Cu2+max prp8]/[Cu2+max PRP8]) and colored blue (worse growth) to yellow (better growth).
Table S2.
| Reporter | Effect | Refs. |
| G1A | Inhibits second step | 24 |
| U2A | Inhibits both steps, lariat intermediate degraded | 18 |
| G5A | Inhibits both steps, activates aberrant 5′SS use | 24 |
| C256A | Inhibits both steps | 24 |
| BrC | Inhibits both steps, limiting for first step | 17, 18 |
| BrG | Inhibits second step | 17 |
| U301G (gAG) | Inhibits second step, lariat intermediate degraded | 18 |
| A302G (UgG) | Inhibits second step, lariat intermediate degraded | 18 |
| G303/4C (UAc/c) | Inhibits second step | 17 |
Numbering is based on the ACT1-CUP1 reporter described in ref. 24.
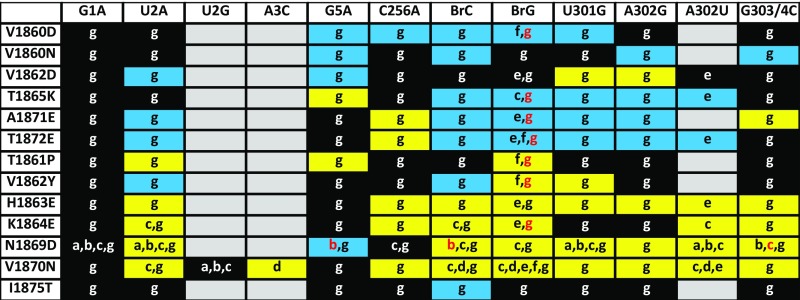
Fig. S1.
Comparison of ACT1-CUP1 growth assays. Data from seven different studies (13, 17–20, 42) are included with the data generated in this work. Background coloring indicates growth of prp8 toggle allele carrying the indicated ACT1-CUP1 splicing reporter, where black indicates WT growth, blue indicates decreased/hyperaccurate growth in at least one reference, yellow indicates increased/error-prone growth in at least one reference, and gray untested. References used to create this table are indicated by letter: a, ref. 20; b, ref. 16; c, ref. 17; d, ref. 18; e, ref. 13; f, ref. 19; and g, this work. Red letter coloring indicates that in the indicated reference the prp8 allele grew as WT. Such differences are likely due to differing strain backgrounds used by different laboratories.
All prp8 toggle alleles grew with efficiency similar to PRP8 when required to splice a WT ACT1-CUP1 reporter. However, individual prp8 toggle alleles exhibited differential growth on ACT1-CUP1 reporters with mutated, nonconsensus splicing sequences (Fig. 2D and Fig. S1). In general, prp8 toggle alleles sorted into two groups: those that exhibited worse growth (blue, Fig. 2D) than PRP8 (indicative of hyperaccurate/inefficient splicing) and those that exhibited better growth (indicative of error-prone splicing) (yellow, Fig. 2D). This division held whether the reporter affected primarily the first step of splicing, the second step, or both (Table S2). Differences in the extent of the effect varied with individual prp8 toggle alleles. For example, V1860D and V1860N both exhibited a hyperaccurate/inefficient phenotype; however, the phenotype was much stronger in prp8 V1860D regardless of reporter. Such variability is not unexpected, because toggle alleles may bias RH domain conformation to differing degrees. These data suggested that prp8 toggle alleles could be broadly classified as exhibiting hyperaccurate/inefficient or error-prone/efficient splicing, associated with the β-hairpin and loop conformations of the RH extension, respectively.
Toggle Alleles Interact Genetically with an ATPase-Deficient prp2 Allele.
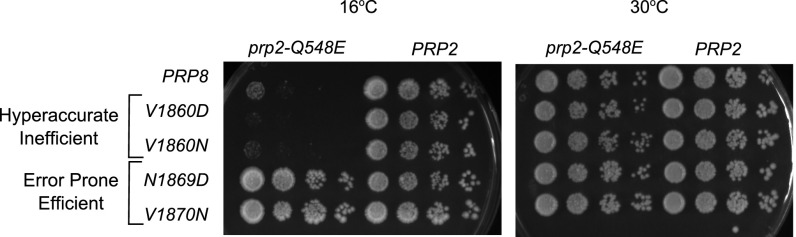
The DEAH-box helicase Prp2 is required for the first catalytic step of splicing (1). Prp2 destabilizes the association between the U2 snRNP and the pre-mRNA before the first step (26) and promotes snRNA rearrangements in preparation for catalysis (27). Strains containing the prp2-Q548E allele are impaired for growth at 16 °C and exhibit defects in pre-mRNA splicing, likely due to deficiencies in ATP binding and/or hydrolysis that lead to inefficient catalytic activation (27). We made double-mutant strains that contained prp2-Q548E in combination with prp8 alleles from each class identified in our ACT1-CUP1 reporter assay: V1860D and V1860N (hyperaccurate/inefficient) and N1869D and V1870N (error-prone/efficient). Hyperaccurate/inefficient prp8 alleles exhibited synthetic sickness at 16 °C when combined with prp2-Q548E whereas error-prone/efficient prp8 toggle alleles rescued the cold sensitivity of the prp2-Q548E strain (Fig. 3). Because Prp2 is required for catalytic activation, these data are consistent with a model in which error-prone/efficient prp8 alleles promote splicing catalysis whereas hyperaccurate/inefficient oppose it.
Fig. 3.
Growth of double-mutant strains carrying WT, V1860D, V1860N, N1869D, and V1870N prp8 alleles in combination with PRP2 or prp2-Q548E at 16 °C (Left) and 30 °C (Right).
In Vitro Characterization of the First-Step Splicing Efficiency of prp8 Toggle Mutants.
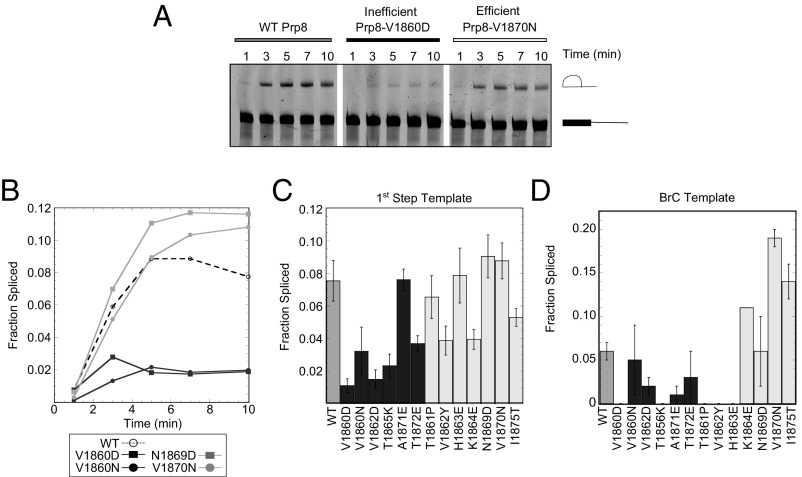
To more directly assess first-step splicing efficiency we performed in vitro splicing assays using a splicing substrate truncated just downstream of the branch site. This substrate is unable to complete the second step because it lacks a 3′SS (19, 28), enabling more robust characterization of the first step. Fig. 4A shows a representative time-course experiment performed in PRP8 extract and extracts made from two toggle alleles (V1860D and V1870N) that, based upon ACT1-CUP1 reporter assays, were expected to have opposite effects on splicing efficiency. Whereas extracts made from error-prone/efficient prp8 toggle alleles spliced the truncated pre-mRNA with efficiency similar to WT, those from hyperaccurate/inefficient prp8 alleles exhibited decreased efficiency.
Fig. 4.
(A) First-step in vitro splicing assay. Gel showing results of a time course of splicing of a fluorescent pre-Act1 truncated template. Pre-mRNA and first-step product indicated. (B) Quantification of representative first-step time course assay. (C) Fraction truncated pre-Act1 spliced after 10 min. (D) Fraction pre-BrC template spliced after 20 min is shown. Hyperaccurate/inefficient alleles colored black, error-prone/efficient gray. Error bars are SEM for three biological replicates.
Because the first-step reaction was essentially complete at 10 min (Fig. 4B), we repeated this analysis with extracts made from all prp8 toggle alleles but focused only on the 10-min time point (Fig. 4C). On the whole, hyperaccurate/inefficient alleles performed the first step of splicing less efficiently than WT, whereas error-prone/efficient alleles performed the first step with similar or increased efficiency. There were a few exceptions: Three of the prp8 toggle alleles classified as error-prone/efficient based on ACT1-CUP1 reporter data (V1862Y, K1864E, and I1875T) spliced with lower efficiency, whereas the hyperaccurate/inefficient prp8 allele A1871E spliced with efficiency similar to WT. This might reflect allele-specific variability in the artificial context of in vitro splicing with a truncated pre-mRNA and hints at additional complexity in the mode of action of the prp8 toggle alleles.
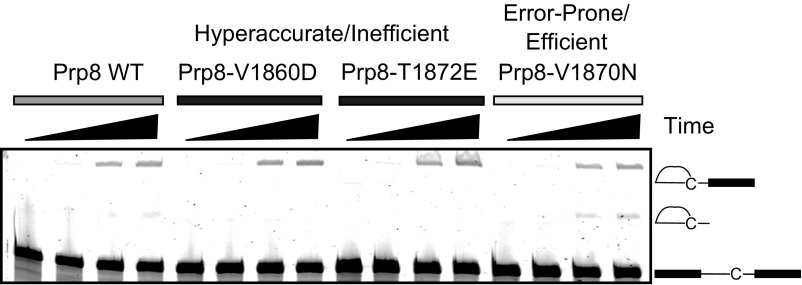
To further characterize first-step catalytic efficiency we used extracts made from prp8 toggle alleles to perform in vitro splicing assays on a full-length ACT1 pre-mRNA harboring a BrC mutation (Fig. 4D and Fig. S2). The BrC mutation decreases the efficiency of both steps, with a particularly strong effect on the first step (17, 29). Extracts from most of the error-prone/efficient prp8 toggle alleles spliced a BrC-containing substrate more efficiently than PRP8. This included extracts from two of the alleles that had spliced the truncated pre-mRNA substrate less efficiently, further indicating potential template-specific effects. All extracts made from hyperaccurate/inefficient prp8 alleles spliced BrC template less efficiently than WT. Some extracts from both prp8 toggle allele classes were unable to splice mutant templates in vitro. Because all of the extracts spliced the WT template, we presume that this inefficiency reflects a specific defect between the mutant template and these extracts.
Fig. S2.
Extracts made from error-prone/efficient prp8 alleles perform both steps of splicing more efficiently than those made from WT PRP8, whereas extracts made from hyperaccurate/inefficient prp8 alleles perform as those made from WT PRP8 or worse. Gel showing results of a time-course experiment comparing splicing of a fluorescent pre-Act1 template carrying a BrC mutation. Pre-mRNA, lariat intermediate first-step product and excised lariat second-step product are indicated.
Lariat Sequencing Reveals Genomewide Alterations to Splicing Fidelity.
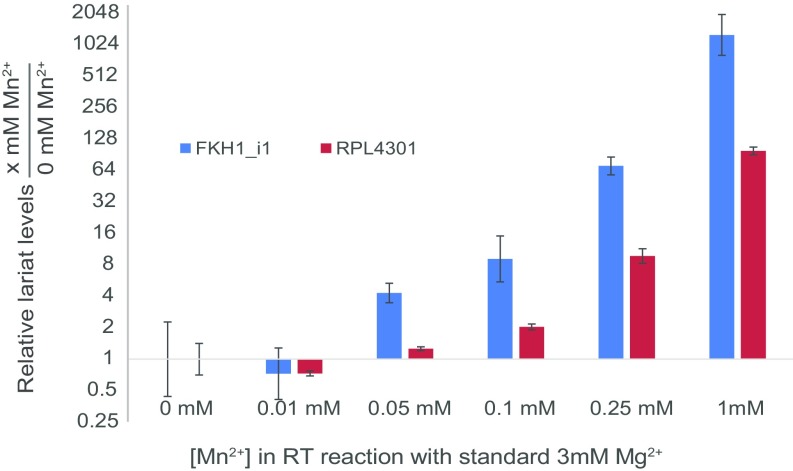
Given the altered fidelity of prp8 RH alleles as revealed by reporter constructs, we sought to assess the global impacts of prp8 RH alleles on the in vivo splice-site selection for native introns. Several groups, including ours, have recently described methods for determining the global locations of splice-site activation by monitoring the lariats generated during each splicing reaction (30–34). Here we developed a modified approach that enables quantitative assessment of splice-site use (SI Materials and Methods). Strains were created carrying the prp8 toggle alleles in combination with a deletion of the DBR1 gene, an essential component of the lariat decay pathway, to facilitate lariat accumulation. Lariat RNAs were enriched from the total RNA by first depleting the sample of rRNAs and then enzymatically degrading linear RNAs. When cDNA generated from lariat RNAs is subjected to high-throughput sequencing, a subset of sequencing reads that traverse the 2′,5′ linkage can be used to extract the specific 5′SS and Br sequences that were activated in the splicing reaction (32). Although reverse transcriptase exhibits a low propensity to transcribe across the 2′,5′ linkage under standard reaction conditions, here we identified conditions that allow for an ∼100-fold increase in the frequency of read-through (Fig. S3).
Fig. S3.
Effect of addition of Mn2+ during cDNA synthesis on relative lariat levels are shown for two Schizosaccharomyces pombe intron lariats, fkh1_Intron1 and rpl4301. Confidence intervals are based on three qPCR technical replicates. cDNA synthesis was performed in standard RT buffer containing 3 mM Mg2+ along with a titration series of MnCl2. The efficiency of RT reading through the 2′,5′ linkage in the lariat branch was measured as a function of number of lariat amplicons available for amplification in a quantitative PCR assay done on the cDNA with primers that were designed such that they faced away from one another in the linear RNA but toward one another in the context of a lariat, as in Fig. 5B. Relative lariat levels were then calculated by normalization with an exon in the corresponding gene under investigation. Then, using the ΔΔCT method of analysis, fold change in relative lariat levels between each sample and the sample made with 0 mM Mn2+ was calculated. Addition of 1 mM Mn2+ in the RT reaction resulted in around a 100-fold increase in relative lariat levels over a sample made with no Mn2+. It is to be noted that the exon levels remained largely unchanged as measured by raw CT values, indicating that the increase in relative lariat levels with increasing [Mn2+] was largely because of an increase in lariat amplicon available for amplification as a result of RT reading through the lariat branch.
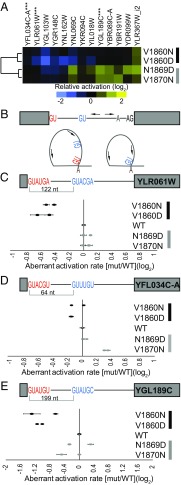
We identified the global locations of splice-site activation in WT PRP8, as well as in two prp8 toggle alleles from each of the hyperaccurate/inefficient (V1860D and V1860N) and error-prone/efficient (N1869D and V1870N) classes (Fig. 5). When considering only the WT PRP8 strain, lariat reads were detected for 287 of the 308 (93%) annotated spliceosomal introns, a level of coverage that exceeded that obtained by other published lariat sequencing approaches in Saccharomyces cerevisiae (33, 34) and established the capacity of this approach to readily capture global sites of splicing activation (Fig. S4 and Datasets S1 and S2). Consistent with previous studies, this analysis also revealed use of alternative splice sites associated with known introns (30–34), herein referred to as aberrant splice sites. We focused further analyses on the aberrant events with sufficient read depth across all strains to enable statistical analyses.
Fig. 5.
(A) Global lariat sequencing identified specific locations of increases (yellow) and decreases (blue) in aberrant 5′SS activation associated with annotated introns. (B) Schematic of primer locations (arrows) that enable discrimination between annotated (red) and aberrant (blue) splice-site activation (Table S4). (C–E) Relative splice-site use as determined by RT-PCR is shown for two biological replicates of each strain. Hyperaccurate/inefficient alleles colored black, error-prone/efficient gray. Error bars are SD for two technical replicates.
Fig. S4.
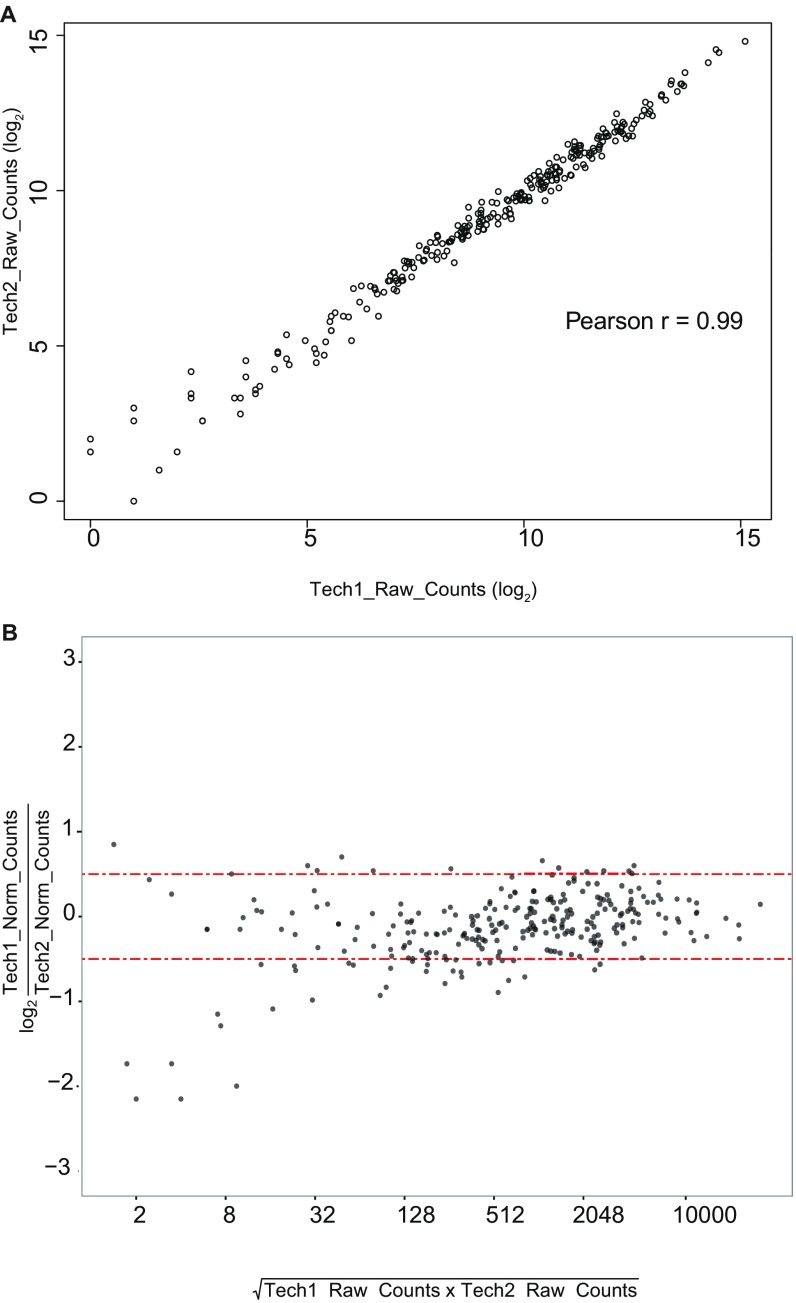
Lariat sequencing datasets are highly reproducible. (A) Raw counts for the 276 annotated introns used in the analysis in Fig. 5A (SI Materials and Methods) for two technical replicates of prp8 N1869D are plotted as an XY scatter. (B) MA plot of the same data depicted in A. The y axis uses raw counts normalized to total raw counts from all annotated introns. Red lines mark changes ±0.5 (log2) between the replicates.
For each event, the frequency of aberrant splice-site activation relative to the frequency of annotated splice-site activation was determined for the selected prp8 toggle alleles. Fig. 5A shows a false-colored representation of the behavior of each of these aberrant splicing events relative to PRP8. When considering the behavior of the different alleles across all of these splicing events, the behaviors of the pair of hyperaccurate/inefficient alleles highly correlated with one another (Pearson r = 0.82), and the behaviors of the pair of error-prone/efficient alleles highly correlated with one another (Pearson r = 0.87). By contrast, the behavior of the hyperaccurate/inefficient pairs was poorly correlated with that of the error-prone/efficient pairs (Pearson r = −0.11), consistent with these classes of alleles having opposing impacts on these aberrant events. Moreover, when considering the behavior of the individual splicing events in the context of prp8 toggle alleles, events largely matched the results seen with ACT1-CUP1 reporters (Fig. 2 and Fig. S1) wherein the hyperaccurate/inefficient alleles showed lower levels of aberrant splice-site activation than the WT, while the error-prone/efficient alleles showed higher levels. As with the reporters, the level of aberrant splice-site activation varied with the different prp8 toggle alleles.
To independently validate the frequencies of specific aberrant splice-site activation events determined from lariat sequencing, we used a PCR-based approach to quantify the different lariat species (Fig. S5). As seen in Fig. 5B, primers were designed such that they faced away from one another in the context of linear RNA but toward one another in the context of a lariat. The amplicons derived from these primer pairs differed in length depending upon the particular 5′SS used in the reaction, allowing the relative abundancies of the two isoforms to be determined by capillary electrophoresis on a Bioanalyzer. The results of these experiments were largely consistent with lariat sequencing results (Fig. 5 C–E). Decreased levels of aberrant splice-site activation were apparent in the hyperaccurate/inefficient alleles relative to WT, with the strongest phenotypes apparent in both assays for the V1860D mutant, consistent with our observations on ACT1-CUP1 reporters. The effects of the error-prone/efficient alleles were more modest, showing subtle yet consistent increases in the use of aberrant splice sites within some of the transcripts and little change from WT within others.
Fig. S5.
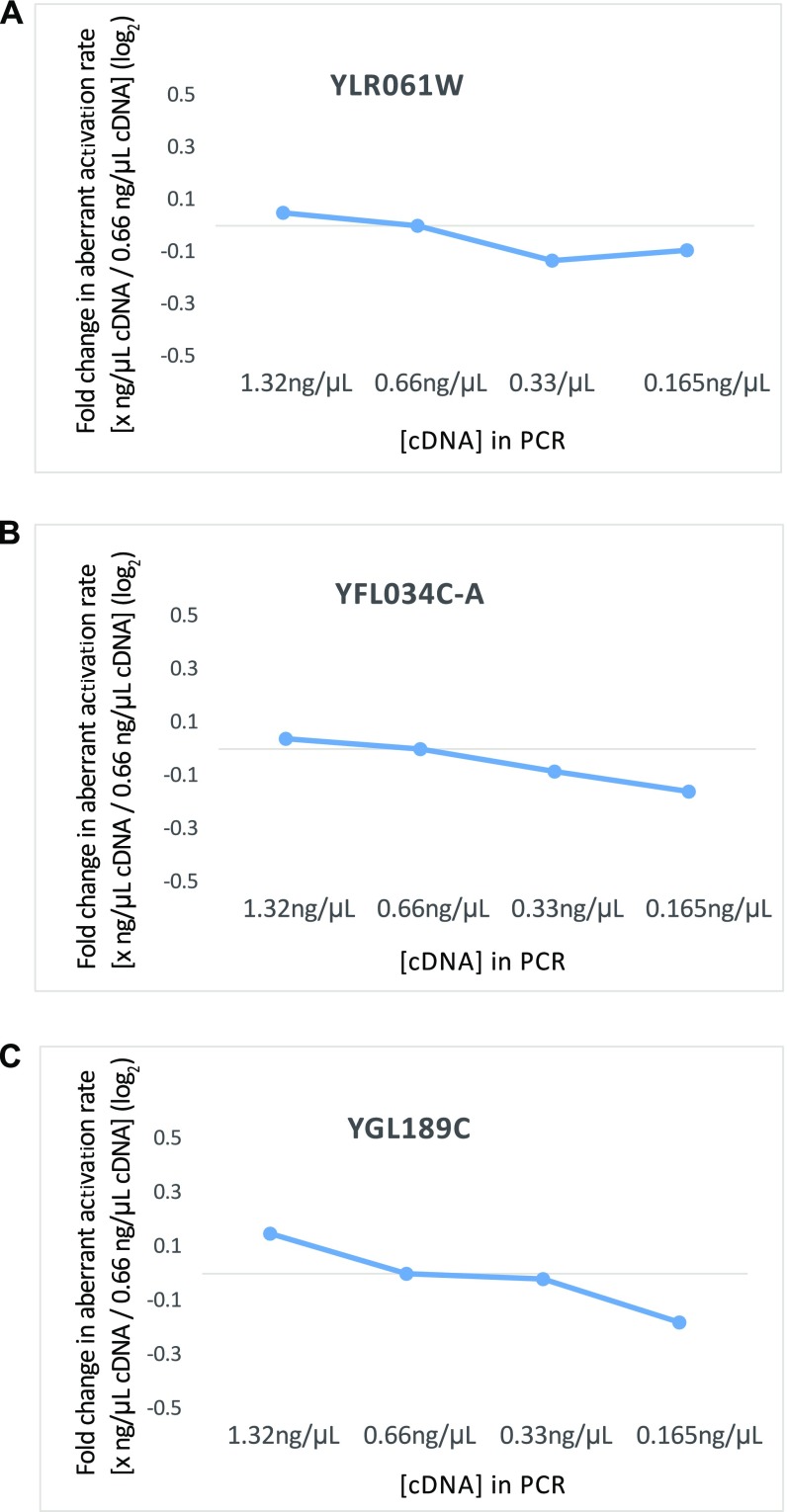
(A–C) Fold change in the measurement of aberrant activation rate with different concentrations of cDNA in the PCR over the concentration (0.66 ng/μL) that was used in the assay in Fig. 5 C–E are shown for the three introns investigated.
SI Materials and Methods
Strains and Plasmids.
See Table S3 for strain genotypes. In vitro splicing extracts were created from strain yTB72 (Mat a prp8∆::LYS2 his3 leu2 ura3 lys2 yCP50-PRP8) (25) with transitional and catalytic allele variants shuffled in. Strain yAMP24 (mat α cup1Δ::ura3-52 prp8Δ::LYS2 ade2 his3 leu2 ura3 lys2 yCP50-PRP8) (40) was used in the ACT1-CUP1 splicing reporter assays. Transitional and catalytic allelic variants of prp8 were created by templated mutagenesis of a pRS313-PRP8 plasmid using the KOD polymerase system (Novagen) and introduced to yTB72 or yAMP24 by 5FOA shuffling (39). The ACT1-CUP1 reporter plasmids have been described previously (25). The strain yTT516 (Mat a cup1Δ::ura3-52 prp2Δ::TRP yCP51-PRP2 prp8Δ::LYS yCP50-PRP8 ade leu his ura) was created by mating stain yTB72 with strain yTY1 (41) and shuffling in plasmids pJPS1910, pJPS1919, and pJPS2501 (27), containing PRP2 or prp2-Q548E encoded on a pRS415 vector (gifts from J. Staley, University of Chicago, Chicago). Strain yTT97 (Mat a prp8∆::LYS2 his3 leu2 ura3 lys2 dbr1::Kan) was created from yTB72 by using homologous recombination to replace the coding sequence of DBR1 with the Kan marker and was used for lariat sequencing analyses. Prp8 plasmids are available through Addgene (www.addgene.org).
Table S3.
Genotype of strains used in this study
| Strain | Genotype |
| yTB72 | mat a prp8∆::LYS2 his3 leu2 ura3 lys2 yCP50-PRP8 |
| yAMP24 | mat α cup1Δ::ura3-52 prp8Δ::LYS2 ade2 his3 leu2 ura3 lys2 yCP50-PRP8 |
| yTT516 | mat a cup1Δ::ura3-52 prp2Δ::TRP yCP51-PRP2 prp8Δ::LYS yCP50-PRP8 ade leu his ura |
| yTT97 | mat a prp8∆::LYS2 his3 leu2 ura3 lys2 dbr1::Kan |
ACT1-CUP1 Reporter Assays.
Strains derivative of yAMP24 containing a PRP8 allele with an ACT1-CUP1 reporter plasmid were grown overnight to saturation, diluted back to OD600 of 0.1, then grown to OD600 of 0.5–0.8. Cultures were then diluted back to an OD600 of 0.1, serially diluted from there in fourfold dilutions, and spotted onto –Leu plates containing varying concentrations of CuSO4 (0, 0.013, 0.025, 0.05, 0.075, 0.1, 0.18, 0.25, 0.3, 0.4, 0.5, 0.75, 1.0, and 1.5 mM). Plates were incubated at 30 °C for 3–4 d. The maximum concentration of copper a specific strain could grow on was assessed for each prp8 mutant in combination with each ACT1-CUP1 reporter and given a score calculated using the formula log2([Cu2+max prp8-catalytic or transitional]/[Cu2+max matched PRP8-WT]) to facilitate comparisons between strains and normalize between replicates (40). To capture minor differences between strains, if two strains of the same set (grown and spotted the same day onto the same batch of plates) were both able to tolerate the same maximum concentration of copper but one was growing clearly better or worse than the other at that concentration we adjusted the “tolerated copper” value up or down (depending on growth relative to multiple other strains on the plate) by 10% before calculating growth relative to WT.
In Vitro Splicing Assays.
Yeast were grown in rich media at 30 °C until they reached an OD600 of ∼1.0. Under these conditions, all mutants grew at a rate similar to PRP8. Extracts and pre-mRNA templates were made as described previously (25). In vitro splicing reactions consisted of a 40% vol/vol splicing extract combined with pre-mRNA template and 3% PEG 8000, 60 mM potassium phosphate, pH 7.0, 2.5 mM MgCl2, 2 mM ATP, and 10 U RNasin (Promega). Reactions were performed as indicated (10 min at 37 °C for first-step analysis, 20 min at 30 °C for BrC template analysis) before being quenched with 2.5% SDS and 1 mM EDTA. Reactions were incubated with 10 U of proteinase K (Invitrogen) at 65 °C for 10 min and loaded [1:1 in formamide loading buffer (96% formamide, 20 mM EDTA, and bromophenol blue or no dye)] onto a 6% polyacrylamide (19:1), 7 M urea, Tris/borate/EDTA gel, visualized on a Typhoon imaging system (GE Healthcare Life Sciences), and quantified using ImageQuant (GE Healthcare Life Sciences). First-step product was quantified as Flariat/(Flariat + Fpre-mRNA), and the fraction spliced in the BrC mutant template assays was quantified as condition (Flariat + Flariat-intermediate + FmRNA)/(Fpre-mRNA + Flariat + Flariat-intermediate + FmRNA). Values reported in Fig. 4 C and D represent the average and SEM for three distinct biological replicates performed using three separate preparations of splicing extract.
Lariat Sequencing: Library Preparation.
Total RNA isolation.
Cells were grown in yeast extract peptone dextrose (YPD) at 30 °C overnight to saturation in a 5-mL culture then back-diluted to an OD600 of 0.05 in a 25-mL YPD culture. When cells reached an OD600 between 0.4 and 0.8, 25 mL of cell culture was harvested by filtration then placed in a 15-mL Falcon tube and flash-frozen using liquid nitrogen and stored until RNA isolation. Total cellular RNA was isolated using hot phenol-chloroform extraction as follows. Into the 15-mL Falcon tube containing filter-collected cells was added 2 mL acid phenol (pH <5.5), followed by 2 mL AES buffer [50 mM sodium acetate (pH 5.3), 10 mM EDTA, and 1% SDS], and the tube was vortexed for 10 s then incubated at 65 °C for 7 min, vortexing for at least 3 s at 1-min intervals. The tube was then incubated on ice for 5 min, then the entire mixture was transferred to a 15-mL PhaseLock Heavy Gel tube (5PRIME) and centrifuged at 4 °C at 5,250 × g for 5 min. Two milliliters of phenol:chloroform:isoamyl alcohol (IAA) (25:24:1) was then added to the supernatant and mixed by shaking the tube up and down vigorously for 5 s. The tube was then centrifuged again in the same manner, and 2 mL chloroform was added to the supernatant, mixing as before. The tube was centrifuged as before, and this time the supernatant was transferred to a new 15-mL Falcon tube, to which 2.5 mL isopropanol and 200 μL 3 M sodium acetate (pH 5.3) were added. The resulting solution was mixed by vortexing and incubated at −20 °C for at least an hour. It was then centrifuged for 30 min at 4 °C at 5,250 × g, then supernatant was removed by decanting and the pellet was transferred to a 1.7-mL centrifuge tube. Then, the pellet was washed with 1.5 mL 70% ethanol twice by centrifuging for 5 min at 4 °C at 18,000 × g. The supernatant was decanted, and the sample was dried using vacuum centrifugation at room temperature. The RNA pellet was then resuspended in RNase-free water.
DNase treatment.
Ten micrograms of total RNA isolated by the above method was then subjected to DNase treatment in a 20-μL reaction with a final composition of 40 mM Tris⋅HCl (pH 8.0), 10 mM MgSO4, 1 mM CaCl2, and 2 U of RQ1 RNase-free DNase (Promega). The reaction was incubated at room temperature for 15 min. The reaction was then cleaned up by phenol-chloroform extraction and ethanol precipitation. All subsequent steps in the protocol used Low Retention tubes (Fisherbrand) to minimize sample loss.
rRNA depletion.
DNased RNA (4.5 μg) was used as input for rRNA depletion using Illumina’s Ribo-Zero Gold rRNA Removal Kit (Yeast) following the manufacturer’s instructions. At the end of the protocol, 85 μL of supernatant was obtained, which was purified by ethanol precipitation. To the 85 μL supernatant, 15 μL of water, 10 μL of 3 M sodium acetate (pH 5.3), 250 μL of ice-cold 200-proof pure ethanol, and 2 μL of glycogen (20 g/L) were added and the solution was incubated at −20 °C for at least an hour and precipitated. The RNA pellet was resuspended in water.
RNaseR treatment.
RNaseR treatment was set up with rRNA-depleted RNA obtained from the previous step in a 50-μL reaction with a final composition of 20 mM Tris⋅HCl (pH 8.0), 100 mM KCl, 0.1 mM MgCl2, and 1 U of RNaseR (Epicentre). The reaction was incubated at 37 °C for 10 min. The reaction was then cleaned up by phenol-chloroform extraction and ethanol precipitation. The RNA pellet was then resuspended in water.
cDNA synthesis.
cDNA synthesis was performed with RNaseR-treated RNA using SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) with modifications to the first-strand cDNA synthesis step. RNA and dN9 primers were mixed in a 10 μL volume and incubated at 65 °C for 10 min followed by cooling on ice for 1 min and on benchtop for 5 min. To this sample, 4 μL of 5× First-Strand Reaction Buffer without MgCl2 [250 mM Tris⋅HCl (pH 8.3) and 375 mM KCl] was added. Then, 2 μL 0.1 M DTT, 1 μL 10 mM dNTP, and 2 μL 30 mM MnCl2 were added and the reaction was equilibrated at 45 °C for 2 min. Then, 1 μL of SuperScript II RT was added and the reaction was incubated at 45 °C for 1 h. As seen in Fig. S3, addition of MnCl2 to a standard cDNA synthesis reaction with 3 mM MgCl2 significantly increases the read-through frequency of the 2′,5′ linkage. Similar read-through efficiency was achieved using 3 mM MnCl2 and no MgCl2. The first-strand reaction was cleaned up with ethanol precipitation to remove MnCl2 from the reaction. The pellet was then resuspended in 105 μL water, 4 μL 5× First-Strand Reaction Buffer with MgCl2 [250 mM Tris⋅HCl (pH 8.3), 375 mM KCl, 15 mM MgCl2], 2 μL 0.1 M DTT, 4 μL 10 mM dNTP, 30 μL 5× Second-Strand Reaction Buffer [100 mM Tris⋅HCl (pH 6.9), 450 mM KCl, 23 mM MgCl2, 0.75 mM β-NAD+, and 50 mM (NH4)2SO4], 4 μL Escherichia coli DNA Polymerase I (10 U/μL), and 1 μL E. coli RNase H (2 U/μL). E. coli DNA Ligase was not added. This reaction was incubated at 16 °C for 2 h. Following this, 2 μL of T4 DNA Polymerase (5 U/μL) was added and the reaction was further incubated at 16 °C for 5 min. The reaction was stopped by addition of 10 μL of 0.5 M EDTA and then ethanol-precipitated. The double-stranded cDNA pellet was then resuspended in water and purified on a 10% native acrylamide gel and sized above 30 bp. The cDNA was then extracted from the gel by soaking the crushed gel pieces in four to five volumes of 0.3 M sodium acetate (pH 5.3) and rotating overnight. The eluate was then precipitated with 2.5 volumes of ethanol and 4 μL gycogen followed by two washes with 70% ethanol. The cDNA pellet was then resuspended in 85 μL water.
End repair.
To 85 μL of double-stranded cDNA from the previous step, 10 μL of 10× NEBNext End Repair Reaction Buffer (500 mM Tris⋅HCl, pH 7.5, 100 mM MgCl2, 100 mM DTT, 10 mM ATP, and 4 mM dNTPs) and 5 μL of NEBNext End Repair Enzyme Mix (NEB End Repair Module) were added and the reaction was incubated at room temperature for 30 min. Cleanup was then done with phenol-chloroform extraction and ethanol precipitation. The RNA pellet was then resuspended in water. Further, the end-repaired DNA was cleaned up with GE Illustra MicroSpin G-25 columns per the manufacturer’s instructions. It was then eluted in 50 μL water.
A-tailing.
To 42 μL of end-repaired DNA, 5 μL of 10× NEBNext dA-Tailing Reaction Buffer (100 mM Tris⋅HCl, pH 7.9, 100 mM MgCl2, 0.5 M NaCl, 10 mM DTT, and 2 mM dATP) and 3 μL of Klenow Fragment (3′→ 5′ exo–) (NEB dA-tailing module) were added and the reaction was incubated at 37 °C for 30 min. Following this, the reaction was cleaned up with phenol-chloroform extraction and ethanol precipitation. Following precipitation, the pellet was resuspended in 16.5 μL water.
Adapter ligation.
Ligation was done with Enzymatics Rapid Ligation kit. To 16.5 μL of A-tailed DNA, 18.5 μL of 2× Rapid Ligation buffer [132 mM Tris⋅HCl (pH 7.6), 20 mM MgCl2, 2 mM DTT, 2 mM ATP, and 15% PEG 6000], 1 μL of T4 DNA Ligase (Rapid) (L6030-HC-L), and 1 μL of Illumina Tru-seq barcoded adapter were added. This reaction was incubated at room temperature for 15 min.
Ligation sizing.
Thirty-seven microliters of 2× denaturing gel loading buffer (95% formamide and 25 mM EDTA) was added to the ligation reaction and this was run on 6% denaturing PAGE. The sample was sized between 150 nt and 330 nt. Ligated DNA was then extracted from the sized gel by soaking the crushed gel pieces in four to five volumes of 0.3 M sodium acetate (pH 5.3) and rotating overnight. The eluate was then precipitated with 2.5 volumes of ethanol and 4 μL glycogen followed by two washes with 70% ethanol and vacuum drying. The pellet was resuspended in 40 μL water.
PCR amplification.
Reactions were performed in 20 μL volume, with 10 μL DNA sample precipitated from gel, 1× Phusion HF buffer (NEB), dNTPs (0.25 mM each), 0.25 μM Illumina P5 PCR primer, 0.25 μM Illumina P7 PCR primer, and Phusion polymerase. The PCR protocol was an initial denaturation step of 98 °C for 30 s, followed by 8–10 cycles of 98 °C for 10 s, 65 °C for 30 s, and 72 °C for 30 s. Final extension was at 72 °C for 5 min. The PCR product was purified on a 6% native acrylamide gel, recovering the material between 150 and 330 bp. The purified products were sequenced on an Illumina NextSeq platform.
Cleanup by phenol-chloroform extraction and ethanol precipitation.
For phenol-chloroform extraction, the sample was made up to 100 μL or 200 μL with water and then an equal volume of phenol:chloroform:IAA (25:24:1) was added, mixed by shaking and centrifuged in a 2-mL PhaseLock Heavy Gel tube (5PRIME) at 18,000 × g for 2 min. Another volume of phenol:chloroform:IAA was again added to the supernatant and centrifuged again and then one volume of chloroform was added to the supernatant. The tube was centrifuged again and the supernatant was transferred to a 1.5-mL tube. To the supernatant, 1/10 volume of 3 M sodium acetate (pH 5.3), 2.5 volumes of ice-cold 200-proof pure ethanol, and 2 μL of glycogen were added and the solution was incubated at −20 °C for at least an hour. This was followed by centrifuging the tube at 18,000 × g for 30 min at 4 °C and removal of the supernatant. This was followed by two washes, each with 1 mL of 70% ethanol and centrifuging at 18,000 × g for 5 min. After the second wash, the supernatant was decanted, and the sample was dried using vacuum centrifugation at room temperature.
Libraries.
Lariat sequencing libraries were prepared for one biological replicate for each strain discussed in Fig. 4. Additionally, to assess the technical reproducibility of the method, two technical replicates were processed separately from the step of rRNA depletion using a single sample of DNased RNA isolated from prp8 N1869D. The method was highly technically reproducible (Pearson r = 0.99) (Fig. S4). The alignment statistics for all libraries are listed in Dataset S2.
Lariat Sequencing: Data Processing.
Lariat sequencing genome alignment.
Illumina sequencing reads were trimmed of any 3′ adapter using Trimmomatic with the following parameters: 3:30:10 and MINLEN:18. Trimmed reads were aligned to the S. cerevisiae genome (Ensembl R64-1-1) using Bowtie2. Parameters of alignment were “score-min L,-0.4,-0.4 –very-sensitive.” End-to-end alignments were done for the initial genome alignment. Paired end alignments on lariat reads were done using “–ff -I 0 -X 5000 –no-mixed –no-discordant.”
Branch-spanning lariat read alignment.
For identification of lariat reads spanning the lariat branch, each read that failed to align to the genome in end-to-end fashion was considered a candidate lariat branch read. These reads were split into mate paired reads at every GT dinucleotide in the forward and reverse complement strands. The subset of mate pairs where each read in the mate pair is at least 10 nucleotides were candidates for alignment. These reads were then assessed for alignment using Bowtie2 in paired-end mode with the fragment containing the GT as mate 1 and the other as mate 2, using the previously noted parameters. Because a single read split in this fashion can yield several possible alignments, the possible GT split alignments were collapsed into a single best available paired-end alignment defined as the alignment that minimizes the number of mismatches.
Identification of annotated introns.
5′SS, Br locations, and strandedness were obtained from each lariat read using the position of GT on the lariat read alignment. Because mapped reads in the SAM/BAM format are represented on the forward strand of the reference genome, if the lariat read showed a GT dinucleotide in the beginning of the left part of the alignment the position of G in the genome was marked as the 5′SS with sense directionality and the last position of the right part was marked as the Br location. If the lariat read showed an AC dinucleotide in the last two positions of the right part of the alignment, the position of C in the genome was marked as the 5′SS with antisense directionality and the first position of the left part was marked as the Br location. Lariat reads with both a GT in the beginning of the left part and AC at the end of the right part were discarded as ambiguous reads (<0.2% of lariat reads). The 5′SS locations were then matched to the annotated 5′SS positions (Ensembl R64-1-1) to identify annotated introns. The list of identified annotated introns (287) along with their Br positions and counts for the PRP8 WT dataset is shown in Dataset S1, A. For the analysis in Fig. 5A, only those annotated intronic reads with Br position mapping to annotated Br locations (based on Br-3′SS distance information from the Ares Intron Database 4.1, intron.ucsc.edu/yeast4.1/) were used (Dataset S1, B) (276 introns in the PRP8 WT dataset). Because reverse transcriptase frequently introduces deletions and mutations when creating the cDNA product that crosses the 5′SS to the Br, to account for errors introduced during reverse transcription the identified Br was allowed to be within ±2 nt of the annotated location to be characterized as a read containing annotated Br. Counts for each annotated intron were then the sum of all reads for that 5′SS with a Br position within ±2 nt of the annotated Br site.
Identification of aberrant 5′ splice sites.
To identify aberrant 5′SSs, only those lariat reads with Br position mapping to ±2 nt of the annotated Br locations were considered for analysis to reduce false-positive rate and restrict the analysis to aberrant 5′SS events in annotated introns. For these reads, if the 5′SS location was found to be more than 5 nt and less than 1,000 nt from the annotated 5′SS location, it was considered an aberrant 5′SS for that intron. The list of identified aberrant 5′SSs and their counts for the PRP8 WT dataset are in Dataset S1, C.
Quantification of aberrant activation rate.
For each annotated intron, reads mapping to all of its aberrant 5′SSs were collected and their counts summed. Introns with collective aberrant counts less than 30 were excluded from further analysis because of the noise associated with low-count events. For the remaining events, the proportion of aberrant site use was calculated as
Validation of Aberrant Splice Sites by RT-PCR.
cDNA was generated from 1 μg of DNase-treated RNA using the methodology described above. PCR was done using primers (Table S4) designed to capture the annotated and most abundant aberrant splicing isoforms as described in Fig. 5B, with 3 min of initial denaturation at 95 °C followed by 24–30 cycles of 95 °C for 10 s, 55 °C for 20 s, and 72 °C for 30 s in a 15-μL reaction containing 10 ng cDNA, 250 nM primers, 10 mM Tris⋅HCl (pH 8.5), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPs (each), 0.25× SyBr Green, 5% vol DMSO, and Taq. Then, 1 μL of the reaction was run on an Agilent Bioanalyzer DNA 1000 assay to resolve the products and they were quantified with Agilent 2100 Bioanalyzer software. For YGL189C, because the aberrant 5′SS isoform was low in abundance relative to the annotated isoform, 10 such PCR reactions were run, pooled, purified, and concentrated into 10-μL volume using DNA Clean & Concentrator-5 kit from Zymo. One microliter of the purified product was then run on Agilent Bioanalyzer DNA 1000 assay. The area under the curve (AUC) for the bands corresponding to the annotated and most abundant aberrant isoform was used for the calculation of aberrant activation rate:
To investigate whether the changes observed in aberrant activation rate in the mutants in Fig. 5 C–E are reflective of actual biological changes and not a result of technical artifacts, the aberrant activation rate for a given intron was calculated across a 16-fold dilution series of cDNA template in the PCR (Fig. S5). Whereas YLR061W and YFL034C-A gave consistent results across a 16-fold cDNA dilution, YGL189C showed a 0.32-fold difference between the highest and lowest dilutions on a log2 scale. However, this change was considerably lower than the decreases in aberrant activation rate observed for V1860D and V1860N in Fig. 5E.
Table S4.
Sequences of RT-PCR Primer primers used
| Primer | Sequence |
| YFL034C-A_FWD_LAR | GGCCAGACATTTTTTCCTCGTCC |
| YFL034C-A_RC_LAR | GTTCGCTGTGAAAGCGGGACTGTTC |
| YLR061W_FWD_LAR | GTAGGCAACTTTGTGGTTTCGGGA |
| YLR061W_RC_LAR | GCAACTAAACACATAGACGCTTC |
| YGL189C_FWD_LAR | CCTTCCCTTTTTGCCACGATC |
| YGL189C_RC_LAR | CAATATTCTTCAATAATAAGTAC |
Discussion
Here we present an analysis of the role of the RNaseH domain (RH) of Prp8 in spliceosomal activity. Specifically we reveal two classes of mutants in the RH extension of Prp8, transitional (inefficient/hyperaccurate) and catalytic (efficient/error-prone). Furthermore, because these transitional and catalytic prp8 alleles stabilize mutually exclusive β-hairpin and loop conformations of the RH extension of Prp8 (Fig. 1 and Table S1) (19), our data directly link a conformational change in the spliceosome to genomewide changes in splicing fidelity.
The Toggle Model.
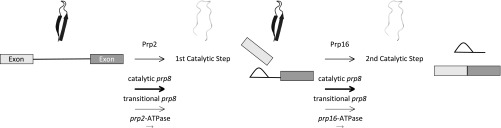
We suggest that the RH extension of Prp8 is a structural toggle, oscillating between mutually exclusive conformations at distinct points in the splicing cycle to promote high-fidelity, high-efficiency splicing. Specifically, we propose that the RH extension adopts the transitional conformation before the first step, toggles to the catalytic for the first step, toggles back to the transitional between catalytic steps, and then again adopts the catalytic conformation for the second step—a “toggle model” (Fig. 6). That the RH extension adopts the catalytic form at both catalytic steps is supported by growth assays where yeast expressing catalytic prp8 alleles promote the splicing of a variety of ACT1-CUP1 reporters in vivo, regardless of which catalytic step is affected by the reporter (Fig. 2 and Fig. S2) (13, 16–19), our in vitro splicing assays (Fig. 4), and the structure of a Bact spliceosome (5). That RH adopts a transitional form before the first and again between the first and second steps is supported by structures of C and C* spliceosomes (7, 8, 10, 11) and by genetic interactions between prp8 toggle alleles and the DEAH-box ATPases Prp2 and Prp16, which promote spliceosome conformational changes required for the first and second steps. Catalytic prp8 alleles suppress growth defects in yeast expressing ATPase-deficient Prp2, whereas transitional prp8 alleles exacerbate them (Fig. 3). These same catalytic alleles also suppress the growth defect of yeast expressing ATPase-deficient Prp16 (17, 18).
Fig. 6.
The toggle model. The Prp8 RH extension toggles between catalytic (error-prone/efficient) and transitional (hyperaccurate/inefficient) forms during the splicing cycle. Prp2 ATPase activity promotes the first catalytic step; Prp16 ATPase activity promotes the second. Catalytic prp8 toggle alleles bias the spliceosome toward the catalytic conformation at both the first and second steps, whereas transitional mutants bias against conversion to the catalytic conformation.
The biological relevance of conformational toggling is well understood for the ribosome, another large, high-fidelity RNP machine that shares numerous similarities with the spliceosome (2). The ribosome repeatedly moves between “open” and “closed” conformations. Proofreading, which allows for the rejection of a near-cognate tRNA, takes place in the open state, whereas peptide bond catalysis occurs in the closed (35). Translational fidelity and ribosome conformation have been directly linked through studies of ribosome mutants known as ribosomal ambiguity (ram) and restrictive. Ram mutants favor the closed, catalytic form of the ribosome and destabilize the proofreading conformation, even in the presence of near-cognate tRNA, resulting in coding errors and more rapid translation. Restrictive mutants have the opposite effect: They favor the open, proofreading conformation and are hyperaccurate (35).
The first-step/second-step model was proposed in 2004 and was the first spliceosomal model to invoke conformational toggling by the ribosome (17). By this model the spliceosome alternates between unique first- and second-step conformations (13, 17–19, 36) wherein first- and second-step alleles of prp8 bias the conformation of the spliceosome toward first- or second-step conformations, respectively. Primer extensions performed on ACT1-CUP1 reporter RNA isolated from strains carrying first- and second-step alleles were key to the development of this model. However, for the subset of prp8 alleles that map to the RH extension, we note that these data are also consistent with the toggle model. Rather than a preference to dwell in a second-step conformation, decreased lariat intermediate and increased mRNA levels exhibited by second-step alleles can also indicate more rapid progression through both steps of splicing, as we propose. Similarly, instead of a bias toward a first-step conformation, decreased pre-mRNA and increased lariat intermediate levels observed in first-step alleles are consistent with a general bias against catalysis; the limited number of spliceosomes that succeed at the first step are unable to complete the second.
The RH domain of Prp8 is not the only spliceosomal component that toggles. Stem II of the U2 snRNA can fold into two mutually exclusive structures: Stem IIa and Stem IIc (37, 38). Alleles that bias toward the Stem IIa conformation have fidelity phenotypes similar to prp8 transitional alleles. These alleles also have genetic and proofreading phenotypes comparable to prp16 ATPase-deficient alleles. In contrast, the U2 Stem IIc conformation has been shown to be necessary for both catalytic steps. Alleles that promote the U2 Stem IIc conformation have phenotypes similar to the prp8 catalytic alleles, as do mutations within the myb-domain of Cef1 (36–38). We speculate that these individual toggles might be linked; the spliceosome as a whole may toggle between catalytic and transitional conformations.
Conclusion.
Although the ribosome and spliceosome are separated by billions of years of evolution, they exhibit many similarities including our demonstration here of a direct coupling of catalytic efficiency with fidelity. Whereas work over the past decade has shed light on the mechanisms by which structural changes in the ribosome enable discrimination of cognate from near-cognate tRNAs, we have only begun to investigate these mechanisms in the spliceosome, owing at least in part to the difficulty in determining whether a splice site should be considered as “cognate” or “near-cognate.” In fact, in the current work probing budding yeast, where splice sites have evolved to conform to a precise consensus sequence, the global locations of aberrant splice-site activation are marked by splice-site sequences that in many instances look far more cognate than do many annotated mammalian splice sites (Fig. 5 C–E). Nevertheless, there are many more locations across the genome where strong potential splice sites exist yet for which we detect no activation. Understanding why the spliceosome activates some of these sites but not others will be key to understanding the relationship between catalytic efficiency and fidelity on the spliceosome. Likewise, it will be crucial to identify structural conformations of the spliceosome that can trigger the interconversion between proofreading and catalytic states, which could contribute to alternative splice-site selection in higher organisms.
Materials and Methods
Standard molecular biology and genetic techniques were used as described previously (24, 25, 39) and are detailed in SI Materials and Methods. Table S3 contains a list of strains used in this work. Plasmids are available through Addgene (www.addgene.org).
For lariat sequencing, enrichment of lariat RNAs from total cellular RNA was accomplished by an initial treatment with Illumina’s Ribo-Zero Gold rRNA Removal Kit (yeast), followed by treatment with RNaseR at 37 °C for 10 min to digest the linear species. First-strand cDNA synthesis was performed on the remaining RNA under otherwise standard conditions but where MgCl2 was replaced with 3 mM MnCl2 to facilitate read-through of the 2′,5′ lariat bond. Libraries for sequencing were generated as previously described (32). A detailed protocol is provided in SI Materials and Methods. Lariat sequencing data can be accessed from the NCBI GEO database (accession no. GSE96891).
Supplementary Material
Acknowledgments
We thank D. Velazquez, A. de Bruyn Kops, K. Patrick, Y. He, Z. Dwyer, and M. Dinglasan for help with experiments and the J. Staley, C. Query, and M. Konarska laboratories for strains and plasmids. We thank J. Burke and H. Madhani for helpful discussions. This work was supported by NIH Grants R01GM2119-42 (to C.G.) and R01GM098634 (to J.A.P.) and a Research Scholar Grant from the American Cancer Society (to J.A.P.). C.G. is an American Cancer Society Research Professor of Molecular Genetics.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the NCBI GEO database (accession no. GSE96891).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701462114/-/DCSupplemental.
References
- 1.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Semlow DR, Staley JP. Staying on message: Ensuring fidelity in pre-mRNA splicing. Trends Biochem Sci. 2012;37:263–273. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen THD, et al. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature. 2016;530:298–302. doi: 10.1038/nature16940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan R, et al. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis. Science. 2016;351:466–475. doi: 10.1126/science.aad6466. [DOI] [PubMed] [Google Scholar]
- 5.Rauhut R, et al. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016;353:1399–1405. doi: 10.1126/science.aag1906. [DOI] [PubMed] [Google Scholar]
- 6.Yan C, Wan R, Bai R, Huang G, Shi Y. Structure of a yeast catalytically activated spliceosome at 3.5 Å resolution. Science. 2016;353:904–11. doi: 10.1126/science.aag0291. [DOI] [PubMed] [Google Scholar]
- 7.Galej WP, et al. Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537:197–201. doi: 10.1038/nature19316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan R, Yan C, Bai R, Huang G, Shi Y. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science. 2016;353:895–904. doi: 10.1126/science.aag2235. [DOI] [PubMed] [Google Scholar]
- 9.Yan C, et al. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science. 2015;349:1182–1191. doi: 10.1126/science.aac7629. [DOI] [PubMed] [Google Scholar]
- 10.Bertram K, et al. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017;542:318–323. doi: 10.1038/nature21079. [DOI] [PubMed] [Google Scholar]
- 11.Fica SM, et al. Structure of a spliceosome remodelled for exon ligation. Nature. 2017;542:377–380. doi: 10.1038/nature21078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grainger RJ, Beggs JD. Prp8 protein: At the heart of the spliceosome. RNA. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang K, Zhang L, Xu T, Heroux A, Zhao R. Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci USA. 2008;105:13817–13822. doi: 10.1073/pnas.0805960105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn AN, Brow DA. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics. 2000;155:1667–1682. doi: 10.1093/genetics/155.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn AN, Reichl EM, Brow DA. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc Natl Acad Sci USA. 2002;99:9145–9149. doi: 10.1073/pnas.102304299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins CA, Guthrie C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev. 1999;13:1970–1982. doi: 10.1101/gad.13.15.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- 18.Liu L, Query CC, Konarska MM. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- 19.Schellenberg MJ, et al. A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation. Nat Struct Mol Biol. 2013;20:728–734. doi: 10.1038/nsmb.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siatecka M, Reyes JL, Konarska MM. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 1999;13:1983–1993. doi: 10.1101/gad.13.15.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fica SM, et al. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcia M, Pyle AM. Visualizing group II intron catalysis through the stages of splicing. Cell. 2012;151:497–507. doi: 10.1016/j.cell.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umen JG, Guthrie C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayerle M, Guthrie C. Prp8 retinitis pigmentosa mutants cause defects in the transition between the catalytic steps of splicing. RNA. 2016;22:798–809. doi: 10.1261/rna.055459.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohrt T, et al. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA. 2012;18:1244–1256. doi: 10.1261/rna.033316.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wlodaver AM, Staley JP. The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic RNA core of the spliceosome. RNA. 2014;20:282–294. doi: 10.1261/rna.042598.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson K, Moore MJ. Bimolecular exon ligation by the human spliceosome bypasses early 3′ splice site AG recognition and requires NTP hydrolysis. RNA. 2000;6:16–25. doi: 10.1017/s1355838200001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fouser LA, Friesen JD. Mutations in a yeast intron demonstrate the importance of specific conserved nucleotides for the two stages of nuclear mRNA splicing. Cell. 1986;45:81–93. doi: 10.1016/0092-8674(86)90540-4. [DOI] [PubMed] [Google Scholar]
- 30.Taggart AJ, DeSimone AM, Shih JS, Filloux ME, Fairbrother WG. Large-scale mapping of branchpoints in human pre-mRNA transcripts in vivo. Nat Struct Mol Biol. 2012;19:719–721. doi: 10.1038/nsmb.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Awan AR, Manfredo A, Pleiss JA. Lariat sequencing in a unicellular yeast identifies regulated alternative splicing of exons that are evolutionarily conserved with humans. Proc Natl Acad Sci USA. 2013;110:12762–12767. doi: 10.1073/pnas.1218353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stepankiw N, Raghavan M, Fogarty EA, Grimson A, Pleiss JA. Widespread alternative and aberrant splicing revealed by lariat sequencing. Nucleic Acids Res. 2015;43:8488–8501. doi: 10.1093/nar/gkv763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gould GM, et al. Identification of new branch points and unconventional introns in Saccharomyces cerevisiae. RNA. 2016;22:1522–1534. doi: 10.1261/rna.057216.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin D, Huang L, Wlodaver A, Andrade J, Staley JP. Sequencing of lariat termini in S. cerevisiae reveals 5′ splice sites, branch points, and novel splicing events. RNA. 2016;22:237–253. doi: 10.1261/rna.052829.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 36.Query CC, Konarska MM. CEF1/CDC5 alleles modulate transitions between catalytic conformations of the spliceosome. RNA. 2012;18:1001–1013. doi: 10.1261/rna.029421.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hilliker AK, Mefford MA, Staley JP. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perriman RJ, Ares M., Jr Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular and Cell Biology. Elsevier; London: 2004. [Google Scholar]
- 40.Price AM, Görnemann J, Guthrie C, Brow DA. An unanticipated early function of DEAD-box ATPase Prp28 during commitment to splicing is modulated by U5 snRNP protein Prp8. RNA. 2013;20:40–60. doi: 10.1261/rna.041970.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edwalds-Gilbert G, et al. Dominant negative mutants of the yeast splicing factor Prp2 map to a putative cleft region in the helicase domain of DExD/H-box proteins. RNA. 2000;6:1106–1119. doi: 10.1017/s1355838200992483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins CA, Guthrie C. Genetic interactions between the 5′ and 3′ splice site consensus sequences and U6 snRNA during the second catalytic step of pre-mRNA splicing. RNA. 2001;7:1845–1854. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.