Significance
The circadian clock in the suprachiasmatic nucleus (SCN) regulates seasonality in physiology and behavior, which is best characterized by the change in the activity time of behavioral rhythms. In nocturnal rodents, the activity time was shortened in long summer days and lengthened in short winter days because of the change in the phase relationship of activity onset and offset, for which different circadian oscillators are predicted. Taking advantage of in vivo monitoring of clock gene expression in freely moving mice, we demonstrated that the circadian rhythms of Per1 and Bmal1 in the SCN are associated differentially with the phase shifts of activity onset and offset, respectively, suggesting the existence of two oscillations with different molecular mechanisms in timing of circadian behavior.
Keywords: clock gene, in vivo recording, suprachiasmatic nucleus, photic phase resetting, E and M oscillators
Abstract
The temporal order of physiology and behavior in mammals is primarily regulated by the circadian pacemaker located in the hypothalamic suprachiasmatic nucleus (SCN). Taking advantage of bioluminescence reporters, we monitored the circadian rhythms of the expression of clock genes Per1 and Bmal1 in the SCN of freely moving mice and found that the rate of phase shifts induced by a single light pulse was different in the two rhythms. The Per1-luc rhythm was phase-delayed instantaneously by the light presented at the subjective evening in parallel with the activity onset of behavioral rhythm, whereas the Bmal1-ELuc rhythm was phase-delayed gradually, similar to the activity offset. The dissociation was confirmed in cultured SCN slices of mice carrying both Per1-luc and Bmal1-ELuc reporters. The two rhythms in a single SCN slice showed significantly different periods in a long-term (3 wk) culture and were internally desynchronized. Regional specificity in the SCN was not detected for the period of Per1-luc and Bmal1-ELuc rhythms. Furthermore, neither is synchronized with circadian intracellular Ca2+ rhythms monitored by a calcium indicator, GCaMP6s, or with firing rhythms monitored on a multielectrode array dish, although the coupling between the circadian firing and Ca2+ rhythms persisted during culture. These findings indicate that the expressions of two key clock genes, Per1 and Bmal1, in the SCN are regulated in such a way that they may adopt different phases and free-running periods relative to each other and are respectively associated with the expression of activity onset and offset.
In mammals, the circadian pacemaker in the hypothalamic suprachiasmatic nucleus (SCN) entrains to a light–dark (LD) cycle and regulates circadian rhythms of behavior and physiology (1, 2). The circadian oscillation in the SCN is autonomous, and the clock genes Per1, Per2, Cry1, Cry2, Clock, and Bmal1 play crucial roles (3). A heterodimer of Clock and Bmal1 proteins (CLOCK/BMAL1) activates the transcription of Per and Cry genes; in turn, the protein products of these genes suppress their own transactivation by CLOCK/BMAL1, closing a feedback loop. One turn of the auto-feedback loop (core loop) takes ∼24 h. On the other hand, Bmal1 expression is enhanced by RAR-related orphan nuclear receptor (ROR) and is repressed by an orphan nuclear receptor in the RevErb family (α, β) through ROR response element (4, 5). The expressions of ROR and the RevErb family are enhanced in turn by a BMAL1/CLOCK heterodimer via an upstream E-box. Thus, the Bmal1 circadian rhythm is auto-regulated by a feedback loop (the Bmal1 loop) which is interlocked with the core loop, maintaining an antiphasic phase relationship with Per1 rhythm. This interlocked Bmal1 loop has been considered to contribute to stabilization and fine tuning of the core loop (4, 6) in addition to the regulation of downstream pathways (7).
The expression of Per genes in the SCN is activated by a timed exposure to light, which phase shifts the circadian pacemaker (8, 9). The phase-dependent phase shifts of clock gene expression are regarded as a key mechanism by which the circadian pacemaker is entrained to a LD cycle. Light signals from the retina stimulate the expression of Per genes, perturbing the core loop dynamics to produce a phase-dependent phase shift (8). However, the mechanisms by which light-induced phase-shift signals from the core loop are transduced to circadian rhythms in physiology and behavior are not well understood.
On the behavioral level, the onset and offset of an activity band (activity onset and offset) of circadian behavioral rhythm are known to respond differentially to a phase-shifted LD cycle (10) and to a single light pulse under continuous darkness (DD) in nocturnal rodents (11). In addition, the phase relation between the activity onset and offset is known to change under different photoperiods (12). Furthermore, the two phases of behavioral rhythm occasionally split under the constant light condition (12). From these findings, the two oscillator hypothesis was advanced to explain behavioral circadian rhythm in nocturnal rodents (13). Namely, one oscillator, designated an evening (E) oscillator, regulates the activity onset, and the other oscillator, designated a morning (M) oscillator, controls the activity offset. Previously, regional differences were reported in the circadian rhythms of clock gene expression in the SCN (14–17), and the phase relation of the two rhythms changed under different photoperiods in parallel with the change in activity time of behavioral circadian rhythm. These findings suggested the existence of the E and M oscillators in the SCN. However, the molecular mechanisms underlying the E and M oscillators are totally unknown. Recently, we developed an in vivo method for monitoring clock gene expression in the SCN of a freely moving mouse (18, 19), enabling us to compare the SCN circadian rhythms with physiological and behavioral rhythms.
In the present study, to obtain insight into the molecular mechanism of light entrainment, we continuously monitored circadian rhythms in clock gene Per1 and Bmal1 expression in the SCN together with behavioral rhythms. Surprisingly, the rhythms of Per1 and Bmal1 expression responded differentially to the light pulse. The dissociation between the two circadian rhythms was confirmed in the cultured SCN slices from double-transgenic mice carrying luciferase reporters for Per1 and Bmal1 expression. These findings indicate that there are at least two circadian pacemakers in the SCN, which have different molecular mechanisms and differentially regulate behavioral outputs.
Results
Differential Responses of Per1-luc and Bmal1-ELuc Circadian Rhythms in the SCN in Vivo.
We continuously measured the expression of the clock genes Per1 and Bmal1 in the SCN of freely moving transgenic mice carrying a bioluminescence reporter (Per1-luc or Bmal1-ELuc) under DD. Spontaneous locomotor activity was monitored simultaneously by an infrared thermal sensor. Bioluminescence emitted from the SCN was collected with an implanted plastic optical fiber connected to a cooled photomultiplier tube (PMT), as described previously (18, 19).
Phase responses of Per1-luc and Bmal1-ELuc circadian rhythms in the SCN were examined when a single light pulse of 9 h duration was given at circadian time (CT) 11.5 to make the largest phase-delay shift according to a previous study (20), in which the activity onset of behavioral circadian rhythm was defined as CT12 (Fig. 1 and Fig. S1A). We also examined the effect of a light pulse of the same duration given at CT21.5, when phase-advance shifts were expected (Fig. 2 and Fig. S1B). Data of the in vivo experiments before a phase-delaying light pulse (n = 4 for Per1 and n = 4 for Bmal1) were used in a previous study (18) to calculate the free-running period and to estimate the circadian and ultradian phases of behavioral and clock gene expression rhythms.
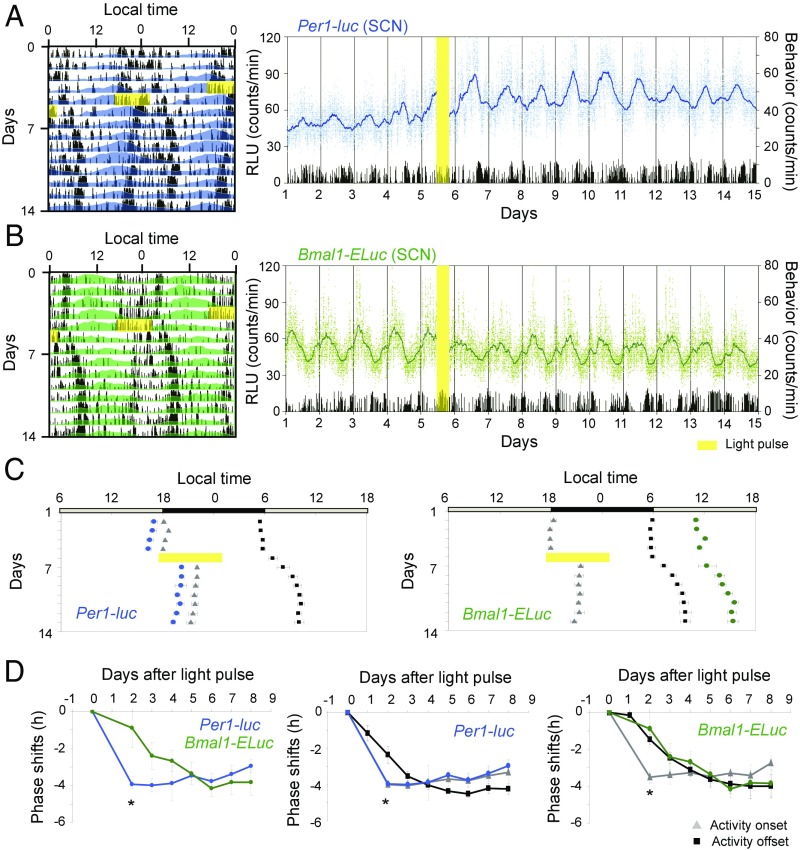
Fig. 1.
Light pulse-induced phase-delay shifts of circadian rhythms in the SCN and behavioral rhythms in freely moving adult mice. (A and B) Typical examples of phase response at CT11.5 are illustrated for the Per1-luc (A) and Bmal1-ELuc (B) rhythms in the SCN with a behavioral rhythm (black histogram) (Left) in double-plotting, in which the colored area indicates bioluminescence larger than the minimum value of a series, and in sequential plotting (Right), in which broken lines indicate raw data and solid lines indicate 4-h moving-averaged values (Per1-luc, blue; Bmal1-ELuc, green). The number of behavioral activities in 1-min intervals is indicated by black vertical bars. A yellow vertical bar indicates the time of the light pulse. (C) Mean acrophases ± SEM (horizontal bar) are illustrated for Per1-luc (blue circles) (Left) and Bmal1-ELuc (green circles) (Right) together with the mean activity onsets (gray triangles) and offsets (black squares). A yellow horizontal bar indicates the time of a light pulse. A horizontal gray and black bar at the top of each panel indicates the LD cycle to which mice had been entrained. (D) Daily phase shifts are reported by mean ± SEM (n = 4) for Per1-luc (blue circles) and Bmal1-ELuc (green circles) (Left) rhythms, for Per1-luc and two phase markers (onset and offset) of behavioral rhythm (Center), and for Bmal1-ELuc and the two phase markers (Right). The abscissa indicates the number of days after a light pulse. An asterisk indicates statistically significant difference (P < 0.05, two-way ANOVA with a post hoc t test) between Per1-luc and Bmal1-ELuc (Left), between Per1-luc and activity offset (Center), and between Bmal1-ELuc and activity onset (Right). RLU, relative luminescence units.
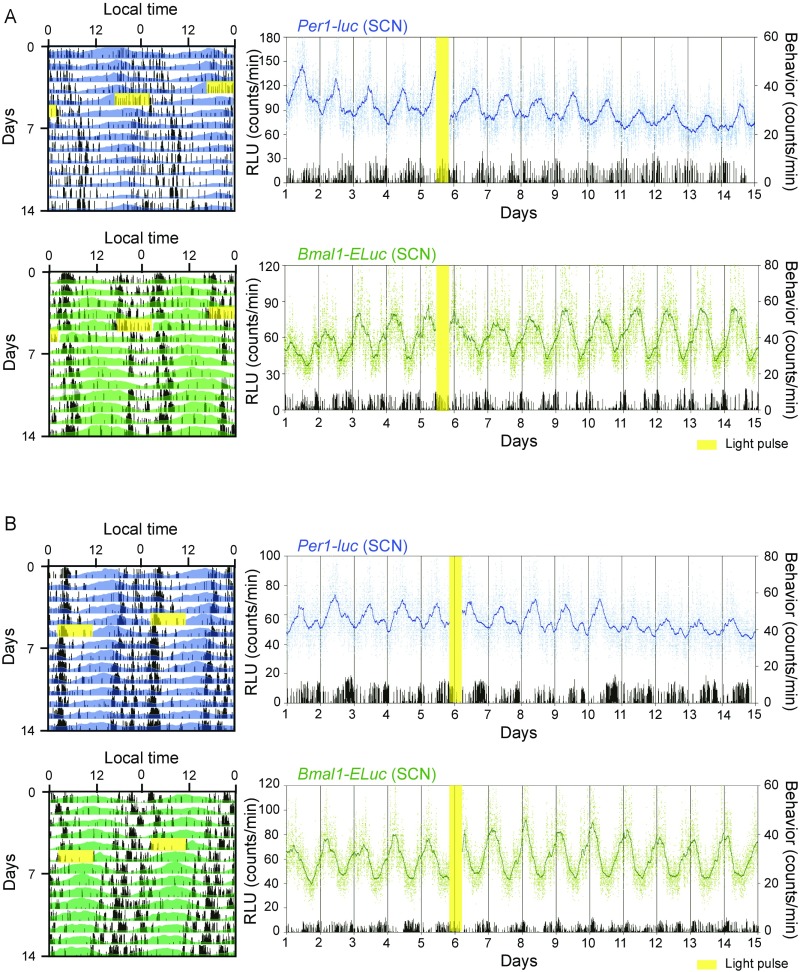
Fig. S1.
Light pulse-induced phase-delay and phase-advance shifts in circadian rhythms in the SCN and in behavioral rhythms in freely moving mice. (A) Other examples of the phase response at CT11.5 are illustrated for the circadian Per1-luc (Upper) and Bmal1-ELuc (Lower) behavioral rhythm plotted in a double-plotted manner (Left) and sequentially (Right). Also see the legends of Fig. 1 A and B. (B) Other examples of the phase response at CT21.5 are illustrated for the Per1-luc (Upper) and Bmal1-ELuc (Lower) circadian rhythm with a behavioral rhythm plotted in a double-plotted manner (Left) and sequentially (Right). Also see Figs. 1 and 2.
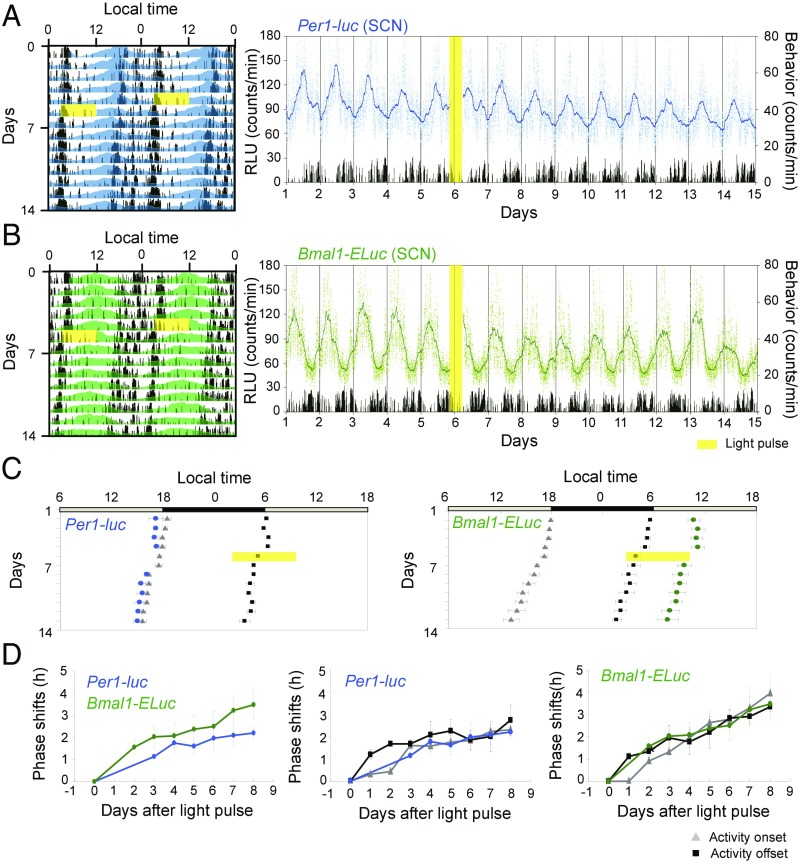
Fig. 2.
Light pulse-induced phase-advance shifts of circadian rhythms in the SCN and behavioral rhythm in freely moving adult mice. (A and B) Typical examples of a light-induced phase response at CT21.5 are illustrated for the Per1-luc (A) and Bmal1-ELuc (B) rhythms in the SCN together with a behavioral rhythm in double plotting (Left) and sequential plot (Right) as in Fig. 1 A and B. (C) Mean acrophases are illustrated for Per1-luc (Left) and Bmal1-ELuc (Right) rhythms together with the mean activity onsets and offsets of behavioral rhythms. Also see the legend of Fig. 1C. (D) The amount of phase shifts after a light pulse are shown as the mean and SEM (n = 3) for Per1-luc (blue circles) and Bmal1-ELuc (green circles) (Left), for Per1-luc and two phase markers (activity onset and offset) of behavioral rhythm (Center), and for Bmal1-ELuc and the phase markers (Right). Also see the legend of Fig. 1D.
In response to a light pulse at CT11.5, the activity onset of the behavioral rhythm was phase delayed immediately by 4.0 ± 0.4 h on average in Per1-luc mice (mean ± SD, n = 4) and by 3.5 ± 0.7 h on average in Bmal1-ELuc mice (n = 4). In contrast, the activity offset was gradually phase delayed over four or five cycles by 4.4 ± 0.3 h in Per1-luc mice and by 3.6 ± 0.8 h in Bmal1-ELuc mice before reaching a free-running steady state (Fig. 1 C and D). The amount of phase shift in either behavioral marker was not significantly different in the two reporter mice. Thus, the activity band was temporarily compressed after the light pulse (Fig. 1C). On the other hand, the circadian peak of Per1-luc rhythm was immediately phase delayed in parallel with the activity onset (Fig. 1 C, Left and D), and the circadian peak of Bmal1-ELuc rhythm was gradually phase delayed in parallel with the activity offset (Fig. 1 C, Right and D). The amount of phase shift in the two bioluminescent rhythms on the second day after the light pulse was significantly different (two-way ANOVA, post hoc t test, P < 0.05) (Fig. 1D, Left). When each bioluminescent rhythm was compared with behavioral rhythms, the amount of phase shift in the Per1-luc rhythm was not different from the phase shift of activity onset but was significantly different from the phase shift of activity offset (two-way ANOVA, post hoc t test, P < 0.05) (Fig. 1D, Center). The amount of phase shift in the Bmal1-ELuc rhythm was not different from the amount of phase shift in activity offset but was significantly different from the amount of phase shift in the activity onset (two-way ANOVA, post hoc t test, P < 0.01) (Fig. 1D, Right).
When a single light pulse was given at CT21.5, the amount of phase shift was small, and the dissociation between the Per1-luc and Bmal1-ELuc circadian rhythms was not detected at statistically significant levels (Fig. 2 and Fig. S1B). Nevertheless, an association of the circadian peak in clock gene expression with the behavioral marker was observed, similar to that observed with a light pulse at CT11.5. These results indicate that the circadian rhythms of Per1 and Bmal1 expression are dissociable when an external perturbation produces a large phase-delay shift in the SCN circadian rhythm in vivo.
Dissociation of Per1 and Bmal1 Oscillations in the SCN Slice.
To test whether the dissociation of Per1 and Bmal1 circadian oscillation really occurs, we made SCN slices from the double-transgenic mice carrying reporters for both Per1 and Bmal1 expression and monitored Per1-luc and Bmal1-ELuc simultaneously from the same SCN slice. Circadian Per1-luc and Bmal1-ELuc rhythms in the cultured SCN were separated successfully, as previously reported (21, 22), and persisted at least for 3 wk (Fig. S2).
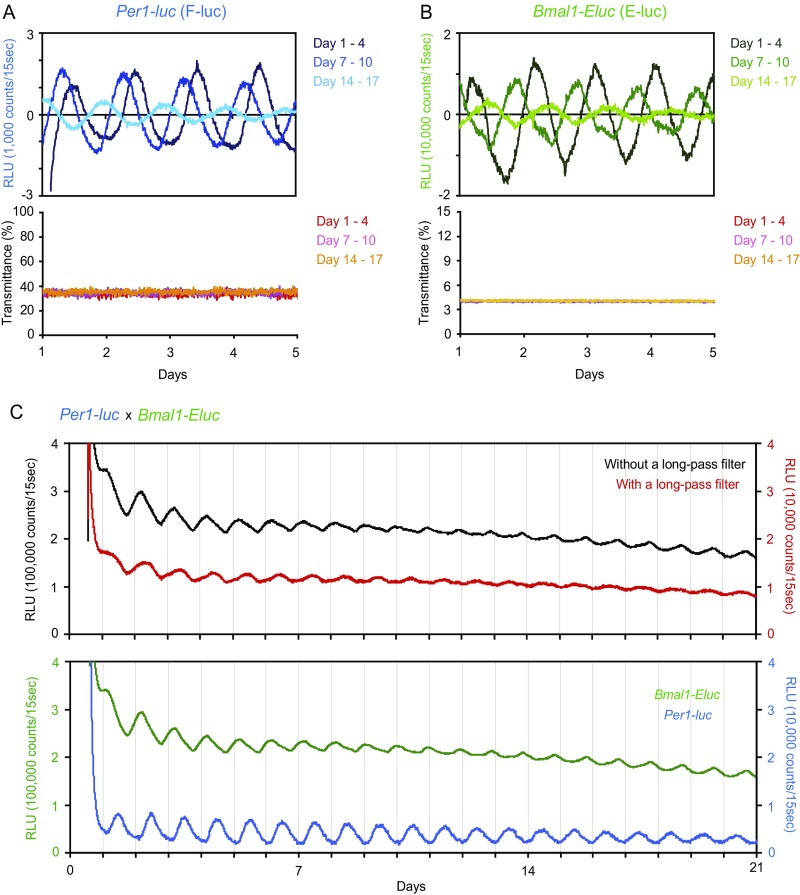
Fig. S2.
Separation of F-luc and E-Luc bioluminescence from the SCN slice. (A and B) Per1-luc (A) or Bmal1-ELuc (B) bioluminescence from a cultured SCN slice from a single-transgenic mouse (Upper) and the percentage of transmittance via a long-pass filter (Lower). Data during days1–4, 7–10, and 14–17 are shown in different colors. Transmittance of Per1-luc and Bmal1-ELuc through a long-pass filter was kept constant during culture for at least 17 d. (C, Upper) Time-series data of bioluminescence intensity in the cultured SCN from a double-transgenic mouse (Per1-luc/Bmal1-ELuc) with or without a long-pass filter. (Lower) Data after separation of Per1-luc and Bmal1-ELuc.
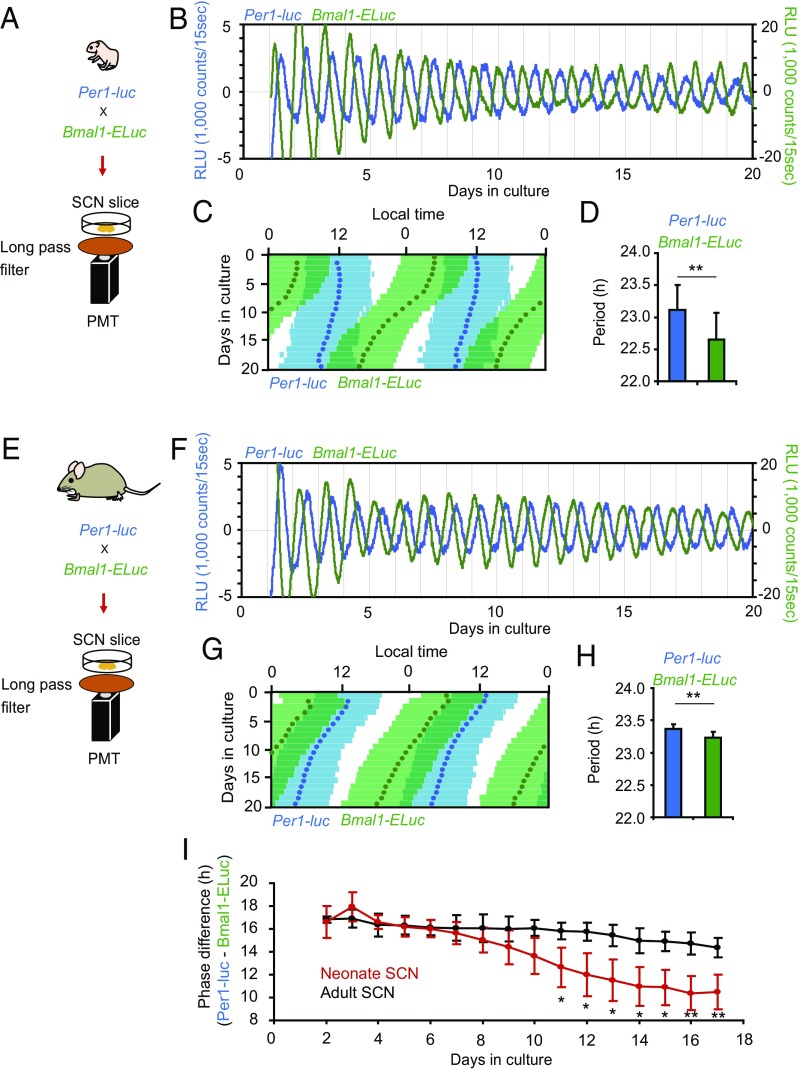
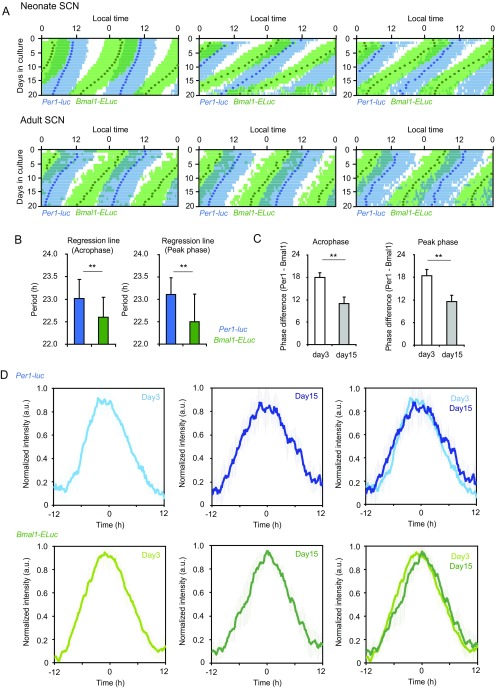
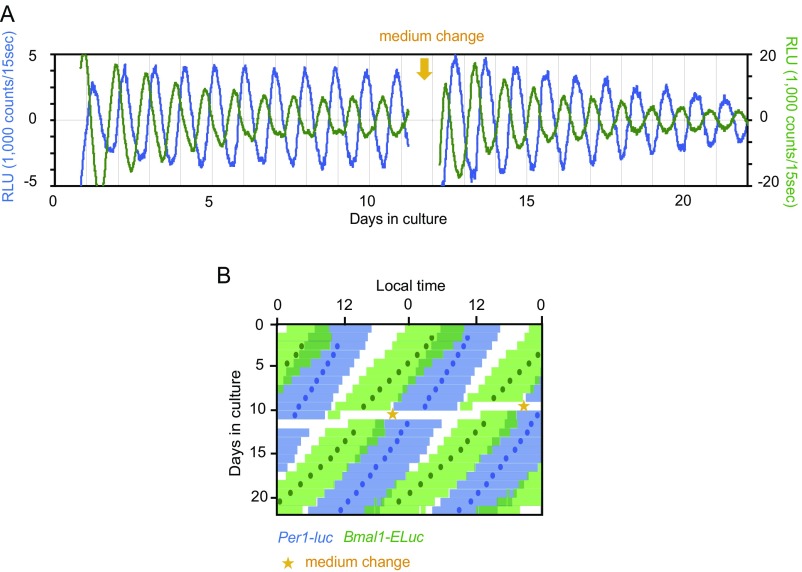
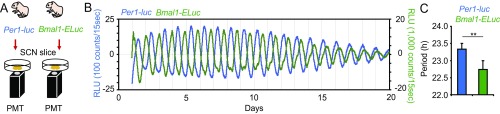
In the neonatal SCN (Fig. 3 A–C), double-plotted circadian rhythms in Per1-luc and Bmal1-ELuc showed internal dissociation, with a pattern similar to relative coordination in which the phase relation of two rhythms changed continuously in the course of free running (23). Similar patterns also were detected in other neonatal SCN slices (Fig. S3A). The mean circadian period of Bmal1-ELuc mice was significantly shorter than that of Per1-luc mice. The circadian period determined by χ2 periodogram was 22.7 ± 0.4 h for Bmal1-ELuc and 23.1 ± 0.4 h for Per1-luc; these periods were significantly different (n = 9, paired t test, P < 0.01) (Fig. 3D). The periods determined by other methods, such as a linear regression line fitted to the consecutive cycle peaks, confirmed the difference (Fig. S3B). As a result, the phase relation between the Per1-luc and Bmal1-ELuc circadian rhythms changed gradually and significantly during culturing (Fig. 3C and Fig. S3A), regardless of the circadian phase marker used [i.e., the acrophase of a fitted cosine curve (Fig. 3I and Fig. S3C, Left) or the peak of a detrended circadian rhythm (Fig. S3C, Right)]. To know whether the changes in the phase relationship between the two circadian rhythms reflected gradual and systematic alterations of the rhythm shape, we compared the shape of rhythmicity on day 3 and day 15 of culture and did not find any difference (Fig. S3D). In addition, the skewness of circadian rhythm was not significantly different between day 3 and day 15 for either Per1-luc or Bmal1-ELuc. Furthermore, to exclude a possible effect of rhythm damping during culture on the determination of rhythm phase, we enhanced the rhythm amplitude by replacing the culture medium with fresh medium; this replacement is known to increase the robustness of rhythmicity (Fig. S4). However, because the medium exchange also is known to shift the circadian rhythm of the neonatal SCN depending on the time of replacement (24), we selected the time to minimize the phase shifts. The phase relation between the two rhythms was kept essentially as it had been after medium exchange, and there was no systematic relation between the rhythm amplitude and phase difference.
Fig. 3.
Simultaneous measurement of Per1-luc and Bmal1-ELuc rhythms in the cultured SCN slice. (A and E) Experimental schemes of simultaneous measurement of Per1-luc and Bmal1-ELuc expression in neonatal (A) and adult (E) SCN slices. (B, C, F, and G) Sequential plots and double plots of Per1-luc (blue) and Bmal1-ELuc (green) circadian rhythms in a neonatal (B and C) and an adult (F and G) SCN slice in culture. In double-plotting, the time zone in which the bioluminescence is higher than the mean value of detrended data in a series is indicated by colored horizontal bars. Colored circles in a double plot (C and G) are acrophases. (D and H) Mean circadian periods calculated by χ2 periodogram (± SD) (21 d) of Per1-luc and Bmal1-ELuc in the neonatal (n = 9) (D) and adult (n = 5) (H) SCN slices are indicated by colored bars. Double asterisks (**) indicate a statistically significant difference between Per1-luc and Bmal1-ELuc (P < 0.01, paired t test). (I) Mean daily phase differences in terms of acrophase between Per1-luc and Bmal1-ELuc circadian rhythms in the neonatal (red, n = 9) and adult (black, n = 5) SCN slices in culture for 17 d. Two-way repeated-measure ANOVA revealed significant differences between the neonatal and adult SCN; **P < 0.01, *P < 0.05 (post hoc t test).
Fig. S3.
Other examples of simultaneous measurement of Per1-luc and Bmal1-ELuc rhythms in the cultured SCN slice. (A) Three additional examples of dissociation between Per1-luc and Bmal1-ELuc rhythms in neonatal (Upper) and adult (Lower) SCN slices. The circadian rhythms are double-plotted. The time zone in which bioluminescence is larger than the mean of detrended data (0 value in the ordinate) is indicated together with acrophases by blue bars and circles, respectively, for Per1-luc and green blue bars and circles, respectively, for Bmal1-ELuc. (B) The mean circadian periods ± SD for Per1-luc and Bmal1-ELuc rhythms calculated by the slope of a regression line fitted to consecutive acrophases (Left) or to daily peak phases (Right). (C) The mean phase differences ± SD in terms of acrophase (Left) and peak phase (Right) between Per1-luc and Bmal1-ELuc rhythms in the cultured SCN slice. (D) Normalized circadian rhythms of Per1-luc or Bmal1-ELuc in the SCN slices on day 3 and day 15 of culture. Data are shown as the mean ± SD. There was no significant difference, in terms of the skewness, in the shapes of Per1-luc and Bmal1-ELuc circadian rhythm in the two stages of culture. Double asterisks indicate a statistically significant difference (P < 0.01, paired t test).
Fig. S4.
Effects of medium exchange on rhythm amplitude and the phase relation between Per1-luc and Bmal1-ELuc rhythms. (A) Representative Per1-luc (blue) and Bmal1-ELuc (green) rhythms in a neonatal SCN slice before and after medium exchange. Medium was replaced with fresh medium at the time indicated by the yellow arrow. (B) Double-plotted rhythms. The time zone in which bioluminescence is larger than the mean of detrended data (0 value in the ordinate) is indicated together with the acrophases by blue bars and circles, respectively, for Per1-luc and green bars and circles, respectively, for Bmal1-ELuc. Medium exchange was done at the time indicated by a yellow star.
A difference in the circadian period of the two clock gene rhythms also was detected in the adult SCN slice (Fig. 3 E–H and Fig. S3). The circadian period of Per1-luc rhythm was 23.4 ± 0.1 h, and that of Bmal1-ELuc rhythm was 23.2 ± 0.1 h (n = 5), a significant difference (paired t test, P < 0.01) (Fig. 3H). However, the amount of dissociation in the adult SCN was less robust, and the phase difference between the two rhythms after day 11 of culture was significantly smaller in the adult than in the neonatal SCN (P < 0.01, two-way repeated-measure ANOVA) (Fig. 3I).
Spontaneous Firing Rhythms in the Cultured SCN Are Dissociated from Per1 and Bmal1 Oscillations.
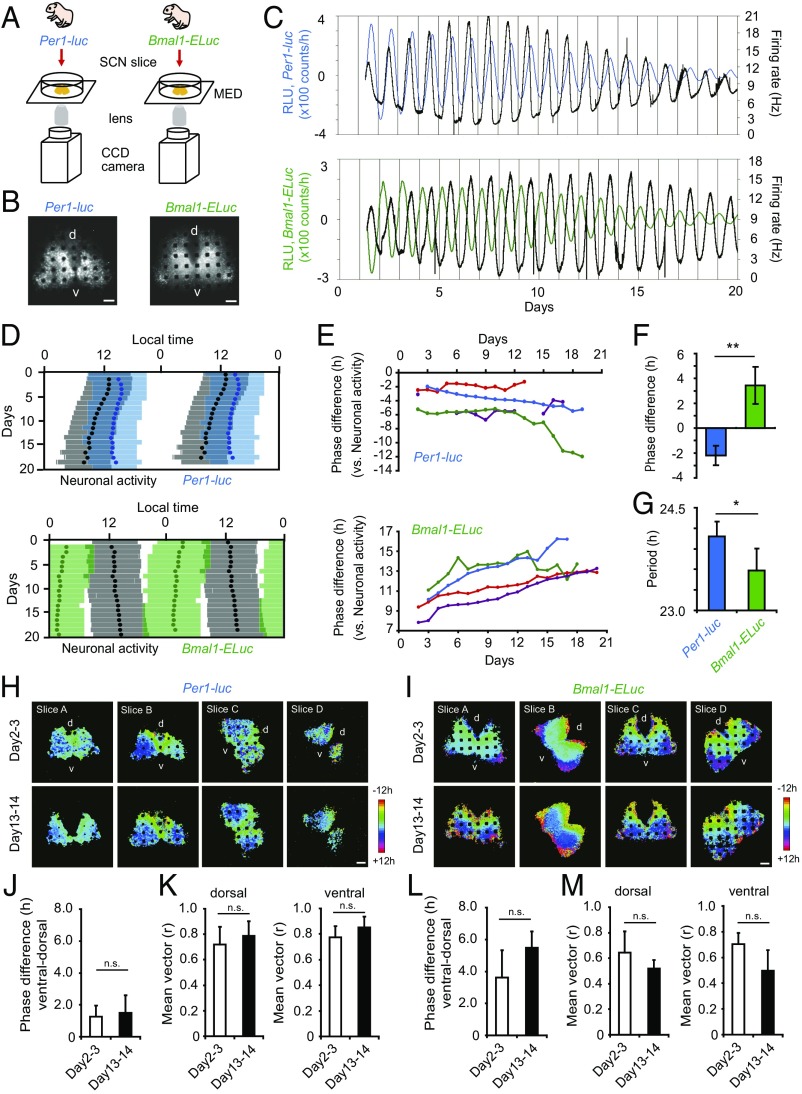
Spontaneous firing is regarded as an important output signal from a core loop in the SCN (25, 26) and is well correlated with behavioral circadian rhythms (27). However, the mechanism by which spontaneous firings in the SCN regulate behavioral circadian rhythms remains unknown. To understand the relationship between the circadian firing rhythm and activity onset or offset, or between spontaneous firing and clock gene expression in the SCN, we measured Per1-luc or Bmal1-ELuc expression simultaneously with spontaneous firing in the neonatal SCN slices using a multielectrode array dish (MED) and a CCD camera (Fig. 4 A and B and Fig. S5 A and B). For this experiment, we used single-transgenic mice carrying a bioluminescence reporter for Per1-luc or Bmal1-ELuc expression. The characteristics of the bioluminescent circadian rhythms in these single-transgenic mice were similar to those in double-transgenic mice (Per1-luc, 23.3 ± 0.1 h, n = 7; Bmal1-ELuc, 22.7 ± 0.3 h, n = 7; Student’s t test, P < 0.01) (Fig. S6).
Fig. 4.
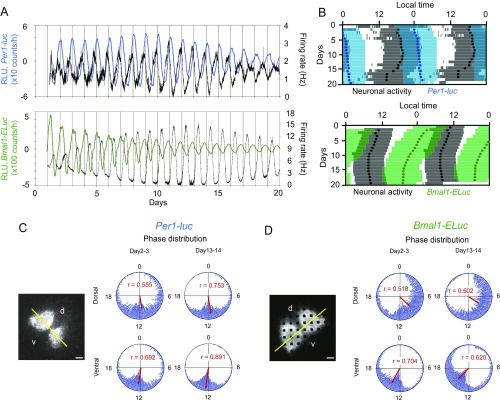
Simultaneous measurement of Per1-luc and Bmal1-ELuc together with spontaneous firing in the cultured SCN. (A) Experimental scheme of simultaneous measurement of Per1-luc, Bmal1-ELuc, and spontaneous firing in the neonatal SCN. (B) Bioluminescence images of Per1-luc (Left) and Bmal1-ELuc (Right) on a MED probe. (Scale bars: 100 µm.) (C) Sequential plots of Per1-luc (Upper, blue) and Bmal1-ELuc (Lower, green) rhythms with firing rhythm (black) from the entire area of the SCN. (D) Double-plotted circadian rhythms of Per1-luc (Upper, blue) and Bmal1-ELuc (Lower, green) with respective acrophases (closed circles) are illustrated together with double-plotted circadian rhythms of spontaneous firing (gray bars) with acrophase (black circles). The colored zones indicate the time when bioluminescence was higher than the mean value of detrended data in a series. Also see the legend of Fig. 3. (E) Daily acrophase differences in Per1-luc (Upper) or Bmal1-ELuc (Lower) rhythms and firing rhythms in four SCNs; SCNs are shown in different colors. One SCN partially lacks firing data. (F) Phase difference (mean ± SD) between days 13–14 and days 2–3 in culture in the Per1-luc (Left) and Bmal1-ELuc (Right) circadian rhythms. Negative values indicate phase delay, and positive values indicate phase advance (**P < 0.01, Student’s t test). (G) Circadian period determined by χ2 periodogram (mean ± SD) using records of Per1-luc and Bmal1-ELuc at day 14 in culture (*P < 0.05, Student’s t test). (H and I) Acrophase maps of Per1-luc (H) and Bmal1-ELuc (I) at days 2–3 (Upper) and 13–14 (Lower) in culture (n = 4). The mean acrophase was adjusted to 0 h. The color key indicates phase distribution in hours. (J–M) The phase difference (mean ± SD) in Per1 (J) or Bmal1 (L) circadian rhythms on the pixel level between the ventral and dorsal regions and the length of the mean vector of the respective rhythm (K and M) in the dorsal and ventral region at days 2–3 and 13–14 in culture.
Fig. S5.
Other examples of simultaneous measurement of Per1-luc and Bmal1-ELuc together with spontaneous firing in the cultured SCN. (A) Sequential plots of Per1-luc (blue, Upper) and Bmal1-ELuc (green, Lower) circadian rhythms together with spontaneous firing rates (black). (B) Double plots of Per1-luc (Upper) and Bmal1-ELuc (Lower) circadian rhythms together with circadian firing rhythms. The time zone in which bioluminescence or firing rates are larger than the mean values of detrended data in a series is indicated by colored bars with respective acrophases. (C and D) Bioluminescence images of Per1-luc (C) and Bmal1-ELuc (D) in the SCN slice. Yellow lines indicate the border between the dorsal and ventral regions in the SCN. Rayleigh plots of acrophase of circadian rhythms in the dorsal (Upper) and ventral (Lower) SCN on days 2–3 (Left) and 13–14 (Right) of culture. The mean vector r is indicated inside the Rayleigh plots.
Fig. S6.
Per1-luc and Bmal1-ELuc rhythms in the cultured neonatal SCN slice. (A) Experimental schemes of Per1-luc or Bmal1-ELuc measurement in the cultured SCN slice. (B) Sequential plots of the Per1-luc and Bmal1-ELuc circadian rhythms of neonatal SCN slices from different mice. Blue and green indicate Per1-luc and Bmal1-ELuc, respectively. (C) Bars indicate the mean circadian periods ± SD of Per1-luc (n = 7) and Bmal1-ELuc (n = 7). The period was measured by χ2 periodogram. Double asterisks indicate a statistically significant difference between Per1-luc and Bmal1-ELuc (P < 0.01, Student’s t test).
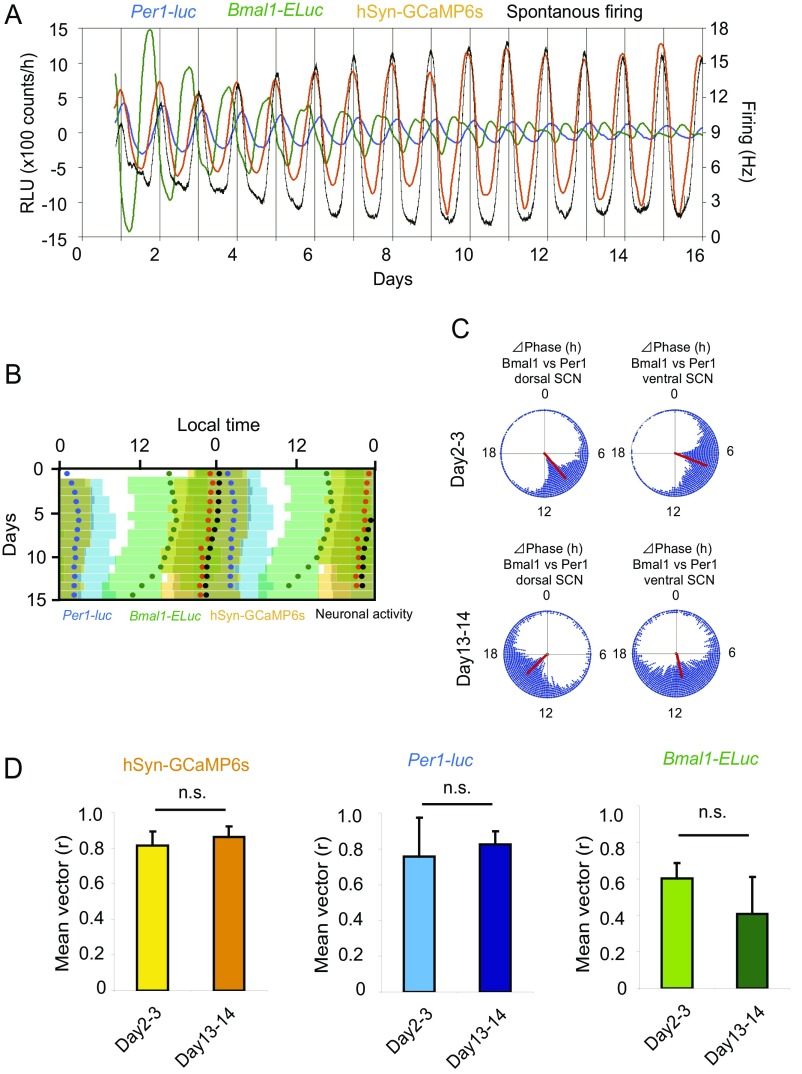
The circadian peak of Per1-luc rhythm in the neonatal SCN slice on the MED was gradually phase delayed relative to that of the spontaneous firing rhythms (Fig. 4 C–E). On the other hand, the circadian peak of Bmal1-ELuc rhythm was gradually phase advanced relative to that of the firing rhythms (Fig. 4 C–E). The phase difference between the firing rhythms and Per1 or Bmal1 expression rhythms was significantly larger at day 15 of culture than that at the beginning of culture (Student’s t test, P < 0.01) (Fig. 4F). The circadian periods of Per1-luc and Bmal1-ELuc rhythms also were significantly different (Student’s t test, P < 0.05) (Fig. 4G). These results indicate that the firing rhythms and Per1 or Bmal1 rhythms in the cultured SCN are dissociable and that the firing rhythms are not a direct consequence of the circadian oscillation of either the core feedback loop or the interlocked Bmal1 loop.
Spatiotemporal Features of Per1 and Bmal1 Expression in the Cultured SCN.
To know whether the dissociation between the Per1 and Bmal1 circadian rhythms is caused by region-specific gene expression in the SCN, we analyzed the spatiotemporal features of Per1-luc and Bmal1-ELuc expression in the SCN during culturing using an automated rhythm analysis with time-series images on the pixel level (28). In both Per1-luc and Bmal1-ELuc rhythms, the mean circadian phase on day 2 or 3 of culture was significantly delayed in the ventral region relative to the dorsal region, by ca. 1 h for Per1-luc (paired t test, P < 0.05) and by ca. 3 h for Bmal1-ELuc (paired t test, P < 0.05) (Fig. 4 J and L). The phase relation between the dorsal and ventral regions was not changed on day 13 or 14 of culture in either the Per1-luc or the Bmal1-ELuc rhythm (paired t test, Per1-luc, P = 0.841; Bmal1-ELuc, P = 0.932) (Fig. 4 J and L). We also analyzed the distribution of circadian peaks in the dorsal and ventral regions of the SCN using Rayleigh plots (Fig. 4 K and M and Fig. S5 C and D). The mean vector of the circadian rhythm was not changed in the dorsal and ventral regions of the SCN during culturing for either Per1-luc (Fig. 4K) or Bmal1-ELuc (Fig. 4M), suggesting that the internal synchrony of cellular circadian rhythms remained unchanged during culture. These results indicate that the dissociation between Per1-luc and Bmal1-ELuc rhythms is not caused by a regional difference in the SCN neural network.
Simultaneous Measurement of Circadian Per1, Bmal1, Calcium, and Spontaneous Firing in the Cultured SCN.
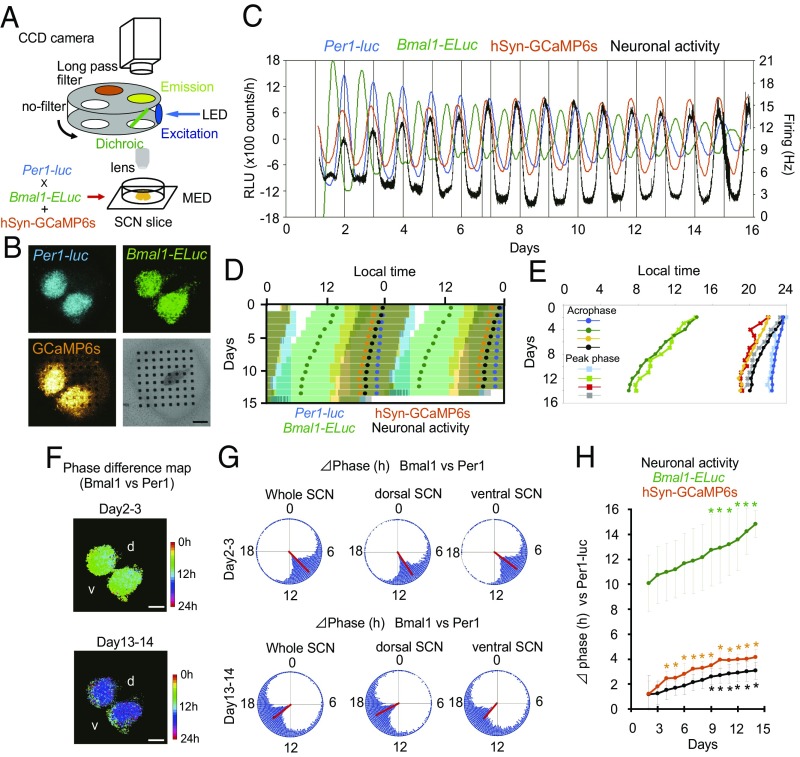
To understand further the relationships among the circadian firing rhythm, the core feedback loop, and the interlocked Bmal1 loop, we simultaneously measured Per1 and Bmal1 gene expression, spontaneous firing, and the intracellular calcium level from single cultured SCN slices. The SCN of double-transgenic mice (Per1-luc and Bmal1-ELuc) expressing the calcium indicator GCaMP6s was cultured on a MED probe, and the bioluminescence of firefly (F)-luc for Per1-luc and of E-Luc for Bmal1-ELuc, the fluorescence of GCaMP6s, and spontaneous firing were monitored (Fig. 5 A and B, Fig. S7, and Movie S1). In addition to an endogenous circadian oscillation as an output of the core feedback loop, intracellular calcium in the SCN is known to be controlled by the input signals from the SCN neural network (29, 30). Plotting of the acrophase or of the circadian peak in each cycle revealed that the circadian rhythms in Bmal1-ELuc, spontaneous firing, and intracellular calcium were gradually phase advanced relative to the Per1-luc circadian rhythms in the course of culturing (one-way repeated-measure ANOVA, P < 0.05 and post hoc Tukey–Kramer test) (Fig. 5 C–H). Interestingly, the calcium circadian rhythm was essentially phase-locked to the circadian firing rhythms, keeping a stable phase difference of about 1 h for more than 10 d (Fig. 5 D and H). Thus, the phase relations of circadian rhythms in Per1-luc, Bmal1-ELuc, and firing changed gradually during culturing. Although the phase relations among these circadian rhythms changed substantially during culture, the phase relation of the Per1-luc and Bmal1-ELuc circadian rhythms (Δphase) in the dorsal and ventral SCN regions did not change (day 2–3, 1.7 ± 0.3 h; day13–14, 2.1 ± 0.9 h; paired t test, P = 0.283) (Fig. 5 F and G and Fig. S7C), suggesting that the change in the phase relationship of the two rhythms occurred at the same rate in both regions. The mean vector in these circadian rhythms was not different between day 2–3 and day 13–14, again suggesting that the internal synchrony of cellular circadian rhythms was maintained during culture (Fig. S7D). On the other hand, the mean vector of the Bmal1-ELuc circadian rhythm was slightly but significantly shorter than those of the other circadian rhythms, suggesting differential internal synchrony and/or sloppiness of the Bmal1-ELuc circadian rhythms.
Fig. 5.
Simultaneous measurement of Per1-luc, Bmal1-ELuc, intracellular calcium, and spontaneous firing in the cultured SCN slice. (A) Experimental scheme of simultaneous multifunctional measurement in the neonate SCN slice. (B) Bioluminescence images of Per1-luc (Upper Left) and Bmal1-ELuc (Upper Right), fluorescence images of GCaMP6s (Lower Left), and a bright field image (Lower Right) of a cultured SCN slice on a MED probe. (Scale bars: 200 µm.) (C) Sequential plots of circadian Per1-luc, Bmal1-ELuc, GCaMP6s, and firing rhythms from the entire area of the SCN. Spontaneous firing was expressed as the mean firing rate from electrodes covered by bilateral SCN. (D) Double plots of circadian rhythms of four measures. Colored circles are acrophases of circadian Per1-luc, Bmal1-ELuc, GCaMP6s, and firing rhythms. Yellow bars indicate the time zone in which GCaMP6s fluorescence is higher than the mean value of detrended data in a series. Also see the legend of Fig. 4D. (E) Longitudinal plotting of daily acrophases and of cycle peaks of the four circadian rhythms demonstrated in C. The features of free running are almost identical regardless of the phase marker. (F) Phase-difference maps (Per1-luc vs. Bmal1-ELuc) at days 2–3 (Upper) and 13–14 (Lower) in culture. The color key indicates the phase difference in hours. (G) Rayleigh plots of phase difference on the pixel level between Per1-luc and Bmal1-ELuc in the whole (Left), dorsal (Center), and ventral (Right) SCN are shown for days 2–3 (Upper) and 13–14 (Lower) in culture. A red line in a Rayleigh circle indicates the mean phase. (H) The mean daily phase difference, in terms of acrophase, between the Per1-luc and three other circadian rhythms (n = 3). (*P < 0.05, vs. day 2, one-way repeated-measure ANOVA with post hoc Tukey–Kramer test).
Fig. S7.
Another example of simultaneous measurement of Per1-luc, Bmal1-ELuc, intracellular calcium, and spontaneous firing in the cultured SCN slice. (A) Sequential plots of circadian Per1-luc, Bmal1-ELuc, GCaMP6s, and firing rhythms in a cultured SCN slice. Spontaneous firing was expressed as the mean firing rate at the electrodes covered by the bilateral SCN. (B) Double plots of the circadian rhythms of four measures. The time zone in which respective measure is larger than the mean of detrended data in a series is indicated by a colored bar: Per1-luc (blue), Bmal1-ELuc (green), GCaMP6s (orange), neural activity (gray). Circles of the respective colors are acrophases of circadian rhythm. (C) Rayleigh plots indicate the distribution of phase difference between Per1-luc and Bmal1-ELuc from the dorsal (Left) and ventral (Right) SCN at days 2–3 (Upper) and days 13–14 (Lower) in culture. The direction and length of a red line in the circle indicates the mean phase difference and its stability. (D) The mean vector r of acrophase for the circadian rhythms of calcium (Left), Per1-luc (Center), and Bmal1-ELuc (Right) on days 2–3 and days 13–14 of culture (n = 3). The mean vector was not significantly different between different days in culture but was significantly different in the circadian rhythms examined [two-way ANOVA; days, not significant (n.s.); rhythms, P < 0.01].
Discussion
In the present study we found that the Per1-luc and Bmal1-ELuc circadian rhythms were transiently dissociated when freely moving mice under DD experienced a phase-shifting light pulse. The gene-expression rhythms were differentially associated with the activity onset and offset of behavioral circadian rhythm. Similar dissociation of Per1 and Bmal1 oscillation was observed in the cultured SCN slices of mice carrying a dual reporter system. Such a transient dissociation between the circadian rhythms was detected only by continuous and simultaneous measurement of multiple rhythms from a single SCN. The dual reporter system used in the present study enabled us to identify the change in the phase relation of two different rhythms. However, because the method is based on luciferin–luciferase reaction and the separation of emitted light by filtering, these findings should be confirmed by more direct measurement of the transcription or translation of clock genes in future. Most of the current direct methods are still difficult to apply because of insufficient time resolution resulting from individual differences and use of a large number of animals.
Interestingly, the circadian rhythm in spontaneous firing was dissociated from the Per1 and Bmal1 circadian rhythms but not from the circadian rhythm of the intracellular calcium level. These findings indicate that the Bmal1 loop has an oscillatory nature similar to that of the core loop and behaves as a constitutional subunit of the circadian system in the SCN.
We demonstrated the dissociation of circadian rhythms in clock gene expression of freely moving mice when the circadian rhythms were on the way to steady-state free running after the light pulse (Fig. 1). A similar dissociation of the circadian rhythms of clock gene expression has been observed previously with shifts in LD schedules (31, 32), but the relevance to behavioral outputs was not clear. Importantly, the phase shifts of Per1 and Bmal1 rhythms in our study were closely associated with the phase shifts of either the activity onset or offset of behavioral rhythm. Previously, Vansteensel et al. (33) reported the dissociation between the Per1-luc circadian rhythm in the SCN and behavioral rhythms that occurred following a shift in the LD cycle. The finding raised the possibility that the Per1-luc circadian rhythm reports only a subset of SCN neurons. In agreement with this report, the present findings suggest not only the existence of two coupled circadian oscillations with different molecular mechanisms but also their differential association with the activity onset and offset of behavioral rhythms. According to Pittendrigh’s hypothesis (13), activity onset and offset of nocturnal rodents are regulated by two different oscillators, the E and M oscillator, respectively. The hypothesis was based on the findings of rhythm splitting under constant light and differential responses to light (12). In agreement with the above hypothesis, Honma et al. (11) demonstrated in rats that activity onset and offset showed different phase responses to a single light pulse. Thus, the Per1-luc circadian rhythm in the SCN seems to associate with the E oscillator, and the Bmal1-ELuc circadian rhythm seems to associate with the M oscillator. Previous reports demonstrated separate regional pacemakers which behaved differently under a long and a short photoperiod in the SCN (14, 15). The pacemaker corresponding to the E oscillator is likely located in the rostral part of SCN, and that corresponding to the M oscillator is likely located in the caudal part. The present findings further suggest that the two circadian oscillations have different molecular mechanisms.
Dissociation between the Per1 and Bmal1 oscillation also was observed in the SCN ex vivo (Figs. 3–5). Because of a significant difference in the circadian period, the phase relation between the Per1 and Bmal1 oscillation changed gradually during culturing; this change was not the result of systematic changes in the shapes of circadian rhythms (Fig. S3 and Fig. S7). The core and interlocked Bmal1 loop have been regarded as the origin of the circadian rhythms of Per1 and Bmal1 expression (3–5). The present results indicate that the two loops are coupled to each other but exhibit ongoing differences in period and relative phase that suggest they may oscillate independently. They are dissociable by a light pulse in vivo and by prolonged free running ex vivo. The theoretical backgrounds of the present findings are a coupling of two oscillating feedback loops and a dependency of the circadian period on the relative strength of interlocked loops as formulated in another associated loop (34). In addition, the relative coordination between of Per1-luc and Bmal1-ELuc rhythms suggests that the core loop and Bmal1 loop are not independent but are mutually interactive even under dissociation. The Bmal1 loop consists of a positive-feedback component of ROR and a negative-feedback component of RevErbα for Bmal1 expression and potentially is capable of oscillating without oscillation of the core loop, at least in theory (35).
The circadian oscillation in the dorsomedial region of the SCN was reported to be faster than that in the ventrolateral region (36). Regional differences also were reported in the intensity of clock gene expression (37). Therefore the dissociation between the two clock gene rhythms could be ascribed to regional differences in circadian oscillation. However, in the present study, the phase relation of the Per1 and Bmal1 expression rhythms was not different in the dorsal and ventral SCN during culturing, bringing into question the possibility that regional differences in respective circadian oscillation might be a cause of dissociation. Recently, we found sharply differentiated clusters of cells that expressed the PER2 circadian rhythm with different periods in the SCN of Cry1,2 double deficient mice (38). The cells in each of these cell clusters showed no regional specificity but instead were distributed diffusely. The finding suggested that in the circadian system of the SCN a specific type of oscillating cell builds up a constitutional oscillator that does not necessarily show regional specificity. The Per1- and Bmal1-specific oscillator cells could be located diffusely throughout the SCN. Alternatively, the dissociation of two clock gene rhythms could be caused by the dissociation of the core loop and the Bmal1 loop in single SCN cells. If so, within a single cell, the core molecular loop would be responsible for the activity onset and the Bmal1 loop would be responsible for the activity offset of behavioral rhythm. In this case, we should assume different output signals from a single cell. In any case, the coupling of two oscillations was less strong in the neonatal SCN than in the adult SCN, suggesting a developmental change in the SCN circadian system.
We had expected that the circadian firing rhythm in the SCN slice might synchronize with either of the two rhythms of clock gene expression or show a splitting into two components. Contrary to our expectation, the simultaneously determined circadian firing rhythms dissociated from both of the circadian gene-expression rhythms (Figs. 4 and 5). However, a coupling between the circadian firing and intracellular calcium rhythms persisted for at least for 2 wk in culture, suggesting that the two overt rhythms are regulated by the same oscillatory mechanism, which could be neither the core loop nor the interlocked Bmal1 loop. The cellular circadian rhythms in the SCN are likely regulated by two circadian oscillations of different origins: the intracellular molecular feedback loop and the SCN network in which the cell is involved. The relative intensities of the two oscillations may determine the nature of the cellular circadian rhythms examined. Circadian firing and calcium rhythms would be affected by feedback loops and the SCN network (39). A strong coherence of the circadian firing rhythm to the intracellular calcium rhythm could be interpreted in terms of causality rather than oscillatory coupling. Neither Ca2+ nor firing rhythm in the SCN is associated directly with the onset or offset of circadian behavioral rhythm. The present findings indicate that the links between the SCN circadian rhythms and behavior are more complicated than previously thought (40).
Taken together, the present findings indicate that external perturbation, such as a single light pulse, transiently uncouples interlocked molecular loops that are separately associated with the onset and offset of behavioral rhythms.
Materials and Methods
Animals.
Male and female Per1-luc (heterozygous or homozygous) (14) and Bmal1-ELuc (heterozygous) (41) reporter mice of C57BL/6j background were used. Per1-luc and Bmal1-ELuc mice were produced in the YS New Technology Institute and the National Institute of Advanced Industrial Science and Technology, respectively, by injecting the reporter construct into fertilized eggs of C57BL/6j background. We back-crossed these mice with C57BL/6j background mice for more than seven generations. Mice were reared in our animal quarters where environmental conditions were controlled (lights-on 6:00–18:00; light intensity ∼100 lx at the cage bottom; humidity 60 ± 10%). Mice had free access to food pellets and water. Experiments were conducted in compliance with the rules and regulations established by the Animal Care and Use Committee of Hokkaido University under the ethical permission of the Animal Research Committee of Hokkaido University (approvals no. 13-0053 for in vivo experiments and no. 13-0064 for ex vivo experiments).
Measurement of Behavioral Activity.
Mice (nine males, four females) were used in the in vivo study at age 2–6 mo. Mice were individually housed in a polycarbonate cage (115 mm wide × 215 mm long × 300 mm high), which was placed in a light-tight and air-conditioned box (40 × 50 × 50 cm). An LED light source with an intensity of 150–250 lx on the ceiling of a box was turned on when a light pulse was given after 5 d exposure to DD. Spontaneous movements were measured by a passive infrared sensor, which detects a change in thermal radiation from an animal caused by body movements (42). The amount of behavioral activity was recorded automatically every minute by a computer (The Chronobiology Kit; Stanford Software System).
Surgery.
Surgery was performed under isoflurane anesthesia as previously described (18, 19). To measure bioluminescence from the SCN in vivo, a handmade guide cannula (i.d. 1.12 mm; o.d. 1.48 mm) was stereotaxically inserted into the brain 0.2 mm posterior to the bregma, 0.2 mm lateral from the midline, and 3.0 mm from the surface of the skull and was fixed to the skull with dental resin. After a recovery period of more than 4 d, a polymethyl methacrylate optical fiber (fiber diameter, 0.5 mm; surface cladding, 0.25 mm thick) was inserted into the guide cannula aimed at the SCN at a 5.8-mm depth from the surface of the skull and was fixed to the skull with dental resin.
More than 4 d after the insertion of the optical fiber, an osmotic pump (model 2002; Alzet; flow speed, 0.5 µL/h, pump volume, 200 µL) containing d-luciferin, a substrate of luciferase, was implanted in the peritoneal cavity. To deliver the substrate, the osmotic pump was filled with d-luciferin K (100 mM) dissolved in physiological saline. After each surgery, penicillin-G (40 units/g of body weight) was administered i.m. to prevent infection. Aspirin (120 mg/kg of body weight, administered orally) or buprenorphine (0.05–0.1 mg/kg of body weight, administered by s.c. injection) was used for postoperative analgesia.
In Vivo Measurement of Bioluminescence.
Three to five days after the implantation of the osmotic pump, bioluminescence from the SCN was measured in freely moving mice under DD. The measurement was performed every minute via an optical fiber connected to a photon-counting device (In vivo Kronos; Atto) (18, 19) equipped with a photomultiplier tube (Hamamatsu Photonics). Recorded data were fed into a computer and analyzed.
Histological Examination.
Once the measurements were completed, mice were anesthetized with ether and were intracardially perfused with physiological saline followed by 4% paraformaldehyde in 0.1 M phosphate buffer (PB). Brains were cryoprotected with 20% sucrose in 0.1 M PB and stored at −80 °C. Serial 30-μm-thick coronal sections of the brain were made using a Leica cryostat and were stained with cresyl violet to identify the placement of the tip of the optical fiber.
Preparation of SCN Slices for Culture.
To measure the bioluminescence from the SCN slice, mice were killed to harvest the SCN between 8:00 and 16:00 h. To measure the level in tissue, 300-μm-thick coronal slices from the SCN of neonatal (postnatal day 7) mice were made with a tissue chopper (Mcllwain), and slices from the SCN of adult (2- to 6-mo-old) mice were made with a microslicer (DTK-1000; Dosaka EM). The SCN tissue was dissected at the mid-rostrocaudal region, and paired SCN slices were cultured on a Millicell-CM culture insert (Millipore Corporation) under culture conditions as described previously (43). Briefly, the slices were cultured in air at 36.5 °C with 1.2 mL DMEM (Invitrogen) with 0.1 mM d-luciferin K and 5% supplement solution.
Measurement of Bioluminescence in SCN Tissue.
Bioluminescence in SCN tissue was measured using a Lumicycle (Actimetrics) or Kronos (Atto) PMT at 10-min intervals with an exposure time of 1 min. For the simultaneous measurement of Per1 and Bmal1 expression, the bioluminescence of Per1-luc and Bmal1-ELuc from the same SCN slice was monitored alternatively by a dish-type luminometer with a turntable for eight recording dishes (Kronos, Atto). Bioluminescence was measured for 15 s in the presence of a 600-nm long-pass filter (R60 filter; Hoya), then in the presence of a 560-nm long-pass filter (O56 filter; Hoya), and then without any filter. The measurement was repeated at 10-min intervals. The intensities of Per1-luc and Bmal1-ELuc bioluminescence were calculated as described previously (21, 22). For bioluminescence calculation, we used a 600-nm long-pass filter.
Simultaneous Recordings of Bioluminescence and Spontaneous Firing from an SCN Slice.
An SCN slice from a 3- to 5-d-old pup was cultured on a MED with 64 electrodes. The size of an electrode was 20 × 20 µm (MED-P210A). Spontaneous firings were recorded using a MED 64 system (Alpha MED Scientific). Spike discharges with a signal-to-noise ratio >2.0 were collected by Spike Detector software (Alpha MED Scientific) as described previously (43). The number of spikes per minute was calculated for each electrode covered by the SCN slice.
The MED was placed in a mini-incubator installed on the stage of a microscope (ECLIPSE TE2000-U or ECLIPSE E1000; Nikon). The culture conditions were as described previously (43). Bioluminescence was recorded with a CCD camera (ORCA-ІІ; Hamamatsu Photonics) cooled at −60 °C. The pixel size was 4.3 × 4.3 μm.
Simultaneous Recordings of Per1-luc, Bmal1-ELuc, Calcium, and Spontaneous Firing from an SCN Slice.
An SCN slice from a 3- to 5-d-old pup of Per1-luc and Bmal1-ELuc pup was cultured on a Millicell-CM culture insert. Aliquots of adeno-associated virus (AAV) serotype rh10 harboring GCaMP6s, a genetically encoded calcium sensor under the control of human synapsin-1 promotor (University of Pennsylvania Gene Therapy Program Vector Core), were inoculated onto the surface of the cultured SCN slice 3–5 d after slice preparation. On the day after AAV infection, the SCN slice was transferred onto the MED probe. Simultaneous measurement of bioluminescence, fluorescence, and neuronal activity began 7–10 d after the SCN was cultured.
The MED was placed in a mini-incubator installed on the stage of a microscope (ECLIPSE-80i; Nikon) equipped with an EM-CCD camera (ImagEM; Hamamatsu Photonics). A fluorescent calcium sensor (GCaMP6s) was excited at cyan color (475/28 nm) with an LED light source (Retra Light Engine; Lumencor) and was visualized with a 495-nm dichroic mirror and 520/35-nm emission filters (SemLock). To measure F-luc and E-Luc bioluminescence (Per1-luc and Bmal1-ELuc) separately, a long-pass filter (AT610, Chroma Technology Corporation) was used. Bioluminescence with or without the filter was measured every hour (exposure time, 29 min for each condition). Per1-luc and Bmal1-ELuc bioluminescence were calculated on the pixel level with the method used for PMT measurement.
Data Analysis.
For the analyses of behavioral rhythms in vivo, spontaneous movements obtained every minute were used. The number of phase shifts was determined on the basis of a double-plotted actograph by visual inspection. A regression line was fitted to the succeeding activity onsets or offsets during a steady-state free-run before and after the light pulse. The circadian period was determined by the slope of the regression line, and the amount of daily phase shift was calculated from the phase difference between the phase on the day of light pulse and that on the following day. Three chronobiology experts (D.O., S.H., and K.H.) participated in the visual inspection.
For the analyses of circadian bioluminescent rhythms in vivo and ex vivo, data obtained with a PMT were smoothed by a 4-h moving average for in vivo data and a 50-min moving average for ex vivo data. The smoothed data then were detrended by a 24-h moving average subtraction method (18, 19). Neuronal activity rhythms also were detrended by a 24-h moving average subtraction method. To compare the peak phases of circadian rhythms in vivo and ex vivo, we used an acrophase obtained by ClockLab software (Actimetrics) or simply the peak phase in a cycle. The circadian period in the SCN slice was calculated by a periodogram or from the slope of a regression line fitted to consecutive peak phases by the least-squares method. The difference in phase angle between two circadian rhythms was calculated using the acrophase of a best-fitted cosine curve or the peak of a circadian cycle. For time-lapse images obtained by a CCD camera, the properties of circadian rhythm in bioluminescence and fluorescence signals were analyzed on the pixel level using custom-made software based on a cosine curve-fitting method as described previously (39, 44). The dorsal and ventral areas of the SCN were separated by a line drawn at the midpoint between the dorsal and ventral edges so that it crossed the third ventricle at a right angle. A Rayleigh plot was made using the Oriana4 software (Kovach Computing Service). Data are expressed as mean ± SD unless otherwise indicated.
Statistics.
Student’s t test was used when two independent group means were compared. A paired t test was used when two dependent group means were compared. A one-way ANOVA with a post hoc Tukey–Kramer test was used to analyze data of a single time series. A two-way ANOVA with a post hoc t test was used when the data of two independent time series were compared (Statview or Statcel 3).
Supplementary Material
Acknowledgments
We thank H. Tei for providing the Per1-luciferase reporter plasmid; M. Shimogawara and H. Kubota for the development of an in vivo bioluminescence measurement system; K. Baba for technical advice; T. Ueda for the analysis program; M. Mieda for advice on AAV purification; M. P. Butler for thorough discussions; and the Genetically-Encoded Neuronal Indicator and Effector (GENIE) Project and the Janelia Farm Research Campus of the Howard Hughes Medical Institute for sharing GCaMP6s constructs. This work was supported in part by the Uehara Memorial Foundation; the Nakajima Foundation; the Project for Developing Innovation Systems of the Ministry of Education, Culture, Sports, Science and Technology (MEXT); the Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program, MEXT; and Japan Society for the Promotion of Science Grants in Aid for Scientific Research (KAKENHI) Grants 15H04679, 26860156, and 15K12763.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613374114/-/DCSupplemental.
References
- 1.Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- 2.Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci USA. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 4.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 5.Triqueneaux G, et al. The orphan receptor Rev-erbalpha gene is a target of the circadian clock pacemaker. J Mol Endocrinol. 2004;33:585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato TK, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Liu AC, et al. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shigeyoshi Y, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 9.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 10.Aschoff J, Hoffmann K, Pohl H, Wever R. Re-entrainment of circadian rhythms after phase-shifts of the Zeitgeber. Chronobiologia. 1975;2:23–78. [PubMed] [Google Scholar]
- 11.Honma K, Honma S, Hiroshige T. Response curve, free-running period, and activity time in circadian locomotor rhythm of rats. Jpn J Physiol. 1985;35:643–658. doi: 10.2170/jjphysiol.35.643. [DOI] [PubMed] [Google Scholar]
- 12.Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. V. Pacemaker structure: A clock for all seasons. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1976;106:333–355. [Google Scholar]
- 13.Pittendrigh CS. Circadian oscillation in cells and the circadian organization of multi-cellular systems. In: Schmitt FO, Worden FG, editors. The Neurosciences, Third Study Program. The MIT Press; Cambridge, MA: 1974. pp. 437–458. [Google Scholar]
- 14.Inagaki N, Honma S, Ono D, Tanahashi Y, Honma K. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci USA. 2007;104:7664–7669. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naito E, Watanabe T, Tei H, Yoshimura T, Ebihara S. Reorganization of the suprachiasmatic nucleus coding for day length. J Biol Rhythms. 2008;23:140–149. doi: 10.1177/0748730408314572. [DOI] [PubMed] [Google Scholar]
- 16.Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Dynamic interactions mediated by nonredundant signaling mechanisms couple circadian clock neurons. Neuron. 2013;80:973–983. doi: 10.1016/j.neuron.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myung J, et al. GABA-mediated repulsive coupling between circadian clock neurons in the SCN encodes seasonal time. Proc Natl Acad Sci USA. 2015;112:E3920–E3929. doi: 10.1073/pnas.1421200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ono D, Honma K, Honma S. Circadian and ultradian rhythms of clock gene expression in the suprachiasmatic nucleus of freely moving mice. Sci Rep. 2015;5:12310. doi: 10.1038/srep12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono D, Honma S, Honma K. Circadian PER2:LUC rhythms in the olfactory bulb of freely moving mice depend on the suprachiasmatic nucleus but not on behaviour rhythms. Eur J Neurosci. 2015;42:3128–3137. doi: 10.1111/ejn.13111. [DOI] [PubMed] [Google Scholar]
- 20.Comas M, Beersma DG, Spoelstra K, Daan S. Phase and period responses of the circadian system of mice (Mus musculus) to light stimuli of different duration. J Biol Rhythms. 2006;21:362–372. doi: 10.1177/0748730406292446. [DOI] [PubMed] [Google Scholar]
- 21.Noguchi T, Ikeda M, Ohmiya Y, Nakajima Y. Simultaneous monitoring of independent gene expression patterns in two types of cocultured fibroblasts with different color-emitting luciferases. BMC Biotechnol. 2008;8:40. doi: 10.1186/1472-6750-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noguchi T, et al. Dual-color luciferase mouse directly demonstrates coupled expression of two clock genes. Biochemistry. 2010;49:8053–8061. doi: 10.1021/bi100545h. [DOI] [PubMed] [Google Scholar]
- 23. Aschoff J (1981) Freerunning and entrained circadian rhythms. Handbook of Behavioral Neurobiology, ed Aschoff J (Plenum, New York), Vol 4, Biological Rhythms, pp 81–93.
- 24.Nishide SY, Honma S, Honma K. The circadian pacemaker in the cultured suprachiasmatic nucleus from pup mice is highly sensitive to external perturbation. Eur J Neurosci. 2008;27:2686–2690. doi: 10.1111/j.1460-9568.2008.06231.x. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz WJ, Gross RA, Morton MT. The suprachiasmatic nuclei contain a tetrodotoxin-resistant circadian pacemaker. Proc Natl Acad Sci USA. 1987;84:1694–1698. doi: 10.1073/pnas.84.6.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones JR, Tackenberg MC, McMahon DG. Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior. Nat Neurosci. 2015;18:373–375. doi: 10.1038/nn.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura W, et al. In vivo monitoring of circadian timing in freely moving mice. Curr Biol. 2008;18:381–385. doi: 10.1016/j.cub.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Yoshikawa T, et al. Spatiotemporal profiles of arginine vasopressin transcription in cultured suprachiasmatic nucleus. Eur J Neurosci. 2015;42:2678–2689. doi: 10.1111/ejn.13061. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda M, et al. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron. 2003;38:253–263. doi: 10.1016/s0896-6273(03)00164-8. [DOI] [PubMed] [Google Scholar]
- 30.Hastings MH, Maywood ES, O’Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol. 2008;18:R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Reddy AB, Field MD, Maywood ES, Hastings MH. Differential resynchronisation of circadian clock gene expression within the suprachiasmatic nuclei of mice subjected to experimental jet lag. J Neurosci. 2002;22:7326–7330. doi: 10.1523/JNEUROSCI.22-17-07326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiessling S, Eichele G, Oster H. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J Clin Invest. 2010;120:2600–2609. doi: 10.1172/JCI41192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vansteensel MJ, et al. Dissociation between circadian Per1 and neuronal and behavioral rhythms following a shifted environmental cycle. Curr Biol. 2003;13:1538–1542. doi: 10.1016/s0960-9822(03)00560-8. [DOI] [PubMed] [Google Scholar]
- 34.Yan J, et al. An intensity ratio of interlocking loops determines circadian period length. Nucleic Acids Res. 2014;42:10278–10287. doi: 10.1093/nar/gku701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uryu K, Tei H. Feedback loops interlocked at competitive binding sites amplify mammalian circadian oscillations. J. Chronobiol. 2016;22:155. doi: 10.1016/j.jtbi.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Koinuma S, et al. Regional circadian period difference in the suprachiasmatic nucleus of the mammalian circadian center. Eur J Neurosci. 2013;38:2832–2841. doi: 10.1111/ejn.12308. [DOI] [PubMed] [Google Scholar]
- 37.Evans JA, et al. Shell neurons of the master circadian clock coordinate the phase of tissue clocks throughout the brain and body. BMC Biol. 2015;13:43. doi: 10.1186/s12915-015-0157-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono D, Honma S, Honma K. Differential roles of AVP and VIP signaling in the postnatal changes of neural networks for coherent circadian rhythms in the SCN. Sci Adv. 2016;2:e1600960. doi: 10.1126/sciadv.1600960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enoki R, et al. Topological specificity and hierarchical network of the circadian calcium rhythm in the suprachiasmatic nucleus. Proc Natl Acad Sci USA. 2012;109:21498–21503. doi: 10.1073/pnas.1214415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramkisoensing A, Meijer JH. Synchronization of biological clock neurons by light and peripheral feedback systems promotes circadian rhythms and health. Front Neurol. 2015;6:128. doi: 10.3389/fneur.2015.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noguchi T, Ikeda M, Ohmiya Y, Nakajima Y. A dual-color luciferase assay system reveals circadian resetting of cultured fibroblasts by co-cultured adrenal glands. PLoS One. 2012;7:e37093. doi: 10.1371/journal.pone.0037093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abe H, et al. Clock gene expressions in the suprachiasmatic nucleus and other areas of the brain during rhythm splitting in CS mice. Brain Res Mol Brain Res. 2001;87:92–99. doi: 10.1016/s0169-328x(00)00295-3. [DOI] [PubMed] [Google Scholar]
- 43.Ono D, Honma S, Honma K. Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nat Communs. 2013;4:1666. doi: 10.1038/ncomms2670. [DOI] [PubMed] [Google Scholar]
- 44.Mieda M, et al. Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron. 2015;85:1103–1116. doi: 10.1016/j.neuron.2015.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.