Abstract
Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3 (APOBEC3) proteins are mammalian-specific cellular deaminases and have a robust ability to restrain lentivirus replication. To antagonize APOBEC3-mediated antiviral action, lentiviruses have acquired viral infectivity factor (Vif) as an accessory gene. Mammalian APOBEC3 proteins inhibit lentiviral replication by enzymatically inserting G-to-A hypermutations in the viral genome, whereas lentiviral Vif proteins degrade host APOBEC3 via the ubiquitin/proteasome-dependent pathway. Recent investigations provide evidence that lentiviral vif genes evolved to combat mammalian APOBEC3 proteins. In corollary, mammalian APOBEC3 genes are under Darwinian selective pressure to escape from antagonism by Vif. Based on these observations, it is widely accepted that lentiviral Vif and mammalian APOBEC3 have co-evolved and this concept is called an “evolutionary arms race.” This review provides a comprehensive summary of current knowledge with respect to the evolutionary dynamics occurring at this pivotal host-virus interface.
Keywords: Vif, APOBEC3, Lentivirus, Mammal, Evolutionary arms race
Overview of lentiviruses and APOBEC3
Classification of lentiviruses
Lentiviruses belong to the family Retroviridae and cause a variety of disorders in several species of mammals [1, 2]. In principle, exogenous lentiviruses are classified into five categories based on the host species: (1) primate lentiviruses (PLVs) in primates, (2) feline immunodeficiency viruses (FIVs) in felids, (3) bovine immunodeficiency virus (BIV) and Jembrana disease virus (JDV) in bovids, (4) Maedi–Visna virus (MVV) and caprine arthritis encephalitis virus (CAEV) in ruminants, and (5) Equine infectious anemia virus (EIAV) in horses (Fig. 1) [1, 2]. Human immunodeficiency virus type 1 (HIV-1) and type 2 (HIV-2) are PLVs and are known as the causative agents of acquired immunodeficiency syndrome (AIDS) in humans (Homo sapiens) (Fig. 1) (http://www.unaids.org/globalreport/). In addition, simian immunodeficiency viruses (SIVs) infect non-human primates including chimpanzees (Pan troglodytes), gorillas (Gorilla gorilla) and more than 40 species of Old World monkeys (OWMs) (Fig. 1).
Fig. 1.

A phylogenetic tree of lentiviral Vif. The names of the viral families (e.g., SIVgor, FIVfca and CAEV) and their strains (shown after under the bars; e.g., CP2135, TM219 and Roccaverano) are labeled on the tips. The hosts of the respective viruses are represented on the right of each branch with an illustration. The estimated CSTs and a recombination of PLVs are indicated with arrows. The circulations of FIVlru/FIVpco (in bobcats and puma) and CAEV/MVV (in goat and sheep as SRLV) in the wild are indicated with the double arrows. The scale bar indicates 0.4 amino acid substitutions per site
Characteristics of APOBEC3 genes
Apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3 (APOBEC3; A3) proteins are cellular cytidine deaminases and are specifically found in mammals but not in other vertebrates [3, 4]. The A3 proteins of mammals, particularly those of primates, are considered to be cell-intrinsic immune factors that combat viruses, including lentiviruses and retrotransposons. To limit the replication of lentiviruses, the A3 proteins expressed in virus-producing cells are packaged into virions released from the cell. Then, A3 proteins are brought into neighboring cells and halt viral replication via enzymatic hypermutation of the viral genome and by blocking reverse transcription directly (for more detail, see references [5, 6]. As a result, lentiviral virions produced in the presence of A3 proteins are dramatically less infectious.
The mammalian A3 genes are duplicated in a chromosomal locus flanked by CBX6 and CBX7 [4, 5, 7]. As summarized in Fig. 2, the number of A3 genes and the history of the A3 duplication process are different in each mammalian lineage (for more detail, see references [4, 5, 7, 8]. For instance, primates, including humans (Homo sapiens), have seven A3 genes, while rodents including mice (Mus musculus) have only one ortholog. Horse (Equus caballus), pig (Sus scrofa), and bovids encode six, two and three A3 genes, respectively (Fig. 2) [7, 8].
Fig. 2.
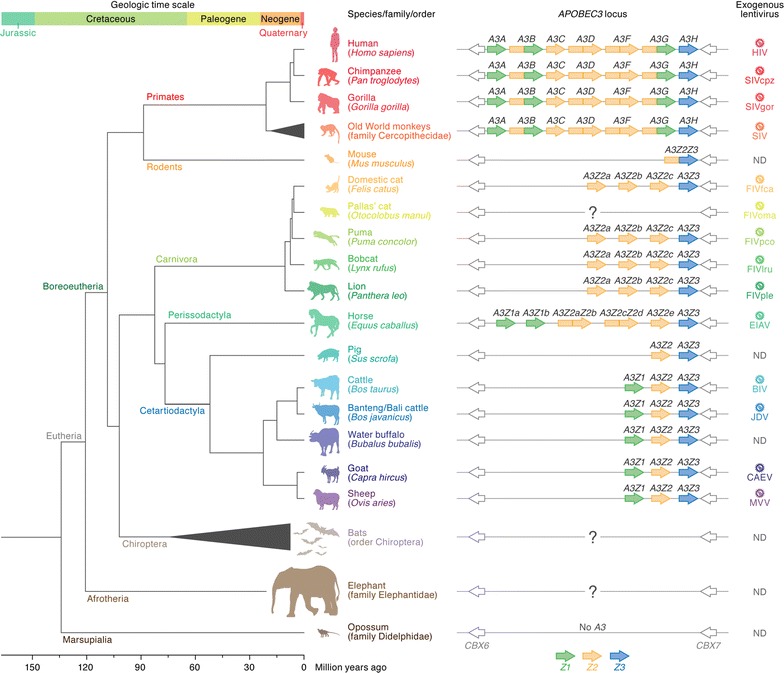
Evolution of mammals and their A3 genes. Left A phylogenetic tree of mammals associated with lentiviruses with the geologic time (top) and age (bottom) scales is shown [14, 125]. In the phylogenetic tree, a subclass (Eutheria), a clade (Boreoeutheria), a superorder (Afrotheria), and some major orders (Primates, Rodents, Carnivora, Perissodactyla, Cetartiodactyla, Chiroptera, and Marsupialia) are labeled on each branch. Representative animals are illustrated with their common names and scientific names (in parentheses). Middle Schematics of the A3 genes in each mammal are shown. The A3 genes encoded in the CBX6-CBX7 loci of respective mammals are summarized. The name of each A3 gene is indicated on each open reading frame. Z1, Z2 and Z3 domains, which are classified by amino acid sequence, are shown in green, yellow, and blue, respectively [4]. Note that A3 genes are not encoded in the genome of opossum [115]. The “?” indicates that the number and composition of A3 genes in these mammals are currently unknown. Right The exogenous lentivirus in each mammal is shown. ND not detected (yet)
Based on the sequence homology of zinc-coordinating (Z) catalytic domains, mammalian A3 proteins are classified into three subsets: Z1, Z2 or Z3. Each A3 protein is composed of single or double Z domains [4]. For instance, human A3A, A3C and A3H encode single Z1, Z2 and Z3 domain proteins, respectively, whereas human A3B, A3D, A3F and A3G encode double Z domain proteins (Fig. 2) [4]. Previous molecular phylogenetic studies have indicated that the mammalian A3 genes are evolving under positive selection [9, 10] and the gene duplications themselves likely result from selection pressures imposed by virus infections [11].
An evolutionary arms race between mammals and lentiviruses
In the field of virology, revealing the co-evolutionary relationship between viruses and their hosts is intriguing and is crucial to understanding how viruses can impact the evolution of their hosts and vice versa. As summarized in Fig. 1, a hallmark of lentivirus ecology is the emergence of new lineages via cross-species transmission (CST) events. Additionally, viral recombination between lentivirus strains occurs frequently. As a result, reconstructing the dynamic relationship between lentiviruses and their hosts is complex.
To gain a better understanding of the evolutionary conflict between lentiviruses and their host species, cell-based virological experiments with a focus on the functional relationship between viral and host proteins have recently been conducted in combination with a molecular phylogenetic approach. This strategy stems from the concept known as the “Red Queen hypothesis [12]”, which proposes that games of cat-and-mouse occur between viral and host proteins as they engage with one another over time [13–15]. Based on this concept, various experiments have been conducted using mammalian A3 proteins and a lentiviral protein, viral infectivity factor (Vif).
Vif-A3 interplay in terms of the CST events and the evolutionary arms race of lentiviruses and mammals
Vif is an accessory protein encoded by all lentiviruses with the exception of EIAV, a lentivirus found in horses [1, 2]. As described above, the replication of lentiviruses lacking a functional Vif protein is robustly impaired by certain A3 proteins in the host. In contrast, lentiviral Vif degrades the A3 proteins expressed in virus-producing cells in an ubiquitin/proteasome-dependent manner to antagonize the A3-mediated antiviral action [5, 6, 16]. As described in Fig. 3, knowledge regarding the functional interaction between Vif and A3 can inform the quest to elucidate the principle of CST events and the subsequent evolutionary arms race between lentiviruses and their hosts. In the following sections, we describe our current understanding of the evolutionary relationships between lentiviruses and their hosts, which has been informed by functional interactions between lentiviral Vif and host A3 proteins.
Fig. 3.
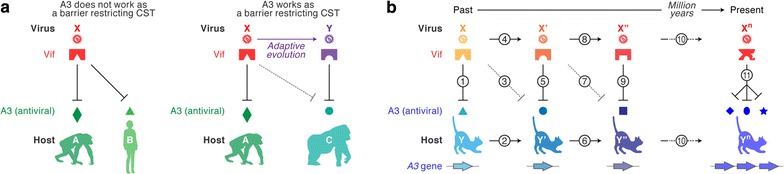
CST and the evolutionary arms race between viruses and hosts. a A3 as a barrier restricting CST. The lentivirus X infects the host A, and the Vif protein of virus X antagonizes the anti-viral A3 protein of host A. a Left When the Vif of virus X antagonizes not only anti-viral A3 of host A but also that of host B, a candidate for the new host, virus X can be successfully transferred from host A to host B without anti-viral restriction mediated by A3 of host B. a Right On the other hand, if the Vif protein of virus X is unable to antagonize anti-viral A3 of host C, another candidate for the new host, the anti-viral A3 of host C plays a role in restricting the CST of virus X from host A to host C. For a successful CST, the virus X evolves to adapt to host C and its Vif acquires the ability to antagonize host C’s anti-viral A3 (“Adaptive evolution” in this panel). As a result, a nascent species virus, the virus Y, has emerged and infects host C. b The concept of an evolutionary arms race between lentiviruses (Vif) and hosts (A3). In the past, an ancestral host (the host Y) was infected with an ancestral pathogenic virus (the virus X) and an anti-viral A3 protein of host Y was antagonized by the Vif protein of virus X (1). To escape from the pathogenic infection of virus X, the host A3 acquires certain mutations to be resistant to the degradation mediated by the virus X Vif, resulting in the emergence of a novel host, the host Y′ (2). Although the anti-viral A3 of the host Y′ is resistant to the virus X Vif (3), the virus X Vif acquires mutations to adapt to the host Y′ (4). Then, a novel virus, the virus X′, emerges and its Vif is able to antagonize an anti-viral A3 of the host Y′ (5). Subsequently, similar to the process of (1) to (5), the selective pressure by the virus X′ produces the host Y″ of which A3 is resistant to the virus X′ Vif (6 and 7), while the virus X′ evolves to antagonize the host Y″ A3 and becomes the virus X″ (8 and 9). Such an “arms race” between lentiviruses and hosts has evolutionarily occurred over a long period of time (10). It is speculated that this process might trigger the duplication of A3 genes in mammals. After n-times arms races, in the present, the host Yn encodes multiple anti-viral A3 proteins, while the Vif of the virus Xn antagonizes them (11)
A3 antagonism by lentiviral Vif proteins
Human A3 versus HIV
In 1983 and 1986, HIV-1 [17, 18] and HIV-2 [19] were isolated as the causative agents of AIDS, respectively. Both viruses likely emerged following independent CST events (Fig. 1), and molecular phylogenetic investigations have indicated that the origin of HIV-1 is SIVcpz (an SIV infecting chimpanzee [Pan troglodytes]) [20] or SIVgor (an SIV infecting gorilla [Gorilla gorilla]) [21], while the origin of HIV-2 is SIVsm (an SIV infecting sooty mangabey [Cercocebus atys]) [22]. Molecular clock analyses have estimated that the CST of SIVcpz (i.e., the birth of HIV-1) occurred ~100 years ago (around A.D. 1908–1933) [23], while the CST of SIVsm took place ~200 years ago (around A.D. 1729–1875) [24]. Moreover, HIV-1 is classified into 4 groups, M (main or major), N (new or non-M-non-O), O (outlier), and P, and each group has emerged following an individual CST event [20, 21, 25, 26] (Fig. 1). It should be noted that the virus causing the worldwide pandemic is HIV-1 group M [27]. The details of HIV pathogenesis and the clinical information through the study of infected individuals are well summarized elsewhere (e.g., see the following references: [2, 28, 29]).
In 2002, Sheehy et al. [30] identified A3G (this gene was originally designated CEM15) as a novel restriction factor against HIV-1 replication. Importantly, the A3G-mediated antiviral action is antagonized by HIV-1 Vif [30]. This report initiated research interest in Vif-A3 interactions and the impact of this interaction on HIV replication and pathogenesis (reviewed in references [5, 6, 16]). Now it is known that human (Homo sapiens) encodes seven A3 genes on chromosome 22 (Fig. 2). Specifically, endogenous A3D, A3F, and A3G expressed in human CD4+ T lymphocytes, a major target cell type for HIV infection, display robust anti-HIV activity in cell culture-based experiments (in vitro) and humanized mice (in vivo) [5, 6, 16, 31–34]. Although other human A3s, such as A3A and A3B, have exhibited anti-HIV-1 activity in overexpression experiments [35, 36], it seems controversial whether endogenously expressed A3A and A3B can elicit anti-HIV effects [37, 38]. Human A3C exhibits weak anti-HIV-1 activity [39, 40], but a recent paper has demonstrated that a single nucleotide polymorphism in human A3C (S188I substitution) results in enhanced HIV-1 inhibition in cell-based overexpression experiments [41].
Intriguingly, OhAinle et al. [42] have reported that A3H is polymorphic in human populations. A follow-up study showed that there are at least seven haplotypes and that three of them, called haplotype II, V, and VII, produce stably expressed proteins and exhibit anti-HIV-1 activity [43]. In contrast, the other four haplotypes (I, III, IV, and VI) do not exhibit stable protein expression [43]. Since chimpanzee A3H appears to be monomorphic, and the protein encoded is stable [42], it appears that the human A3H protein has developed reduced stability since the divergence from chimpanzee. Furthermore, the frequency of each haplotype is different among human populations, with a higher frequency of stable A3H in the African-descendant population and a lower frequency in other populations such as European, American and Asian populations [42, 43].
Although the ability of HIV-1 Vif to antagonize antiviral human A3s including A3D, A3F, and A3G is highly conserved [44], the ability of Vif to degrade stable A3H is not; the Vif proteins of some HIV-1 isolates (e.g., strains LAI, 93RW037 and 93BR029) are capable of degrading stable A3H (e.g., haplotype II), while other isolates (e.g., strains NL4-3, 92TH026, AD.MDR01 and 93TH305) are not [45]. Mutagenesis experiments determined that the Vif residues at positions 39 and 48 are closely associated with the ability to counteract stable human A3H [35, 46, 47]. These observations suggest that the extent of anti-HIV-1 ability elicited by endogenous A3H varies among humans, and the gain and loss of the function of HIV-1 Vif to counteract stable A3H has repeatedly occurred during viral spread in vivo and human population.
Non-human primate A3 versus SIV
The non-human primate hosts that are naturally infected with PLV in the wild can be divided into two groups: the OWMs in sub-Saharan Africa and the great apes, including chimpanzees (Pan troglodytes) and gorillas (Gorilla gorilla) (Fig. 1) [1]. Similar to humans, both OWMs and great apes encode seven A3 genes in their genomes.
As summarized in Fig. 1, a variety of OWM species reside in Africa, and most of them harbor a species-specific SIV that is not overtly pathogenic [48–51] suggesting that SIVs have co-existed with their OWM hosts for a very long time [52]. In contrast, there is also evidence for more recent CST events that give rise to new lentivirus infections. Bailes et al. [26] have reported that SIVcpz emerged through the recombination of two SIV lineages: SIVrcm (an SIV infecting red-capped mangabeys [Cercocebus torquatus]) and SIVgsn/mon/mus (SIVs infecting greater spot-nosed monkeys [Cercopithecus nictitans], mona monkeys [Cercopithecus mona] and mustached monkeys [Cercopithecus cephus]) (Fig. 1), and this recombination event is assumed to have occurred ~500 years ago (around A.D. 1266–1685) [24]. SIVgor appears to have emerged through the CST of SIVcpz [21]. In sharp contrast to SIV infection in OWMs in Africa, exogenous PLVs have not been found in the New World.
As briefly described in the introduction, Vif-A3 interactions can provide clues to elucidate virus-host (i.e., lentivirus-mammal) evolutionary relationships and history. Here, we introduce three major examples suggesting that host A3 proteins may represent a barrier to inhibit CST: (1) rhesus macaque (Macaca mulatta) A3G is resistant to degradation mediated by HIV-1 Vif (strain NL4-3) [53, 54]; (2) rhesus macaque A3G is resistant to degradation mediated by SIVagm (an SIV infecting African green monkeys [Chlorocebus aethiops]; strain TAN-1) Vif [53]; and (3) gorilla A3G is resistant to degradation mediated by the Vif proteins of SIVcpz (e.g., strains LB715, MT145, EK505 and Gab2), the putative ancestor of SIVgor [25]. In contrast to example (3), SIVgor Vif is able to degrade and counteract gorilla A3G [25], suggesting that this virus has gained the ability to counteract gorilla A3G through adaptation to gorilla following CST from chimpanzee. Moreover, Compton and Emerman have recently demonstrated that the Vif proteins of SIVs naturally infecting OWMs, such as SIVagm, SIVmus, SIVsm, SIVdeb (an SIV infecting De brazza’s monkeys [Cercopithecus neglectus]) and SIVolc (an SIV infecting colobus monkeys [Procolobus verus]) (Fig. 1), counteract A3G proteins in a species-specific manner [55]. The species specificity of Vif action arises because many species encode point mutations in the region of A3G recognized by Vif [55]. These findings suggest that each SIV Vif has evolutionarily acquired the ability to overcome the restriction imposed by A3G of their respective hosts. Furthermore, three codons have been inserted into the A3G gene of the Colobinae primate subfamily, rendering it resistant to most SIV Vif proteins. The recurrent mutation of A3G at sites important for sensitivity to Vif suggests that Vif or Vif-like viral proteins must have applied selective pressure in ancestral primates. This implies that these ancient lentiviruses caused lethal pathogenesis and/or reproductive defects that drove evolutionary change in primate hosts. By dating the appearance of adaptive mutations in primate A3G, it has been inferred that SIV has been infecting non-human primates for at least 12 million years.
Similar to the case of human A3H (described above), A3G polymorphism has been observed in OWMs and is believed to affect viral transmission within and between species [55–58]. Furthermore, the antiviral potency can be divergent between different A3 proteins in the same host. For example, human A3F exhibits higher antiviral activity than human A3D, while chimpanzee A3F exhibits lower antiviral activity than chimpanzee A3D [59]. Altogether, the following issues make unraveling the functional interaction of PLV Vif and primate A3s and its consequence a challenge: the multiplicity of primate A3 genes, the difference in anti-viral activity among A3 genes and their hosts, inter- and intra-species heterogeneity of A3 gene sequences, and divergence in PLV vif genes. Therefore, future investigations addressing the Vif-A3 interaction using experimental and comprehensive approaches through the techniques of molecular phylogenetic, evolutionary biology and mathematic modeling will be important to elucidate detail of the evolutionary arms race between PLVs and mammals.
Carnivore A3 versus FIV
FIV was first isolated in 1987 from domestic cats (Felis catus) with chronic AIDS-like disorders [60]. Epidemiological studies by ELISA, immunoblotting, and/or PCR techniques have revealed that FIV is detectable in a broad range of wild animals in the family Felidae [61–63]. FIV has been detected in felids residing in the Old World, including lions (Panthera leo), cheetahs (Acinonyx jubatus) and leopards (Panthera pardus), and those in the New World, including pumas (Puma concolor), jaguars (Panthera onca), and ocelots (Leopardus pardalis) [61]. FIV appears to be distributed worldwide, since a felid species in Asia, Pallas’ cat (Otocolobus manul), has been shown to be positive for FIV by PCR [61, 64, 65]. Furthermore, one study has confirmed that a FIV-related virus is present in spotted hyenas (Crocuta crocuta), a related carnivore family belonging to Hyaenidae [61]. In contrast to the disorders seen in FIV-infected domestic cats, it appears that FIV infections in wild felids are relatively benign and apathogenic because of less divergence in the viral sequences and no obvious symptoms in the infected felids [66]. Therefore, it has been assumed that FIV has co-existed with felids for a long period of time, as in the case of SIV infection in OWMs (see above). The higher FIV pathogenicity in domestic cat is reminiscent of the observation of SIV infection in rhesus macaques (Macaca mulatta): SIVmac (an SIV infecting rhesus macaque [Macaca mulatta]) is highly pathogenic in this monkey and emerged from a CST of SIVsm from sooty mangabeys in 1985 (Fig. 1) [67]. Therefore, similar to the situation of SIVmac emergence, FIVfca might also have emerged from a relatively recent CST event from wild felids.
Because surveys designed to detect FIV have largely utilized serological methods (e.g., ELISA and immunoblot), FIV vif sequences are only available for the following viruses: FIVfca (an FIV infecting domestic cats [Felis catus]), FIVoma (an FIV infecting Pallas’ cats [Otocolobus manul]), FIVpco (an FIV infecting pumas [Puma concolor]), FIVlru (an FIV infecting bobcats [Lynx rufus]), and FIVple (an FIV infecting lions [Panthera leo]) (Fig. 1). Although each FIV has adapted to a specific host felid, recent studies have suggested that FIVlru and a subclass of FIVpco co-circulate in both puma and lynx in North America (Fig. 1) [68, 69].
Felids, including domestic cats (Felis catus), pumas (Puma concolor), bobcats (Lynx rufus) and lions (Panthera leo), encode multiple A3 proteins: three A3Z2 proteins (A32a, A32b and A32c; the orthologs of primate A3C) and a single A3Z3 (the ortholog of primate A3H) (Fig. 2) [70, 71]. In addition, a two-domain A3Z2Z3 hybrid protein is generated through alternative splicing [70, 71]. Similar to the relationship between primate A3s and PLVs (see above), previous studies revealed that the feline A3Z3 and A3Z2Z3 proteins potently suppress the infectivity of vif-deleted FIVfca [70–75] and that the Vif proteins of FIVfca (strains Petaluma, C36 and Shizuoka) counteract the antiviral action of domestic cat A3Z3 and A3Z2Z3 [71, 74].
Interestingly, de Castro et al. [76] have reported that there are at least seven A3Z3 haplotypes in domestic cats (haplotypes I to VII) containing four nonsynonymous polymorphisms at codons 65, 68, 94, and 96, and a synonymous substitution at codon 100. A follow-up molecular phylogenetic analysis determined that codon 65 is under positive selection [74]. Intriguingly, cell-based experiments revealed that domestic cat A3Z3 haplotype V (A65I substitution compared to haplotype I) is resistant to degradation mediated by the FIVfca Vif proteins (strains Petaruma, C36 and Shizuoka) [74]. BEAST (Bayesian evolutionary analysis by sampling trees) analysis further implied that domestic cat A3Z3 haplotype V emerged approximately 60,000 years ago [74]. These observations strongly suggest that domestic cat A3Z3 haplotype V has been naturally selected to escape from an ancestral FIV and that an evolutionary arms race between FIVfca (or its ancestor) and domestic cats has taken place.
In addition to the interaction between FIVfca and domestic cat A3s, a previous paper has revealed that FIVfca (strain Petaluma) Vif is able to antagonize A3Z3 and A3Z2Z3 of other felids, including pumas, lions and tigers [71, 75]. In contrast, FIVpco (strain PLV-1695) Vif is unable to degrade domestic cat A3Z3 [75]. Most intriguingly, FIVpco (strain PLV-1695) Vif is incapable of counteracting A3Z3 of pumas, which are its natural host [75]. This is the first report demonstrating that a non-primate lentiviral Vif protein is unable to degrade an antiviral A3 protein of its natural host. Then, how is it possible that FIVpco Vif does not counteract A3 of its natural host? One possibility is that this may result in the elimination of this virus from circulation. However, a recent study by Lee et al. [77] have demonstrated an on-going cross-species circulation of FIVpco in puma and bobcat in North America. Additionally, as a higher viremia has been detected in the pumas infected with FIVpco [77], these observations argue against this possibility. Another possibility is that puma A3Z3 may provide only partial protection. Since the expression levels of endogenous A3 genes in puma remain unassessed, it may be plausible to speculate that the anti-viral effect of endogenous puma A3Z3 is negligible. However, endogenous anti-viral A3 proteins are expressed in domestic cats and exhibit robust activity against FIVfca [78], Yoshikawa et al. [79] have recently demonstrated that the Vif proteins of a subtype of FIVfca (subtype B) lose their ability to antagonize domestic cat A3-mediated anti-viral effect despite the worldwide spread of this subtype. These findings imply that the Vif-A3 interaction is more complicated than expected, at least in the interplay between FIV Vif and feline A3. Therefore, future investigations should focus on elucidating the role of feline A3 in controlling feline lentiviruses and their cross-species circulation in the wild.
As described above, several FIV lineages have been isolated from various felids; however, the molecular interaction between their Vif proteins and the A3 proteins of their hosts remains unclear. Also, the importance of Vif antagonism of A3 proteins may depend on the host-virus context, and may suggest differences in lentivirus replication and pathogenesis in vivo. Therefore, investigation of the interplay of FIV Vif and the A3 proteins of Felidae will lead to a better understanding of an intriguing virus-host co-evolutionary episode.
Artiodactyla A3 versus their lentiviruses
To evaluate the antiviral activity of A3 proteins of Artiodactyla such as cattle and sheep, cell-based experiments have been performed using a viral vector system based on HIV-1 and murine leukemia virus and expression plasmids for the A3Z3 and A3Z2Z3 proteins derived from cattle (Bos taurus) and sheep (Ovis aries) [80–82]. These studies revealed that the A3Z3 and A3Z2Z3 proteins of both cattle and sheep have the ability to suppress the infectivity of vif-deleted HIV-1 and murine leukemia virus (a prototype retrovirus in mice) [80–82]. Cattle A3Z3 is degraded by the Vif proteins of BIV (strain BIM127), MVV (strain Iceland), and SIVmac (strain 239) but not by those of HIV-1 (strain LAI) and FIV (strain NCSU) [80, 81]. On the other hand, sheep A3Z3 is degraded by the Vif proteins of MVV (strain Iceland) and HIV-1 (strain LAI) but not by those of SIVmac (strain 239), BIV (strain BIM127) and FIV (strain NCSU) [80, 81]. These findings imply that the Vif-A3Z3 interaction can be promiscuous and that each lentiviral Vif protein is optimized to antagonize the A3Z3 protein of its mammalian host.
Bovine A3 versus BIV and JDV
At least 2 lineages of lentivirus have been isolated in bovids: BIV and JDV. BIV was isolated in 1972 in the United States from cattle (Bos taurus) displaying persistent lymphocytosis [83]. However, subsequent studies reported that the disorders caused by BIV infection in cattle seem relatively mild compared to those that occur in humans as a result of HIV-1 infection [1, 84, 85]. In contrast, sporadic outbreaks of JDV infection that result in severe disorders and high mortality have occurred in Bali cattle and domesticated banteng (Bos javanicus), in certain islands of Indonesia since 1964 [86]. JDV was subsequently identified as the causative agent of severe disorders in Bali cattle in 1993 [87]. Serosurveillance studies reported that BIV infection appears worldwide in cattle [88–94], while JDV infection is endemic in Bali cattle, banteng, and cattle of the islands of Southeast Asia including Indonesia and Malaysia [86, 87, 95, 96].
To address the co-evolutionary dynamics of bovine A3 and the two bovine lentiviruses, BIV and JDV, a recent study determined the sequences of three A3 genes (i.e., A3Z1, A3Z2, and A3Z3) of various bovids belonging to the tribe Bovini, which includes the genera Bos and Bison: cattle (Bos taurus), zebu (Bos indicus), banteng (Bos javanicus), gaur (Bos gaurus), yak (Bos grunniens), European bison (Bison bonasus) and American bison (Bison bison), as well as the genus Bubalus (water buffalo [Bubalus bubalis]) [97]. Molecular phylogenetic analyses inferred that bovine A3Z3 is under strong positive selection and that the amino acids at positions 32, 62, and 92 have been positively selected [97]. Intriguingly, cell-based assays using a vif-deleted HIV-1-based reporter system, BIV Vif (strain R27) and JDV Vif (strain Tabanan/87) expression plasmids and respective bovine A3Z3 proteins revealed that gaur A3Z3 is specifically resistant to JDV-mediated degradation [97]. The resistance of gaur A3Z3 to JDV Vif is attributed to 3 codons at position 32, 62, and 92, all of which show evidence of positive selection [97]. As the A3Z3 protein of the MRCA (most recent common ancestor) of cattle, banteng, zebu and gaur, which has been estimated using a molecular phylogenetic method, is sensitive to JDV Vif-mediated degradation [97], the resistance of gaur A3Z3 was found to be acquired after diverging from the other bovids approximately 2.6 million years ago [98]. Investigations on paleontology [99] and molecular phylogenetic [98] have suggested that the tribe Bovini originated in Asia. Because JDV is an endemic lentivirus in Southeastern Asia [86, 87, 95, 96], these findings provide the first evidence suggesting that an evolutionary arms race between lentivirus (JDV or its ancestral virus) and mammals (bovids) occurred in Asia in the past.
Ovine and caprine A3 versus SRLV
During the 1930s through the 1950s, a pathogenic agent was identified in sheep (Ovis aries) in Iceland [100], and subsequent investigations in the early 1970s led to the isolation of MVV as the first lentivirus [101, 102]. CAEV was isolated from goats (Capra hircus) [1, 103, 104]. Despite their initial classification as distinct viruses subsequent genetic analyses of MVV and CAEV indicated that these viruses cluster closely [1, 105–107]. Therefore, MVV and CAEV have recently been classified into a single group called SRLV.
Sheep A3Z3 and A3Z2Z3 have the ability to impair vif-deleted HIV-1 infection, and these proteins are degraded by MVV Vif (Icelandic strain 1514) [80, 81]. In addition, both the sheep and goat A3Z2Z3 proteins are degraded by the Vif proteins of MVV (strain 1514) and CAEV (strains Cork, 1GA, and Roccaverano) [108]. Both sheep and goats are ruminants that belong to the subfamily Caprinae, and current evidence indicates that sheep split from goats approximately 4 million years ago [109]. Therefore, in contrast to felids and bovids, SRLV may have co-evolved with ruminants.
Vif co-factors
Throughout lentiviral lineages, the vif gene serves the same functional purpose of degrading antiviral A3 proteins. However, there is only ~25% conservation of its genetic sequence [81]. Despite the significant divergence of this genetic sequence, all lentiviral Vifs are capable of hijacking the cullin-RING ubiquitin ligase (CRL) complex, which consists of cullin E3 ubiquitin ligase (CUL; CUL2 or CUL5), ring box protein 2 (RBX2) and elongin B/C (ELOB/C) [81, 110, 111]. Although the vif sequence is highly divergent, the S/TLQ motif is highly conserved in all lentiviral Vif proteins, and Vif interacts with the CRL complex in an S/TLQ motif-dependent manner [81].
In 2012, two groups identified core binding factor subunit β (CBFB) as the co-factor necessary for PLV Vif to degrade host A3 proteins [110, 111]. PLV Vif, CBFB and ELOB/C form a substrate adaptor for CUL5 and RBX2, which allows the Vif interaction with suitable and susceptible A3 proteins [110]. Vif is the adaptor between the A3 proteins and the CRL complex [111]. CBFB is required for PLV Vif to degrade host A3s but is dispensable for other lentiviral Vif proteins [81, 110–112].
A comparative approach combining proteomic, biochemical, structural, and virological techniques conducted by Kane et al. [81] identified cyclophilin A (CYPA; also known as peptidylprolyl isomerase A) as the co-factor for MVV Vif. The requirement of CYPA is specific for MVV Vif (strain Iceland) [81], and a subsequent study has recently revealed that CAEV (strains Cork, 1GA, and Roccaverano), in addition to MVV Vif (strain 1514), also bind to CYPA [108]. These observations indicate that the CYPA requirement for host A3 degradation is a common feature of SRLV Vif proteins, although PLV Vif does not appear to need CYPA. Moreover, a combination approach of molecular phylogenetic and structural techniques has suggested that mammalian CBFB and CYPA are evolutionarily and structurally conserved [108]. Therefore, lentiviral Vif may have evolved to utilize evolutionarily and structurally stable proteins to degrade host A3 proteins [108]. In contrast to PLV and SRLV, it is intriguing that BIV Vif activity does not appear to be reliant on any co-factors, and no co-factors have yet been identified for FIV Vif activity [81]. These insights further suggest that each lentiviral Vif has evolved an individual strategy to adapt to each host mammal.
Future perspective: when and how did the A3 and vif genes emerge?
Here, we described the co-evolutionary relationship between mammalian A3 genes and exogenous lentiviruses. However, many intriguing questions remain: when and how was the A3 gene acquired in mammals? When and how did lentiviruses acquire the vif gene? Was vif gene acquired for the purpose of combating host A3s? Or did Vif perform other functions in lentivirus replication that were supplanted? Why is vif gene lacking in EIAV?
Regarding the origin of A3 genes; A3 is known as a component of AID/APOBEC family. AID (activation-induced cytidine deaminase) is a nucleotide mutator contributing to the somatic hypermutation of immunoglobulin genes in B cells, while APOBEC1 edits the mRNA encoding apolipoprotein B that is expressed in small intestine (reviewed in [113, 114]). Since both AID and APOBEC1 are commonly encoded in all mammals as well as birds and reptiles [5], it is plausible to speculate that these genes can be the origin of A3 genes in mammals. On the other hand, it is known that A3 genes are not present in opossums (Fig. 2) [115], suggesting that A3 genes were acquired after (in which family/order of mammals?) divergence with Marsupialia. However, it is unclear whether A3 gene acquisition occurred in the common ancestor of Eutheria or Boreoeutheria (see Fig. 2). Moreover, we still do not know how many A3 genes are encoded in the other mammals such as bats and elephants (Fig. 2). Particularly, the mammals belonging to the order Chiroptera (e.g., microbats and megabats) are highly divergent [116]. Therefore, it is plausible to assume that Chiroptera A3 genes exhibit a high level of diversity. This information will be useful for considering the evolutionary scenario of A3 acquisition/duplication, which is likely to depend on the evolutionary relationship between lentiviruses and these hosts. Furthermore, Ikeda et al. [117] have recently demonstrated that opossum A1 possesses the activity to impair the replication of lentiviruses and retroelements, suggesting that certain marsupial AID/APOBEC family proteins potently exhibit compensatory activity to limit lentiviral replication instead of A3. Nevertheless, it is still intriguing that the duplications of AID and A1 genes have not been found and that the gene duplication of AID/APOBEC family in mammals is specifically occurred in A3 genes.
The driving force and moment of A3 duplication are still both unclear. In contrast to the Z1 and Z2 domains, it is intriguing that the duplication of Z3 domain has not been observed in any mammals (Fig. 2). This is reminiscent of the “Kondrashov hypothesis” (also known as “deterministic mutation hypothesis”), which assumes that the majority of deleterious mutations are of small effect and that each subsequent mutation has an increasingly large effect on host fitness [118]. According to this concept, duplication of the A3Z3 gene may be evolutionarily constrained due to deleterious effects for the host organism. The possibility of host A3Z3 toxicity is further supported by findings related to the A3H (the ortholog to A3Z3 in primates) gene of human and chimpanzee. Human has seven A3H haplotypes, but four are not expressed at the protein level [42]. In contrast, chimpanzee A3H is monomorphic, and this protein is expressed [42], suggesting that there is a cost to maintain multiple copies of functional A3H. In fact, a recent paper has revealed that human A3H likely contributes to cancer mutagenesis [119]. Therefore, functional A3H has been evolutionarily lost in humans after the divergence from chimpanzees due to its toxicity.
Similar to mammalian A3 genes, the origin of lentiviral vif genes is also unclear. In this regard, endogenous lentiviruses have been detected in the genomes of various mammals, including European rabbits (Oryctolagus cuniculus) [120], lemurs belonging to two different genera (Microcebus and Cheirogaleus) [121, 122], and ferrets (Mustela putorius furo) [123, 124]. More intriguingly, all endogenous lentiviruses ever detected appear to encode putative vif sequences [121–123]. Considering that all exogenous lentiviruses, with the exception of EIAV in horses, also possess this accessory gene, vif gene is an ancient lentiviral component with crucial importance to maintaining lentivirus infections worldwide.
Authors’ contributions
YN, HA, AS, EY, MM, GJ-F, YK and KS wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank Robert Gifford (University of Glasgow, United Kingdom), Alex Compton (National Cancer Institute, USA) and Terumasa Ikeda (University of Minnesota, USA) for providing crucial comments, and Mss. Naoko Misawa and Kotubu Misawa for their dedicated support.
Competing interests
The authors declare that they have no competing interests.
Funding
This study was supported in part by CREST, JST (to KS); Takeda Science Foundation (to KS); Mochida Memorial Foundation for Medical and Pharmaceutical Research (to KS); Salt Science Research Foundation (to KS); Smoking Research Foundation (to KS); Grants-in-Aid for Scientific Research C 15K07166 (to KS), Scientific Research B (Generative Research Fields) 16KT0111 (to KS), and Scientific Research on Innovative Areas 17H05813 (to KS) and 24115008 (to YK) from JSPS; the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Dynamic Approaches to Living System) from AMED (to EY); and the JSPS Core-to-Core program, A. Advanced Research Networks (to YK); and Research on HIV/AIDS 16fk0410203h0002 from AMED (to YK).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yusuke Nakano, Email: nakano.yusuke.6r@kyoto-u.ac.jp.
Hirofumi Aso, Email: aso.hirofumi.75r@st.kyoto-u.ac.jp.
Andrew Soper, Email: beatlesno1fan9@gmail.com.
Eri Yamada, Email: eyamada@virus.kyoto-u.ac.jp.
Miyu Moriwaki, Email: mmoriwak@virus.kyoto-u.ac.jp.
Guillermo Juarez-Fernandez, Email: gukle.juarez@gmail.com.
Yoshio Koyanagi, Email: ykoyanag@virus.kyoto-u.ac.jp.
Kei Sato, Email: ksato@virus.kyoto-u.ac.jp.
References
- 1.Evans DT, Elder JH, Desrosiers RC. Nonhuman lentiviruses. In: Knipe DM, Howley PM, editors. Fields virology, vol. 2. 6. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1584–1612. [Google Scholar]
- 2.Freed EO, Martin MA. Human immunodeficiency viruses: replication. In: Knipe DM, Howley PM, editors. Fields virology, vol. 2. 6. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1502–1560. [Google Scholar]
- 3.Koito A, Ikeda T. Intrinsic restriction activity by AID/APOBEC family of enzymes against the mobility of retroelements. Mob Genet Elements. 2011;1:197–202. doi: 10.4161/mge.1.3.17430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479–480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 7.LaRue RS, Jonsson SR, Silverstein KA, Lajoie M, Bertrand D, El-Mabrouk N, et al. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zielonka J, Bravo IG, Marino D, Conrad E, Perkovic M, Battenberg M, et al. Restriction of equine infectious anemia virus by equine APOBEC3 cytidine deaminases. J Virol. 2009;83:7547–7559. doi: 10.1128/JVI.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallender EJ, Lahn BT. Positive selection on the human genome. Hum Mol Genet. 2004;13(Spec No 2):R245–R254. doi: 10.1093/hmg/ddh253. [DOI] [PubMed] [Google Scholar]
- 10.Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2:E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. doi: 10.1016/S0169-5347(03)00033-8. [DOI] [Google Scholar]
- 12.Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc Lond B Biol Sci. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- 13.Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol. 2012;12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gifford RJ. Viral evolution in deep time: lentiviruses and mammals. Trends Genet. 2012;28:89–100. doi: 10.1016/j.tig.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Kirchhoff F. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe. 2010;8:55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 18.Gallo RC, Sarin PS, Gelmann EP, Robert-Guroff M, Richardson E, Kalyanaraman VS, et al. Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS) Science. 1983;220:865–867. doi: 10.1126/science.6601823. [DOI] [PubMed] [Google Scholar]
- 19.Clavel F, Guyader M, Guetard D, Salle M, Montagnier L, Alizon M. Molecular cloning and polymorphism of the human immune deficiency virus type 2. Nature. 1986;324:691–695. doi: 10.1038/324691a0. [DOI] [PubMed] [Google Scholar]
- 20.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 21.Plantier JC, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemee V, et al. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15:871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 22.Gao F, Yue L, White AT, Pappas PG, Barchue J, Hanson AP, et al. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature. 1992;358:495–499. doi: 10.1038/358495a0. [DOI] [PubMed] [Google Scholar]
- 23.Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wertheim JO, Worobey M. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLoS Comput Biol. 2009;5:e1000377. doi: 10.1371/journal.pcbi.1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’arc M, Ayouba A, Esteban A, Learn GH, Boue V, Liegeois F, et al. Origin of the HIV-1 group O epidemic in western lowland gorillas. Proc Natl Acad Sci USA. 2015;112:E1343–E1352. doi: 10.1073/pnas.1502022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailes E, Gao F, Bibollet-Ruche F, Courgnaud V, Peeters M, Marx PA, et al. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- 27.Merson MH, O’Malley J, Serwadda D, Apisuk C. The history and challenge of HIV prevention. Lancet. 2008;372:475–488. doi: 10.1016/S0140-6736(08)60884-3. [DOI] [PubMed] [Google Scholar]
- 28.Kuritzkes DR, Koup RA. HIV-1: pathogenesis, clinical manifestations, and treatment. In: Knipe DM, Howley PM, editors. Fields virology, vol. 2. 6. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1561–1583. [Google Scholar]
- 29.Tebit DM, Arts EJ. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect Dis. 2011;11:45–56. doi: 10.1016/S1473-3099(10)70186-9. [DOI] [PubMed] [Google Scholar]
- 30.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 31.Krisko JF, Begum N, Baker CE, Foster JL, Garcia JV. APOBEC3G and APOBEC3F act in concert to extinguish HIV-1 replication. J Virol. 2016;90:4681–4695. doi: 10.1128/JVI.03275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krisko JF, Martinez-Torres F, Foster JL, Garcia JV. HIV restriction by APOBEC3 in humanized mice. PLoS Pathog. 2013;9:e1003242. doi: 10.1371/journal.ppat.1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato K, Izumi T, Misawa N, Kobayashi T, Yamashita Y, Ohmichi M, et al. Remarkable lethal G-to-A mutations in vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice. J Virol. 2010;84:9546–9556. doi: 10.1128/JVI.00823-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato K, Takeuchi JS, Misawa N, Izumi T, Kobayashi T, Kimura Y, et al. APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. PLoS Pathog. 2014;10:e1004453. doi: 10.1371/journal.ppat.1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ooms M, Krikoni A, Kress AK, Simon V, Munk C. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. J Virol. 2012;86:6097–6108. doi: 10.1128/JVI.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger G, Durand S, Fargier G, Nguyen XN, Cordeil S, Bouaziz S, et al. APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 2011;7:e1002221. doi: 10.1371/journal.ppat.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Refsland EW, Hultquist JF, Harris RS. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 2012;8:e1002800. doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitamura S, Ode H, Iwatani Y. Structural features of antiviral APOBEC3 proteins are linked to their functional activities. Front Microbiol. 2011;2:258. doi: 10.3389/fmicb.2011.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol. 2012;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 41.Wittkopp CJ, Adolph MB, Wu LI, Chelico L, Emerman M. A Single nucleotide polymorphism in human APOBEC3C enhances restriction of lentiviruses. PLoS Pathog. 2016;12:e1005865. doi: 10.1371/journal.ppat.1005865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.OhAinle M, Kerns JA, Li MM, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, Abudu A, Son S, Dang Y, Venta PJ, Zheng YH. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol. 2011;85:3142–3152. doi: 10.1128/JVI.02049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Binka M, Ooms M, Steward M, Simon V. The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. J Virol. 2012;86:49–59. doi: 10.1128/JVI.06082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Refsland EW, Hultquist JF, Luengas EM, Ikeda T, Shaban NM, Law EK, et al. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014;10:e1004761. doi: 10.1371/journal.pgen.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ooms M, Brayton B, Letko M, Maio SM, Pilcher CD, Hecht FM, et al. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe. 2013;14:411–421. doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Klatt NR, Silvestri G, Hirsch V. Nonpathogenic simian immunodeficiency virus infections. Cold Spring Harb Perspect Med. 2012;2:a007153. doi: 10.1101/cshperspect.a007153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.VandeWoude S, Apetrei C. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin Microbiol Rev. 2006;19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Locatelli S, Peeters M. Cross-species transmission of simian retroviruses: how and why they could lead to the emergence of new diseases in the human population. AIDS. 2012;26:659–673. doi: 10.1097/QAD.0b013e328350fb68. [DOI] [PubMed] [Google Scholar]
- 52.McCarthy KR, Kirmaier A, Autissier P, Johnson WE. Evolutionary and functional analysis of old world primate TRIM5 reveals the ancient emergence of primate lentiviruses and convergent evolution targeting a conserved capsid interface. PLoS Pathog. 2015;11:e1005085. doi: 10.1371/journal.ppat.1005085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mariani R, Chen D, Schröfelbauer B, Navarro F, König R, Bollman B, et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/S0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 54.Hatziioannou T, Princiotta M, Piatak M, Jr, Yuan F, Zhang F, Lifson JD, et al. Generation of simian-tropic HIV-1 by restriction factor evasion. Science. 2006;314:95. doi: 10.1126/science.1130994. [DOI] [PubMed] [Google Scholar]
- 55.Compton AA, Emerman M. Convergence and divergence in the evolution of the APOBEC3G-Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog. 2013;9:e1003135. doi: 10.1371/journal.ppat.1003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Compton AA, Hirsch VM, Emerman M. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe. 2012;11:91–98. doi: 10.1016/j.chom.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Compton AA, Malik HS, Emerman M. Host gene evolution traces the evolutionary history of ancient primate lentiviruses. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120496. doi: 10.1098/rstb.2012.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krupp A, McCarthy KR, Ooms M, Letko M, Morgan JS, Simon V, et al. APOBEC3G polymorphism as a selective barrier to cross-species transmission and emergence of pathogenic SIV and AIDS in a primate host. PLoS Pathog. 2013;9:e1003641. doi: 10.1371/journal.ppat.1003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Etienne L, Bibollet-Ruche F, Sudmant PH, Wu LI, Hahn BH, Emerman M. The role of the antiviral APOBEC3 gene family in protecting chimpanzees against lentiviruses from monkeys. PLoS Pathog. 2015;11:e1005149. doi: 10.1371/journal.ppat.1005149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pedersen NC, Ho EW, Brown ML, Yamamoto JK. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235:790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- 61.Troyer JL, Pecon-Slattery J, Roelke ME, Johnson W, VandeWoude S, Vazquez-Salat N, et al. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J Virol. 2005;79:8282–8294. doi: 10.1128/JVI.79.13.8282-8294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olmsted RA, Langley R, Roelke ME, Goeken RM, Adger-Johnson D, Goff JP, et al. Worldwide prevalence of lentivirus infection in wild feline species: epidemiologic and phylogenetic aspects. J Virol. 1992;66:6008–6018. doi: 10.1128/jvi.66.10.6008-6018.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carpenter MA, Brown EW, Culver M, Johnson WE, Pecon-Slattery J, Brousset D, et al. Genetic and phylogenetic divergence of feline immunodeficiency virus in the puma (Puma concolor) J Virol. 1996;70:6682–6693. doi: 10.1128/jvi.70.10.6682-6693.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sutton CA, Gordnier PM, Avery RJ, Casey JW. Comparative replication kinetics of two cytopathic feline lentiviruses ex vivo. Virology. 2005;332:519–528. doi: 10.1016/j.virol.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 65.Barr MC, Zou L, Long F, Hoose WA, Avery RJ. Proviral organization and sequence analysis of feline immunodeficiency virus isolated from a Pallas’ cat. Virology. 1997;228:84–91. doi: 10.1006/viro.1996.8358. [DOI] [PubMed] [Google Scholar]
- 66.Pecon-Slattery J, Troyer JL, Johnson WE, O’Brien SJ. Evolution of feline immunodeficiency virus in Felidae: implications for human health and wildlife ecology. Vet Immunol Immunopathol. 2008;123:32–44. doi: 10.1016/j.vetimm.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Daniel MD, Letvin NL, King NW, Kannagi M, Sehgal PK, Hunt RD, et al. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 68.VandeWoude S, Troyer J, Poss M. Restrictions to cross-species transmission of lentiviral infection gleaned from studies of FIV. Vet Immunol Immunopathol. 2010;134:25–32. doi: 10.1016/j.vetimm.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee JS, Bevins SN, Serieys LE, Vickers W, Logan KA, Aldredge M, et al. Evolution of puma lentivirus in bobcats (Lynx rufus) and mountain lions (Puma concolor) in North America. J Virol. 2014;88:7727–7737. doi: 10.1128/JVI.00473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Münk C, Beck T, Zielonka J, Hotz-Wagenblatt A, Chareza S, Battenberg M, et al. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 2008;9:R48. doi: 10.1186/gb-2008-9-3-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zielonka J, Marino D, Hofmann H, Yuhki N, Löchelt M, Münk C. Vif of feline immunodeficiency virus from domestic cats protects against APOBEC3 restriction factors from many felids. J Virol. 2010;84:7312–7324. doi: 10.1128/JVI.00209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang J, Zhang W, Lv M, Zuo T, Kong W, Yu X. Identification of a Cullin5–ElonginB–ElonginC E3 complex in degradation of feline immunodeficiency virus Vif-mediated feline APOBEC3 proteins. J Virol. 2011;85:12482–12491. doi: 10.1128/JVI.05218-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stern MA, Hu C, Saenz DT, Fadel HJ, Sims O, Peretz M, et al. Productive replication of Vif-chimeric HIV-1 in feline cells. J Virol. 2010;84:7378–7395. doi: 10.1128/JVI.00584-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoshikawa R, Izumi T, Yamada E, Nakano Y, Misawa N, Ren F, et al. A naturally occurring domestic cat APOBEC3 variant confers resistance to FIV infection. J Virol. 2015;90:474–485. doi: 10.1128/JVI.02612-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoshikawa R, Nakano Y, Yamada E, Izumi T, Misawa N, Koyanagi Y, et al. Species-specific differences in the ability of feline lentiviral Vif to degrade feline APOBEC3 proteins. Microbiol Immunol. 2016;60:272–279. doi: 10.1111/1348-0421.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Castro FL, Junqueira DM, de Medeiros RM, da Silva TR, Costenaro JG, Knak MB, et al. Analysis of single-nucleotide polymorphisms in the APOBEC3H gene of domestic cats (Felis catus) and their association with the susceptibility to feline immunodeficiency virus and feline leukemia virus infections. Infect Genet Evol. 2014;27:389–394. doi: 10.1016/j.meegid.2014.08.024. [DOI] [PubMed] [Google Scholar]
- 77.Lee J, Malmberg JL, Wood BA, Hladky S, Troyer R, Roelke M, et al. Feline immunodeficiency virus cross-species transmission: implications for emergence of new lentiviral infections. J Virol. 2017;91. doi:10.1128/JVI.02134-16. [DOI] [PMC free article] [PubMed]
- 78.Troyer RM, Thompson J, Elder JH, VandeWoude S. Accessory genes confer a high replication rate to virulent feline immunodeficiency virus. J Virol. 2013;87:7940–7951. doi: 10.1128/JVI.00752-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoshikawa R, Takeuchi JS, Yamada E, Nakano Y, Misawa N, Kimura Y, et al: Feline immunodeficiency virus evolutionarily acquires two proteins, Vif and protease, capable of antagonizing feline APOBEC3. J Virol. 2017. doi:10.1128/JVI.00250-17. [DOI] [PMC free article] [PubMed]
- 80.LaRue RS, Lengyel J, Jonsson SR, Andresdottir V, Harris RS. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J Virol. 2010;84:8193–8201. doi: 10.1128/JVI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kane JR, Stanley DJ, Hultquist JF, Johnson JR, Mietrach N, Binning JM, et al. Lineage-specific viral hijacking of non-canonical E3 ubiquitin ligase cofactors in the evolution of Vif anti-APOBEC3 activity. Cell Rep. 2015;11:1236–1250. doi: 10.1016/j.celrep.2015.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jonsson SR, Hache G, Stenglein MD, Fahrenkrug SC, Andresdottir V, Harris RS. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 2006;34:5683–5694. doi: 10.1093/nar/gkl721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Van der Maaten MJ, Boothe AD, Seger CL. Isolation of a virus from cattle with persistent lymphocytosis. J Natl Cancer Inst. 1972;49:1649–1657. doi: 10.1093/jnci/49.6.1649. [DOI] [PubMed] [Google Scholar]
- 84.Hirai N, Xuan S, Ochiai K, Onuma M. Alteration of immune responses of rabbits infected with bovine immunodeficiency-like virus. Microbiol Immunol. 1994;38:943–950. doi: 10.1111/j.1348-0421.1994.tb02151.x. [DOI] [PubMed] [Google Scholar]
- 85.Onuma M, Koomoto E, Furuyama H, Yasutomi Y, Taniyama H, Iwai H, et al. Infection and dysfunction of monocytes induced by experimental inoculation of calves with bovine immunodeficiency-like virus. J Acquir Immune Defic Syndr. 1992;5:1009–1015. [PubMed] [Google Scholar]
- 86.Wilcox GE. Jembrana disease. Aust Vet J. 1997;75:492–493. doi: 10.1111/j.1751-0813.1997.tb14379.x. [DOI] [PubMed] [Google Scholar]
- 87.Kertayadnya G, Wilcox GE, Soeharsono S, Hartaningsih N, Coelen RJ, Cook RD, et al. Characteristics of a retrovirus associated with Jembrana disease in Bali cattle. J Gen Virol. 1993;74(Pt 9):1765–1778. doi: 10.1099/0022-1317-74-9-1765. [DOI] [PubMed] [Google Scholar]
- 88.Horzinek M, Keldermans L, Stuurman T, Black J, Herrewegh A, Sillekens P, et al. Bovine immunodeficiency virus: immunochemical characterization and serological survey. J Gen Virol. 1991;72:2923–2928. doi: 10.1099/0022-1317-72-12-2923. [DOI] [PubMed] [Google Scholar]
- 89.McNab WB, Jacobs RM, Smith HE. A serological survey for bovine immunodeficiency-like virus in Ontario dairy cattle and associations between test results, production records and management practices. Can J Vet Res. 1994;58:36–41. [PMC free article] [PubMed] [Google Scholar]
- 90.Polack B, Schwartz I, Berthelemy M, Belloc C, Manet G, Vuillaume A, et al. Serologic evidence for bovine immunodeficiency virus infection in France. Vet Microbiol. 1996;48:165–173. doi: 10.1016/0378-1135(95)00138-7. [DOI] [PubMed] [Google Scholar]
- 91.Cavirani S, Donofrio G, Chiocco D, Foni E, Martelli P, Allegri G, et al. Seroprevalence to bovine immunodeficiency virus and lack of association with leukocyte counts in Italian dairy cattle. Prev Vet Med. 1998;37:147–157. doi: 10.1016/S0167-5877(98)00099-3. [DOI] [PubMed] [Google Scholar]
- 92.Meas S, Ohashi K, Sugimoto C, Onuma M. Phylogenetic relationships of bovine immunodeficiency virus in cattle and buffaloes based on surface envelope gene sequences. Arch Virol. 2001;146:1037–1045. doi: 10.1007/s007050170134. [DOI] [PubMed] [Google Scholar]
- 93.Meas S, Nakayama M, Usui T, Nakazato Y, Yasuda J, Ohashi K, et al. Evidence for bovine immunodeficiency virus infection in cattle in Zambia. Jpn J Vet Res. 2004;52:3–8. [PubMed] [Google Scholar]
- 94.Gonzalez ET, Licursi M, Vila Roza V, Bonzo E, Mortola E, Frossard JP, et al. Evidence of bovine immunodeficiency virus (BIV) infection: serological survey in Argentina. Res Vet Sci. 2008;85:353–358. doi: 10.1016/j.rvsc.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 95.Desport M, Lewis J. Jembrana disease virus: host responses, viral dynamics and disease control. Curr HIV Res. 2010;8:53–65. doi: 10.2174/157016210790416370. [DOI] [PubMed] [Google Scholar]
- 96.Desport M, Stewart ME, Mikosza AS, Sheridan CA, Peterson SE, Chavand O, et al. Sequence analysis of Jembrana disease virus strains reveals a genetically stable lentivirus. Virus Res. 2007;126:233–244. doi: 10.1016/j.virusres.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 97.Yamada E, Yoshikawa R, Nakano Y, Misawa N, Kobayashi T, Ren F, et al. A naturally occurring bovine APOBEC3 confers resistance to bovine lentiviruses: implication for the co-evolution of bovids and their lentiviruses. Sci Rep. 2016;6:33988. doi: 10.1038/srep33988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hassanin A, Ropiquet A. Molecular phylogeny of the tribe Bovini (Bovidae, Bovinae) and the taxonomic status of the Kouprey, Bos sauveli Urbain 1937. Mol Phylogenet Evol. 2004;33:896–907. doi: 10.1016/j.ympev.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 99.Bibi F, Bukhsianidze M, Gentry AW, Geraads D, Kostopoulos DS, Vrba ES. The fossil record and evolution of Bovidae: state of the field. Palaeontol Electronica. 2009;12:10A. [Google Scholar]
- 100.Sigurdsson B, Palsson P, Grimsson H. Visna, a demyelinating transmissible disease of sheep. J Neuropathol Exp Neurol. 1957;16:389–403. doi: 10.1097/00005072-195707000-00010. [DOI] [PubMed] [Google Scholar]
- 101.Haase AT, Garapin AC, Faras AJ, Varmus HE, Bishop JM. Characterization of the nucleic acid product of the visna virus RNA dependent DNA polymerase. Virology. 1974;57:251–258. doi: 10.1016/0042-6822(74)90125-1. [DOI] [PubMed] [Google Scholar]
- 102.Haase AT, Varmus HE. Demonstration of a DNA provirus in the lytic growth of visna virus. Nat New Biol. 1973;245:237–239. doi: 10.1038/newbio245237a0. [DOI] [PubMed] [Google Scholar]
- 103.Adams DS, Crawford TB, Banks KL, McGuire TC, Perryman LE. Immune responses of goats persistently infected with caprine arthritis–encephalitis virus. Infect Immun. 1980;28:421–427. doi: 10.1128/iai.28.2.421-427.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Adams DS, Crawford TB, Klevjer-Anderson P. A pathogenetic study of the early connective tissue lesions of viral caprine arthritis–encephalitis. Am J Pathol. 1980;99:257–278. [PMC free article] [PubMed] [Google Scholar]
- 105.Minardi da Cruz JC, Singh DK, Lamara A, Chebloune Y. Small ruminant lentiviruses (SRLVs) break the species barrier to acquire new host range. Viruses. 2013;5:1867–1884. doi: 10.3390/v5071867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ramirez H, Reina R, Amorena B, de Andres D, Martinez HA. Small ruminant lentiviruses: genetic variability, tropism and diagnosis. Viruses. 2013;5:1175–1207. doi: 10.3390/v5041175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Valas S, Benoit C, Guionaud C, Perrin G, Mamoun RZ. North American and French caprine arthritis–encephalitis viruses emerge from ovine maedi–visna viruses. Virology. 1997;237:307–318. doi: 10.1006/viro.1997.8800. [DOI] [PubMed] [Google Scholar]
- 108.Yoshikawa R, Izumi T, Nakano Y, Yamada E, Moriwaki M, Misawa N, et al. Small ruminant lentiviral Vif proteins commonly utilize cyclophilin A, an evolutionarily and structurally conserved protein, to degrade ovine and caprine APOBEC3 proteins. Microbiol Immunol. 2016;60:427–436. doi: 10.1111/1348-0421.12387. [DOI] [PubMed] [Google Scholar]
- 109.Jiang Y, Xie M, Chen W, Talbot R, Maddox JF, Faraut T, et al. The sheep genome illuminates biology of the rumen and lipid metabolism. Science. 2014;344:1168–1173. doi: 10.1126/science.1252806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jäger S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, et al. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang W, Du J, Evans SL, Yu Y, Yu XF. T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 112.Hultquist JF, Binka M, LaRue RS, Simon V, Harris RS. Vif proteins of human and simian immunodeficiency viruses require cellular CBFβ to degrade APOBEC3 restriction factors. J Virol. 2012;86:2874–2877. doi: 10.1128/JVI.06950-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 115.Fujino T, Navaratnam N, Jarmuz A, von Haeseler A, Scott J. C → U editing of apolipoprotein B mRNA in marsupials: identification and characterisation of APOBEC-1 from the American opossum Monodelphus domestica. Nucleic Acids Res. 1999;27:2662–2671. doi: 10.1093/nar/27.13.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mayer F, Dietz C, Kiefer A. Molecular species identification boosts bat diversity. Front Zool. 2007;4:4. doi: 10.1186/1742-9994-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ikeda T, Shimoda M, Ebrahimi D, VandeBerg JL, Harris RS, Koito A, et al. Opossum APOBEC1 is a DNA mutator with retrovirus and retroelement restriction activity. Sci Rep. 2017;7:46719. doi: 10.1038/srep46719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kondrashov AS. Deleterious mutations and the evolution of sexual reproduction. Nature. 1988;336:435–440. doi: 10.1038/336435a0. [DOI] [PubMed] [Google Scholar]
- 119.Starrett GJ, Luengas EM, McCann JL, Ebrahimi D, Temiz NA, Love RP, et al. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat Commun. 2016;7:12918. doi: 10.1038/ncomms12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc Natl Acad Sci USA. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gilbert C, Maxfield DG, Goodman SM, Feschotte C. Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 2009;5:e1000425. doi: 10.1371/journal.pgen.1000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gifford RJ, Katzourakis A, Tristem M, Pybus OG, Winters M, Shafer RW. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc Natl Acad Sci USA. 2008;105:20362–20367. doi: 10.1073/pnas.0807873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Han GZ, Worobey M. Endogenous lentiviral elements in the weasel family (Mustelidae) Mol Biol Evol. 2012;29:2905–2908. doi: 10.1093/molbev/mss126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cui J, Holmes EC. Endogenous lentiviruses in the ferret genome. J Virol. 2012;86:3383–3385. doi: 10.1128/JVI.06652-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Springer MS, Stanhope MJ, Madsen O, de Jong WW. Molecules consolidate the placental mammal tree. Trends Ecol Evol. 2004;19:430–438. doi: 10.1016/j.tree.2004.05.006. [DOI] [PubMed] [Google Scholar]


