Significance
Plant development depends on the Polycomb Group (PcG) pathway, which ensures an epigenetic memory of gene repression. PcG proteins form complexes that decorate histones of target genes with signature modifications. Decorated histones are diluted during DNA replication, therefore compensatory mechanisms must ensure their renewal during cell division. We show that a protein related to a yeast replication factor plays an important role in maintaining a PcG-associated chromatin modification in dividing plant cells.
Keywords: Ctf4, epigenetic inheritance, ENHANCER OF LHP1 1, Polycomb repressive complex 2, replication
Abstract
Polycomb Repressive Complex (PRC) 2 catalyzes the H3K27me3 modification that warrants inheritance of a repressive chromatin structure during cell division, thereby assuring stable target gene repression in differentiated cells. It is still under investigation how H3K27me3 is passed on from maternal to filial strands during DNA replication; however, cell division can reinforce H3K27me3 coverage at target regions. To identify novel factors involved in the Polycomb pathway in plants, we performed a forward genetic screen for enhancers of the like heterochromatin protein 1 (lhp1) mutant, which shows relatively mild phenotypic alterations compared with other plant PRC mutants. We mapped enhancer of lhp1 (eol) 1 to a gene related to yeast Chromosome transmission fidelity 4 (Ctf4) based on phylogenetic analysis, structural similarities, physical interaction with the CMG helicase component SLD5, and an expression pattern confined to actively dividing cells. A combination of eol1 with the curly leaf (clf) allele, carrying a mutation in the catalytic core of PRC2, strongly enhanced the clf phenotype; furthermore, H3K27me3 coverage at target genes was strongly reduced in eol1 clf double mutants compared with clf single mutants. EOL1 physically interacted with CLF, its partially redundant paralog SWINGER (SWN), and LHP1. We propose that EOL1 interacts with LHP1–PRC2 complexes during replication and thereby participates in maintaining the H3K27me3 mark at target genes.
Polycomb group (PcG) proteins function as transcriptional repressors by establishing a compacted chromatin status that is less accessible to transcription factors and RNA polymerase II complexes (1). Two chromatin modifications are hallmarks of PcG actions: Histone H2A lysine monoubiquitination and histone H3 lysine 27 trimethylation (H3K27me3) deposited respectively by Polycomb Repressive Complexes (PRC) 1 and 2 (2, 3).
In Arabidopsis thaliana, small multigene families encode three of the four components of the evolutionary conserved PRC2 (2, 4). MSI1 is part of a multigene family, but the only one member copurifying with PRC2 (5, 6). In contrast, three genes encoding relatives of the Drosophila melanogaster Su(Z)12 are reported to define distinct PRC2 complexes showing functional specialization, e.g., in gametophyte development, delay of reproductive development, and response to vernalization in the sporophyte (2, 4). The SET domain containing catalytic core of PRC2 is encoded by three genes, which are the partially redundant CLF and SWN acting mainly in the sporophyte and MEDEA (MEA), which is functional in gametophyte development (2, 4). Several proteins associate with PRC2 to modulate its function, such as VEL/VIL PHD-finger proteins (7), BLISTER (BLI) (8), LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) (5, 6), EMBRYONIC FLOWER (EMF) 1 (5), and ANTAGONIST OF LHP1 (ALP) 1 (5). PRC1 is less defined than PRC2 and likely exists in several forms (2, 4). In A. thaliana, PRC1 variants contain seven proteins, including LHP1, EMF1, and five RING-domain proteins (AtRING1A/B and AtBMI1A/B/C) (2, 9). RING domain proteins are responsible for H2A ubiquitination and interact with LHP1, whose chromodomain binds to H3K27me3 (10, 11).
Mutation in the single-copy gene LHP1 causes relatively mild phenotypes compared with strong PcG mutants (11–16), suggesting that the role of LHP1 as interface between PRC1 and PRC2 is backed up by redundant mechanisms. For example, the maize VIVIPAROUS 1 and A. thaliana ABA INSENSITIVE 3-like (VAL) subgroup of B3-domain transcription factors recruit AtBMI1 proteins to target genes regulating seed maturation (10, 11). Other transcription factors and cis-elements such as GA(A)GA(A) repeats and telobox cis elements have been linked to PRC2 recruitment (17, 18). An interaction between RING1A and CLF explains PRC2 recruitment by PRC1 components (19). Finally, PRC2 has affinity to its own target modification through a binding pocket in its ESC homologs (20). Binding of H3K27me3 causes allosteric changes that increase the catalytic activity of PRC2 to modify H3 tails of an adjacent nucleosome (21).
Requirements for PRC2 catalytic activity and maintenance of H3K27me3 are different in dividing and resting cells, which form the majority of plant tissues (22, 23). Based on animal and yeast models, nucleosomes are fully removed from the maternal strand before passage of the replication fork and, therefore, need to be reassembled and complemented on both daughter strands, first for H3-H4 and then for H2A-H2B dimers (22, 23). In animals, PRC2 associates with the replication fork, thereby facilitating stable inheritance of the H3K27me3 mark (24). Communication between mother and daughter strands, and between daughter strands in close vicinity of the replication fork, maintains epigenetic information (22).
Three DNA-polymerases (DNA-pol) participate in leading and lagging DNA strand synthesis, and all three have been connected to PcG-mediated gene repression in A. thaliana. Mutations in INCURVATA (ICU) 2 and EARLY IN SHORT DAYS (ESD) 7, respectively encoding homologs of the priming DNA-polα and the leading strand-specific DNA-polε, were shown to enhance both lhp1 and clf mutant phenotypes (25–27). Similar genetic interaction was suggested by the observation that a temperature-conditional DNA-polδ mutant, gigantea suppressor (gis) 5, expresses increased levels of SEPPALATA (SEP) 3 and FLOWERING LOCUS T (FT), which are also up-regulated in lhp1 and clf mutants (28). Physical interaction between LHP1 and ESD7 or ICU2 was shown by in vitro pull-down (25, 26), but the latter could not be confirmed in a later study (27). Nevertheless, the genetic data suggest that there exists a tight link between DNA-pols and an LHP1-associated PRC2.
Here, we identified a mutation in a previously uncharacterized gene as causal for an enhancer of lhp1 (eol) phenotype. We provide evidence that the gene encodes the A. thaliana homolog of yeast Chromosome transmission fidelity 4 (Ctf4), which acts in a trimeric complex to couple DNA-Polα and the DNA helicase complex during DNA replication (29). We show that the function of EOL1 within the PcG pathway depends on H3K27me3 activity and contributes to H3K27me3 inheritance during cell division. EOL1 interacts physically with PcG components CLF, SWN, and LHP1. We propose that EOL1 recruits LHP1-PRC2 to ensure faithful inheritance of the H3K27me3 modification during replication.
Results
Mapping an eol Mutation.
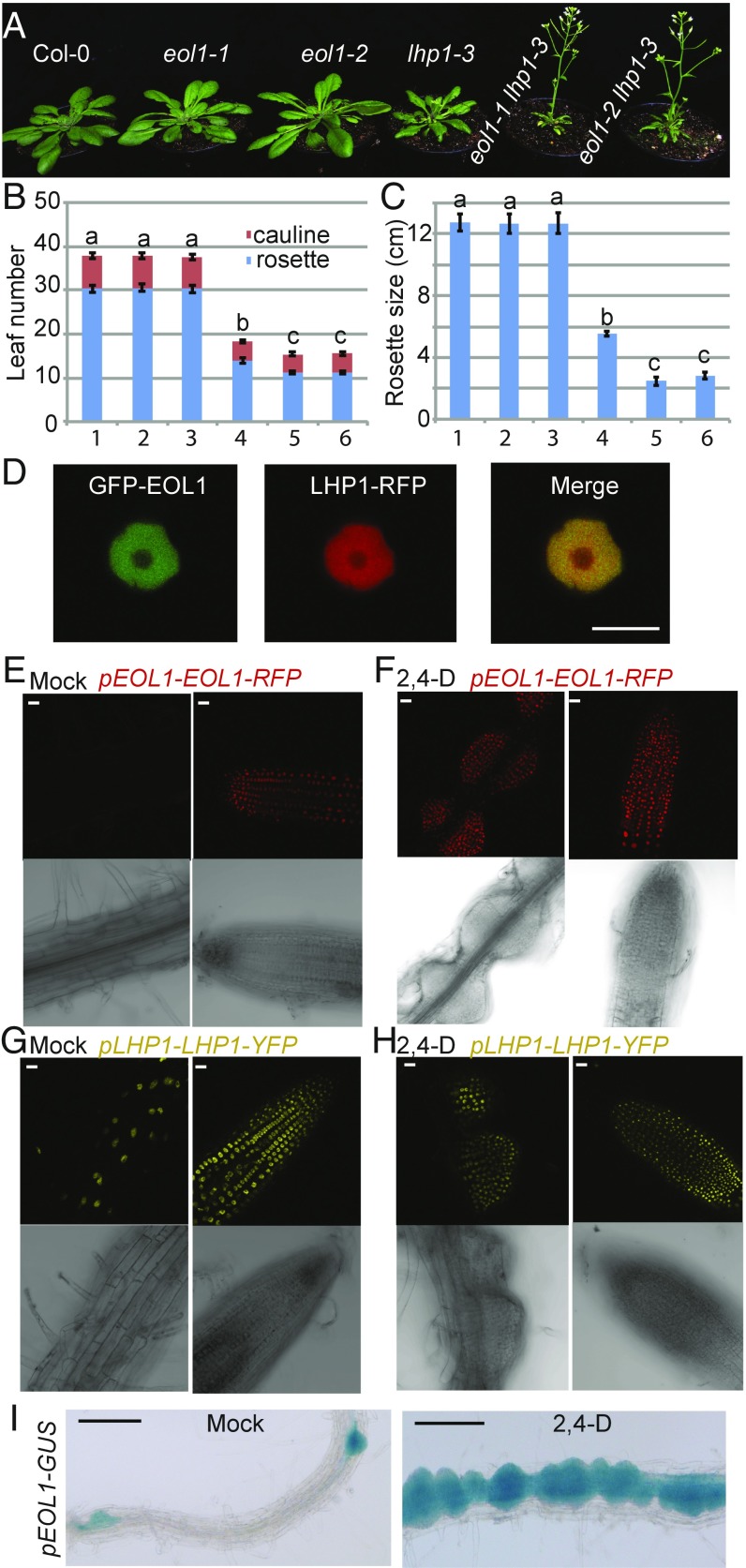
To identify novel genes participating in the plant PcG pathway, we induced mutations in the loss of function lhp1-3 mutant background (ref. 30, from now lhp1) by ethyl methane-sulfonate (EMS) and screened for genetic enhancers of the lhp1 phenotype, which consists of reduced plant size, early flowering in all photoperiod conditions, and altered leaf morphology compared with wild type (12, 31). In the M2 generation, plants that were smaller and flowered earlier than lhp1 under short day (SD) conditions were selected and their offspring grown at 16 °C in 12-h light/dark (MD) photoperiod. A recessive eol1 showed reduced rosette size and earlier flowering compared with lhp1, and did not segregate additional phenotypes after outcrossing to wild-type Col-0, suggesting that a single mutated locus affected development in the lhp1 background but not alone (Fig. 1 A–C). We used fast isogenic mapping (32) in an F2 population generated after two backcrosses to lhp1 to identify a single nucleotide polymorphism, causing conversion of a tryptophan (TGG) to a premature stop codon (TGA) in exon 1 of At3g42660 as candidate for the causative mutation (SI Appendix, Fig. S1A and Table S1). The identity of eol1 was confirmed by crossing lhp1 to an independent T-DNA insertion allele in At3g42660 (GABI_382A06; eol1-2), which showed no transcription across the insertion site (SI Appendix, Fig. S1B). F2 individuals homozygous for both the T-DNA allele and lhp1 showed phenotypic enhancement, whereas all plants carrying at least one functional LHP1 allele were undistinguishable from Col-0 (Fig. 1 A–C). Last, an 8-kb genomic fragment containing At3g42660 and 2-kbp and 1.5-kbp upstream and downstream regions complemented the eol1-1 lhp1 phenotype (SI Appendix, Fig. S2).
Fig. 1.
Loss-of-function alleles of eol1 enhance the lhp1 mutant phenotype. (A) Phenotype of Col-0 (1), eol1-1 (2), eol1-2 (3), lhp1-3 (4), eol1-1 lhp1-3 (5), and eol1-2 lhp1-3 (6) plants at day 30 after germination (DAG). Plants were grown at 16 °C in MD (12 h light/12 h dark) photoperiod. (B) Flowering time of genotypes grown as in A as number of leaves. Error bars indicate mean ± SE (n = 9). Significance determined by one-way ANOVA with multiple comparison correction by Tukey honest significant difference (HSD). Letters indicate significance groups (P < 0.001). (C) Rosette diameter of plants as in A, significance tested as above. (D) Nuclear colocalization of fluorescent GFP-EOL1 and LHP1-RFP transiently expressed in N. benthamiana leaf epidermis. (E–H) EOL1 and LHP1 protein levels in dividing tissues. Localization of fluorescent fusion protein in stable pEOL1-EOL1-RFP line (E and F) and pLHP1-LHP1-YFP (G and H) in absence (Mock) or presence of 2,4-D in the differentiation zone (Left) and the meristematic zone (Right) of the primary root. (I) EOL1 response to 2,4-D is transcriptional. Stably transformed promoter EOL1-GUS lines were grown on MS medium in absence (Mock, Left) or presence of 2,4-D (Right). Histochemical GUS detection shows signals in emerging side roots and in 2,4-D-induced calli. (Scale bars: A, 1 cm; D–H, 10 μm; I, 100 μm.).
EOL1 Encodes a Nuclear Protein Expressed in Dividing Cells.
An amino-terminal fusion of green fluorescent protein (GFP) to EOL1 was transiently expressed in tobacco and showed fluorescence exclusively in the nucleus with a notable depletion in the nucleolar region, similar to the localization of a carboxyl-terminal fusion of RFP to LHP1 (Fig. 1D and SI Appendix, Fig. S3). To determine the spatial expression pattern of EOL1 at cellular resolution, we inserted the codons for tagRFP before the EOL1 stop codon within a genomic fragment that included 2-kb promoter and the full structural gene. Red fluorescent signal was detected in the primary root of seedlings within the meristematic zone but not in the differentiation zone (Fig. 1E). This pattern suggested that EOL1 was present in cells undergoing active cell division. To confirm this link to cell division, we induced cell divisions by treating roots with the auxin analog 2,4-D. After 3 d of 2,4-D treatment, proliferating cell clusters were detected and nuclei within these showed red fluorescence (Fig. 1F). In contrast, LHP1 was detected in all root nuclei independent of 2,4-D treatment (Fig. 1 G and H).
To evaluate whether EOL1 was predominantly regulated at the transcriptional or posttranscriptional level, we fused the 2-kb EOL1 promoter to a β-glucuronidase (GUS) reporter gene. Histochemical detection of GUS activity confirmed that EOL1 expression was restricted to tissues undergoing active cell division, such as the root meristematic zone, initiating lateral roots, young leaves, and the shoot apex (SI Appendix, Fig. S4). Furthermore, GUS activity was increased in 2,4D-treated roots (Fig. 1I). Last, EOL1 GFP fusion protein expressed under the control of a constitutive promoter did accumulate in roots irrespective of cell division (SI Appendix, Fig. S5). Taken together, EOL1 is regulated at the transcriptional level to express in cells undergoing active cell division.
EOL1 Is Related to Ctf4/AND1/WDHM1.
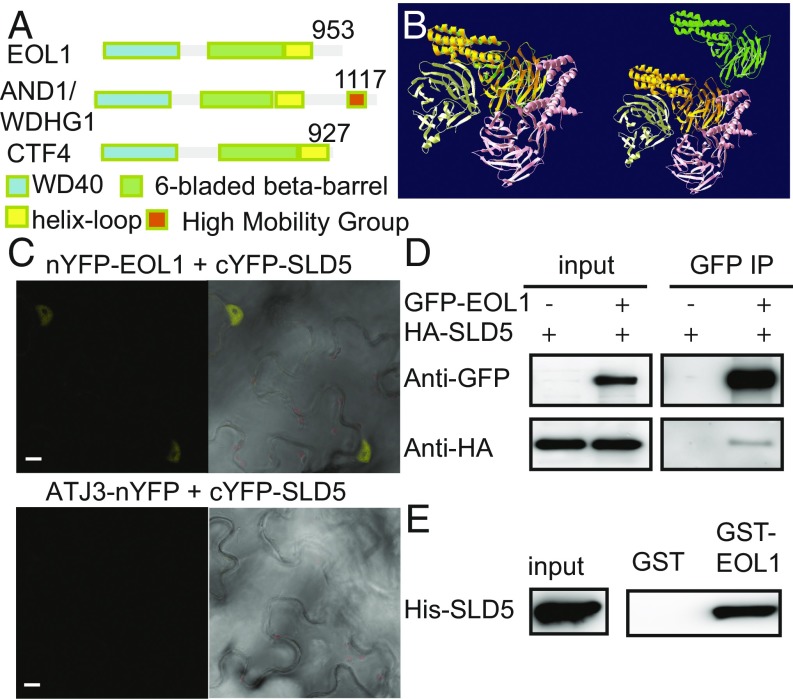
Although EOL1 is annotated as an unknown protein in The Arabidopsis Information Resource (TAIR), analysis of the primary sequence by SMART (33) predicts the presence of an amino-terminal WD40 domain and a centrally located domain of unknown function (DUF) 3639. This domain structure is found in yeast Ctf4 and metazoan Actinodin 1 (AND1)/WD-and-High-mobility-group-domain protein 1 (WDHD1), components of the nuclear DNA replisome (34, 35). A recently solved crystal structure of Ctf4 showed that DUF3639 is part of a novel WD40-related domain that forms a β-propeller with six blades (β-6-prop), forming a stable trimer (29). The carboxyl-terminal end of Ctf4 extends as α-helical fold that can interact with a motif found in both DNA-Polα and Sld5 (29). Sld5 is part of the GINS complex associated with the replicative MCM2-7 helicase complex, which unwinds the parental DNA double strand. Despite its proposed role as an important adaptor between essential components of the replisome, the role of Ctf4 in replication is not yet entirely clear. Presence of Ctf4 is essential for replication in D. melanogaster, but not in yeast, which show a reduced growth phenotype only in the presence of DNA-damaging agents (36, 37).
We probed the expression of DNA damage markers BREAST CANCER SUSCEPTIBILIY 1 (BRCA1), RAD51, and Poly-ADP-RIBOSE POLYMERASE 2 (PARP2) in wild-type and eol1 mutant seedlings in the absence and presence of the DNA damaging agents Bleocin and Zebularine (38). The marker genes were induced by both agents, irrespective of the genotype (SI Appendix, Fig. S6A). Furthermore, whereas both chemicals reduced root growth, the response was the same in eol1 and wild-type seedlings (SI Appendix, Fig. S6B).
Alignment of the primary sequences of representative Ctf4 homologs showed high conservation between kingdoms for the amino-terminal WD40 and the β-6-prop domains whereas the carboxyl-terminal extension and interdomain sequences were more variable (Fig. 2A and SI Appendix, Fig. S7). Phylogenetic neighbor joining trees based on the amino acid sequence alignment of the β-6-prop domain corroborated the classification of plant homologs as Ctf4 related (SI Appendix, Fig. S8). Furthermore, an in silico model of the A. thaliana protein generated a structure that fitted well to both the β-6-prop and carboxyl-terminal extension (39) (Fig. 2B).
Fig. 2.
EOL1 encodes the A. thaliana homolog of Ctf4. (A) Domain structure of A. thaliana EOL1, mouse AND1/WDHG1 and yeast Ctf4. (B) Overlay of structural prediction of EOL1 (green) with the published crystal structure of the yeast Ctf4 trimer (yellow, orange, pink), all without the WD40 domain, the lower left subunit also without carboxyl-terminal α-helical fold. At Right, EOL1 is translocated to the upper right. (C) BiFC analysis of the physical association between EOL1 and SLD5. Plasmid pairs, as indicated, were coinfiltrated into N. benthamiana leaves by using Agrobacterium tumefaciens. ATJ3 was used as a negative control. (Scale bars: 10 µm.) (D) CoIP assay of EOL1 and SLD5 transiently expressed in A. thaliana mesophyll protoplasts as indicated. Inputs and precipitates from anti-GFP trap beads were analyzed by Western blotting with anti-HA or anti-GFP antibodies. (E) Protein pulldown assay with GST-EOL1 or GST as bait. Bacterial protein extracts containing His-SLD5 were trapped with bait proteins bound to glutathione-linked resins and analyzed by Western blotting with anti-His antibodies.
We tested interaction between EOL1 and A. thaliana homologs of the GINS-complex components Sld5 and Psf1 to corroborate EOL1’s association with the replication machinery. Bimolecular fluorescence complementation (BiFC) (40) assays were performed in tobacco leaves by transiently coexpressing fusions of the amino-terminal half of yellow fluorescent protein to EOL1 (nYFP-EOL1) and its carboxyl-terminal half to SLD5 (cYFP-SLD5) or PSF1 (cYFP-PSF1). A. thaliana DNAJ HOMOLOG 3 (ATJ3, mainly localized in the nucleus) was used as a negative control. Respectively strong and weak YFP signals were detected for nYFP-EOL1/cYFP-SLD5 and nYFP-EOL1/cYFP-PSF1 (Fig. 2C and SI Appendix, Fig. S9A). Coimmunoprecipitation (Co-IP) experiments from Col-0 protoplasts transiently coexpressing GFP-EOL1 and HA-SLD5 or HA-PSF1 showed that GFP-EOL1 copurified HA-SLD5 (Fig. 2D) but not HA-PSF1 (SI Appendix, Fig. S9B). The interaction between EOL1 and SLD5 seemed direct because in vitro pull-down assays using bacterially expressed GST fusions to EOL1 purified His-SLD5 (Fig. 2E) but not His-PSF1 (SI Appendix, Fig. S9C). Thus, EOL1 shows a conserved molecular interaction with SLD5 in the GINS complex.
The Molecular Basis of the eol Phenotype.
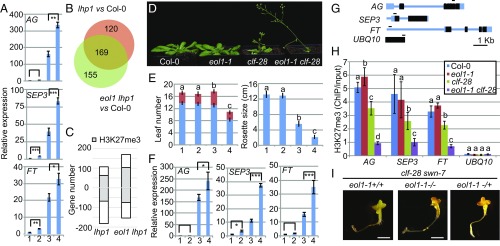
The small leaf phenotype of lhp1 mutants is linked to ectopic expression of floral meristem identity genes in leaves (41), whereas early flowering is due to up-regulation of FT (31). We tested expression of FT and the floral meristem identity genes AGAMOUS (AG) and SEP3 in eol1 lhp1 and eol1 mutants. Seedlings were grown in MD photoperiod and expression was evaluated on material collected at Zeitgeber (ZT) 12 h, when FT levels are expected to be highest in wild-type plants grown in this light regime. In the presence of the eol1-1 allele, SEP3 and FT expressed at approximately twofold higher levels compared with the respective controls, Col-0 and lhp1 (Fig. 3A). For AG, expression was not increased in eol1 single mutants compared with wild-type Col-0 controls but in eol1-1 lhp1 double mutants over lhp1.
Fig. 3.
EOL1 impacts H3K27me3 marks through PRC2. (A) qRT-PCR analysis of AG, SEP3, and FT expression at 10 DAG of Col-0 (1), eol1-1 (2), lhp1-3 (3), and eol1-1 lhp1-3 (4) seedlings grown at 16 °C in MDs. Values were normalized to PP2A and are displayed as fold difference to Col-0. Error bars indicate mean ± SD (three biological replicates). Significance analysis by Student’s t test for two group comparisons. (B) Venn diagram of genes differentially expressed in lhp1-3 and eol1-1 lhp1-3 seedlings compared with Col-0 based on RNA-seq data (biological replicates n = 3, GLM test with FDR correction on cpm values, threshold FDR < 0.05, log2 FC ≥ 2). (C) Number of genes significantly up- or down-regulated and the relative proportion of H3K27me3-positive genes. (D) Phenotype 21 DAG of Col-0 (1), eol1-1 (2), clf-28 (3) and eol1-1 clf-28 (4) plants grown at 22 °C in LDs. (E) Flowering time (Left) and rosette size (Right) of genotypes grown as in D scored as number of leaves and rosette diameter, respectively. Error bars indicate mean ± SE (n = 9). Significance determined by one-way ANOVA with multiple comparison correction by Tukey HSD. Letters indicate significance groups (P < 0.001). (F) qRT-PCR analysis of expression levels of AG, SEP3, and FT. RNA extracted from 10 DAG seedlings of genotypes as in D grown at 22 °C in LDs. Values were normalized to PP2A and analyzed as in A. (G) Structure of AG, SEP3, FT, and UBQ10 loci. Exons and untranslated regions are represented by black and blue boxes, respectively, and introns and other regions by blue lines. Fragments for ChIP PCR are indicated. (H) ChIP analysis of H3K27me3 levels at AG, SEP3, FT, and UBQ10 in genotypes as in D at 10 DAG. UBQ10 was amplified as negative control for H3K27me3. Error bars indicate mean ± SD calculated (three biological replicates). Significance determined by one-way ANOVA with multiple comparison correction by Tukey HSD. Letters indicate significance groups (P < 0.05). (I) Phenotype at 20 DAG of clf-28 swn-7 eol1-1+/+, clf-28 swn-7 eol1-1−/− and clf-28 swn-7 eol1−/+ plants grown at 22 °C on MS plates in a growth chamber. (Scale bars: 1 cm.)
RNA-seq transcriptome comparisons of Col-0, lhp1, and eol1 lhp1 seedlings detected a similar number of misregulated genes in the lhp1 and eol1 lhp1 mutants compared with Col-0 with a large overlap between both (Fig. 3B, 169 genes, FDR < 0.05; at least threefold change in expression; Datasets S1 and S2). We interrogated the proportion of H3K27me3-positive genes by intersecting differentially expressed gene sets with a list of H3K27me3 target genes in seedlings determined in a previous ChIP-seq analysis (18). More H3K27me3-positive genes showed increased expression in eol1 lhp1 over lhp1 mutants (Fig. 3C). Furthermore, H3K27me3 target genes up-regulated in lhp1 showed a clear trend toward a further increase in expression in the eol1 lhp1 double mutant, whereas H3K27me3-negative genes did not (SI Appendix, Fig. S10). Genes down-regulated in lhp1 did not show an H3K27me3-related pattern in the double eol1 lhp1 mutant (SI Appendix, Fig. S10). In sum, lack of EOL1 function further increases misexpression of a number of H3K27me3 target genes that are already up-regulated in lhp1 single mutants.
EOL1 Impacts H3K27me3 Inheritance.
Considering the exclusive expression of EOL1 in dividing cells (Fig. 1) (31), increased ectopic expression of floral meristem identity genes in nondividing cells in the eol1 lhp1 compared with lhp1 mutants suggests an epigenetic contribution of EOL1 in the regulation of typical LHP1 target genes. The PcG epigenetic memory is linked to H3K27me3 inheritance (22). Mutation of SWN does not lead to an obvious morphological phenotype, which was also the case for eol1-1 swn-7 double mutants (SI Appendix, Fig. S11). However, introducing the eol-1 allele into the clf-28 mutant background enhanced many aspects of the clf phenotype, such as smaller rosette size and earlier flowering (Fig. 3 D and E). Similar to eol1-1 lhp1, eol1-1 clf-28 mutants showed a further increase in expression of AG, SEP3, and FT compared with clf-28 single mutants (Fig. 3F). We performed chromatin immunoprecipitation in single and double mutants of clf and eol1 to evaluate H3K27me3 levels at target genes. H3K27me3 levels were decreased at FT, AG, and SEP3 in clf-28 single mutants compared with Col-0 controls and further decreased in the eol1-1 clf-28 background (Fig. 3 G and H). In eol1 single mutants, H3K27me3 levels at all genes were as in wild-type seedlings. As a control for chromatin quality, all extracts were also precipitated with antibodies against histone H3, which enriched equal amounts of DNA from all genotypes (SI Appendix, Fig. S12).
The genetic data suggested that the effect of EOL1 on H3K27me3 inheritance could be particular important to ensure functionality of PRC2 complexes that contain SWN at the catalytic core (clf mutant) while being less crucial for CLF-containing PRC2 complexes (swn mutant). Despite the presence of MEA, H3K27me3 cannot be detected in clf swn mutants (42, 43). Because clf swn double mutants were epistatic to clf swn eol1 triple mutants, EOL1 function seems to depend on PRC2 activity (Fig. 3I). In sum, EOL1 function is required to maintain H3K27me3 marks at target genes already affected in the inheritance of this mark in clf mutants. In wild-type plants, EOL1 seems dispensable for H3K27me3 inheritance.
EOL1 Physically Interacts with LHP1, CLF, and SWN.
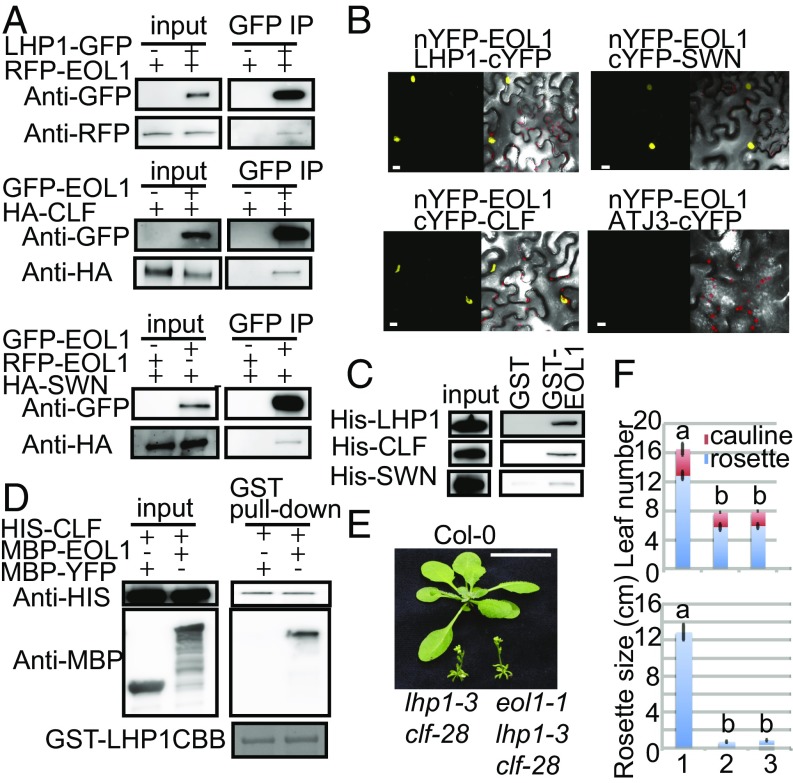
Because the genetic interaction suggested that EOL1 required PRC2 activity, we tested whether this observation was explained by direct protein interaction. Because physical associations between LHP1 and PRC2 are described (5, 6), we also interrogated whether EOL1 could be part of an LHP1–PRC2 complex. Co-IP assays performed from transiently transfected Col-0 protoplasts showed that GFP-EOL1 copurified HA-tagged CLF and SWN and that GFP-LHP1 copurified RFP-EOL1 (Fig. 4A). These interactions were confirmed by BiFC (40) using nYFP-EOL1 and cYFP-CLF, cYFP-SWN, or LHP1-cYFP. A fluorescent signal as indicator of molecular interaction was detected in nuclei of tobacco epidermis cells expressing nYFP-EOL1 together with cYFP-CLF, cYFP-SWN, or LHP1-cYFP but not ATJ3-cYFP (Fig. 4B). Furthermore, cYFP-CLF, cYFP-SWN, or LHP1-cYFP did not show fluorescent signal in combination with ATJ3-nYFP (SI Appendix, Fig. S13).
Fig. 4.
EOL1 binds to LHP1, CLF, and SWN in vitro and in vivo. (A) CoIP assay of EOL1 with LHP1, CLF, or SWN transiently expressed in A. thaliana mesophyll protoplasts. LHP1-GFP or GFP-EOL1 were immunoprecipitated with anti-GFP trap beads from protoplasts cotransfected with RFP-EOL1, HA-CLF, or HA-SWN as indicated, the precipitates were analyzed by Western blotting with anti-HA, anti-RFP, or anti-GFP antibodies. (B) BiFC analysis of the physical association of EOL1 with LHP1, CLF, and SWN. Plasmid pairs, as indicated, were coinfiltrated into Nicotiana benthamiana leaves by using A. tumefaciens. ATJ3 was used as a negative control. (C) Protein pull down assay with GST-EOL1 or GST as bait. Total protein extracts of bacteria expressing His-LHP1, or Sf21 cells expressing His-CLF or His-SWN were incubated with bait proteins bound to glutathione-linked resins. Proteins associated with the resins were analyzed by Western blotting with an anti-His antibody or stained with Coomassie Brilliant Blue (CBB). (D) Proteins pull down assay with GST-LHP1 as bait. Total protein extracts of insect cells expressing His-CLF were mixed with bacterial extracts expressing MBP-YFP or MBP-EOL1 and incubated with GST-LHP1 bound to glutathione-linked resin. Proteins associated with the resins were analyzed by Western blotting with anti-His or anti-MBP antibodies. (E) Phenotype of Col-0 (1), lhp1-3 clf-28 (2), and eol1-1 lhp1-3 clf-28 (3) plants at 20 DAG. Plants were grown at 22 °C in LDs. (F) Flowering time and rosette size of genotypes grown as in D scored as number of leaves and rosette diameter, respectively. Error bars indicate mean ± SE (n = 9). Significance determined by one-way ANOVA with multiple comparison correction by Tukey HSD. Letters indicate significance groups (P < 0.001). (Scale bars: B, 10 µm; E, 1 cm.)
These assays suggested that EOL1 associates with LHP1 and CLF/SWN inside nuclei but could not resolve whether these interactions were direct. We therefore performed in vitro pull-down assays by using GST to EOL1 fusions produced in bacteria. Prey proteins, which were either bacterially expressed His-LHP1 or His-CLF and His-SWN expressed in Sf21 insect cells, were separately mixed with recombinant bait proteins. Analysis of pull-down products on immunoblots with anti-His antibodies showed that EOL1 pulled down LHP1, CLF, and SWN (Fig. 4C). Taken together, the data suggested that interaction between EOL1 and LHP1 or the PRC2 components CLF and SWN is direct.
Because LHP1 was previously shown to copurify with CLF and MSI1 (5, 6), it seems that EOL1 interacts with an LHP1–PRC2 complex through two protein interfaces that directly bind EOL1. We tested whether EOL1 mediated the previously described interaction between PRC2 and LHP1. However, in vitro pulldown experiments indicated that LHP1 and CLF interact in the absence of EOL1 (Fig. 4D). Because LHP1 copurified equal amounts of CLF irrespective of the presence or absence of EOL1, the interaction between these partners seems noncompetitive (Fig. 4D). Genetic data supported the idea that EOL1 acts through an LHP1–PCR2 complex because the addition of the nonfunctional eol1-1 allele to a clf lhp1 background did not enhance the lhp1 clf double mutant phenotype (Fig. 4 E and F). In sum, EOL1 interacts with an LHP1–PRC2 supercomplex through nonexclusive interactions with LHP1 and CLF or SWN.
Discussion
Based on several lines of evidence, we propose that EOL1 is the plant homolog of yeast Ctf4 and mammalian WDHD1/AND1 (Fig. 2 and SI Appendix, Fig. S7). EOL1 was restricted to narrow domains undergoing active cell division, domains that massively expanded in auxin-induced roots (Fig. 1). E2F transcription factors regulate genes required for DNA replication and cell cycle in all eukaryotes, and EOL1 was identified in a previous genome-wide study of E2F target genes in A. thaliana, supporting the notion that EOL1 is part of a gene network coordinated during cell division (44).
Although the overall sequence similarity for the proposed functional homologs from different kingdoms is low, structural predictions identify three common domains (Fig. 2 and SI Appendix, Fig. S7). Mammalian WDHD1/AND1 features a High Mobility Group (HMG) domain in a carboxyl-terminal extension that is absent from yeast and plant homologs. Yeast Ctf4 forms a functional trimer that acts as a multivalent interface between the MCM2-7 helicase-associated GINS complex and DNA-polα during replication (29). This interaction is mediated through specific recognition of a peptide motif found in DNA-polα and Sld5 with the α-helical fold of Ctf4 (29, 34, 35). We confirmed an interaction between plant EOL1 and SLD5 (Fig. 2), despite the fact that the EOL1 α-helical fold is poorly conserved and that the signature for the interacting peptide is undetectable in ICU2 or SLD5.
Different from the phenotype observed in yeast Ctf4 null mutants, eol1 mutants were insensitive to treatment with DNA damaging drugs (SI Appendix, Fig. S6). Although EOL1 is a single copy gene in A. thaliana, the presence of functionally redundant proteins could attenuate the requirement for EOL1 in DNA damage control. Alternatively, plants may deal with DNA damage in an EOL1/Ctf4 independent way.
Mutation of EOL1 enhances the phenotypes displayed by lhp1 or clf but not lhp1 clf or clf swn double mutants, indicating that EOL1 interacts with an LHP1–PRC2 (Figs. 1, 3, and 4). Accordingly, EOL1 physically interacts with both, LHP1 and CLF, in a noncompetitive manner (Fig. 4). Although EOL1 mutants show no developmental aberrations in our experimental growth conditions, we detected a reduction of H3K27me3 levels in eol1 clf double mutant compared with clf mutant (Fig. 3). We suggest that LHP1–PRC2 complexes containing SWN (present in clf mutants) depend on EOL1 function, whereas they seem less affected if CLF is present (in swn mutants) at their catalytic core.
Mutations in the three DNA-pols enhance the abnormal phenotype of PcG mutants or cause deregulated expression of PcG target genes (25–28). Although evidence for a direct interaction of LHP1 with ICU2 is slightly conflicting (25, 27), it is conceivable that these proteins, which engage in higher-order protein complexes, possess several interfaces to other proteins. Complexes may also share subunits, as reported for the WD40 domain protein MSI1, which is an integral part of PRC2 but also of Chromatin assembly factor 1 (CAF1), the histone H3-H4 chaperone that assists nucleosome assembly during replication (45). Like EOL1, MSI1 features a WD40 domain and reportedly binds to both CLF and LHP1 (Fig. 4 and ref. 6). Furthermore, in animals it has been speculated that association of PRC2 with the replication fork may be promoted by affinity of the E(z) component to single-stranded DNA that is generated upon unwinding by the helicase complex (46, 47).
In conclusion, we propose that maintenance of epigenetic information during replication requires a specialized network of interactions between the replication apparatus and PRC2 and that these networks include LHP1 and EOL1.
Materials and Methods
The eol1-1 mutant was derived from lhp1-3 background by EMS mutagenesis. Details of experimental procedures, such as mapping of EOL1, quantitative PCR, Co-IP, pull-down, BiFC, microscopic analyses, GUS staining, and ChIP are described in SI Appendix, SI Materials and Methods. See SI Appendix, Table S2 for the primers used in this study.
Supplementary Material
Acknowledgments
We thank Petra Taenzler and Leon Langereis for excellent technical help, Nicolas Thomä (Friedrich Miescher Institute for Biomedical Research, Switzerland) for providing pNT74i Vector and technical advice, Jane Parker for critical reading of the manuscript, and Ales Pecinka and Andreas Finke for helping us to test phenotype and marker gene expression during treatments with DNA-damaging drugs. We thank the Max Planck Society for funding.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the European Nucleotide Archive (ENA) database, www.ebi.ac.uk/ena (accession no. PRJEB20122).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620955114/-/DCSupplemental.
References
- 1.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 2.Förderer A, Zhou Y, Turck F. The age of multiplexity: Recruitment and interactions of Polycomb complexes in plants. Curr Opin Plant Biol. 2016;29:169–178. doi: 10.1016/j.pbi.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 3.Grossniklaus U, Paro R. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb Perspect Biol. 2014;6:a019331. doi: 10.1101/cshperspect.a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mozgova I, Hennig L. The polycomb group protein regulatory network. Annu Rev Plant Biol. 2015;66:269–296. doi: 10.1146/annurev-arplant-043014-115627. [DOI] [PubMed] [Google Scholar]
- 5.Liang SC, et al. Kicking against the PRCs - A domesticated transposase antagonises silencing mediated by polycomb group proteins and is an accessory component of polycomb repressive complex 2. PLoS Genet. 2015;11:e1005660, and erratum (2016) 12:e1005812. doi: 10.1371/journal.pgen.1005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derkacheva M, et al. Arabidopsis MSI1 connects LHP1 to PRC2 complexes. EMBO J. 2013;32:2073–2085. doi: 10.1038/emboj.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA. 2008;105:16831–16836. doi: 10.1073/pnas.0808687105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schatlowski N, Stahl Y, Hohenstatt ML, Goodrich J, Schubert D. The CURLY LEAF interacting protein BLISTER controls expression of polycomb-group target genes and cellular differentiation of Arabidopsis thaliana. Plant Cell. 2010;22:2291–2305. doi: 10.1105/tpc.109.073403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merini W, Calonje M. PRC1 is taking the lead in PcG repression. Plant J. 2015;83:110–120. doi: 10.1111/tpj.12818. [DOI] [PubMed] [Google Scholar]
- 10.Yang C, et al. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr Biol. 2013;23:1324–1329. doi: 10.1016/j.cub.2013.05.050. [DOI] [PubMed] [Google Scholar]
- 11.Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol. 2010;20:1853–1859. doi: 10.1016/j.cub.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 12.Gaudin V, et al. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development. 2001;128:4847–4858. doi: 10.1242/dev.128.23.4847. [DOI] [PubMed] [Google Scholar]
- 13.Xu L, Shen WH. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol. 2008;18:1966–1971. doi: 10.1016/j.cub.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez R, Kim MY, Calonje M, Moon YH, Sung ZR. Temporal and spatial requirement of EMF1 activity for Arabidopsis vegetative and reproductive development. Mol Plant. 2009;2:643–653. doi: 10.1093/mp/ssp004. [DOI] [PubMed] [Google Scholar]
- 15.Chanvivattana Y, et al. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131:5263–5276. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- 16.Chen D, Molitor A, Liu C, Shen WH. The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 2010;20:1332–1344. doi: 10.1038/cr.2010.151. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, et al. Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of polycomb group proteins in association with distinct sets of cis-regulatory elements. PLoS Genet. 2016;12:e1005771. doi: 10.1371/journal.pgen.1005771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Hartwig B, James GV, Schneeberger K, Turck F. Complementary activities of telomere repeat binding proteins and polycomb group complexes in transcriptional regulation of target genes. Plant Cell. 2016;28:87–101. doi: 10.1105/tpc.15.00787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molitor AM, Bu Z, Yu Y, Shen WH. Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 2014;10:e1004091. doi: 10.1371/journal.pgen.1004091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350:aac4383. doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 22.Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15:340–356. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- 23.Iglesias FM, Cerdán PD. Maintaining epigenetic inheritance during DNA replication in plants. Front Plant Sci. 2016;7:38. doi: 10.3389/fpls.2016.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petruk S, et al. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrero JM, González-Bayón R, del Pozo JC, Ponce MR, Micol JL. INCURVATA2 encodes the catalytic subunit of DNA polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell. 2007;19:2822–2838. doi: 10.1105/tpc.107.054130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.del Olmo I, et al. EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J. 2010;61:623–636. doi: 10.1111/j.1365-313X.2009.04093.x. [DOI] [PubMed] [Google Scholar]
- 27.Hyun Y, et al. The catalytic subunit of Arabidopsis DNA polymerase α ensures stable maintenance of histone modification. Development. 2013;140:156–166. doi: 10.1242/dev.084624. [DOI] [PubMed] [Google Scholar]
- 28.Iglesias FM, et al. The arabidopsis DNA polymerase δ has a role in the deposition of transcriptionally active epigenetic marks, development and flowering. PLoS Genet. 2015;11:e1004975. doi: 10.1371/journal.pgen.1004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon AC, et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature. 2014;510:293–297. doi: 10.1038/nature13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsson AS, Landberg K, Meeks-Wagner DR. The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics. 1998;149:597–605. doi: 10.1093/genetics/149.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotake T, Takada S, Nakahigashi K, Ohto M, Goto K. Arabidopsis TERMINAL FLOWER 2 gene encodes a heterochromatin protein 1 homolog and represses both FLOWERING LOCUS T to regulate flowering time and several floral homeotic genes. Plant Cell Physiol. 2003;44:555–564. doi: 10.1093/pcp/pcg091. [DOI] [PubMed] [Google Scholar]
- 32.Hartwig B, James GV, Konrad K, Schneeberger K, Turck F. Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol. 2012;160:591–600. doi: 10.1104/pp.112.200311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letunic I, Doerks T, Bork P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka H, et al. Ctf4 coordinates the progression of helicase and DNA polymerase alpha. Genes Cells. 2009;14:807–820. doi: 10.1111/j.1365-2443.2009.01310.x. [DOI] [PubMed] [Google Scholar]
- 35.Kang YH, et al. Interaction between human Ctf4 and the Cdc45/Mcm2-7/GINS (CMG) replicative helicase. Proc Natl Acad Sci USA. 2013;110:19760–19765. doi: 10.1073/pnas.1320202110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gosnell JA, Christensen TW. Drosophila Ctf4 is essential for efficient DNA replication and normal cell cycle progression. BMC Mol Biol. 2011;12:13. doi: 10.1186/1471-2199-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kouprina N, et al. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5736–5747. doi: 10.1128/mcb.12.12.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pecinka A, Liu CH. Drugs for plant chromosome and chromatin research. Cytogenet Genome Res. 2014;143:51–59. doi: 10.1159/000360774. [DOI] [PubMed] [Google Scholar]
- 39.Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis. 2009;30:S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 40.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez-Vernaza M, et al. Antagonistic roles of SEPALLATA3, FT and FLC genes as targets of the polycomb group gene CURLY LEAF. PLoS One. 2012;7:e30715. doi: 10.1371/journal.pone.0030715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lafos M, et al. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet. 2011;7:e1002040. doi: 10.1371/journal.pgen.1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kinoshita T, Yadegari R, Harada JJ, Goldberg RB, Fischer RL. Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell. 1999;11:1945–1952. doi: 10.1105/tpc.11.10.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verkest A, et al. A generic tool for transcription factor target gene discovery in Arabidopsis cell suspension cultures based on tandem chromatin affinity purification. Plant Physiol. 2014;164:1122–1133. doi: 10.1104/pp.113.229617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hennig L, Bouveret R, Gruissem W. MSI1-like proteins: An escort service for chromatin assembly and remodeling complexes. Trends Cell Biol. 2005;15:295–302. doi: 10.1016/j.tcb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Krajewski WA, Nakamura T, Mazo A, Canaani E. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol Cell Biol. 2005;25:1891–1899. doi: 10.1128/MCB.25.5.1891-1899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.