Significance
Many clinically significant human viral and bacterial pathogens use dynamin-dependent endocytosis to initiate infection or deliver toxin into host cells. Owing to the complex nature of this cellular process, the molecular mechanisms that regulate this pathway remain to be fully elucidated. Here, we use rotavirus (RV) as a model and identify drebrin as a regulatory protein that restricts the cell entry of multiple viruses. We demonstrate that genetic depletion or chemical inhibition of drebrin leads to enhanced RV infection in vitro and increased diarrhea incidence and virus shedding in vivo. Our current study provides insights into endocytosis regulation in general and highlights the potential broad application of blocking drebrin to augment the uptake of viruses and other dynamin-mediated cargo.
Keywords: drebrin, rotavirus, endocytosis
Abstract
Despite the wide administration of several effective vaccines, rotavirus (RV) remains the single most important etiological agent of severe diarrhea in infants and young children worldwide, with an annual mortality of over 200,000 people. RV attachment and internalization into target cells is mediated by its outer capsid protein VP4. To better understand the molecular details of RV entry, we performed tandem affinity purification coupled with high-resolution mass spectrometry to map the host proteins that interact with VP4. We identified an actin-binding protein, drebrin (DBN1), that coprecipitates and colocalizes with VP4 during RV infection. Importantly, blocking DBN1 function by siRNA silencing, CRISPR knockout (KO), or chemical inhibition significantly increased host cell susceptibility to RV infection. Dbn1 KO mice exhibited higher incidence of diarrhea and more viral antigen shedding in their stool samples compared with the wild-type littermates. In addition, we found that uptake of other dynamin-dependent cargos, including transferrin, cholera toxin, and multiple viruses, was also enhanced in DBN1-deficient cells. Inhibition of cortactin or dynamin-2 abrogated the increased virus entry observed in DBN1-deficient cells, suggesting that DBN1 suppresses dynamin-mediated endocytosis via interaction with cortactin. Our study unveiled an unexpected role of DBN1 in restricting the entry of RV and other viruses into host cells and more broadly to function as a crucial negative regulator of diverse dynamin-dependent endocytic pathways.
Endocytosis is a complex and tightly regulated process pivotal to uptake of nutrients, neurotransmitters, and hormones into cells (1). Endocytosis is also central to the host innate immune response because the entry and hence detection of various bacterial toxins and viral pathogens are dependent on their internalization and access into endocytic vesicles (2, 3). Endocytosis can be grossly categorized into dynamin-dependent and dynamin-independent pathways, based on the reliance on dynamins, the host GTPases essential to the scission reaction of nascent vesicles from plasma membranes (2). Dynamin-dependent pathways can be further divided into clathrin- or caveolae-mediated endocytosis (4), whereas dynamin-independent pathways involve phagocytosis by specialized immune cells, macropinocytosis, direct fusion, and other routes, such as the nonclathrin/noncaveolae-dependent carrier and lipid raft pathway (5).
Rotavirus (RV), a nonenveloped, segmented, double-stranded RNA virus, is the leading cause of severe dehydrating diarrhea in infants, accounting for 215,000 deaths annually worldwide (6). RV infections mostly impact the young but can also affect the immunocompromised, the elderly, and healthy adults (7). Therefore, RV is a significant global health threat, and a deeper understanding of the RV–host interaction is needed to identify cellular pathways that could serve as drug targets to prevent or limit infection. RV efficiently replicates in the mature intestinal epithelial cells (IECs) in the small bowel and its entry into host cells relies primarily on the viral outer capsid spike protein VP4 (8, 9). After VP4 binds to its cognate receptors on cellular surfaces, it undergoes a marked conformational change that allows the RV particles to be taken up by the host cells via endocytosis. Multiple studies including two recent genome-wide siRNA screens suggest that RV enters via a dynamin-2–dependent endocytosis (10, 11). RV infection of polarized IECs from the apical side is also shown to depend on clathrin (12). Despite these advances, the intricate molecular mechanisms of how VP4 interacts with the host proteins at a postattachment step remain unclear. Here, we used tandem immunoprecipitation (IP) coupled with high-resolution mass spectrometry (MS) to systematically construct a VP4–host proteome interactive network, which revealed that a large number of actin-binding proteins are conducive or inhibitory to RV infection. Notably, we found that drebrin (encoded by DBN1), known to stabilize actin filaments (13), inhibits RV infection in a virus strain-independent manner at the entry step. Genetic depletion or chemical inhibition of DBN1 significantly facilitates the endocytosis of dynamin-dependent cargo, including transferrin, cholera toxin, vesicular stomatitis virus (VSV), as well as RV. Our interrogation of the RV–host protein interactome unveiled a role of DBN1 as a master regulator of both clathrin- and caveolin-dependent endocytic pathways.
Results
VP4 Interactome Analysis Reveals Multiple Host Cytoskeleton-Associated Proteins That Regulate RV Infection.
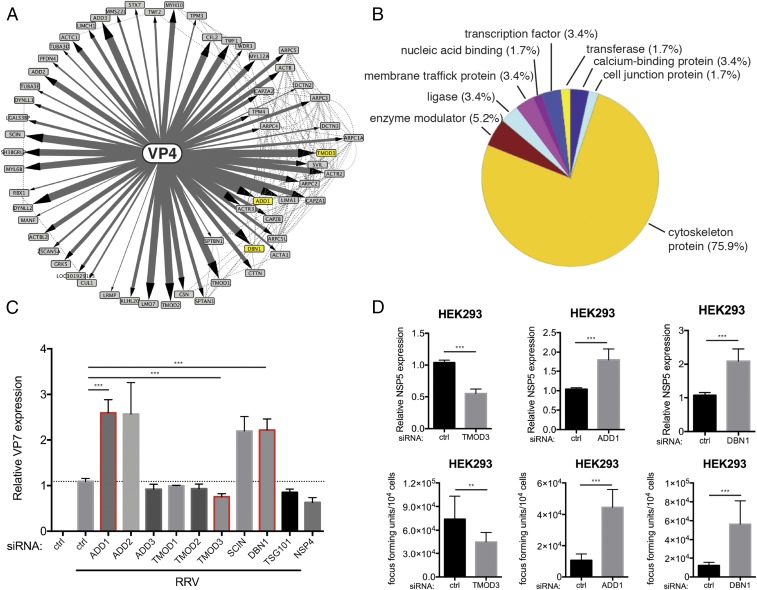
To comprehensively identify novel host proteins involved in RV entry, we focused our attention on VP4, the viral hemagglutinin that, along with VP7, constitutes the outermost layer of the infectious RV particle. We adopted a tandem IP–MS approach as previously described (14, 15). In brief, we generated an HEK293 stable cell line that, upon doxycycline treatment, expresses VP4 derived from a rhesus rotavirus (RRV) strain fused at the N terminus with a localization and affinity purification (LAP) tag (EGFP-TEV-S). A large-scale culture of this cell line was produced and used for immunoprecipitation. Based on the IP–MS results (Dataset S1), we constructed a systems view of high-confidence VP4-interacting partners (Fig. 1A). We identified 59 cellular proteins that coprecipitated with VP4. Gene ontology analysis indicated that the majority are associated with the cytoskeleton (Fig. 1B). Such strong interaction with the host cytoskeleton was not observed for the other RV proteins such as NSP1 that we previously analyzed (15). We also noticed several protein complexes/families specifically enriched in our VP4 interactome, such as the actin-related protein 2/3 complex (ARPC; subunits 1A, 2, 3, 4, 5, and 5L), F-actin–capping protein (CAPZ; subunits A1, A2, and B), adducin (ADD1–3), and tropomyosin (TMOD1–3) (Fig. 1A).
Fig. 1.
VP4 proteomic network reveals cytoskeleton-binding proteins that modulate RV infection. (A) Interactome of the bait viral protein (VP4) and high-confidence host binding proteins. Solid lines with arrowheads represent interactions identified in this study. The width of lines corresponds to the strength of interaction detected in the IP–MS experiment. Dotted lines represent publicly curated protein–protein interactions. The proteins studied in this paper are highlighted by yellow nodes. (B) Pie chart of PANTHER functional classification of VP4-interacting proteins shown in A. (C) HEK293 cells were transfected with indicated siRNA for 48 h and infected with simian RV RRV strain at MOI = 1 for 24 h. Levels of RV replication were measured by RT-qPCR examining the expression of viral gene VP7, normalized to that of GAPDH. The genes studied in this paper are outlined by red boxes. (D) Same experiment as in C except that viral gene NSP5 was measured instead of VP7 by RT-qPCR and virus titer in the supernatants was determined by a focus-forming unit (FFU) assay. For C and D, experiments were repeated at least five times. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (**P ≤ 0.01; ***P ≤ 0.001).
To functionally study the role of these cytoskeleton binding proteins in RV infection, we used small interfering RNA (siRNA) to knock down the expression of select proteins and quantified RV replication by measuring viral VP7 RNA levels with RT-qPCR at 24 h postinfection (p.i.) (Fig. 1C). Other than tumor susceptibility gene 101 (TSG101), none of the other siRNAs induced obvious cytopathic effect (SI Appendix, Fig. S1A). TSG101 and NSP4 served as positive controls because these proteins were previously shown to be important for promoting RV infection (11, 16). Interestingly, depletion of TMOD3 led to a decrease in VP7 expression comparable to that induced by the positive controls (>20%), suggesting that TMOD3 might facilitate simian RV RRV strain infection. In contrast, siRNA-mediated silencing of ADD1, ADD2, SCIN, and DBN1 (which encodes a protein named drebrin) resulted in enhanced RV infection.
Based on the physical interaction strength with VP4 revealed by IP–MS and the effect on RV replication following siRNA depletion, we chose to further examine TMOD3, ADD1, and DBN1. We found that TMOD3 siRNA reduced both intracellular viral RNA, as measured by NSP5 expression, and virus yield in the cell supernatants at 24 h p.i. (Fig. 1D). Further, the effect of TMOD3 on RV infectivity was virus strain-specific (SI Appendix, Fig. S1B) and RRV infectivity was enhanced in HEK293 cells overexpressing TMOD3 (SI Appendix, Fig. S1C), suggesting that TMOD3 indeed facilitates RRV infection. In contrast, siRNA knockdown of ADD1 and DBN1 increased intracellular viral RNA levels and extracellular virus titers (Fig. 1D), indicative of inhibitory roles of ADD1 and DBN1 on RV infection. The effect of ADD1 on RV infection was also strain dependent (SI Appendix, Fig. S2A). To further examine the anti-RV replication role of ADD1, we used CRISPR-Cas9 technology to generate ADD1−/− HEK293 cells (SI Appendix, Fig. S2 B and C). ADD1−/− cells supported higher levels of RV replication and virus release than did control cells (SI Appendix, Fig. S2 D and E), confirming that ADD1 acts as a host restriction factor of RV infection. Of note, the effects of both TMOD3 and ADD1 on RV replication appeared to be strain specific although the mechanistic basis for this strain specificity was not further examined. Taken together, our interrogation of the VP4–host protein interactome revealed several host cytoskeleton proteins and identified TMOD3 as proviral factor and ADD1 and DBN1 as antiviral factors.
DBN1 Interacts with VP4 and Restricts RV Entry.
We next narrowed our focus to more fully examine the role of DBN1 in regulating RV replication because little is known concerning its potential role during virus infections. DBN1 is a cytoskeletal protein whose aberrant expression has been implicated in several malignancies and neurological disorders (17, 18). We first validated the physical interaction between DBN1 and VP4 during both exogenous expression and active RV infection. Using the HEK293 cells stably expressing VP4, we observed that GFP-tagged VP4 specifically coprecipitated with endogenous DBN1 upon doxycycline induction (Fig. 2A). In addition, consistent with previous observation (19), we found DBN1 at the plasma membrane and it strongly colocalized with VP4 and actin filaments during RV infection (Fig. 2B). We further validated the specific colocalization of DBN1 and VP4, but not two other RV structural proteins (VP6 and VP7), using a proximity ligation assay (SI Appendix, Fig. S3 A and B). Consistent with our confocal results, DBN1–VP4 interaction was detected at the periphery of RV-infected cells (SI Appendix, Fig. S3A). Importantly, this interaction also took place during active RV entry into host cells, although the number of foci was much lower (SI Appendix, Fig. S3 C and D). Because VP4 is naturally cleaved by trypsin-like enzymes in the small intestine into N-terminal VP8* and C-terminal VP5* fragments, we next asked which region is responsible for DBN1 interaction. Using an in vitro transcription and translation system, we expressed full-length VP4, VP8*, and VP5* and performed immunoprecipitation with recombinant GST-tagged DBN1 protein. We found that in contrast to VP4 and C-terminal VP5* coprecipitation with DBN1, the N-terminal VP8* region responsible for the sialic acid binding and cell attachment was unable to pull down DBN1 (SI Appendix, Fig. S4).
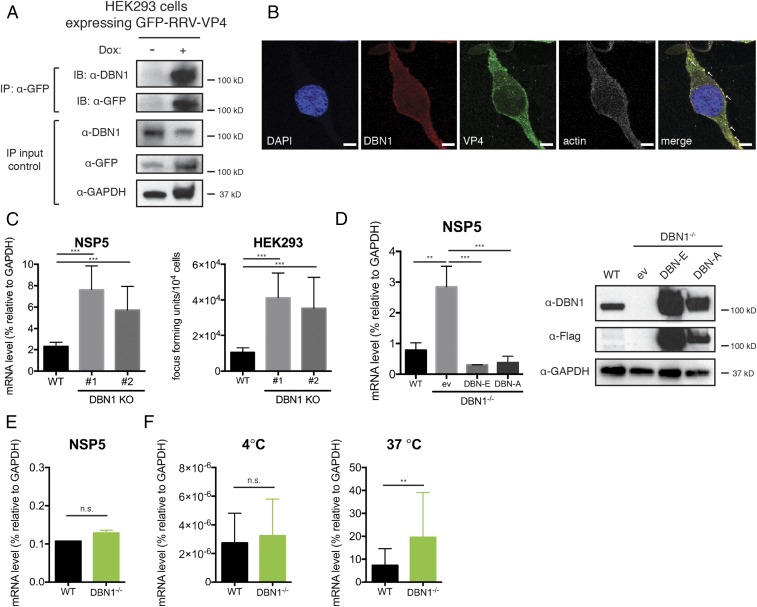
Fig. 2.
Drebrin deficiency results in enhanced RV infection. (A) Lysates of HEK293 cells stably expressing a GFP-tagged RRV VP4 were stimulated with or without doxycycline (Dox, 1 μg/mL) for 24 h, then subjected to IP using α-GFP antibody, and analyzed with Western blot using indicated antibodies. Bottom panels are 10% input. (B) HEK293 cells were infected with RRV at MOI = 3 for 24 h and analyzed by confocal microscopy for the localization of VP4 (green), DBN1 (red), actin (phalloidin, white), and nucleus (DAPI, blue). Colocalization (yellow) is highlighted by white arrowheads. Panels are single z slices. (Scale bar, 8 μm.) (C) WT and two individual clones of DBN1 knockout (KO) HEK293 cells were infected with RRV at MOI = 1 for 24 h. Viral gene NSP5 expression was measured by RT-qPCR and normalized to that of GAPDH (Left). Virus particles in the supernatants were titrated by an FFU assay (Right). (D) DBN1 KO HEK293 cells were transfected with pCMV6-entry empty vector (ev), and Flag-tagged full-length DBN-A or DBN-E, and infected with RRV at MOI = 1 for 24 h. Viral gene NSP5 level was measured by RT-qPCR and normalized to that of GAPDH (Left) and lysates were harvested for Western blot using indicated antibodies (Right). (E) WT and DBN1 KO HEK293 cells were transfected with RV double-layered particles (DLPs) for 16 h and NSP5 expression was measured by RT-qPCR. (F) WT and DBN1 KO HEK293 cells were infected with RRV (MOI = 50) and then incubated at either 4 °C for 1 h or incubated at 4 °C for 1 h and then shifted to 37 °C for an additional 1 h. The level of viral gene NSP5 was measured by RT-qPCR and normalized to that of GAPDH, as an indicator of input viral genomes. For all figures, experiments were repeated at least three times. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (**P ≤ 0.01; ***P ≤ 0.001; n.s., not significant).
To obtain a clean background to better study DBN1 function, we generated clonal DBN1−/− HEK293 cells using the CRISPR-Cas9 system. We isolated two individual colonies and confirmed complete deletion of DBN1 by Sanger sequencing and Western blot (SI Appendix, Fig. S5A). Importantly, consistent with our siRNA results, DBN1−/− cells exhibited increased intracellular viral RNA and production of RV progeny in the supernatants compared with wild-type (WT) HEK293 cells (Fig. 2C). Unlike effects of depletion of TMOD3 and ADD1, the effect of DBN1 deletion was strain independent (SI Appendix, Fig. S5B). Adding back two major isoforms of Flag-tagged DBN1 (DBN-A and DBN-E), but not the empty vector, completely rescued resistance to RV infection in the DBN1−/− cells (Fig. 2D), whereas DBN1 overexpression had minimal effect on RV infection (SI Appendix, Fig. S5 C–E). Collectively, these findings indicate that endogenous DBN1 coprecipitates and colocalizes with VP4 and functions to restrict RV infection in HEK293 cells. This restriction could not be recapitulated by actin polymerization inhibitors, suggestive of a novel function of DBN1 other than stabilization of actin filaments (SI Appendix, Fig. S6A).
To mechanistically determine how DBN1 antagonizes RV infection, we took advantage of RV double-layered particles (DLPs), which are transcriptionally active virus subparticles lacking the VP4 and VP7 surface proteins and cannot directly infect cells unless transfected into the cytoplasm to bypass entry (20). Transfected DLPs resulted in comparable levels of virus replication in WT and DBN1−/− cells (Fig. 2E), suggesting that the DBN1-mediated inhibition occurs at the virus entry step. In addition, time-course experiments revealed that levels of intracellular input viral RNA were already elevated at 1 h postinfection in DBN1−/− cells compared with WT cells (SI Appendix, Fig. S5D, Left), a further indication that RV entry is inhibited by DBN1.
RV entry relies on VP4 binding to surface receptors, triggering conformational changes that allow the virus particles to be internalized into the cells in a dynamin-dependent route (21). To examine whether the initial RV attachment to host cells is affected by the loss of DBN1, we performed a standard virus adsorption assay. At 4 °C, energy-dependent endocytosis is inhibited, and at this temperature similar amounts of RV particles were found to bind to cellular surfaces in WT and DBN1−/− cells (Fig. 2F, Left). Importantly, when cells preincubated with virus at 4 °C were shifted to 37 °C for 1 h, virus uptake into DBN1−/− cells was higher than into WT cells (Fig. 2F, Right), suggesting that DBN1-mediated RV inhibition occurs at a postattachment step. Using a conformation-specific antibody that only recognizes trimeric VP7 on incoming virus particles (22), we also observed that in contrast to the peripheral VP7 staining in WT cells during early infection, in DBN1−/− cells VP7 staining was diffuse throughout the cytoplasm (SI Appendix, Fig. S7), consistent with the enhanced rate of virus particle trafficking into the cytoplasm of DBN1-deficient cells. Remarkably, the replication of vesicular stomatitis virus (VSV), a completely unrelated RNA virus from the Rhabdoviridae family, was also restricted by the presence of DBN1 early in infection (SI Appendix, Fig. S6B). Altogether, these data indicate that endogenous DBN1 broadly restricts cell entry of VSV and multiple strains of RV.
DBN1 Negatively Regulates the Endocytosis of Dynamin-Dependent Cargo.
Our findings that DBN1 deficiency results in an increase in the entry of RV and VSV, both of which are known to depend on the host GTPase dynamin for endocytosis (21, 23) led us to ask whether DBN1 might play a more general role in regulating dynamin-mediated endocytosis. Consistent with this hypothesis, the uptake of fluorescently labeled transferrin, a well-studied ligand of classical clathrin-mediated endocytosis (24), was enhanced in the absence of DBN1: Whereas no transferrin was detected in WT cells, cytoplasmic transferrin signal was visible at 10 min and became more evident at 20 min postincubation in DBN1-deficient cells (Fig. 3A and SI Appendix, Fig. S8A). To complement the immunofluorescence staining experiments, we quantitatively measured transferrin uptake by flow cytometry and observed a concordantly weaker transferrin endocytosis in WT cells than in DBN1−/− counterparts 10 min posttransferrin addition (SI Appendix, Fig. S8B).
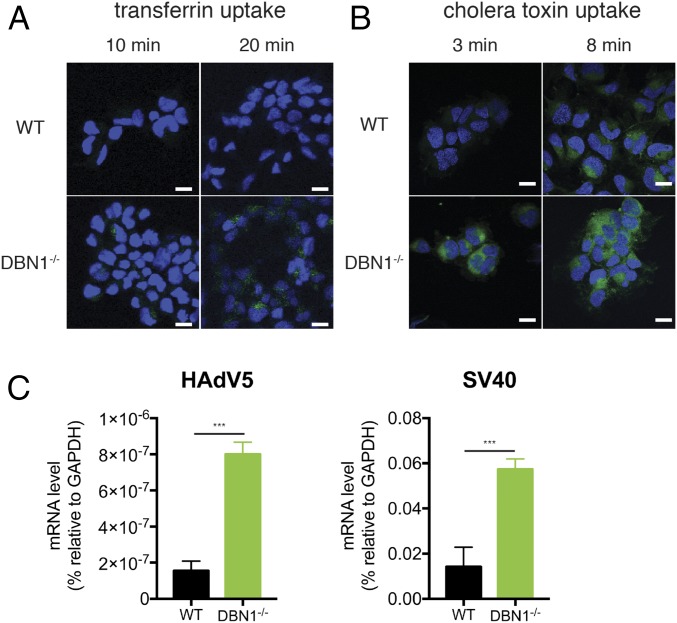
Fig. 3.
Loss of drebrin leads to enhanced endocytosis of dynamin-dependent cargo. (A) WT and DBN1 KO HEK293 cells were incubated with FITC-conjugated transferrin (Tfn, 20 μg/mL) and fixed at indicated time points for confocal microscopy to examine intracellular transferrin (green) and nucleus (DAPI, blue). (Scale bar in panels and single z slices, 20 μm.) (B) WT and DBN1 KO HEK293 cells were incubated with FITC-conjugated cholera toxin subunit B (CTxB, 10 μg/mL) and fixed at indicated time points for confocal microscopy for internalized toxin (green) and nucleus (DAPI, blue). (Scale bar in panels and single z slices, 20 μm.) (C) WT and DBN1 KO HEK293 cells were infected with human adenovirus serotype 5 (HAdV5) or simian vacuolating virus 40 (SV40) at MOI = 0.1 for 1 h. Expression levels of hexon (HAdV5) and large T antigen (SV40) were measured by RT-qPCR and normalized to that of GAPDH. For all figures, experiments were repeated at least three times. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (***P ≤ 0.001).
In addition to transferrin, we tested cholera toxin subunit B (CTxB), which mediates the internalization of the holotoxin in a caveolae-mediated, dynamin-dependent manner (24). CTxB uptake was also markedly enhanced at early time points in DBN1-deficient cells (Fig. 3B and SI Appendix, Fig. S8C). This result was further verified by single-cell analysis (SI Appendix, Fig. S8D). In contrast, dextran, a high molecular weight molecule internalized via dynamin-independent macropinocytosis (24), was detected at similar levels in WT and DBN1−/− cells at all time points (SI Appendix, Fig. S8 E–G). In summary, loss of DBN1 resulted in increased endocytosis of RV, VSV, transferrin, and CTxB, but not dextran, highlighting a specific and critical role of DBN1 in dynamin-controlled endocytic events that include both the clathrin- and caveolae-dependent pathways.
To further investigate how DBN1 influences viral endocytosis, we next examined uptake of various other DNA and RNA viruses in the presence and absence of DBN1. Higher amounts of human adenovirus serotype 5 (HAdV5) and simian vacuolating virus 40 (SV40), reliant on clathrin-dependent and caveolae-dependent endocytosis, respectively (5), were observed in DBN1−/− cells than in WT cells (Fig. 3C). In contrast, uptake of Vaccinia virus and Sendai virus, neither requiring dynamin (5), was similar in DBN1−/− and WT cells (SI Appendix, Fig. S8H). These data support our hypothesis that DBN1 specifically inhibits dynamin-dependent endocytic pathways. Interestingly, two important mosquito-borne flaviviruses, Zika virus and dengue virus, with undefined endocytic pathways, infected WT and DBN1−/− cells at the same efficiency (SI Appendix, Fig. S8H), suggesting that these two viruses likely enter cells independently of dynamin.
DBN1 Inhibits Dynamin-2 Function via Cortactin.
Next, to define the mechanism by which DBN1 blocks dynamin function, we preincubated WT and DBN1−/− cells with dynasore, an inhibitor that blocks the activity of all three isoforms of dynamin (25), before addition of RV. Consistent with the previous report (22), dynasore treatment only modestly inhibited (1.5-fold) infection in WT cells (Fig. 4A). By contrast, dynasore substantially reduced (15-fold) RV infectivity in the DBN1−/− cells (Fig. 4A). A similar pattern was observed with VSV infection (SI Appendix, Fig. S9A). These data, together with the spectrum of cargo regulated by DBN1, support the possibility that DBN1 acts to inhibit the function of dynamin, thereby limiting the potency of additional chemical dynamin inhibitors such as dynasore. We next examined the effect of Dyngo-4a, a more specific small-molecule inhibitor for dynamin-2 (DNM2) (26). RV infection was more significantly inhibited by Dygno-4a in DBN1−/− cells (18-fold) than in WT cells (1.3-fold), suggesting that DBN1 likely functions through inhibition of DNM2 (SI Appendix, Fig. S9B). To directly test this hypothesis, we knocked down the expression of each of the three dynamins in WT and DBN1−/− cells and evaluated RV infectivity in these cells. Importantly, only the siRNA targeting DNM2, and not those that reduced levels of DNM1 or DNM3 expression, resulted in reduction of RV infection, and the inhibition was more pronounced in DBN1−/− cells than in WT cells (SI Appendix, Fig. S9C).
Fig. 4.
Drebrin inhibits dynamin-2–mediated endocytosis via interaction with cortactin. (A) WT and DBN1 KO HEK293 cells were treated with either vehicle control (DMSO) or dynasore (100 μM) for 30 min before RRV infection (MOI = 1). Total RNA was harvested at 1 hpi and NSP5 level was measured by RT-qPCR and normalized to that of GAPDH. (B) WT HEK293 cells were transfected with indicated siRNA for 48 h before infection with RRV at MOI = 1 for 1 h. Viral NSP5 level was measured by RT-qPCR and normalized to that of GAPDH. Dotted line denotes NSP5 level of cells transfected with ctrl siRNA (set at 0.984); cortactin (encoded by CTTN) is outlined by a red box. (C) DBN1 KO HEK293 cells were transfected with indicated siRNA (gelsolin, encoded by GSN) for 48 h and infected with RRV at MOI = 1 for 1 h. The level of viral NSP5 was measured by RT-qPCR and normalized to that of GAPDH. (D) HEK293 cells were transfected with the first siRNA for 48 h and then transfected with second siRNA for an additional 48 h before RRV infection (MOI = 1) for 1 h. The input viral genome was measured by NSP5 levels using RT-qPCR and normalized to that of GAPDH. For all figures, experiments were repeated at least three times. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; n.s., not significant).
Because there are no reports of DBN1–DNM2 interaction and that DBN1 did not directly interfere with the GTPase activity of DNM2 (SI Appendix, Fig. S9D), we interrogated their possible connections by combining available proteomics data of known DBN1 binding proteins from publicly curated protein–protein interaction databases (27) with an siRNA screen. We knocked down the expression of all reported strong DBN1 interactors in HEK293 cells and then examined how individual siRNA depletion affects RV entry. Among these, siRNA-mediated silencing of genes encoding ARP3 actin-related protein 3 (ACTR3), spectrin beta nonerythrocytic 1 (SPTBN1), and cortactin (CTTN) resulted in a statistically significant decrease in RV infectivity (Fig. 4B). Intriguingly, CTTN has been reported to assist DNM2-mediated endocytosis by mediating an association between DNM2-tethered vesicles and ACTR3; this interaction promotes actin filament growth and pushes the cargo-containing vesicles further into the cytoplasm (28). Therefore, we hypothesized that CTTN might be involved in DBN1-mediated inhibition of DNM2 activity. Indeed, siRNA knockdown of CTTN impaired RV infection in DBN1−/− cells to an extent similar to chemical inhibition and siRNA-mediated silencing of DNM2 (Fig. 4C). Inhibition of expression of another DBN1 interacting protein gelsolin (GSN), previously shown to be antiviral through regulation of actin dynamics (29), had minimal effect either in WT or DBN1−/− cells (Fig. 4 B and C). Efficient knockdown of GSN and CTTN was validated by RT-qPCR analysis (SI Appendix, Fig. S9E). Further genetic dissection revealed that, unlike the enhancement of RV entry in WT cells by DBN1 siRNA, silencing of both DBN1 and CTTN did not enhance RV infection compared with transfection with CTTN siRNA alone. This finding suggests that these two proteins function in the same pathway. In addition, CTTN deficiency abrogated enhanced endocytosis in cells previously transfected with DBN1 siRNA (Fig. 4D), suggesting that CTTN likely acts upstream of DBN1 in the pathway.
DBN1 Colocalizes and Coprecipitates with Cortactin.
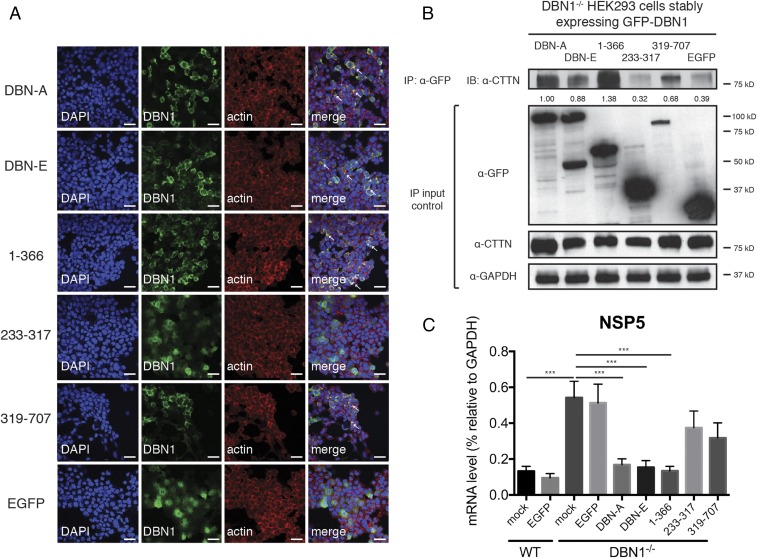
To further delineate the molecular mechanisms of DBN1-mediated inhibition of DNM2, we used a set of previously described DBN1 mutants (30) to identify the domain within DBN1 that is responsible for inhibiting DNM2-mediated endocytosis. Stable cell lines were constructed from DBN1−/− HEK293 cells reconstituted with two isoforms of full-length DBN1 or mutants that encode the N-terminal domain (amino acids 1–366), middle region (amino acids 233–317), or the C-terminal domain (amino acids 319–707). Interestingly, the N-terminal fragment had peripheral localization similar to that of the full-length DBN1 and colocalized with actin filaments (Fig. 5A). In contrast, the middle region of DBN1 was observed diffusely throughout the cytoplasm and the C-terminal mutant only partially colocalized with the cytoskeleton (Fig. 5A). Importantly, the full-length DBN1 and N-terminal domain colocalized with and coimmunoprecipitated with endogenous CTTN (Fig. 5B and SI Appendix, Fig. S10A) and were able to fully rescue DBN1 deficiency as shown by restricted RV infection in cells expressing these constructs (Fig. 5C). Despite comparable expression levels (SI Appendix, Fig. S10B), the C-terminal fragment and the middle region of DBN1 had an intermediate phenotype and did not inhibit RV entry as effectively as the other DBN1 mutants (Fig. 5C). Further mutagenesis analysis with DBN1 mutants (31) revealed that the most N-terminal ADF-H domain within DBN1 is responsible for restriction of viruses and other cargo (SI Appendix, Fig. S10C). Collectively, our data suggest that DBN1’s localization to the actin cytoskeleton and interaction with CTTN, mediated by its N-terminal region, are likely necessary for its ability to block dynamin function.
Fig. 5.
N-terminal domain of drebrin localizes to the actin cytoskeleton and coprecipitates with cortactin. (A) DBN1 KO HEK293 cells stably expressing indicated GFP-tagged DBN1 constructs or control EGFP were analyzed by confocal microscopy for the localization of DBN1 (green), actin (red), and nucleus (DAPI, blue). Colocalization (yellow) is highlighted by white arrowheads. (Full-length: DBN isoforms A and E; N terminus: amino acid 1–366; middle region: 233–317; C terminus: 319–707). (Scale bar in panels and single z slices, 40 μm.) (B) DBN1 KO HEK293 cells stably expressing indicated GFP-tagged DBN1 constructs or control EGFP were subject to IP using α-GFP antibody and analyzed by Western blot using indicated antibodies. The IP band intensities were normalized to endogenous CTTN levels in IP input and compared with that of DBN-A (lane 1), which was set as 1.00. Bottom panels are 10% input. (C) Reconstituted DBN1 KO HEK293 cells were infected with RRV at MOI = 1 for 24 h and examined by RT-qPCR for viral NSP5 expression, normalized to that of GAPDH. For all figures, experiments were repeated at least three times. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (***P ≤ 0.001).
Loss of DBN1 Enhances RV Infection in Vivo and in Human Intestinal Enteroids.
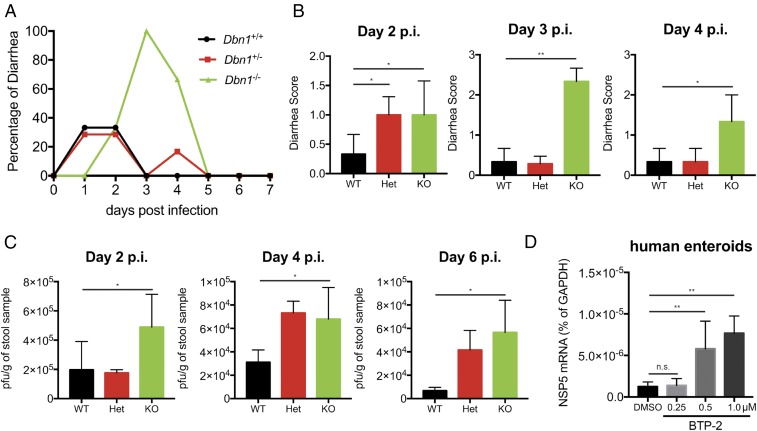
We next extended our analysis of DBN1 regulation of RV infection from in vitro cell culture systems to a suckling mouse model of RV infection. Five-day-old pups born from Dbn1 heterozygous (Het) breeding pairs were orally inoculated with 106 plaque-forming units (pfus) of the simian RRV strain. We monitored both diarrhea occurrence and fecal shedding of viral antigens. Notably, although the overall gut homeostasis and permeability was not altered by the loss of Dbn1 (SI Appendix, Fig. S11), Dbn1 knockout (KO) pups exhibited increased diarrhea compared with both WT and Het animals (Fig. 6 A and B). In addition, we detected significantly more infectious virus particles in the stool samples harvested from Dbn1−/− mice than their WT littermates on days 2, 4, and 6 postinfection (Fig. 6C), further supporting an important role of DBN1 in restricting RV infection in vivo.
Fig. 6.
Drebrin knockout results in increased RV infection in vivo and in human enteroids. (A) Five-day-old C57BL/6 pups of indicated Dbn1 genotypes (WT, n = 3; Het, n = 7; KO, n = 3) were orally inoculated with 106 pfu of the simian RV RRV strain and monitored for the incidence of diarrhea for 7 d. (B) Quantification of A for the indicated days postinfection based on diarrheal severity parameters. (C) Fecal specimens were collected on the indicated days postinfection and subject to a standard plaque forming unit (pfu) assay to determine infectious virus particles per gram of stool samples. (D) Human intestinal organoids were treated with either vehicle control (DMSO) or BTP-2 at indicated concentrations for 30 min before infection with the human RV Wa strain (MOI = 1). Total RNA was harvested at 1 hpi and NSP5 level was measured by RT-qPCR and normalized to that of GAPDH. For D, experiments were repeated at least three times. Data are represented as mean ± SEM. Statistical significance is determined by Student’s t test (*P ≤ 0.05; **P ≤ 0.01; n.s., not significant).
To determine the physiological relevance of DBN1 in the human small intestines, we tested the effect of DBN1 inhibition in a primary 3D human intestinal enteroid system, also known as the “mini-gut” that recapitulates many important features of normal intestinal epithelium and supports robust human RV infection (32). Because it is technically challenging to directly deplete DBN1 expression in the enteroids, we used BTP-2, a well-characterized DBN1 inhibitor (33), as an alternative approach. BTP-2 treatment of HEK293 cells mimicked CRISPR-mediated DBN1 depletion: RV entry was increased by BTP-2 in a dose-dependent manner at concentrations that did not induce obvious cytotoxicity or loss of tight junctions (SI Appendix, Fig. S12 A–C). Importantly, inhibition of DBN1 by BTP-2 also significantly promoted RV infection in human enteroids (Fig. 6D), suggesting that DBN1 broadly functions to restrict RV entry in a cell type-independent fashion in vitro as well as in vivo.
Discussion
Despite the widespread availability of several safe and effective RV vaccines, RVs remain the leading cause for severe diarrheal diseases in infants and young children. Many fundamental aspects of RV pathogenesis and virus–host interaction are inadequately understood, thus hindering the development of improved vaccines and effective antiviral therapeutics. In particular, RV entry into target host cells, primarily IECs in the small intestine, is a well-coordinated yet complex event that warrants further investigation. We report here a comprehensive set of studies to identify and examine some of the host protein factors that interact with VP4, the viral outmost capsid protein that mediates RV attachment and internalization into target cells. Of note, we identified several cytoskeleton-associated host proteins that either inhibit or facilitate RV infection. One of these VP4 interacting proteins, drebrin (encoded by DBN1), broadly functions to dampen dynamin-dependent endocytosis, including the early entry steps of several viruses including RV, VSV, HAdV5, and SV40. Our current study, while providing specific mechanistic insights into how DBN1 serves as a general gatekeeper for dynamin-mediated endocytic pathways, exemplifies how the study of viral protein/host protein interactions can facilitate the general interrogation of host protein function and signal transduction.
Viruses, as obligate intracellular pathogens, have to gain access through the plasma membrane to use intracellular resources for efficient replication and progeny production. During virus infection, the host cell cytoskeleton constitutes an early line of defense by posing a physical barrier to exogenous agents, before the induction of innate immune responses. Virtually all viruses have to devise ways to overcome the interconnected meshwork to facilitate entry and/or egress. In the present study, using an unbiased tandem IP–MS approach (Fig. 1A), we identified the actin-binding protein DBN1 as a restriction factor for RV infection (Fig. 1D). DBN1 coprecipitated with RV VP4 and colocalized with actin filaments and VP4 during both ectopic expression and virus infection (Fig. 2 A and B and SI Appendix, Fig. S4). The presence of DBN1 restricted RV replication in a strain-independent manner by specifically inhibiting virus entry into target host cells (SI Appendix, Fig. S5). Importantly, DBN1 expression also served to inhibit the entry of several other DNA and RNA viruses that rely on dynamin for cell entry, including VSV, HAdV5, and SV40 (SI Appendix, Fig. S6B and Fig. 3C). Recently, Zika virus (ZIKV) has emerged as a global public health threat and has been clearly identified as an important cause of microcephaly and rarely, Guillain-Barre syndrome (34). The cell entry pathway of ZIKV has yet to be defined. Our data suggest that, in contrast to the previously mentioned viruses, ZIKV and dengue virus cell entry is not regulated by DBN1 (SI Appendix, Fig. S8H) and likely occurs by a dynamin-independent endocytosis mechanism. In this aspect, DBN1−/− cells may be a useful tool to study viruses and other cargo with unknown entry pathways.
Using an siRNA screen of DBN1-interacting proteins, we found that DBN1 regulates dynamin-2 (DNM2)-dependent endocytosis via an association with cortactin (CTTN) (Fig. 4B). The C-terminal SH3 domain of CTTN binds to the proline-rich domain of dynamin-2, promoting actin filament polymerization and vesicle fission (35). Knocking down CTTN in DBN1−/− cells paralleled the effect of Dygno-4a, a small-molecule DNM2 inhibitor, and significantly abrogated the enhanced RV entry in these knockout cells (Fig. 4C and SI Appendix, Fig. S9B). Immunofluorescence and IP analysis pinpointed the N terminus of DBN1 as responsible for its colocalization and interaction with CTTN at the plasma membrane (Fig. 5 and SI Appendix, Fig. S10A). Based on these findings, we hypothesize that, at steady state in WT cells, DBN1 blocks DNM2 function by interfering with CTTN–ACTR3 interaction, disconnecting CTTN from the actin bundle and preventing vesicle-associated filament growth. However, in DBN1-deficient cells, CTTN–DNM2 complex is able to directly establish the connection between DNM2-bound vesicles with Arp2/3 complex, thereby facilitating the internalization of DNM2-dependent cargo.
Given the broad spectrum of cargos regulated by DNM2-dependent endocytic pathways, we expect that DBN1−/− cells will also be altered in many other aspects of cell biology and immune responses beyond the specific circumstance of RV infection. Because the endocytosis of a large number of cytokines, growth factors, and hormones is dependent on DNM2, it will be intriguing to examine whether DBN1 deficiency affects the uptake of these molecules as well. It is worth noting that interleukin-2 (IL-2) enters cells via a clathrin- and caveolae-independent but dynamin-mediated pathway (36) and it plays a crucial role in regulating T-cell homeostasis. Whether T cells in Dbn1−/− mice are hyperresponsive to IL-2 stimulation remains to be tested. In addition, Wnt signaling, critical for maintaining the stem cell niche in the crypts of small intestines, is dependent on caveolae and dynamin (37). One might predict, based on our findings, that chemical inhibition of DBN1 by the small-molecule inhibitor BTP-2 in human enteroid culture will significantly reshape the dynamics of IEC subpopulations and affect how these cells respond to viral or bacterial challenge. Finally, it is tempting to speculate whether short-term suppression of DBN1 by genetic depletion or chemical inhibition, might serve to boost the DNM2-dependent cell entry pathway in a manner that might augment drug delivery or immunization. For example, one of the most commonly used viral vectors for gene therapy, adeno-associated virus (AAV), enters via both dynamin-dependent and -independent routes (38, 39). Specific and transient DBN1 inhibition could enhance AAV uptake into target tissues and increase immunogenicity.
In summary, we initiated a systems-view analysis to identify the host proteins that interact with RV VP4, the viral receptor. As an initial result of this screen, we discovered and then investigated a function of DBN1: the ability to restrict the entry of multiple viral pathogens including RV into host cells. Our results suggest DBN1, via colocalization and interaction with CTTN at the actin filaments, specifically inhibits multiple examples of DNM2-mediated endocytosis. These findings further highlight the important role of cytoskeleton proteins in virus infection and shed light on the delicate regulatory mechanisms by which DNM-dependent endocytic events are controlled. The current study also provides a mechanistic basis for the potential design of DBN1 agonists and antagonists for therapeutic purposes to enhance or inhibit cell entry, including entry of multiple viruses/viral vectors.
Materials and Methods
Additional procedures are described in detail in SI Appendix, SI Materials and Methods.
Cells and Viruses.
Human embryonic kidney fibroblast HEK293 cells (CRL-1573) were obtained from American Type Culture Collection (ATCC) and cultured in DMEM supplemented with 10% FBS, 2 mM l-glutamine, 100 IU/mL of penicillin, and 100 μg/mL of streptomycin. African Green Monkey kidney MA104 cells (CRL-2378.1) were obtained from ATCC and cultured in complete M199 medium. HEK293 cells stably expressing VP4 were cultured in complete DMEM in the presence of puromycin (0.5 μg/mL). Expression of VP4 was induced by doxycycline (1 μg/mL) treatment for 24 h. HEK293 cells overexpressing Flag-tagged TMOD3, overexpressing Flag-tagged DBN1, and reconstituted with DBN1 constructs stable cell lines were screened and cultured in complete DMEM in the presence of G418 (0.5 mg/mL).
Human RV Wa and DS1 strain, simian RV RRV and SA11 strain, bovine RV United Kingdom strain, porcine RV SB1A, OSU strain, and murine RV ETD strains were propagated in MA104 cells as previously described (40). Viruses were activated by trypsin (5 μg/mL) at 37 °C for 20 min before infection. Cells were washed with serum-free medium (SFM) twice and incubated with virus at different multiplicity of infections (MOIs) at 37 °C for 1 h. After removal of virus inoculum, complete medium or SFM was added back to cells. Lysates and cell supernatants were harvested at different time points for virus quantification. RV TLPs, DLPs, and VLPs were prepared as previously described (20).
Recombinant VSV (strain Indiana) expressing GFP was previously characterized (41) and was propagated and titrated in BHK-21 cells. ZIKV (strain P6-740) was provided by Robert Tesh, University of Texas Medical Branch, Galveston, TX and propagated in C6/36 cells. DENV-1 (strain 276RKI) was obtained through NIH Biodefense and Emerging Infections Research Resources Repository, National Institute of Allergy and Infectious Diseases, NIH, and propagated in C6/36 cells. Vaccinia virus (strain MVA) and SV40 (strain EK) were purchased from ATCC. Human adenovirus serotype 5 and Sendai virus (strain Cantell) were kindly provided by Xin Wang, Cleveland Clinic, Cleveland, OH.
Virus Infections.
For assays of the virus entry, WT and DBN1−/− HEK293 cells were incubated with RV at MOI = 10 at 37 °C for 1 h. After removal of virus inoculum, cells were washed with PBS twice and harvested for virus quantification. Strand-specific RT-PCR was performed to ensure that viral RNA (mRNA level of NSP5) detected by qPCR was derived from input viral genomes and not newly transcribed RNA. For assays of the virus binding, cells were washed with DMEM two times and infected with RRV (MOI = 50) at 4 °C for 1 h. Cells were washed twice with DMEM and harvested for detection of viral RNA. For VSV, HAdV5, SV40, VV, SeV infection, WT, and DBN1−/− HEK293 cells were infected at MOI of 0.1 for 1 h. For ZIKV and DENV-1 infection, WT and DBN1−/− HEK293 cells were infected at MOI of 0.01 for 3 h. Cell lysates were directly harvested using RLT lysis buffer and the copy number of viral genome present in the cells was quantified by RT-qPCR.
Mice Infection.
Six-week-old sex-matched DBN1 heterozygous mice on a C57BL/6NJ background (C57BL/6NJ-Dbn1em1J/J, 027202) were purchased from The Jackson Laboratory. Mice were specific pathogen free, maintained under a strict 12 h light cycle (lights on at 7:00 AM and off at 7:00 PM), and given a regular chow diet (Harlan, diet 2018) ad libitum. All mice were maintained in Veterinary Medical Unit of VA Palo Alto Health Care System (VAPAHCS). The Institutional Animal Care Committee at the VAPAHCS approved these studies.
To obtain DBN1 KO mice, we crossed DBN1 heterozygous male mice with DBN1 heterozygous female mice. The offspring were born at a Mendelian ratio: three WT, seven heterozygous, and three KO. Five-day-old suckling mice were orally inoculated with 106 pfu of the simian RRV strain. From 1 to 7 dpi, littermates were examined daily for the occurrence of diarrheal disease. The percentage and severity of diarrhea among the littermates during the course of infection was recorded as previously described (42). In brief, diarrhea was scored based on color, consistency, and amount, and numbered as follows: 0 = normal; 1 = pasty; 2 = semiliquid; 3 = liquid, and score ≥2 considered as diarrhea. Fecal specimens were collected into preweighed Eppendorf tubes and stored at −80 °C before measurement by a standard plaque assay. At 7 dpi, mouse tails and intestinal tissues were harvested for genotyping by PCR and Western blot, respectively. The primers used for genotyping are listed in SI Appendix, Table S1. Small intestinal tissues were collected from WT, heterozygous, and knockout mice at 5 d postbirth and processed for hematoxylin and eosin staining at Histo-Tec Laboratory.
Lipofection of DLPs.
Infection of WT and DBN1−/− HEK293 cells with RRV DLPs using lipofectin-mediated transfection was carried out as described (20). Briefly, DLPs were diluted in DMEM and incubated with a mixture of Lipofectamine (Life Technologies) in DMEM (15% vol/vol) for 40 min at room temperature. A total of 50 μL of this mixture was added to the cells for 4 h at 37 °C and 5% CO2, and then cells were washed with DMEM and harvested at different time points for measuring viral RNA.
RNA Isolation and Real-Time Quantitative PCR.
Total RNA was extracted from the cell lysates using RNeasy kit (Qiagen) as previously described (43). Virus cDNA was generated by reverse transcription using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qPCR was performed using the Stratagene Mx3005P (Agilent) with each reaction composed of cDNA reverse transcribed from 50 ng of total RNA, 12.5 µL of Power SYBR Green Master Mix (Applied Biosystems), and 200 nM both forward and reverse primers in a total volume of 25 µL (44). SYBR Green primers used in this paper are listed in SI Appendix, Table S1.
Focus Forming Unit Assay.
WT and DBN1−/− HEK293 cells were infected with RRV at MOI of 0.1 in 24-well plates. At different time points after infection, supernatants were collected and added to MA104 cells in a 10× dilution series. After 1 d, cells were fixed, incubated with primary rabbit antibodies against DLP at 37 °C for 60 min, secondary incubation was performed with anti-rabbit IgG at 37 °C for 60 min, AEC substrate was added (SK-4200, Vector Laboratories) and observed for color developing, and fluorescent colonies were counted. Two independent experiments were performed with triplicate infections and one representative experiment is shown.
Human Intestinal Enteroids.
Primary human intestinal enteroids were kindly provided by Calvin Kuo, Stanford University, Stanford, CA. The methods for enteroid culture and RV infection were similar to previous publication (32). In brief, enteroids were treated with TrypLE (Gibco) into single cell suspension, stimulated with BTP-2 (0.25, 0.5, or 1.0 μM) for 30 min, and infected with Wa (MOI = 10) for 1 h. RNA was harvested from infected enteroids for measuring RV NSP5 levels as an indicator of input viral genomes.
Mass Spectrometry and Network Analysis.
Tandem-affinity purification and mass spectrometry were carried out as previously described (15) at the Stanford MS core facility. For each LC/MS experiment, spectral counts were transformed into normalized spectral abundance factors. Using a panel of 66 other experiments also conducted in HEK293 cells, lognormal probability distributions were inferred for the observation of RV VP4. For each observed gene product, a Z test was conducted against that background distribution. The Benjamini–Hochberg method was used to correct P values so obtained, and gene products were accepted below a false discovery rate of 0.12. Interactions between gene products identified in this way were obtained by cross-reference with BioGrid (27). Networks were visualized and figures rendered with Cytoscape (45).
Statistical Analysis.
The results were shown as means ± SEM. Statistical significance was determined by Student’s t test using Prism 7 (GraphPad Software). Significant differences are indicated in the figures (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
Supplementary Material
Acknowledgments
We thank all members of the H.B.G. laboratory for their support; Dr. Jan E. Carette (Stanford University), Dr. Xin Wang (Cleveland Clinic), and Dr. Stefan Linder (University of Hamburg) for kindly providing valuable reagents and useful suggestions; and Drs. Chris Adams and Ryan Leib (Stanford Mass Spectrometry core facility) for their assistance. This work is supported by NIH Grants R01 AI021362, R56 AI021362, U19 AI116484, and by US Department of Veterans Affairs Merit Review Grant GRH0022 (to H.B.G.). S.D. is supported by the Walter V. and Idun Berry Postdoctoral Fellowship Program; the Stanford Institute for Immunity, Transplantation, and Infection Young Investigator Award; and an Early Career Award from the Thrasher Research Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619266114/-/DCSupplemental.
References
- 1.Marsh M, editor. 2001. Endocytosis, Frontiers in Molecular Biology (Oxford University Press, Oxford)
- 2.Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 4.Cossart P, Helenius A. Endocytosis of viruses and bacteria. Cold Spring Harb Perspect Biol. 2014;6:6. doi: 10.1101/cshperspect.a016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercer J, Schelhaas M, Helenius A. Virus entry by endocytosis. Annu Rev Biochem. 2010;79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 6.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD. World Health Organization–Coordinated Global Rotavirus Surveillance Network Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin Infect Dis. 2016;62:S96–S105. doi: 10.1093/cid/civ1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estes MK, Greenberg HB. 2013. Rotaviruses. Lippincott Williams & Wilkins, Philadelphia.
- 8.Méndez E, López S, Cuadras MA, Romero P, Arias CF. Entry of rotaviruses is a multistep process. Virology. 1999;263:450–459. doi: 10.1006/viro.1999.9976. [DOI] [PubMed] [Google Scholar]
- 9.Ludert JE, et al. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J Virol. 1996;70:487–493. doi: 10.1128/jvi.70.1.487-493.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green VA, Pelkmans L. A systems survey of progressive host-cell reorganization during rotavirus infection. Cell Host Microbe. 2016;20:107–120. doi: 10.1016/j.chom.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Silva-Ayala D, et al. Genome-wide RNAi screen reveals a role for the ESCRT complex in rotavirus cell entry. Proc Natl Acad Sci USA. 2013;110:10270–10275. doi: 10.1073/pnas.1304932110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cevallos Porta D, López S, Arias CF, Isa P. Polarized rotavirus entry and release from differentiated small intestinal cells. Virology. 2016;499:65–71. doi: 10.1016/j.virol.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa R, et al. Drebrin, a development-associated brain protein from rat embryo, causes the dissociation of tropomyosin from actin filaments. J Biol Chem. 1994;269:29928–29933. [PubMed] [Google Scholar]
- 14.Torres JZ, Miller JJ, Jackson PK. High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics. 2009;9:2888–2891. doi: 10.1002/pmic.200800873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding S, et al. Comparative proteomics reveals strain-specific β-TrCP degradation via rotavirus NSP1 hijacking a host Cullin-3-Rbx1 complex. PLoS Pathog. 2016;12:e1005929. doi: 10.1371/journal.ppat.1005929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parr RD, et al. The rotavirus enterotoxin NSP4 directly interacts with the caveolar structural protein caveolin-1. J Virol. 2006;80:2842–2854. doi: 10.1128/JVI.80.6.2842-2854.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kojima N, Shirao T. Synaptic dysfunction and disruption of postsynaptic drebrin-actin complex: A study of neurological disorders accompanied by cognitive deficits. Neurosci Res. 2007;58:1–5. doi: 10.1016/j.neures.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Xu SQ, et al. A novel role for drebrin in regulating progranulin bioactivity in bladder cancer. Oncotarget. 2015;6:10825–10839. doi: 10.18632/oncotarget.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keon BH, Jedrzejewski PT, Paul DL, Goodenough DA. Isoform specific expression of the neuronal F-actin binding protein, drebrin, in specialized cells of stomach and kidney epithelia. J Cell Sci. 2000;113:325–336. doi: 10.1242/jcs.113.2.325. [DOI] [PubMed] [Google Scholar]
- 20.Bass DM, et al. Liposome-mediated transfection of intact viral particles reveals that plasma membrane penetration determines permissivity of tissue culture cells to rotavirus. J Clin Invest. 1992;90:2313–2320. doi: 10.1172/JCI116119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Díaz-Salinas MA, et al. The spike protein VP4 defines the endocytic pathway used by rotavirus to enter MA104 cells. J Virol. 2013;87:1658–1663. doi: 10.1128/JVI.02086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf M, Vo PT, Greenberg HB. Rhesus rotavirus entry into a polarized epithelium is endocytosis dependent and involves sequential VP4 conformational changes. J Virol. 2011;85:2492–2503. doi: 10.1128/JVI.02082-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johannsdottir HK, Mancini R, Kartenbeck J, Amato L, Helenius A. Host cell factors and functions involved in vesicular stomatitis virus entry. J Virol. 2009;83:440–453. doi: 10.1128/JVI.01864-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Roy C, Wrana JL. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol. 2005;6:112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- 25.Macia E, et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Harper CB, et al. Dynamin inhibition blocks botulinum neurotoxin type A endocytosis in neurons and delays botulism. J Biol Chem. 2011;286:35966–35976. doi: 10.1074/jbc.M111.283879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stark C, et al. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schafer DA, et al. Dynamin2 and cortactin regulate actin assembly and filament organization. Curr Biol. 2002;12:1852–1857. doi: 10.1016/s0960-9822(02)01228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irving AT, et al. Regulation of actin dynamics by protein kinase R control of gelsolin enforces basal innate immune defense. Immunity. 2012;36:795–806. doi: 10.1016/j.immuni.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi K, et al. Domain analysis of the actin-binding and actin-remodeling activities of drebrin. Exp Cell Res. 1999;253:673–680. doi: 10.1006/excr.1999.4663. [DOI] [PubMed] [Google Scholar]
- 31.Rehm K, Panzer L, van Vliet V, Genot E, Linder S. Drebrin preserves endothelial integrity by stabilizing nectin at adherens junctions. J Cell Sci. 2013;126:3756–3769. doi: 10.1242/jcs.129437. [DOI] [PubMed] [Google Scholar]
- 32.Finkbeiner SR, et al. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio. 2012;3:e00159-12. doi: 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mancini A, et al. Regulation of myotube formation by the actin-binding factor drebrin. Skelet Muscle. 2011;1:36. doi: 10.1186/2044-5040-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petersen LR, Jamieson DJ, Powers AM, Honein MA. Zika virus. N Engl J Med. 2016;374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 35.Kessels MM, Qualmann B. Extending the court for cortactin: From the cortex to the Golgi. Nat Cell Biol. 2005;7:448–449. doi: 10.1038/ncb0505-448. [DOI] [PubMed] [Google Scholar]
- 36.Lamaze C, et al. Interleukin 2 receptors and detergent-resistant membrane domains define a clathrin-independent endocytic pathway. Mol Cell. 2001;7:661–671. doi: 10.1016/s1097-2765(01)00212-x. [DOI] [PubMed] [Google Scholar]
- 37.Blitzer JT, Nusse R. A critical role for endocytosis in Wnt signaling. BMC Cell Biol. 2006;7:28. doi: 10.1186/1471-2121-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan D, et al. Dynamin is required for recombinant adeno-associated virus type 2 infection. J Virol. 1999;73:10371–10376. doi: 10.1128/jvi.73.12.10371-10376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nonnenmacher M, Weber T. Adeno-associated virus 2 infection requires endocytosis through the CLIC/GEEC pathway. Cell Host Microbe. 2011;10:563–576. doi: 10.1016/j.chom.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoshino Y, Wyatt RG, Greenberg HB, Flores J, Kapikian AZ. Serotypic similarity and diversity of rotaviruses of mammalian and avian origin as studied by plaque-reduction neutralization. J Infect Dis. 1984;149:694–702. doi: 10.1093/infdis/149.5.694. [DOI] [PubMed] [Google Scholar]
- 41.Dalton KP, Rose JK. Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology. 2001;279:414–421. doi: 10.1006/viro.2000.0736. [DOI] [PubMed] [Google Scholar]
- 42.Ball JM, Tian P, Zeng CQ, Morris AP, Estes MK. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272:101–104. doi: 10.1126/science.272.5258.101. [DOI] [PubMed] [Google Scholar]
- 43.Ding S, Khoury-Hanold W, Iwasaki A, Robek MD. Epigenetic reprogramming of the type III interferon response potentiates antiviral activity and suppresses tumor growth. PLoS Biol. 2014;12:e1001758. doi: 10.1371/journal.pbio.1001758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolen CR, Ding S, Robek MD, Kleinstein SH. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 2014;59:1262–1272. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.