Abstract
RNA-binding proteins (RBPs) are central players in post-transcriptional regulation and immune homeostasis. The ribonuclease and RBP Regnase-1 exerts critical roles in both immune cells and non-immune cells. Its expression is rapidly induced under diverse conditions including microbial infections, treatment with inflammatory cytokines and chemical or mechanical stimulation. Regnase-1 activation is transient and is subject to negative feedback mechanisms including proteasome-mediated degradation or mucosa-associated lymphoid tissue 1 (MALT1) mediated cleavage. The major function of Regnase-1 is promoting mRNA decay via its ribonuclease activity by specifically targeting a subset of genes in different cell types. In monocytes, Regnase-1 downregulates IL-6 and IL-12B mRNAs, thus mitigating inflammation, whereas in T cells, it restricts T-cell activation by targeting c-Rel, Ox40 and Il-2 transcripts. In cancer cells, Regnase-1 promotes apoptosis by inhibiting anti-apoptotic genes including Bcl2L1, Bcl2A1, RelB and Bcl3. Together with up-frameshift protein-1 (UPF1), Regnase-1 specifically cleaves mRNAs that are active during translation by recognizing a stem-loop (SL) structure within the 3′UTRs of these genes in endoplasmic reticulum-bound ribosomes. Through this mechanism, Regnase-1 rapidly shapes mRNA profiles and associated protein expression, restricts inflammation and maintains immune homeostasis. Dysregulation of Regnase-1 has been described in a multitude of pathological states including autoimmune diseases, cancer and cardiovascular diseases. Here, we provide a comprehensive update on the function, regulation and molecular mechanisms of Regnase-1, and we propose that Regnase-1 may function as a master rapid response gene for cellular adaption triggered by microenvironmental changes.
Keywords: Autoimmune diseases, Regnase-1, ribonuclease, RNA-binding proteins, microenvironment
Introduction
RNA splicing, editing, decay and translation are four core post-transcriptional processes that determine the activation, plasticity and tolerance of the immune system.1 These processes are tightly regulated by RNA-binding proteins (RBPs) that recognize specific RNA sequences or structures (generally called ‘elements'), which are commonly localized at untranslated RNA regions such as 3′UTRs.2 The 3′UTRs are also hotspots for miRNA binding and miRNA-mediated gene regulation.3 Therefore, RBPs and miRNAs, or the miRNA loaded RNA-induced silencing complex (RISC), may bind to the same 3′UTR.4 The interaction between RBPs and 3′UTRs may inhibit miRNA function either through competition for the same binding motif or by alteration of the RNA structure, thus rendering it inaccessible to RISC complexes.4 In contrast, the binding between RBPs and 3′UTRs may also modify the structure of the 3′UTR, thereby favoring miRNA binding and gene regulation.4 Thus, a given RBP can function either as an inhibitor or an enhancer of a particular miRNA.4 Beyond the cooperative roles of RBPs and miRNAs in gene regulation, RBPs are also involved in many aspects of miRNA processing and maturation.5 Given that RBPs function as key components in RNA processing pathways, increased knowledge of particular RBPs in immune cells would greatly enhance understanding of immune regulation.
RBPs are abundant in mammalian cells and comprise more than 30 families and 1000 members encoded by the human genome.6 The biological functions of the majority of RBPs in the immune system are largely unknown, but genetic mouse models of several RBP family members have clearly demonstrated their important functions.1 Genetic knockouts of ZFP36 (TTP), ELAVL1 (HuR), hnRNPD (AUF1) or L13a in mice result in acute or chronic inflammation.7, 8, 9, 10 Genetic ablation of RC3H1 (Roquin-1), RC3H2 (Roquin-2), ASF (SRSF1) or ZC3H12A (MCPIP-1 or Regnase-1) triggers systemic autoimmunity,11, 12, 13 whereas mice lacking ARID5A, CIRBP (hnRNPA18) or EIF4E are resistant to infection and inflammation.14, 15, 16, 17 RBPs recognize mRNAs through binding to RNA elements such as AU-rich elements (AREs) or constitutive decay elements (CDEs).18, 19 The ARE is the most studied sequence in the 3′UTR and is characterized by an enrichment in A and U as well as in at least one copy of the core sequence AUUUA.18 AREs reside within more than 4000 transcripts and may be extensively occupied by RBPs. TTP, AUF1 and Roquin are a few of the RBPs that have been studied in the immune system. TTP and AUF1 recognize AU-rich elements, whereas Roquin recognizes CDEs.1 Recent studies have also indicated that secondary structures such as ‘stem-loop' (SL) are also important for recognition by RBPs.20, 21 To date only select RBPs such as Regnase-1, Roquin and ARID5A have been shown to bind to SL structures. Unlike TTP and Roquin, Regnase-1 is unique in that it possesses endoribonuclease activity in addition to its RNA-binding domain.
Regnase-1 (Regulatory RNase 1), also known as ZC3H12A or MCPIP-1, was originally detected in monocyte chemoattractant protein-1 (MCP-1) treated human peripheral blood monocytes through microarrays, and the full length gene was subsequently cloned from these cells.13, 22 At that time, owing to the structural prediction of a potential DNA binding zinc finger domain, Regnase-1 had been proposed to be a transcription factor that induces the expression of apoptotic genes and contributes to ischemic heart disease (IHD).22 Accumulating evidence suggests that Regnase-1 has four domains: an N-terminal domain (NTD), a PilT N-terminus like (PIN) domain, a zinc finger (ZF) domain and a C-terminal domain (CTD)23, 24 (Figure 1g). Among these, the PIN domain contains the RNase catalytic center, but it requires an intramolecular interaction with NTD for full enzymatic activity.23, 24, 25 The ZF domain is responsible for the recognition and direct binding of mRNAs.23, 24, 25 With the cooperation of its domains, Regnase-1 degrades target mRNAs by recognizing a SL structure at the 3′UTRs of these genes.26 To date, dozens of mRNA targets have been identified and verified through various experimental approaches.26, 27 Similarly to non-sense mediated mRNA decay (NMD), Regnase-1 degrades mRNAs in a manner dependent on the helicase UPF1. Through this mechanism, Regnase-1 directly regulates mRNAs independently of mRNA de novo synthesis, thus endowing Regnase-1 with the ability to immediately and efficiently regulate gene expression in response to microenvironmental changes and other stimuli. Nevertheless, the expression of Regnase-1 is also tightly controlled by multiple feedback systems (Figure 1) and is induced by several stimuli such as TNFα, lipopolysaccharide (LPS) and IL-1β.13, 28 Therefore, Regnase-1 is not only rapidly promotes cellular adaptation to changes in the microenvironment but also facilitates homeostasis. Here, we provide a comprehensive review of the functions and regulation of Regnase-1. We also emphasize that Regnase-1 is a rapidly responsive ribonuclease that has critical roles in diverse physiological processes, and its dysregulation contributes to numerous pathological diseases.
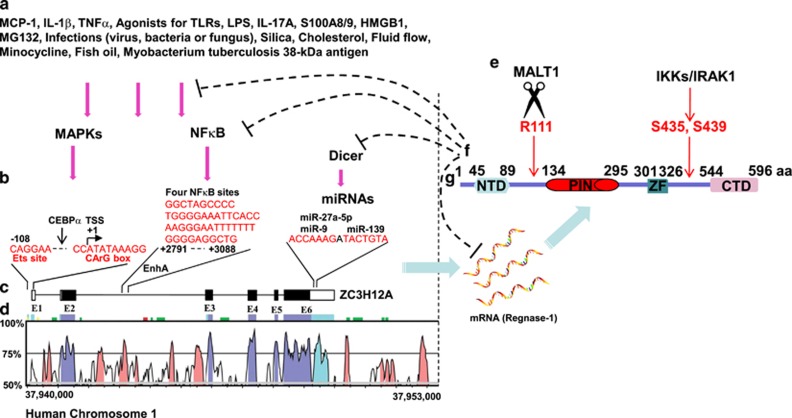
Figure 1.
Dynamic and tight regulation of Regnase-1. Regnase-1 expression is rapidly induced by diverse stimuli through MAPKs, AKT and NF-κB pathways (a). In the promoter and second intron as well as 3′UTR regions of Regnase-1, there are several conserved DNA elements that are important for its transcriptional and post-transcriptional regulation (b). The Regnase-1 coding gene ZC3H12A is shown to comprise 6 exons (c), to be located on human chromosome 1 and to have several highly conserved regions in humans and mice, according to the VISTA program (http://genome.lbl.gov/vista/index.shtml) (d). The Regnase-1 protein is post-translationally inactivated by MALT1 and IKKs/IRAK1 (e). Regnase-1 inhibits its own mRNA stability, NF-κB activity, Dicer function and microbial infections (f). Mouse Regnase-1 protein has 596 amino acids with four domains (g).
Rapidly induced expression of regnase-1 after challenges
Human Regnase-1, encoded by the ZC3H12A gene, is located on chromosome 1 and includes six exons22 (Figures 1c and d). Both its protein and mRNA expression are kept at low levels in normal tissues at resting states. Its mRNA expression, however, is rapidly induced by inflammatory cytokines, microbial infection and chemical or mechanical treatments (Figure 1a).
Inflammatory cytokines (for example, IL-1β and TNFα) and the chemokine MCP-1 enhance Regnase-1 expression through different mechanisms. MCP-1 is the first factor found to robustly induce Regnase-1 expression.22 MCP-1 (also known as CCL2) binds to its cognate receptor CCR2 and then activates ERK or AKT pathways, thus upregulating the expression of Regnase-1. After stimulation with IL-1β, ERK is activated and further promotes the activation of the transcription factor ELK1, which in turn binds to the promoter of ZC3H12A.29 In the promoter region between −76 bp and +60 bp, there is an ETS binding site and a CArG box (Figure 1b), which is recognized by ELK1 and the ELK1 partner serum response factor (SRF). Mutation of either site abolishes the responsiveness of the ZC3H12A promoter to the ELK-VP16 fusion protein.29 Another IL-1β-responsive region, located in the second intron of ZC3H12A, contains four functional NF-κB-binding sites (Figure 1b), which contribute to rapid induction after IL-1β stimulation.30 Moreover, the expression of Regnase-1 is induced by viral infections such as hepatitis C virus (HCV), as well as by bacterial and fungal infections.31 LPS, the major outer membrane component of Gram-negative bacteria, and the mycobacterium tuberculosis 38-kDa antigen both trigger Toll-like receptor 2/4 signaling and consequently increase Regnase-1 expression.32, 33, 34, 35, 36, 37 Notably, the signaling-induced expression of Regnase-1 exerts feedback inhibition on the same pathways that induce its expression.37, 38
In addition to these well-recognized inducers, new molecules that upregulate Regnase-1 expression have recently been described. The signature cytokine of Th17 cells, IL-17A, rapidly increases Regnase-1 expression through JAK/STAT3 signaling.39 IL-17 also promotes the formation of a complex between CIKS and DDX3X, which stabilizes Regnase-1 mRNA.40 Several extracellular proteins such as the S100 family members S100A8 and S100A9 also enhance Regnase-1 expression in cancer cells through an unknown mechanism.41 Extracellular high mobility group box 1 (HMGB-1) increases the expression of Regnase-1 in microglia, whereas Regnase-1 negatively regulates HMGB-1-mediated neuroinflammation and neuronal toxicity.42 Interestingly, several chemical treatments, such as MG-132, SAHA, silica, cholesterol and fish oil, stimulate the expression of Regnase-1. The proteasome inhibitor MG-132 induces the de novo mRNA synthesis of Regnase-1 through mechanisms that are independent of protein-degradation and dependent on ERK 1/2 and p38 kinase activation.43 However, in vascular muscle cells, MG-132 stimulation activates AKT and p38 kinases, thus rapidly increasing Regnase-1 expression.44 Histone deacetylase inhibitor SAHA induces the expression of Regnase-1 in chondrocytes via enhancing the recruitment of CEBPα to the ZC3H12A promoter.45 Silica, a common inhaled agent responsible for silicosis, stimulates the expression of Regnase-1, which in turn increases the cell migration of human pulmonary fibroblasts.46, 47 However, ectopic expression of Regnase-1 has been shown to impede cell migration through the induction of p53.48 Cholesterol treatment of human umbilical vein endothelial cells (HUVECs) elevates the expression of Regnase-1 and consequently contributes to cholesterol-induced damage.49 Fish oil significantly increases Regnase-1 mRNA expression.50 Finally, mechanical stimulation of cells with fluid flow also markedly increases the expression of Regnase-1 in osteocytic cells.51
Although a number of stimuli and transcription factors induce the expression of Regnase-1, the underlying molecular mechanisms are still poorly understood. Which intracellular signaling pathways are predominantly involved downstream of various stimuli remain to be fully elucidated. MAPKs and NF-κB signaling pathways are likely to be important in Regnase-1 induction, but it still remains unclear whether other signaling pathways are also involved. Furthermore, it is still largely unknown which cis elements in the ZC3H12A regulatory regions are key for its stimulus-dependent induction. Most importantly, a fundamental question is why Regnase-1 is induced by such a diverse panel of stimuli. What is the precise role of Regnase-1 in response to these stimuli?
Functional termination of regnase-1 through multiple mechanisms
Regnase-1 is rapidly and robustly induced by many stimuli, but the induction is transient and can be terminated via multiple mechanisms. After receiving signals from TLR or IL-1R, the IκB kinase (IKK) complex cooperates with the kinase IRAK1 to phosphorylate Regnase-1 at its DSGXXS motif13 (Figure 1e). Phosphorylated Regnase-1 is inherently unstable and is subject to proteasomal degradation.13 In T cells, TCR signaling activates MALT1, the only paracaspase in mammalian cells, which then cleaves at arginine 111 and inactivates Regnase-1 52 (Figure 1e). The MALT1 inhibitor MI-2 rescues the expression of Regnase-1 and consequently selectively eliminates HIV-1 latently infected CD4+ T cells.53 Thus, the Regnase-1 protein is tightly controlled by signal-induced degradation or cleavage. Further studies to understand the precise mechanisms of Regnase-1 turnover may provide useful strategies for the treatment of Regnase-1 associated diseases.
In addition to Regnase-1 protein, Regnase-1 mRNA levels are also tightly regulated through a variety of mechanisms, including miRNAs. MiR-9 was the first miRNA reported to control Regnase-1 expression. There is a putative miR-9 binding site in the 3′UTR of ZC3H12A mRNA54 (Figure 1b). The platelet-derived growth factor-BB (PDGF-BB)-mediated upregulation of miR-9 in turn downregulates its target gene ZC3H12A.55 MiR-9 also upregulates IL-6 expression by targeting Regnase-1 in human chondrocytes.56 The miR-9/Regnase-1 axis also has a role in rat acute spinal cord injury.57 Influenza A virus (IAV) hijacks the miR-9/Regnase-1 axis, thereby benefitting the viral life cycle.58 It has been reported that miR-139 and miR-27a-5P promote IL-6 expression by targeting Regnase-1 in human OA chondrocytes59, 60 (Figure 1b). In contrast, Regnase-1 acts as a broad suppressor of miRNA synthesis by counteracting Dicer, a central ribonuclease in miRNA processing.61, 62 Regnase-1 suppresses miRNA biosynthesis via cleavage of the terminal loops of precursor miRNAs (pre-miRNAs).61 Previous studies have shown that miRNA-146a and miR-3613-3p are specifically down-regulated by Regnase-1, but the mechanisms have not been identified.63, 64 Moreover, Regnase-1 is also involved in the degradation of both non-uridylated and uridylated pre-let-7.65
Together, these findings indicate that ZC3H12A is a rapid response gene that can be swiftly induced under different conditions through both transcriptional and post-transcriptional mechanisms. Several identified feedback systems including those activated by microbial infections, NF-κB signaling and miRNAs ensure that the expression of Regnase-1 can be precisely modulated in different contexts (Figure 1f). Therefore, Regnase-1 expression is tightly controlled by post-transcriptional and post-translational mechanisms. Dysregulated expression of Regnase-1 can lead to disease states, which will be summarized in the following sections.
Regnase-1 acts as a ribonuclease that degrade mRNA
As mentioned above, the original identification of Regnase-1 had been reported in 2006, but its true function was not elucidated until 2009. Akira's group has provided the first evidence that Regnase-1 is a ribonuclease, which not only binds the 3′UTR of the IL-6 gene but also degrades IL-6 mRNA as well as other mRNAs without sequence specificity in vitro.66 In contrast to the in vitro findings, Regnase-1 recognizes a specific secondary RNA structure to degrade target mRNAs in vivo. Regnase-1 negatively regulates the mRNA stability of IL-6 and IL-12B, but not that of TNFα. The 3′UTR of IL-6 contains five ARE elements and one SL structure. However, the SL structure, but not the AREs, is important for Regnase-1-mediated RNA decay in vivo, as demonstrated by serial deletions of the 3′UTR of IL-6 mRNA66 (Figure 2a). The SL structure, recognized by Regnase-1, is highly sequence-specific; UAU and UGU loops are the favored structures recognized by Regnase-1, whereas ACA, AAA and UCU loops are not recognized by Regnase-1.26 No common stem sequence has been identified among Regnase-1 targets.26 However, a recent report has indicated that the SL surrounding sequences affect Regnase-1 recognition;67 these results are controversial and need to be further examined.
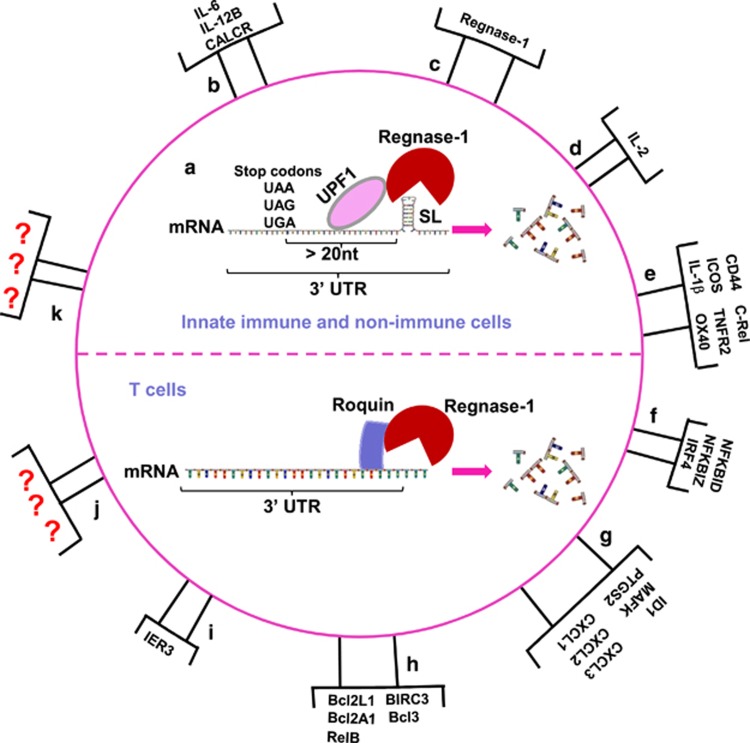
Figure 2.
Regnase-1 is a ribonuclease that degrades mRNAs. In innate immune and non-immune cells, Regnase-1 and Roquin cooperatively downregulate several common genes, but are functionally independent. Together with up-frameshift protein-1 (UPF1), Regnase-1 specifically cleaves mRNAs active during translation by recognizing a SL structure within the 3′UTR of these genes in endoplasmic reticulum-bound ribosomes. The SL structure at the 3′UTR must be located more than 20 nucleotides away from the stop codon for Regnase-1-mediated degradation. Conversely, Roquin localizes at processing bodies or stress granules and controls translationally inactive mRNAs. In T cells, Regnase-1 and Roquin are functionally dependent and cooperatively downregulate a common set of genes via the RNA-binding domain of Roquin and RNase activity of Regnase-1 (a). Regnase-1 identified targets are summarized: IL-6, IL-12B, CALCR (b); Regnase-1 (c); IL-2 (d); CD44, ICOS, IL-1β, c-Rel, TNFR2, OX40 (e); NFKBID, NFKBIZ, IRF4 (f); ID1, MAFK, PTGS2, CXCL1, CXCL2, CXCL3 (g); Bcl2L1, Bcl2A1, RelB, BIRC3, Bcl3 (h); IER3 (i). There are likely many unidentified Regnase-1 targets (j and k).
Although the SL structure recognized by Regnase-1 has been defined, the mechanisms that mediate this recognition structure and the subsequent degradation of SL-containing mRNAs have yet to be fully determined. A recent study of Regnase-1 in innate immune and non-immune cells has revealed that Regnase-1 recognizes and degrades mRNAs depending on the location and the status of mRNAs. Regnase-1 localizes at the ribosome/endoplasmic reticulum (ER), but not at processing bodies (PBs) or stress granules (SGs). The ribosome protein UPF1 associates with Regnase-1, and its helicase activity is critical for Regnase-1-mediated mRNA decay26 (Figure 2a). Only mRNAs active during translation are targeted for Regnase-1-mediated degradation, because the protein translation inhibitor cycloheximide (CHX) is able to rescue mRNA degradation from Regnase-1.26 Furthermore, the SL structure at the 3′UTR must be located more than 20 nucleotides away from the stop codon for Regnase-1-mediated degradation. In this case, Regnase-1 and Roquin, another CCCH type zinc finger member, cooperatively downregulate several common genes, although the mechanisms appear to be distinct26 (Figure 2a). Roquin localizes at processing bodies/stress granules and controls mRNAs that are inactive during translation.26 However, a study in T cells has shown that Regnase-1 and Roquin are functionally dependent; they cooperatively downregulate a common set of genes dependent on the RNA-binding domain of Roquin and the RNase activity of Regnase-1.52 In Regnase-1-deficient T cells, Roquin fails to downregulate its targets. Furthermore, in Roquin-deficient T cells, Regnase-1 fails to degrade its targets.52 Although this cooperation may be cell-type specific, Regnase-1 and Roquin have widely overlapping mRNA targets. In different cell types, the intracellular distribution, the identified targets and associated degradation pathways of Regnase-1 may be different.
Identified targets of regnase-1 ribonuclease activity
The Regnase-1 ribonuclease targets specific mRNAs independently of de novo mRNA synthesis. Much effort has been dedicated to identify the targets of Regnase-1. By comparing mRNA expression profiles between Regnase-1 knockout T cells to wild type controls, it has been shown that 424 genes are upregulated more than 10-fold in Regnase-1-deficient cells.28 To further determine the direct targets, a RNA-immunoprecipitation (IP) sequencing (RIP-seq) analysis has been performed, and 68 mRNAs have been found to be significantly enriched in Regnase-1 IPs.26 These large scale and global techniques have greatly narrowed the pool of potential targets, but true targets of Regnase-1 still need to be experimentally validated under different conditions.
To date, several cytokines, chemokines, co-stimulatory receptors and transcription factors have been defined as Regnase-1 targets, although these vary among different cell types and conditions (Figure 2). The first identified target of Regnase-1 was IL-6, as confirmed by single-molecule RNA FISH.68 In addition to IL-6, other cytokines such as IL-2, IL-1β and IL-12B have been identified as direct targets of Regnase-1.23, 69, 70 The chemokines CXCL1, CXCL2 and CXCL3 serve as Regnase-1 direct targets.26 Regnase-1 also targets mRNAs encoding T-cell co-stimulatory receptors, which consist of ICOS, TNFR2 and OX40, as well as T-cell activation marker CD44.28 In addition, transcription factors including NFKBID, NFKBIZ, RelB and IRF4 have been reported to be Regnase-1 targets.52 Finally, the gene encoding the anti-apoptotic immediate early response 3 (IER3) protein is a recently identified Regnase-1 target, as demonstrated by single-molecule RNA FISH; IER3 mRNA is clearly co-localized with Regnase-1 protein in single cells.71
Recently, the number of identified Regnase-1 targets has increased dramatically (Figure 2). There is no doubt that additional targets of Regnase-1 will be found. However, a provocative question is why the identified Regnase-1 targets vary dramatically among different cells and conditions. The process of Regnase-1 target recognition is likely to be determined by multiple factors and distinct layers of regulation that may differ among cell types. Although the identification and determination of functional important Regnase-1 targets under different conditions can be very challenging, it will probably provide insight into diverse physiological and pathological processes. Furthermore, these investigations will further increase knowledge of the molecular mechanisms of Regnase-1 regulation.
The role of regnase-1 in physiological and pathological conditions
The functional importance of Regnase-1 has been highlighted by genetic knockout mouse models. ZC3H12A-deficient mice display growth retardation and severe anemia, and most mice die within 12 weeks of birth.66, 72, 73, 74 Total knockout mice also show severe splenomegaly and lymphadenopathy. Conditional deficiency of Regnase-1 in bone marrow results in severe multi-organ inflammation.75 These phenotypes support the notion that Regnase-1 has critical roles in restricting inflammation and that its function is not redundant with those of other family members (Table 1).
Table 1. The physiological and pathological roles of Regnase-1.
| Category of biological process | Name of biological process | Role of Regnase-1 | References |
|---|---|---|---|
| T-cell activation | Inhibiting T-cell activation | 28, 52, 97, 98 | |
| Monocyte activation | Inhibiting monocytes activation | 34, 93 | |
| Apoptosis | Promoting apoptosis | 22, 32, 49, 56, 89, 101 | |
| Physiology | Cell differentiation | Promoting cell differentiation | 85 |
| Angiogenesis | Enhancing angiogenesis | 83, 84 | |
| Adipogenesis | Promoting or inhibiting adipogenesis | 86, 87, 88 | |
| Autoimmune gastritis | Preventing autoimmune gastritis | 72 | |
| Rheumatoid arthritis | Promoting rheumatoid arthritis | 96 | |
| Myasthenia gravis | Unknown | 94 | |
| Infections | Inhibiting viral infections | 31, 36, 37, 91 | |
| Pathology | Psoriasis | Unknown | 39, 95 |
| Silicosis | Promoting silicosis | 47 | |
| Atherogenesis | Promoting atherogenesis | 75 | |
| Ischemia | Promoting the development of ischemia-associated diseases but inhibiting tissue remodeling | 22, 35, 76, 77, 78, 79, 80, 81, 82 | |
| Pernicious anemia | Preventing anemia | 66, 72, 73, 74 | |
| Carcinogenesis | Inhibiting carcinogenesis | 64, 99, 100, 101 |
In the mouse ischemic heart disease model established by cardiomyocyte-targeted expression of MCP-1, the mRNA level of Regnase-1 is significantly elevated compared with that in controls.22, 76, 77 Moreover, Regnase-1 mRNA is dramatically increased in human ischemic heart samples.22 Mechanistically, Regnase-1 elicits cardiac myocyte apoptosis and ventricular dysfunction but attenuates endotoxin-induced myocardial dysfunction by suppressing cardiac NF-κB activation.78 Furthermore, Regnase-1 expression in the ischemic myocardium protects against adverse cardiac remodeling and dysfunction after myocardial infarction, possibly through mitigating NF-κB signaling and suppressing inflammation-associated microRNA expression such as miR-126, -146a, -155 and -199a.79 Regnase-1 also promotes endothelial and cardiac differentiation from mesenchymal stem cells (MSCs) and thus enhances myocardial repair and regeneration of ischemic tissues.80 Interestingly, Regnase-1 also protects from ischemic brain damage through the downregulation of proinflammatory cytokines35 and is thus implicated in ischemia tolerance.81, 82 Regnase-1 also mediates minocycline-induced neuroprotection against cerebral ischemia/reperfusion injury.82 Collectively, these findings clearly indicate a critical role of Regnase-1 in ischemia-associated diseases, but the exact mechanism and function are still unknown (Table 1). Accordingly, Regnase-1 may have distinct functions in different stages of ischemic disease. In the early phase of ischemia, Regnase-1 may accelerate disease progression by promoting cell apoptosis; in the late phase, it probably restricts inflammation and thus impedes organ remodeling and dysfunction. In addition, Regnase-1 enhances angiogenesis by inducing the expression of CDH12 and CDH19.83, 84 It has also been reported that the expression of Regnase-1 in neuroprogenitor cells promotes glial cell differentiation.85
Adipogenesis is a key process that contributes to obesity and type 2 diabetes and is controlled by several key transcription factors, such as peroxisome proliferator-activated receptor gamma (PPARγ).86 Strikingly, Regnase-1 is induced earlier than C/EBP family transcription factors and (PPARγ) with adipogenesis-inducing medium, thus indicating that Regnase-1 promotes adipogenesis independently of PPARγ86 (Table 1). A further study has reported that Regnase-1 enhances adipogenesis via oxidative ER stress and autophagy.87 In contrast, another group has shown that Regnase-1 impairs adipogenesis by promoting the decay of C/EBPβ mRNA.88 Because high expression of Regnase-1 promotes cell apoptosis,89 the published inconsistencies may be due to the expression level of Regnase-1. It is also possible that Regnase-1 target specificity may be altered by different levels of Regnase-1 expression.
After viral infection, the expression of Regnase-1 in patients with the CC genotype at the IL-28B SNP rs12979860 is higher than that in patients with the TT genotype.90 In addition, the CC genotype at IL-28B SNP rs12979860 is a marker of sustained viral response, thus suggesting that Regnase-1 may have a protective role in antiviral responses91 (Table 1). Indeed, Regnase-1 has been shown to degrade viral RNAs by directly binding and subsequently inhibiting single positive-stranded RNA virus replication, such as Japanese encephalitis virus (JEV) and dengue virus (DEN).37 Regnase-1, with its wide antiviral spectrum, efficiently inhibits positive-sense RNA viruses (sindbis virus and encephalomyocarditis virus), negative-sense RNA viruses (influenza A virus), and DNA viruses (adenovirus) through its RNase, RNA binding and oligomerization activities.37 Regnase-1 also restricts HIV-1 and HCV infections by directly targeting the viral RNAs.31, 36 Notably, the antiviral effects of Regnase-1 on HIV-1 and simian immunodeficiency virus (SIV) cannot be suppressed by Vpx, an accessory protein that enhances viral replication by degrading the cellular restriction factor SAMHD1.92 The direct antiviral effects of Regnase-1 by degrading viral genomes may be a promising strategy to prevent or treat viral infections. Efforts aimed at increasing Regnase-1 expression levels may prove effective in combating chronic viral infections.
Activation of monocytes and macrophages by LPS induces a robust expression of Regnase-1, which in turn antagonizes the LPS-induced production of inflammatory cytokines and NO.34 In addition to LPS, all other TLR agonists, with the exception of CpG DNA, markedly promote Regnase-1 expression.93 Furthermore, bacterial, viral and fungal infections all strongly enhance Regnase-1 expression,93 thus suggesting that Regnase-1 has a broad effect in innate cells infected with diverse pathogens. Keratinocytes are the predominant cell type in the epidermis and provide the first barrier against dangerous environmental stimuli such as pathogenic bacteria, fungi, parasites, viruses, heat, UV radiation and dehydration. In keratinocytes, IL-17A-induced Regnase-1 expression is dependent on JAK/STAT3 signaling.39 Moreover, patients with autoimmune diseases such as myasthenia gravis (MG) exhibit elevated levels of Regnase-1.94 Aberrant expression of Regnase-1 is also associated with the inflammatory skin disease psoriasis.39, 95 Regnase-1 also causes endothelial dysfunction in rheumatoid arthritis.96 Therefore, Regnase-1 modifies the function of innate cells and thus shapes antiviral responses, and it additionally directly targets viral RNAs. A more detailed understanding of the different Regnase-1 functions may provide new strategies to treat infectious diseases.
Beyond its functions in innate immune cells, Regnase-1 also regulates the activation of adaptive immune cells (Table 1). Mice with the specific deletion of Regnase-1 in T cells begin to die at 8 weeks after birth, and most die within 17 weeks.28 These mice also develop severe splenomegaly. The total numbers of splenic T cells from Regnase-1-deficient mice are elevated, most of which are CD62L− CD44hi effector/memory T cells. Splenic T cells from Regnase-1-deficient mice retain the ability to be polarized to various helper T-cell subsets but display elevated expression of Ki67, thus indicating accelerated cell cycle progression.28 In addition, the nuclease activity of Regnase-1 functions together with the RNA-binding activity of Roquin in inhibiting Th17 cell differentiation by repressing its target mRNAs, which encode for Th17 cell-promoting factors such as IL-6, ICOS, c-Rel, IRF4, IκBNS and IκBζ.52, 97 Interestingly, Regnase-1 haploinsufficient mice show enhanced resistance to disseminated Candida albicans infection but show exacerbated IL-17-dependent pathology in EAE and pulmonary inflammation models.98 In the early phase of TCR activation, the inhibitory function of Regnase-1 in T cells is relieved by MALT1-mediated cleavage.28 Further investigation will be needed to clarify Regnase-1 regulation and the potential distinctive roles of Regnase-1 in different T-cell subsets.
Recent studies of Regnase-1 in different cancers have indicated that Regnase-1 may either inhibit or promote tumorigenesis (Table 1). Primary neuroblastomas have been shown to lack Regnase-1 expression.99 Established neuroblastoma cell lines also express low levels of Regnase-1, and enforced Regnase-1 expression in these cells diminishes cell viability and proliferation.99 Overexpression of Regnase-1 in neuroblastoma cells significantly downregulates choline transporter-CTL1 and miRNA-3613-3p.64 Regnase-1 overexpression also antagonizes MCF7 breast cancer cell growth by stabilizing the RGS2 tumor suppressor100 and stimulates the death of breast cancer cells by selectively enhancing the mRNA decay of anti-apoptotic gene transcripts, such as Bcl2L1, Bcl2A1, RelB, Birc3 and Bcl3.101 Low Regnase-1 expression in tumor samples from breast cancer patients is strongly associated with poor survival 13 years after diagnosis.101 However, in colon cancer cells, S100A8/A9 activation upregulates Regnase-1, which may instead promote cancer formation.41 Therefore, the role of Regnase-1 in different cancers appears to be complex, and understanding of Regnase-1 in carcinogenesis is largely incomplete. There is clear evidence that Regnase-1 may act as a tumor suppressor, but studies from large datasets including multiple cancer types are still not available. Furthermore, Regnase-1 mRNA is directly monitored and regulated by its ribonuclease, as mentioned above. Because of this feedback regulating mechanism, it is insufficient or even misleading to research Regnase-1 at only the mRNA level. Systematic analysis of both Regnase-1 transcripts and protein levels may provide more insight into its role in tumorigenesis. Regnase-1 may also have indirect roles in cancer by regulating inflammation, which is known to fuel the growth of certain cancers. Investigations using Regnase-1-deficient and transgenic mouse models will be essential to clarify the exact roles of Regnase-1 in cancer development and/or progression.
Ribonuclease-independent functions of regnase-1
It is now well established that Regnase-1 functions as a ribonuclease. However, besides the ribonuclease-related NTD and PIN domains, Regnase-1 also has a zinc finger (ZF) domain and a C-terminal domain (CTD). Thus, in addition to its role as a ribonuclease, Regnase-1 may also have other functions such as acting as a transcription factor and/or a deubiquitinase. Regnase-1 was originally identified as a transcription factor through its ZF domain, which might regulate transcription.22 A GAL4-Regnase-1 fusion protein has been found to induce luciferase activity driven by GAL4-binding sites, thus suggesting that Regnase-1 may regulate transcription by recruiting the transcription initiation complex.22 Furthermore, the transcription regulatory function of Regnase-1 has been implicated in angiogenesis.83 In HEK 293 T cells, overexpressed Regnase-1 binds to cdh12 and cdh19 genes and upregulates the expression of both cdh12 and cdh19. This interaction thus supports the idea that Regnase-1 may have DNA binding capability and may activate gene transcription.83 The ZF domain of Regnase-1 belongs to the CCCH type zinc finger, which is a known motif for RNA binding. Thus, in addition to its ribonuclease activity used to degrade mRNAs, Regnase-1, as an RBP, may be extensively involved in RNA processing.
Regnase-1 also has been proposed to act as a deubiquitinase because it removes ubiquitin moieties from signaling proteins, including TRAF2, TRAF3 and TRAF6.102, 103 Wild type, but not C306R mutated, Regnase-1 protein purified from HEK293T cells is able to cleave linear tetra-ubiquitin.102 Regnase-1 also inhibits genotoxic stress-induced NF-κB activation by the deubiquitination of NEMO and TRAF6.102, 103 Both the deubiquitinase and RNase activities of Regnase-1 are required for its promotion of M2 macrophage polarization by the sequential induction of ROS, ER stress and autophagy.104 Although protein domain analysis coupled with in vivo and in vitro data support the notion that Regnase-1 has both transcriptional and deubiquitinase functions, further studies are needed to firmly establish these roles. A genetic knock-in mouse model with mutations in the individual functional domains will provide valuable information to clarify the physiological function of each domain. Of course, it is certainly possible that these two functions are minor activities in comparison to its ribonuclease activity, or perhaps the function that Regnase-1 acquires is dependent on binding partners and/or cellular stimulations. Precise dissection of these functional domains may provide opportunities to manipulate Regnase-1's individual functions instead of its collective functions, thus potentially resulting in fewer side effects in targeting Regnase-1 in clinical settings.
Perspectives
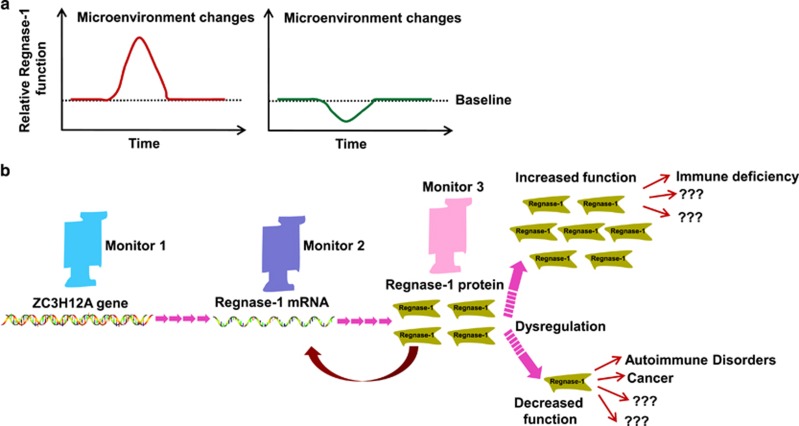
The generation of mouse models and key mechanistic experiments has provided important insights into the function of Regnase-1, and as such, knowledge regarding this important regulator has dramatically increased in the past several years. Genetic mouse models have clearly demonstrated that mice lacking proper function of Regnase-1 exhibit multi-systemic disorders involving multiple organs. Therefore, the dysregulation of Regnase-1 leads to multiple diseases, and the function of Regnase-1 must be tightly controlled (Figures 1 and 3). However, current studies regarding the pathological roles of Regnase-1 are limited. Regarding its physiological functions, Regnase-1 has several important interesting regulatory features: first, its transcription is rapidly induced by multiple stimuli/conditions (Figures 1a and 3a); second, Regnase-1 rapidly regulates mRNA profiles by targeting a subset of genes without mRNA de novo synthesis (Figure 2); third, its function can be inactivated through distinct mechanisms in different cell types (Figures 1e and 3a); fourth, Regnase-1 mRNA is a direct target of its own ribonuclease, thus providing an efficient feedback system to control Regnase-1 activation (Figures 1f and 3b). These features suggest that Regnase-1 has the ability to rapidly respond to microenvironmental changes. Several questions, however, remain unclear and need to be addressed in future studies. How does Regnase-1 rapidly facilitate cell adaptation to diverse stressors? How do different cell types use distinct mechanisms to modulate Regnase-1 activation in various stress responses? How does Regnase-1 recognize different SL structures within target genes with base-pair differences? SL structures have been identified in thousands of mRNAs or non-coding RNAs, yet it is unlikely that all of these serve as targets of Regnase-1. In this regard, there may only be a few targets of Regnase-1 in response to a given specific stimulus. The identification of stimulus-dependent Regnase-1 targets may prove difficult but will greatly benefit understanding of the biological functions of Regnase-1 and may also provide useful information in understanding how Regnase-1 recognizes stimulus-specific mRNAs or non-coding RNAs. Furthermore, it is likely that Regnase-1 dynamically interacts with its partners, and these interactions are probably crucial in Regnase-1 functions. However, Regnase-1 binding proteins are still largely unidentified. Addressing these key questions will be important in future studies. In this review, we have emphasized that Regnase-1 is a potentially important stress responsive gene, which can be either functionally induced or terminated in response to microenvironmental changes (Figure 3a). Regnase-1 function is tightly controlled at multiple levels, and its dysregulation leads to many diseases (Figure 3b). Targeting its ribonuclease activity as well as its regulatory pathways may provide a useful strategy in the treatment of human diseases such as autoimmunity, cancers and cardiovascular diseases.
Figure 3.
Regnase-1 may be a master stress responsive gene that, when dysregulated, can lead to disease. Regnase-1 has been demonstrated to be a rapid response gene. We propose that Regnase-1 may function as a master regulator that facilitates cell adaptation to microenvironmental changes. After encountering microenvironmental changes, cells respond appropriately either by upregulating Regnase-1 to ‘start the engine' or by downregulating Regnase-1 to ‘release the brake' (a). To ensure the proper function of Regnase-1, cells tightly regulate the precise Regnase-1 levels at each step. Both gain and loss of Regnase-1 function result in various diseases, including immune related diseases and other yet to be identified diseases (b).
Acknowledgments
This work was supported by the Distinguished Professorship Program of Jiangsu Province to YF; the National Natural Science Foundation of China (81641164; 81600386; 81471539 and 30801350); and the Natural Science Foundation of Jiangsu Province (BK20141236). EWH is supported by NIH grants RO1CA135362 and R21AI112763. We apologize to the many scientists who made contributions to the field but have not been cited because of space limitations.
Footnotes
The authors declare no conflict of interest.
References
- Kafasla P, Skliris A, Kontoyiannis DL. Post-transcriptional coordination of immunological responses by RNA-binding proteins. Nat Immunol 2014; 15: 492–502. [DOI] [PubMed] [Google Scholar]
- Ufer C. The biology of the RNA binding protein guanine-rich sequence binding factor 1. Curr Protein Pept Sci 2012; 13: 347–357. [DOI] [PubMed] [Google Scholar]
- Moreno-Moya JM, Vilella F, Simon C. MicroRNA: key gene expression regulators. Fertil Steril 2014; 101: 1516–1523. [DOI] [PubMed] [Google Scholar]
- Connerty P, Ahadi A, Hutvagner G. RNA binding proteins in the miRNA pathway. Int J Mol Sci 2016; 17: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer 2011; 11: 644–656. [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet 2014; 15: 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 1996; 4: 445–454. [DOI] [PubMed] [Google Scholar]
- Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G et al. HuR as a negative posttranscriptional modulator in inflammation. Mol Cell 2005; 19: 777–789. [DOI] [PubMed] [Google Scholar]
- Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev 2006; 20: 3174–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poddar D, Basu A, Baldwin WM, Kondratov RV, Barik S, Mazumder B. An extraribosomal function of ribosomal protein L13a in macrophages resolves inflammation. J Immunol 2013; 190: 3600–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratama A, Ramiscal RR, Silva DG, Das SK, Athanasopoulos V, Fitch J et al. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity 2013; 38: 669–680. [DOI] [PubMed] [Google Scholar]
- Moulton VR, Grammatikos AP, Fitzgerald LM, Tsokos GC. Splicing factor SF2/ASF rescues IL-2 production in T cells from systemic lupus erythematosus patients by activating IL-2 transcription. Proc Natl Acad Sci USA 2013; 110: 1845–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Takeuchi O, Teraguchi S, Matsushita K, Uehata T, Kuniyoshi K et al. The IkappaB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat Immunol 2011; 12: 1167–1175. [DOI] [PubMed] [Google Scholar]
- Qiang X, Yang WL, Wu R, Zhou M, Jacob A, Dong W et al. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nat Med 2013; 19: 1489–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K, Ripley B, Nishimura R, Mino T, Takeuchi O, Shioi G et al. Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo. Proc Natl Acad Sci USA 2013; 110: 9409–94014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov SS, Roy CR. Pathogen signatures activate a ubiquitination pathway that modulates the function of the metabolic checkpoint kinase mTOR. Nat Immunol 2013; 14: 1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognese AC, Sharma A, Yang WL, Nicastro J, Coppa GF, Wang P. Cold-inducible RNA-binding protein activates splenic T cells during sepsis in a TLR4-dependent manner. Cell Mol Immunol 2016. [DOI] [PMC free article] [PubMed]
- Zhang T, Kruys V, Huez G, Gueydan C. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem Soc Trans 2002; 30: 952–958. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Lu M, Rattenbacher B, Moroni C. A constitutive decay element promotes tumor necrosis factor alpha mRNA degradation via an AU-rich element-independent pathway. Mol Cell Biol 2003; 23: 3506–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullner EW, Kuhn LC. A stem-loop in the 3' untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell 1988; 53: 815–825. [DOI] [PubMed] [Google Scholar]
- Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell 2013; 153: 869–881. [DOI] [PubMed] [Google Scholar]
- Zhou L, Azfer A, Niu J, Graham S, Choudhury M, Adamski FM et al. Monocyte chemoattractant protein-1 induces a novel transcription factor that causes cardiac myocyte apoptosis and ventricular dysfunction. Circ Res 2006; 98: 1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Peng W, Sun Y, Wang X, Xu Y, Li X et al. Structural study of MCPIP1 N-terminal conserved domain reveals a PIN-like RNase. Nucleic Acids Res 2012; 40: 6957–6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa M, Tsushima T, Noda NN, Kumeta H, Enokizono Y, Yamashita K et al. Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions. Sci Rep 2016; 6: 22324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Fu S, Peng W, Rao Z. MCP-1-induced protein-1, an immune regulator. Protein Cell 2012; 3: 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mino T, Murakawa Y, Fukao A, Vandenbon A, Wessels HH, Ori D et al. Regnase-1 and roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell 2015; 161: 1058–1073. [DOI] [PubMed] [Google Scholar]
- Uehata T, Akira S. mRNA degradation by the endoribonuclease Regnase-1/ZC3H12a/MCPIP-1. Biochim Biophys Acta 2013; 1829: 708–713. [DOI] [PubMed] [Google Scholar]
- Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K et al. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell 2013; 153: 1036–1049. [DOI] [PubMed] [Google Scholar]
- Kasza A, Wyrzykowska P, Horwacik I, Tymoszuk P, Mizgalska D, Palmer K et al. Transcription factors Elk-1 and SRF are engaged in IL1-dependent regulation of ZC3H12A expression. BMC Mol Biol 2010; 11: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalniak L, Mizgalska D, Zarebski A, Wyrzykowska P, Koj A, Jura J. Regulatory feedback loop between NF-kappaB and MCP-1-induced protein 1 RNase. FEBS J 2009; 276: 5892–5905. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Chu JS, Chien HL, Tseng CH, Ko PC, Mei YY et al. MCPIP1 suppresses hepatitis C virus replication and negatively regulates virus-induced proinflammatory cytokine responses. J Immunol 2014; 193: 4159–4168. [DOI] [PubMed] [Google Scholar]
- Lim YJ, Choi JA, Lee JH, Choi CH, Kim HJ, Song CH. Mycobacterium tuberculosis 38-kDa antigen induces endoplasmic reticulum stress-mediated apoptosis via toll-like receptor 2/4. Apoptosis 2015; 20: 358–370. [DOI] [PubMed] [Google Scholar]
- Schott J, Reitter S, Philipp J, Haneke K, Schafer H, Stoecklin G. Translational regulation of specific mRNAs controls feedback inhibition and survival during macrophage activation. PLoS Genet 2014; 10: e1004368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wang J, Azfer A, Song W, Tromp G, Kolattukudy PE et al. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem 2008; 283: 6337–6346. [DOI] [PubMed] [Google Scholar]
- Liang J, Wang J, Saad Y, Warble L, Becerra E, Kolattukudy PE. Participation of MCP-induced protein 1 in lipopolysaccharide preconditioning-induced ischemic stroke tolerance by regulating the expression of proinflammatory cytokines. J Neuroinflammation 2011; 8: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Qiu C, Miao R, Zhou J, Lee A, Liu B et al. MCPIP1 restricts HIV infection and is rapidly degraded in activated CD4+ T cells. Proc Natl Acad Sci USA 2013; 110: 19083–19088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Chien HL, Lin SY, Chang BL, Yu HP, Tang WC et al. MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res 2013; 41: 3314–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Miao R, Zhou Z, Wang T, Liu J, Liu G et al. MCPIP1 negatively regulates toll-like receptor 4 signaling and protects mice from LPS-induced septic shock. Cell Signal 2013; 25: 1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Romeu E, Ferran M, Gimenez-Arnau A, Bugara B, Lipert B, Jura J et al. MCPIP1 RNase is aberrantly distributed in psoriatic epidermis and rapidly induced by IL-17A. J Invest Dermatol 2016; 136: 1599–1607. [DOI] [PubMed] [Google Scholar]
- Somma D, Mastrovito P, Grieco M, Lavorgna A, Pignalosa A, Formisano L et al. CIKS/DDX3X interaction controls the stability of the Zc3h12a mRNA induced by IL-17. J Immunol 2015; 194: 3286–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res 2011; 9: 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XX, Wang C, Huang SF, Chen Q, Hu YF, Zhou L et al. Regnase-1 in microglia negatively regulates high mobility group box 1-mediated inflammation and neuronal injury. Sci Rep 2016; 6: 24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalniak L, Koj A, Jura J. Proteasome inhibitor MG-132 induces MCPIP1 expression. FEBS J 2013; 280: 2665–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Gao J, Shi Z, Tai S, Chan LL, Yang Y et al. MG132 induces expression of monocyte chemotactic protein-induced protein 1 in vascular smooth muscle cells. J Cell Physiol 2017; 232: 122–128. [DOI] [PubMed] [Google Scholar]
- Makki MS, Haqqi TM. Histone deacetylase inhibitor vorinostat (SAHA, MK0683) perturb miR-9-MCPIP1 axis to block IL-1beta-induced IL-6 expression in human OA chondrocytes. Connect Tissue Res 2016; 12: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang Y, Zhang W, Liu H, Zhou Z, Dai X et al. MCPIP1 regulates alveolar macrophage apoptosis and pulmonary fibroblast activation after in vitro exposure to silica. Toxicol Sci 2016; 151: 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Dai X, Cheng Y, Fang S, Zhang Y, Wang X et al. MCPIP1 mediates silica-induced cell migration in human pulmonary fibroblasts. Am J Physiol Lung Cell Mol Physiol 2016; 310: 121–132. [DOI] [PubMed] [Google Scholar]
- Chao J, Dai X, Pena T, Doyle DA, Guenther TM, Carlson MA. MCPIP1 regulates fibroblast migration in 3-D collagen matrices downstream of MAP kinases and NF-kappaB. J Invest Dermatol 2015; 135: 2944–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da J, Zhuo M, Qian M. MCPIP is induced by cholesterol and participated in cholesterol-caused DNA damage in HUVEC. Int J Clin Exp Pathol 2015; 8: 10625–10634. [PMC free article] [PubMed] [Google Scholar]
- Monk JM, Liddle DM, De Boer AA, Brown MJ, Power KA, Ma DW et al. Fish-oil-derived n-3 PUFAs reduce inflammatory and chemotactic adipokine-mediated cross-talk between co-cultured murine splenic CD8+ T cells and adipocytes. J Nutr 2015; 145: 829–838. [DOI] [PubMed] [Google Scholar]
- Govey PM, Kawasawa YI, Donahue HJ. Mapping the osteocytic cell response to fluid flow using RNA-Seq. J Biomech 2015; 48: 4327–4332. [DOI] [PubMed] [Google Scholar]
- Jeltsch KM, Hu D, Brenner S, Zoller J, Heinz GA, Nagel D et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat Immunol 2014; 15: 1079–1089. [DOI] [PubMed] [Google Scholar]
- Li H, He H, Gong L, Fu M, Wang TT. Short communication: preferential killing of HIV latently infected CD4(+) T Cells by MALT1 inhibitor. AIDS Res Hum Retroviruses 2016; 32: 174–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Ma R, Yang L, Hu G, Chen X, Duan M et al. MiR-9 promotes microglial activation by targeting MCPIP1. Nat Commun 2014; 5: 4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Chao J, Kook YH, Gao Y, Yao H, Buch SJ. Involvement of miR-9/MCPIP1 axis in PDGF-BB-mediated neurogenesis in neuronal progenitor cells. Cell Death Dis 2013; 4: e960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makki MS, Haseeb A, Haqqi TM. MicroRNA-9 promotion of interleukin-6 expression by inhibiting monocyte chemoattractant protein-induced protein 1 expression in interleukin-1beta-stimulated human chondrocytes. Arthritis Rheumatol 2015; 67: 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, An BY, Xi XB, Li ZW, Li FY. MicroRNA-9 controls apoptosis of neurons by targeting monocyte chemotactic protein-induced protein 1 expression in rat acute spinal cord injury model. Brain Res Bull 2016; 121: 233–240. [DOI] [PubMed] [Google Scholar]
- Dong C, Sun X, Guan Z, Zhang M, Duan M. Modulation of influenza A virus replication by microRNA-9 through targeting MCPIP1. J Med Virol 2017; 89: 41–48. [DOI] [PubMed] [Google Scholar]
- Makki MS, Haqqi TM. miR-139 modulates MCPIP1/IL-6 expression and induces apoptosis in human OA chondrocytes. Exp Mol Med 2015; 47: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Du L, Jiao H, Zhu H, Xu K, Guo S et al. Mmu-miR-27a-5p-Dependent Upregulation of MCPIP1 Inhibits the Inflammatory Response in LPS-Induced RAW264.7 Macrophage Cells. Biomed Res Int 2015; 2015: 607692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell 2011; 44: 424–436. [DOI] [PubMed] [Google Scholar]
- Choudhury NR, Michlewski G. Terminal loop-mediated control of microRNA biogenesis. Biochem Soc Trans 2012; 40: 789–793. [DOI] [PubMed] [Google Scholar]
- Qu B, Cao J, Zhang F, Cui H, Teng J, Li J et al. Type I interferon inhibition of microRNA-146a maturation through up-regulation of monocyte chemotactic protein-induced protein 1 in systemic lupus erythematosus. Arthritis Rheumatol 2015; 67: 3209–3218. [DOI] [PubMed] [Google Scholar]
- Boratyn E, Nowak I, Horwacik I, Durbas M, Mistarz A, Kukla M et al. Monocyte chemoattractant protein-induced protein 1 overexpression modulates transcriptome, including microRNA, in human neuroblastoma cells. J Cell Biochem 2016; 117: 694–707. [DOI] [PubMed] [Google Scholar]
- Suzuki HI, Katsura A, Miyazono K. A role of uridylation pathway for blockade of let-7 microRNA biogenesis by Lin28B. Cancer Sci 2015; 106: 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 2009; 458: 1185–1190. [DOI] [PubMed] [Google Scholar]
- Wawro M, Kochan J, Kasza A. The perplexities of the ZC3H12A self-mRNA regulation. Acta Biochim Pol 2016; 63: 411–415. [DOI] [PubMed] [Google Scholar]
- Kochan J, Wawro M, Kasza A. Simultaneous detection of mRNA and protein in single cells using immunofluorescence-combined single-molecule RNA FISH. Biotechniques 2015; 59: 209–212. [DOI] [PubMed] [Google Scholar]
- Li M, Cao W, Liu H, Zhang W, Liu X, Cai Z et al. MCPIP1 down-regulates IL-2 expression through an ARE-independent pathway. PLoS ONE 2012; 7: e49841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizgalska D, Wegrzyn P, Murzyn K, Kasza A, Koj A, Jura J et al. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. Febs J 2009; 276: 7386–7399. [DOI] [PubMed] [Google Scholar]
- Kochan J, Wawro M, Kasza A. IF-combined smRNA FISH reveals interaction of MCPIP1 protein with IER3 mRNA. Biol Open 2016; 5: 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Miao R, Huang S, Elder B, Quinn T, Papasian CJ et al. MCPIP1 deficiency in mice results in severe anemia related to autoimmune mechanisms. PLoS ONE 2013; 8: e82542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S. Regnase-1, a ribonuclease involved in the regulation of immune responses. Cold Spring Harb Symp Quant Biol 2013; 78: 51–60. [DOI] [PubMed] [Google Scholar]
- Miao R, Huang S, Zhou Z, Quinn T, Van Treeck B, Nayyar T et al. Targeted disruption of MCPIP1/Zc3h12a results in fatal inflammatory disease. Immunol Cell Biol 2013; 91: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Du F, Wang Y, Huang S, Miao R, Major AS et al. Bone marrow deficiency of MCPIP1 results in severe multi-organ inflammation but diminishes atherogenesis in hyperlipidemic mice. PLoS One 2013; 8: e80089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan NI, Goldschmidt-Clermont PJ, Parker-Thornburg J, Shapiro SD, Kolattukudy PE. Contribution of monocytes/macrophages to compensatory neovascularization: the drilling of metalloelastase-positive tunnels in ischemic myocardium. Circ Res 2000; 87: 378–384. [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE, Quach T, Bergese S, Breckenridge S, Hensley J, Altschuld R et al. Myocarditis induced by targeted expression of the MCP-1 gene in murine cardiac muscle. Am J Pathol 1998; 152: 101–111. [PMC free article] [PubMed] [Google Scholar]
- Niu J, Wang K, Graham S, Azfer A, Kolattukudy PE. MCP-1-induced protein attenuates endotoxin-induced myocardial dysfunction by suppressing cardiac NF-small ka, CyrillicB activation via inhibition of Ismall ka, CyrillicB kinase activation. J Mol Cell Cardiol 2011; 51: 177–186. [DOI] [PubMed] [Google Scholar]
- Niu J, Jin Z, Kim H, Kolattukudy PE. MCP-1-induced protein attenuates post-infarct cardiac remodeling and dysfunction through mitigating NF-kappaB activation and suppressing inflammation-associated microRNA expression. Basic Res Cardiol 2015; 110: 26. [DOI] [PubMed] [Google Scholar]
- Labedz-Maslowska A, Lipert B, Berdecka D, Kedracka-Krok S, Jankowska U, Kamycka E et al. Monocyte chemoattractant protein-induced protein 1 (MCPIP1) enhances angiogenic and cardiomyogenic potential of murine bone marrow-derived mesenchymal stem cells. PLoS ONE 2015; 10: e0133746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Liang J, Wang J, Kolattukudy PE. Delayed brain ischemia tolerance induced by electroacupuncture pretreatment is mediated via MCP-induced protein 1. J Neuroinflammation 2013; 10: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Liang J, Wang J, Kolattukudy PE. MCP-induced protein 1 mediates the minocycline-induced neuroprotection against cerebral ischemia/reperfusion injury in vitro and in vivo. J Neuroinflammation 2015; 12: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J, Azfer A, Zhelyabovska O, Fatma S, Kolattukudy PE. Monocyte chemotactic protein (MCP)-1 promotes angiogenesis via a novel transcription factor, MCP-1-induced protein (MCPIP). J Biol Chem 2008; 283: 14542–14551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J, Wang K, Zhelyabovska O, Saad Y, Kolattukudy PE. MCP-1-induced protein promotes endothelial-like and angiogenic properties in human bone marrow monocytic cells. J Pharmacol Exp Ther 2013; 347: 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrotsos EG, Kolattukudy PE, Sugaya K. MCP-1 involvement in glial differentiation of neuroprogenitor cells through APP signaling. Brain Res Bull 2009; 79: 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younce CW, Azfer A, Kolattukudy PE. MCP-1 (monocyte chemotactic protein-1)-induced protein, a recently identified zinc finger protein, induces adipogenesis in 3T3-L1 pre-adipocytes without peroxisome proliferator-activated receptor gamma. J Biol Chem 2009; 284: 27620–27628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younce C, Kolattukudy P. MCP-1 induced protein promotes adipogenesis via oxidative stress, endoplasmic reticulum stress and autophagy. Cell Physiol Biochem 2012; 30: 307–320. [DOI] [PubMed] [Google Scholar]
- Lipert B, Wegrzyn P, Sell H, Eckel J, Winiarski M, Budzynski A et al. Monocyte chemoattractant protein-induced protein 1 impairs adipogenesis in 3T3-L1 cells. Biochim Biophys Acta 2014; 1843: 780–788. [DOI] [PubMed] [Google Scholar]
- Qi D, Huang S, Miao R, She ZG, Quinn T, Chang Y et al. Monocyte chemotactic protein-induced protein 1 (MCPIP1) suppresses stress granule formation and determines apoptosis under stress. J Biol Chem 2011; 286: 41692–41700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Hayes CN, Ochi H, Tsuge M, Miki D, Hiraga N et al. Inverse association of IL28B genotype and liver mRNA expression of genes promoting or suppressing antiviral state. J Med Virol 2011; 83: 1597–1607. [DOI] [PubMed] [Google Scholar]
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009; 461: 399–401. [DOI] [PubMed] [Google Scholar]
- Li H, Wang TT. MCPIP1/regnase-I inhibits simian immunodeficiency virus and is not counteracted by Vpx. J Gen Virol 2016; 97: 1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazusiak E, Florczyk D, Jura J, Potempa J, Koziel J. Differential regulation by Toll-like receptor agonists reveals that MCPIP1 is the potent regulator of innate immunity in bacterial and viral infections. J Innate Immun 2013; 5: 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KH, Jung J, Lee JH, Hong YH. Blood transcriptome profiling in myasthenia gravis patients to assess disease activity: a pilot RNA-seq study. Exp Neurobiol 2016; 25: 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Chen Z, Wang Q, Song X, Zhang L. Comparisons of gene expression in normal, lesional, and non-lesional psoriatic skin using DNA microarray techniques. Int J Dermatol 2014; 53: 1213–1220. [DOI] [PubMed] [Google Scholar]
- He M, Liang X, He L, Wen W, Zhao S, Wen L et al. Endothelial dysfunction in rheumatoid arthritis: the role of monocyte chemotactic protein-1-induced protein. Arterioscler Thromb Vasc Biol 2013; 33: 1384–1391. [DOI] [PubMed] [Google Scholar]
- Jeltsch KM, Heissmeyer V. Regulation of T cell signaling and autoimmunity by RNA-binding proteins. Curr Opin Immunol 2016; 39: 127–135. [DOI] [PubMed] [Google Scholar]
- Garg AV, Amatya N, Chen K, Cruz JA, Grover P, Whibley N et al. MCPIP1 endoribonuclease activity negatively regulates interleukin-17-mediated signaling and inflammation. Immunity 2015; 43: 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalniak A, Boratyn E, Tyrkalska SD, Horwacik I, Durbas M, Lastowska M et al. Expression of the monocyte chemotactic protein-1-induced protein 1 decreases human neuroblastoma cell survival. Oncol Rep 2014; 31: 2385–2392. [DOI] [PubMed] [Google Scholar]
- Lyu JH, Park DW, Huang B, Kang SH, Lee SJ, Lee C et al. RGS2 suppresses breast cancer cell growth via a MCPIP1-dependent pathway. J Cell Biochem 2015; 116: 260–267. [DOI] [PubMed] [Google Scholar]
- Lu W, Ning H, Gu L, Peng H, Wang Q, Hou R et al. MCPIP1 selectively destabilizes transcripts associated with an antiapoptotic gene expression program in breast cancer cells that can elicit complete tumor regression. Cancer Res 2016; 76: 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J, Shi Y, Xue J, Miao R, Huang S, Wang T et al. USP10 inhibits genotoxic NF-kappaB activation by MCPIP1-facilitated deubiquitination of NEMO. Embo J 2013; 32: 3206–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Huang X, Xin HB, Fu M, Xue A, Wu ZH. TRAF family member-associated NF-kappaB activator (TANK) inhibits genotoxic nuclear factor kappaB activation by facilitating deubiquitinase USP10-dependent deubiquitination of TRAF6 ligase. J Biol Chem 2015; 290: 13372–13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N, Niu J, Saad Y, Kumar S, Sirakova T, Becerra E et al. Transcription factors STAT6 and KLF4 implement macrophage polarization via the dual catalytic powers of MCPIP. J Immunol 2015; 194: 6011–6023. [DOI] [PMC free article] [PubMed] [Google Scholar]