Abstract
Hepatocellular carcinoma (HCC) is the most common type of liver cancer in humans. The focal adhesion tyrosine kinase (FAK) is often over-expressed in human HCC and FAK inhibition may reduce HCC cell invasiveness. However, the anti-oncogenic effect of FAK knockdown in HCC cells remains to be clarified. We found that FAK depletion in HCC cells reduced in vitro and in vivo tumorigenicity, by inducing G2/M arrest and apoptosis, decreasing anchorage-independent growth, and modulating the expression of several cancer-related genes. Among these genes, we showed that FAK silencing decreased transcription and nuclear localization of enhancer of zeste homolog 2 (EZH2) and its tri-methylation activity on lysine 27 of histone H3 (H3K27me3). Accordingly, FAK, EZH2 and H3K27me3 were concomitantly upregulated in human HCCs compared to non-tumor livers. In vitro experiments demonstrated that FAK affected EZH2 expression and function by modulating, at least in part, p53 and E2F2/3 transcriptional activity. Moreover, FAK silencing downregulated both EZH2 binding and histone H3K27me3 levels at the promoter of its target gene NOTCH2. Finally, we found that pharmacological inhibition of FAK activity resembled these effects although milder. In summary, we demonstrate that FAK depletion reduces HCC cell growth by affecting cancer-promoting genes including the pro-oncogene EZH2. Furthermore, we unveil a novel unprecedented FAK/EZH2 crosstalk in HCC cells, thus identifying a targetable network paving the way for new anticancer therapies.
Hepatocellular carcinoma (HCC) is one of the most common human cancers, accounting for 70–85% of primary liver malignancies, and the third leading cause of cancer-related death worldwide.1, 2
Despite progress in cancer research and treatment, HCC carries an extremely poor prognosis since it is often diagnosed at advanced stages, restricting efficient therapeutic options to either surgical resection or transplantation.3, 4 Hence, it is urgent to develop new and more effective therapeutic strategies to defeat HCC. To achieve this goal, it is crucial to deeply dissect molecular signaling pathways that drive and/or control the development and progression of HCC.
It is widely accepted that hepatocarcinogenesis is characterized by the accumulation of genetic and epigenetic changes that occur during onset, promotion and progression of HCC.5 Comparative genomic hybridization studies and whole-genome sequencing have started to reveal the genetic landscape of HCC, disclosing the most important oncogenic pathways.6, 7, 8 Mutations in TP53 and β-catenin gene (CTNNB1) frequently occur in HCC.6 Kan et al.8 found that 26.1% of HCCs harbor amplification of the human protein tyrosine kinase 2 (PTK2) gene. This gene is located on 8q24 chromosome and codes for a 125-kDa non-receptor tyrosine kinase named focal adhesion kinase (FAK).9, 10
FAK is placed at the crossroads of many signaling pathways that can promote cancer growth and metastasis. Indeed, FAK is able to control, in a kinase-dependent manner, tumor cell motility and invasion.11, 12 Moreover, FAK can facilitate tumor cell survival and regulate evasion of anti-tumor immunity through kinase-independent scaffolding functions.13, 14
In HCC, FAK over-expression has been even associated with promotion of portal venous invasion and consequent intrahepatic metastasis.15, 16, 17, 18 Although liver homeostasis is not affected in a mouse model with a specific deletion of FAK in hepatocytes, FAK is required for c-Met/β-catenin-driven hepatocarcinogenesis.19 Moreover, FAK is found to be a Met direct target responsible for Met-induced invasion and metastasis.19, 20 Other promising studies showed that (i) pharmacological inhibition of FAK reduces HCC invasion in vitro and growth and metastasis in vivo; and (ii) silencing of FAK decreases cell invasion and viability in vitro.21, 22 Altogether these findings corroborate the hypothesis of a master role of FAK in the growth of HCC. However, the effect of a specific genetic reduction of FAK levels in HCC cells in vivo remains to be clarified and the potential crosstalk of FAK with master genes involved in HCC is yet to be unveiled.
Here, we found that stable silencing of FAK strongly reduced both in vitro and in vivo HCC growth,confirming the key role of this protein in hepatocarcinogenesis. We further demonstrated that FAK depletion critically affected the transcription, nuclear localization and activity on histone trimethylation at lysine 27 (H3K27me3) of enhancer of zeste homolog 2 (EZH2), an epigenetic repressor and putative oncogene in HCC.23, 24, 25 In addition, FAK, EZH2 and H3K27me3 were concomitantly upregulated in human HCCs compared to non-tumor livers. Importantly, we found that p53 and E2F2/3 were key mediators of FAK-dependent effects on EZH2 expression/activity.
In conclusion, here we describe a new molecular nexus between FAK and EZH2 that controls HCC growth.
Results
FAK silencing reduces cell growth and induces apoptosis of HCC cells in vitro
To explore the functional role of FAK in the control of HCC cell homeostasis, we established a model of stable silencing of FAK by lentivirus-mediated co-delivery of two short hairpin RNAs (shRNAs) specifically targeting both 3′UTR and coding sequence of PTK2. HCC cell lines (HepG2 and Huh6) silenced for FAK (shFAK) exhibited a reduction of FAK mRNA and protein compared to the cells infected with a non-targeting shRNA vector (Control) (Supplementary Figures S1a and b). The upregulation of the FAK-target gene E-Cadherin (CDH1) confirmed the reduction of FAK activity (Supplementary Figures S1a and b).
HCC cells transduced with FAK shRNAs had a slower growth rate and a decreased DNA synthesis with respect to control cells (Supplementary Figure S1c, Figure 1a). Moreover, as a counterweight to the decrease of cells populations in G0/G1 and S phase, the proportion of cells in G2/M phase was higher in shFAK than in control HCC cells (Figure 1b) coherently with an increased early apoptosis (Figure 1c). HCC cells stably silenced for FAK also exhibited a significant decrease in colony formation with respect to control cells (Supplementary Figure S1d).
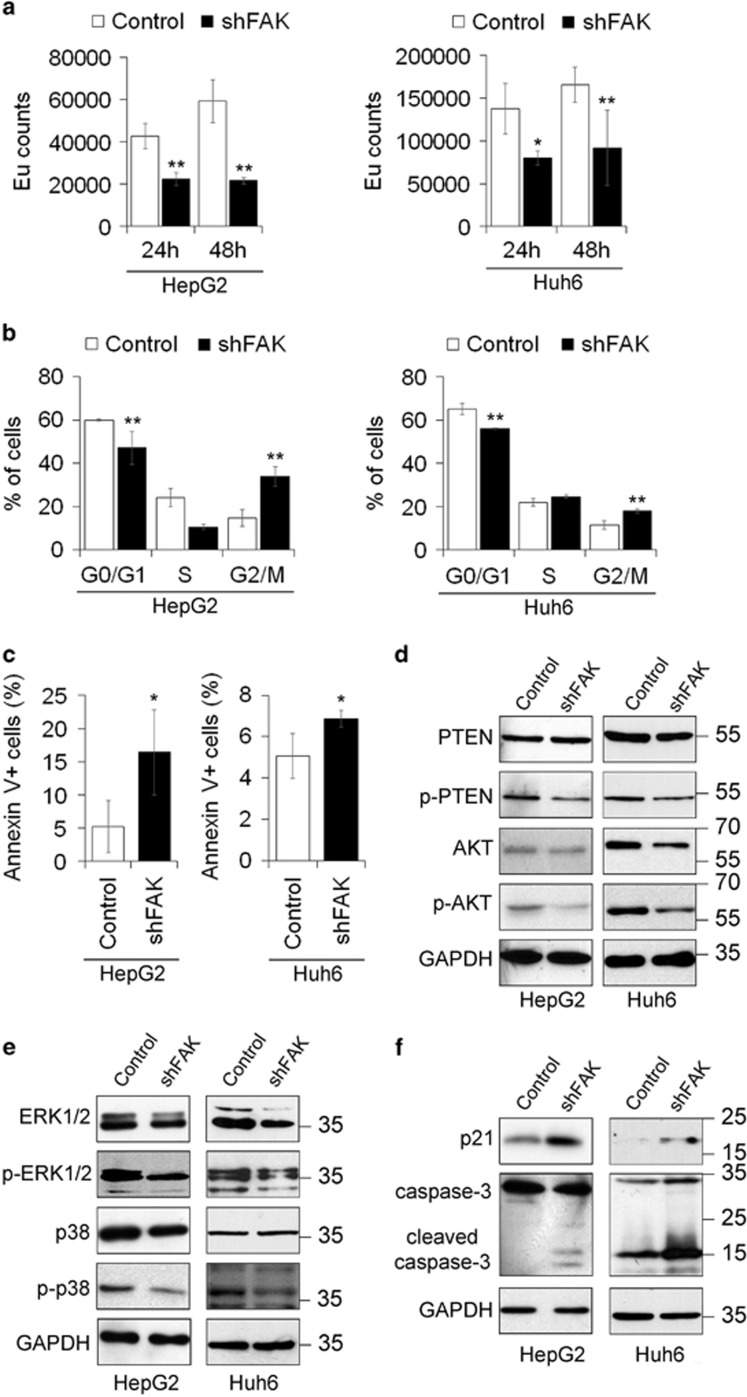
Figure 1.
FAK depletion inhibits growth and promotes apoptosis in HCC cells. (a) Cell proliferation assayed via a BrdU incorporation kit and expressed as Europium (Eu) counts in Control and shFAK HepG2 and Huh6 cells. Values are mean±SD (*P< 0.05;**P<0.01; n=4). (b) Distribution of Control and shFAK HepG2 and Huh6 cells in G0/G1, S and G2/M phase of the cell cycle by propidium iodide (PI) staining and flow cytometric analysis. Values are plotted as mean±SD (**P<0.01; n=3). (c) Percentage of apoptotic Control and shFAK HepG2 and Huh6 cells measured by Annexin V and flow cytometry. Values are plotted as mean±SD (*P<0.05; n=3). (d) Representative western blots (WB) for PTEN and p-PTEN, and for AKT and p-AKT in Control and shFAK HepG2 and Huh6 cells. (e) Representative WB for ERK1/2 and p-ERK1/2, and for p38 and p-p38 in Control and shFAK HepG2 (n=3) and Huh6 cells (n=2). (f) Representative WB for p21 and caspase-3 in Control and shFAK HepG2 (n = 2) and Huh6 cells (n = 2). GAPDH is reported as a loading control
Reduced proliferation rate and apoptosis induction in HCC cells silenced for FAK were confirmed by the decreased activity of signaling pathways commonly involved in these processes. In fact, the expression of the serine 380 phosphorylated/inactive form of phosphatase and tensin homolog (PTEN) and of its target serine 473 phosphorylated/active form of AKT decreased in shFAK cells (Figure 1d,Supplementary Figure S1e). Moreover, the protein expression levels of total and phosphorylated/active form of ERK1/2 and p38 mitogen activated protein kinase were reduced by knockdown of FAK as well (Figure 1e, Supplementary Figure S1f). Finally, p21 increase and the appearance of the cleaved form of caspase-3 were also observed in shFAK cells, confirming G2/M arrest and apoptosis reported before (Figure 1f).
FAK silencing reduces growth of human HCC in mouse xenografts
The role of FAK in in vivo HCC growth was tested in orthotopic and heterotopic tumor-xenograft models, using HepG2 cells engineered with a luciferase reporter (HepG2-luc).26
HepG2-luc-CTRL and HepG2-luc-shFAK cells were injected intramuscularly in nude mice or directly into the liver of NOD/SCID mice, and tumor growth was monitored at different time points by bioluminescence imaging. Notably, NOD/SCID mice showed a very significant reduction of tumor growth generated through intrahepatic injection of HepG2-luc-shFAK as compared to HepG2-luc-CTRL xenografts (Figures 2a and b, Supplementary Figure S2a). The anti-tumor effect of FAK silencing was also confirmed by the intramuscular injection of HCC cells in nude mice (Supplementary Figure S2b).
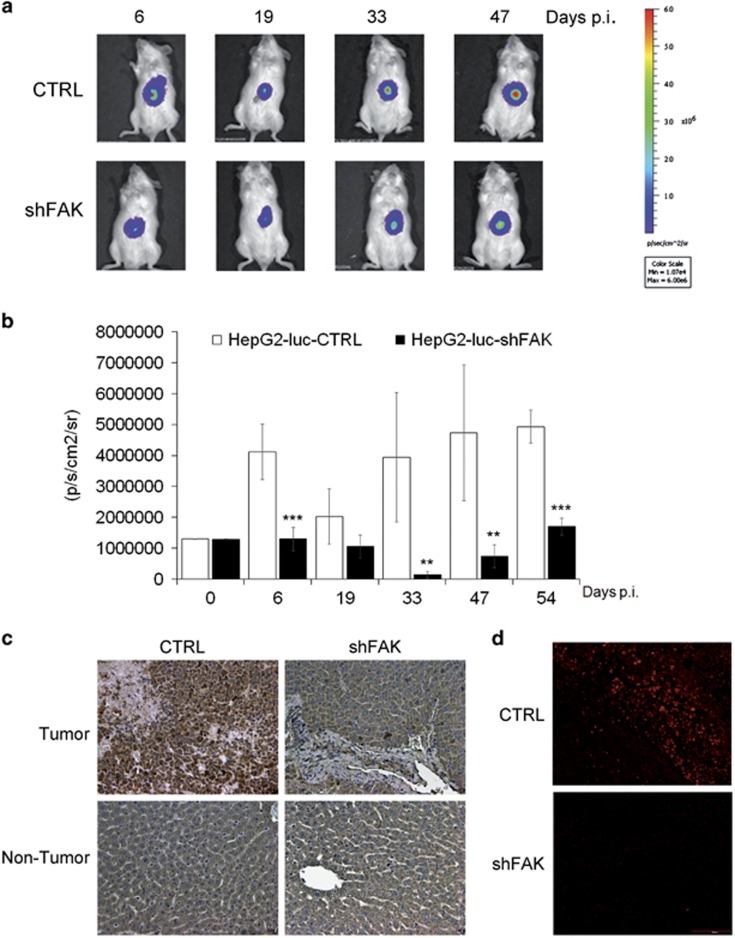
Figure 2.
FAK depletion impairs HCC growth in mouse xenograft models. (a) Representative imaging of NOD/SCID mice bearing intrahepatic tumors from respectively HepG2-luc-CTRL (CTRL) and HepG2-luc-shFAK cells (shFAK). (b) Quantitative analysis of luciferase activity in vivo at various time points. The luminescence signals are expressed as mean±SD of total flux of photon/sec/cm2/steradian (p/s/cm2/sr) (**P<0.01; ***P<0.001; n=6). (c) Representative IHC of FAK expression in liver samples (tumor and non-tumor) from CTRL and shFAK xenografts at 54 days p.i. (×20 magnification). (d) Representative IF of PCNA expression in intrahepatic tumor samples from CTRL and shFAK xenografts at 54 days p.i. (×20 magnification)
Tissue analysis confirmed that the expression of FAK at 54 days post injection (p.i.) was drastically reduced in intrahepatic tumors derived from FAK-silenced HepG2 cells (Figure 2c). Concomitantly, FAK knockdown caused, in the tumor area, downregulation of the expression of proliferating cell nuclear antigen (Figure 2d), a marker of cell proliferation in HCC,27 upregulation of caspase-3 (Supplementary Figure S2c), and over-expression of E-Cadherin protein and mRNA (Supplementary Figures S2d and e).
FAK regulates the expression of cancer-related genes in HCC cells
To gain mechanistic insights into the role of FAK in HCC, we sought to determine FAK-dependent expression of cancer genes in HepG2 cells silenced or over-expressing PTK2 gene, respectively called shFAK and PTK2. To this end, we performed the analysis of 624 cancer genes with an Open Array Real-Time PCR platform. Gene expression profile analysis led to the identification of differentially up- or downregulated genes upon FAK depletion or over-expression (Figures 3a and b, Supplementary Tables S1 and S2).
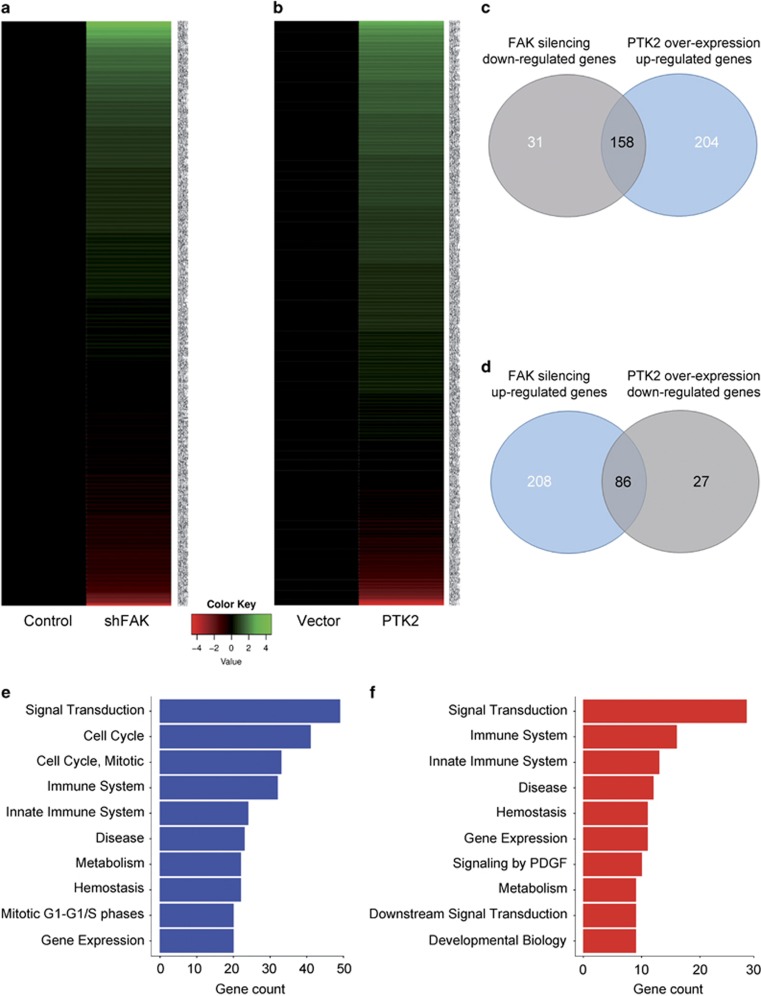
Figure 3.
Cancer-associated genes are influenced by FAK expression. (a) Heatmap representation of the expression of cancer-related genes in HepG2 cells upon FAK silencing compared to control (shFAK versus Control). (b) Heatmap representation of the expression of cancer-related genes in HepG2 cells upon FAK over-expression compared to empty vector (PTK2 versus Vector). (c) Venn diagram showing the overlapping of genes downregulated upon FAK depletion with those upregulated upon FAK over-expression. (d) Venn diagram showing the overlapping of genes upregulated upon FAK depletion with those downregulated upon FAK over-expression. (e) Bar plot of the 10 most abundant pathways for genes downregulated upon FAK silencing. (f) Bar plot of the 10 most abundant pathways for genes upregulated upon FAK silencing
As shown by Venn diagram (Figure 3c), among the genes downregulated after FAK knockdown, we identified a set of 158 potential direct/indirect specific targets being also upregulated upon FAK over-expression. Moreover, we found a set of 86 genes that are either upregulated by FAK silencing or downregulated under FAK over-expression (Figure 3d). These genes are involved in the control of several cellular processes, comprising signal transduction, immune response, cell cycle and gene expression regulation, metabolism and hemostasis (Figures 3e and f). Moreover, from the analysis of genes downregulated by FAK silencing and upregulated by FAK over-expression, some epigenetic regulators of gene expression, such as EZH2, have emerged for the first time as direct/indirect potential targets of FAK.
FAK modulates EZH2 expression and trimethylation of histone H3 at lysine 27 in HCC cells
A previous study demonstrated that over-expression of EZH2 correlated with upregulation of total or phosphorylated FAK predicting a more aggressive behavior in endometrial carcinoma.28 In HCC, the relationship between FAK and EZH2 has never been investigated before. Thus, we evaluated the mRNA and protein expression of EZH2 either in a condition of FAK depletion or over-expression in HCC cells. Our results confirmed that FAK knockdown reduced both EZH2 mRNA and protein levels (Figures 4a and b). Conversely, FAK over-expression increased EZH2 mRNA levels (Supplementary Figure S3a).
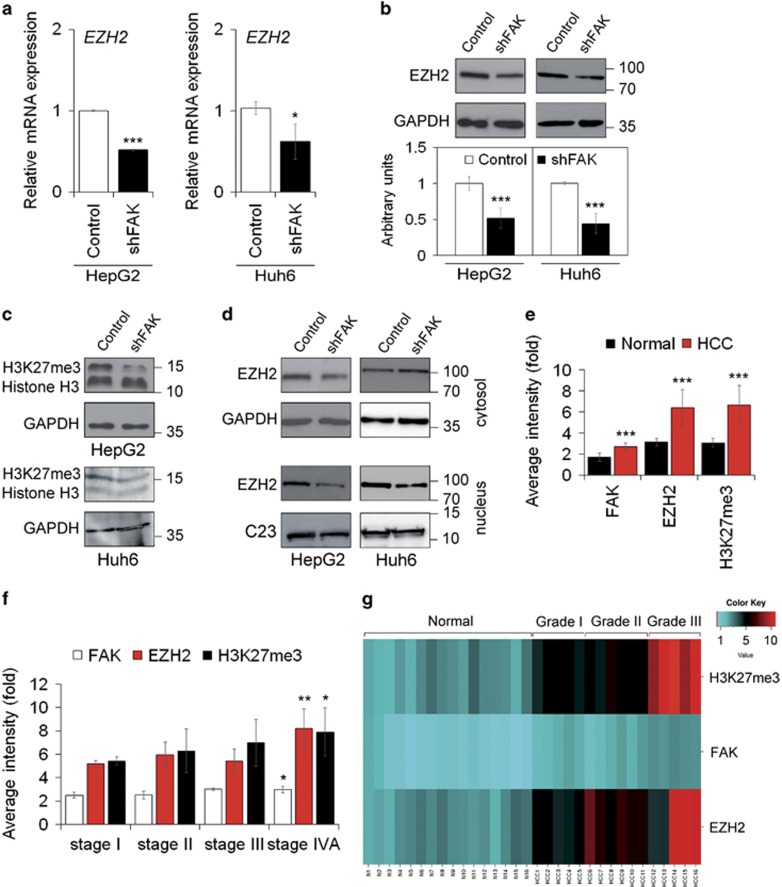
Figure 4.
FAK silencing influences EZH2 expression and activity in HCC cells. (a) Relative mRNA expression of EZH2 gene as measured by quantitative Real-Time PCR (qRT-PCR) in Control and shFAK HepG2 and Huh6 cells. Values are expressed as fold mean±SD (*P<0.05; ***P<0.001; n=3). (b) Representative WB and densitometric quantification for EZH2 protein in Control and shFAK HepG2 and Huh6 cells. GAPDH is reported as a loading control (***P<0.001; n=3). (c) Representative WB of H3K27me3 in Control and shFAK HepG2 and Huh6 cells. GAPDH is reported as a loading control (n=2). (d) Representative WB of cytosolic and nuclear EZH2 protein in Control and shFAK HepG2 and Huh6 cells. GAPDH and C23 are reported as loading controls (n=2). (e) Average fluorescence intensity calculated for FAK, EZH2 and H3K27me3 in liver tumor (HCC) and non-tumor (Normal) samples from the same patients (n=16). Values are plotted as mean±SD (***P<0.001). (f) Average fluorescence intensity calculated for FAK, EZH2 and H3K27me3 in HCC samples according to tumor staging. Values are plotted as mean±SD (*P<0.05; **P<0.01). (g) Heatmap representation of the expression of FAK, EZH2 and H3K27me3 in Normal tissues and HCC according to grade of disease (I, II, II). This image was generated by using the online tools provided by CIMminer (http://discover.nci.nih.gov/cimminer/home.do)
EZH2 is the core catalytic component of the Polycomb repressive complex 2 (PRC2), responsible for gene repression through trimethylation of lysine 27 on histone H3 (H3K27me3) of target gene promoters.29 We found that shFAK HCC cells exhibited a strong reduction of H3K27me3 total levels associated with a decrease of nuclear EZH2 compared to control cells (Figures 4c and d). As expected, miR-200b, a well-described target of EZH2,30 was upregulated by FAK silencing (Supplementary Figure S3b).
According to in vitro data, immunostaining for EZH2 and H3K27me3 was substantially reduced (Supplementary Figures S3c and d) in tumors from HepG2-luc-shFAK NOD/SCID mice with respect to HepG2-luc-CTRL animals. Furthermore, the analysis of tissue arrays, which contained 16 cases of HCC, in duplicates, with corresponding normal non-tumor zone from the same patients as controls, revealed concomitant upregulation of FAK, EZH2 and H3K27me3 in tumor specimens (Figure 4e). Noteworthy, the increase of all three proteins correlate with the severity of the disease in terms of tumor staging, differentiation and presence of metastasis (Figure 4e–g, Supplementary Figures S3e and f,Supplementary Table S3).
p53 mediates FAK-dependent effects on EZH2 expression and activity
To unveil the molecular link connecting FAK to EZH2, we investigated different pathways that have previously been described as regulators of EZH2 expression and activity.
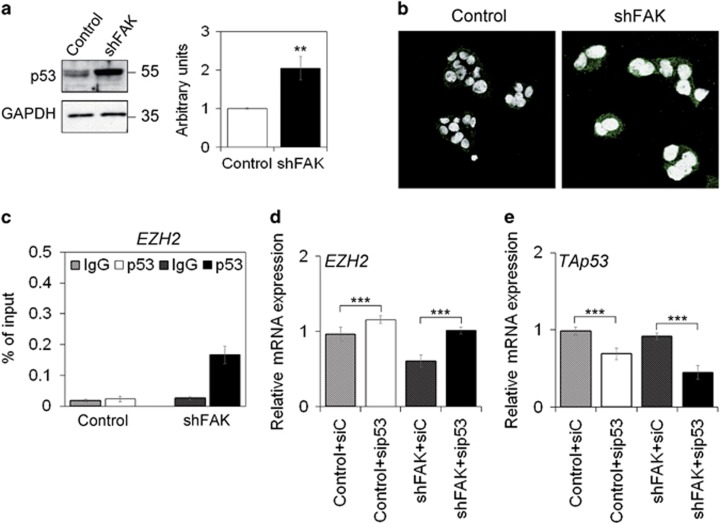
FAK depletion did not affect expression levels of miR-101 (Supplementary Figure S4a), which is known to act as a repressor of EZH2 transcription.31 Interestingly, silencing of FAK in HepG2 cells resulted in upregulation of both total and phosphorylated p53 (Figures 5a and b). A previously published study demonstrated that p53 over-expression resulted in the repression of EZH2 transcription.32 To test the hypothesis of a p53-driven EZH2 regulation in our model, we performed a Chromatin Immunoprecipitation (ChIP) assay using a specific anti-p53 antibody to precipitate chromatin fragments from either control or FAK-depleted HepG2 cells. To this purpose, specific primers have been designed around the CpG Island in the promoter region of EZH2 (Supplementary Figures S4b and c). Increased p53 binding to the EZH2 promoter in a region without p53 binding motifs confirmed our hypothesis (Figure 5c and Supplementary Figure S4d). The role of p53 in mediating FAK-dependent regulation of EZH2 was confirmed by transient silencing experiments showing that p53 silencing in a condition of FAK depletion caused upregulation of EZH2 gene expression (Figures 5d and e).
Figure 5.
p53 is a mediator of FAK silencing effects on EZH2 in HepG2 cells. (a) Representative WB and densitometric quantification for p53 in Control and shFAK HepG2 cells. GAPDH is reported as a loading control. Values are mean±SD of at least three independent experiments (**P<0.01; n=3). (b) Representative IF of serine 15 phosphorylated form of p53 in Control and shFAK HepG2 cells. DRAQ5 was used to stain nuclei ( × 60 magnification). (c) ChIP assays for p53 at the EZH2 promoter in Control and shFAK HepG2 cells. Values are expressed as percentage of input DNA of three independent experiments. IgG was used as negative ChIP control. (d) Relative mRNA expression of EZH2 gene as measured by qRT-PCR in Control and shFAK HepG2 cells after transient silencing with siC or sip53. Values are expressed as fold mean±SD (***P<0.001; n=3). (e) Relative mRNA expression of TAp53 gene as measured by qRT-PCR in Control and shFAK HepG2 cells after transient silencing with siC or sip53. Values are expressed as fold mean±SD (***P<0.001; n=3)
Experiments performed in Huh7.5 p53 functionally defective cells further confirmed the relevance of p53 in shFAK-dependent reduction of HCC cell growth and EZH2 regulation. The efficiency of FAK silencing was similar to that observed in HepG2 cells (Supplementary Figures S5a and b), but its effect on EZH2 mRNA and protein was less evident than in HepG2 cells (Supplementary Figures S5c and d). Moreover, FAK depletion also induced a slight reduction of proliferation rate and apoptosis in Huh7.5 cells (Supplementary Figures S5e and f).
FAK silencing affects EZH2 transcription/activity by E2F
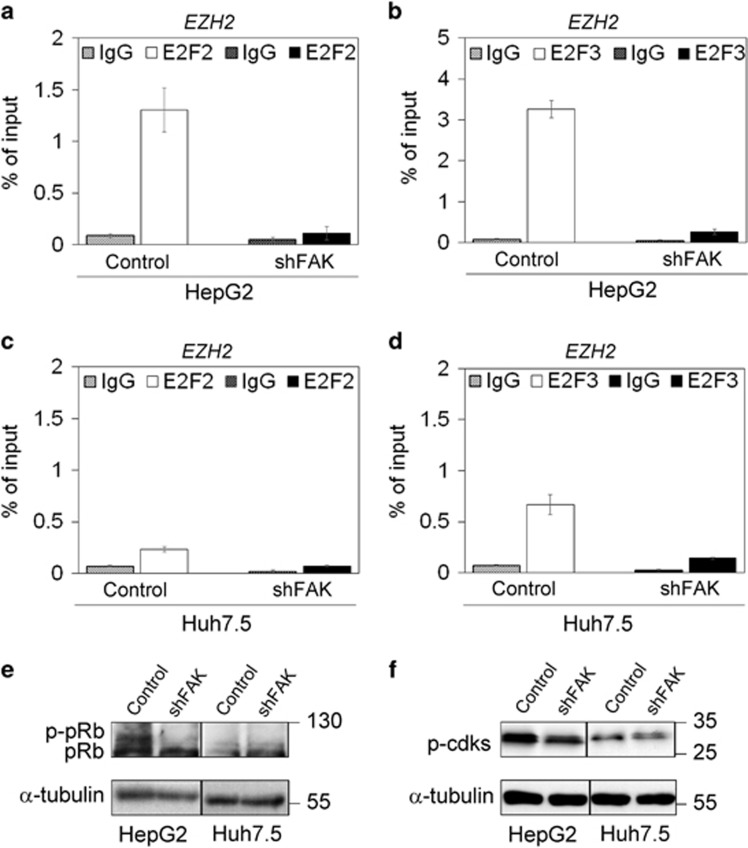
To characterize additional mediators of FAK-related EZH2 transcriptional downregulation, we tested the potential role of E2F, which is able to bind EZH2 promoter to turn on its transcription.33 ChIP assay demonstrated that FAK-depleted HepG2 cells displayed a decrease in the binding of both E2F2 and E2F3 to EZH2 promoter with respect to control HepG2 cells (Figures 6a and b). These results were confirmed in Huh7.5 cells (Figures 6c and d), thus suggesting that FAK/E2F/EZH2 is totally p53 independent.
Figure 6.
FAK silencing in HCC cells affects E2F2 and E2F3 binding to EZH2 promoter. ChIP assay of E2F2 (a) and E2F3 (b) on the EZH2 promoter in Control and shFAK HepG2 cells. ChIP assays of E2F2 (c) and E2F3 (d) on the EZH2 promoter in Control and shFAK Huh7.5 cells. Values are expressed as percentage of input DNA of three independent experiments. IgG was used as negative ChIP control. (e) Representative WB for pRb in Control and shFAK HepG2 and Huh7.5 cells (n=3). (f) Representative WB for phosphorylated cdks (p-cdks) in Control and shFAK HepG2 and Huh7.5 cells. α-tubulin is reported as a loading control (n=3)
It is well known that E2F activity on target genes, such as EZH2, was shown to be constrained by the binding to retinoblastoma protein (pRb).33, 34 We found that FAK depletion in HCC cells increased the expression of hypophosphorylated pRb (Figure 6e). This effect of FAK silencing on pRb could depend on the well-known ERK1/2-mediated relationship between FAK and G1-Phase regulators, such as cyclin D1/cdk4-6 complex and p16INK4, which may influence pRb phosphorylation status.35, 36 Accordingly, under FAK depletion we found a downregulation of phosphorylated cdk level (Figure 6f), and an increase of p16INK4 gene (CDKN2A) expression (Supplementary Figure S6a).
As we previously observed that FAK silencing causes a reduction of EZH2 nuclear localization and activity, we supposed that a physical interaction between these two proteins might also occur. Our preliminary data showed that FAK co-precipitated with EZH2, and that FAK knockdown in HepG2 cells reduced this interaction, suggesting also a possible functional physical interaction. (Supplementary Figures S6b and c).
NOTCH2 is a target gene for the FAK/EZH2 network in HCC cells
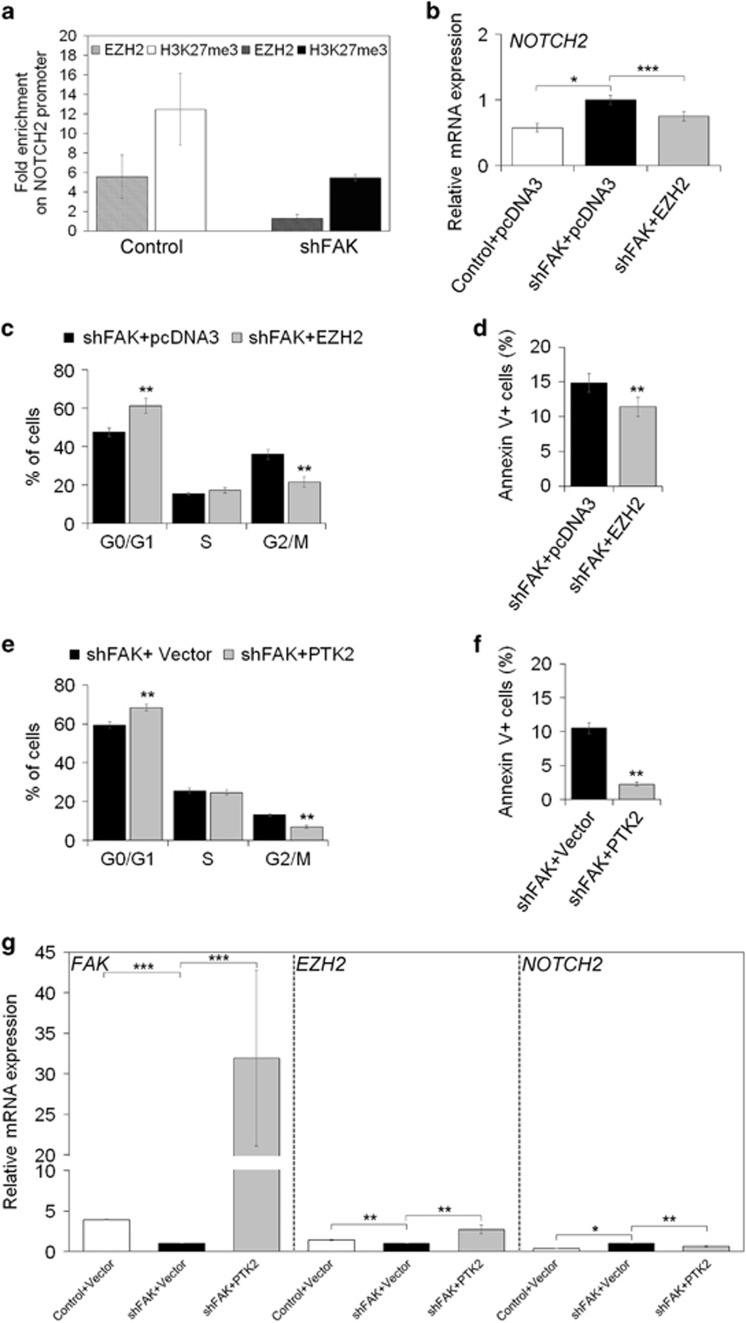
Among all the genes that could be controlled by FAK/EZH2 network, we explored NOTCH2, which has already been reported to be under the control of EZH2 in HCC.37 As previously observed with gene expression profile by OpenArray, FAK silencing caused upregulation of NOTCH2 mRNA levels in HepG2 cells (Supplementary Tables S1). Coherently, upon FAK silencing, we found a reduction of EZH2 and H3K27me3 enrichment on NOTCH2 promoter (Figure 7a). Transient over-expression of an exogenous EZH2 in FAK-silenced HepG2 cells (Supplementary Figure S7a) rescued the expression levels of NOTCH2 (Figure 7b). In addition, EZH2 forced upregulation reverted the expression of other genes modulated by FAK depletion (Supplementary Figures S7b and c,Supplementary Table S4). From a functional point of view, however, EZH2 rescue was able to reduce G2/M phase arrest and apoptosis (Figures 7c and d), and to partially counteract the anchorage-independent growth impaired by FAK silencing (Supplementary Figure S7d).
Figure 7.
FAK silencing affects EZH2 dependent repression of NOTCH2 transcription. (a) ChIP assays for EZH2 and H3K27me3 at the NOTCH2 promoter in Control and shFAK HepG2 cells. Data are expressed as fold enrichment of three independent experiments repeated in duplicate. (b) Relative mRNA expression of NOTCH2 gene as measured by qRT-PCR in Control+pcDNA3, shFAK+pcDNA3 and shFAK+EZH2 HepG2 after 72 h from plasmid transfection. Values are expressed as fold mean±SD of three independent experiments repeated in triplicate (*P<0.05; ***P<0.001). (c) Distribution of shFAK+pcDNA3 and shFAK+EZH2 HepG2 in G0/G1, S, G2/M and SubG1 phase of the cell cycle by PI staining and flow cytometric analysis. Values are plotted as mean±SD (**P<0.01; n=3). (d) Percentage of apoptotic shFAK+pcDNA3 versus shFAK+EZH2 HepG2 cells measured by Annexin V and flow cytometry. Values are plotted as mean±SD (**P< 0.01; n =5). (e) Distribution of shFAK+Vector and shFAK+PTK2 HepG2 in G0/G1, S, G2/M and SubG1 phase of the cell cycle by PI staining and flow cytometric analysis. Values are plotted as mean±SD (**P<0.01; n=3). (f) Percentage of apoptotic of shFAK+Vector and shFAK+PTK2 HepG2 cells measured by Annexin V and flow cytometry. Values are plotted as mean±SD (**P< 0.01; n=3). (g) Relative mRNA expression of FAK, EZH2 and NOTCH2 genes as measured by qRT-PCR after 48 h from plasmid transfection (empty or PTK2 vector) in Control and shFAK HepG2 cells (**P<0.01; *P<0.05; n=3)
A further confirmation that FAK/EZH2 network is crucial for the regulation of HCC cell homeostasis and the effect on target genes, such as NOTCH2, was obtained by re-expressing FAK in HCC cells. In fact, FAK rescue decreased G2/M phase arrest and apoptosis, and caused upregulation of EZH2 mRNA and downregulation of NOTCH2 mRNA in FAK-depleted HepG2 (Figures 7e–g) and Huh7.5 cells (Supplementary Figures S7e–g).
Inhibition of FAK activation induces cell cycle arrest at the G0/G1 phase in HCC cells
In order to evaluate if the observed pro-apoptotic effects depend on FAK activity we treated HepG2 and Huh7.5 cells with PND-1186 that was able to inhibit Tyr-397 phosphorylation of FAK.38
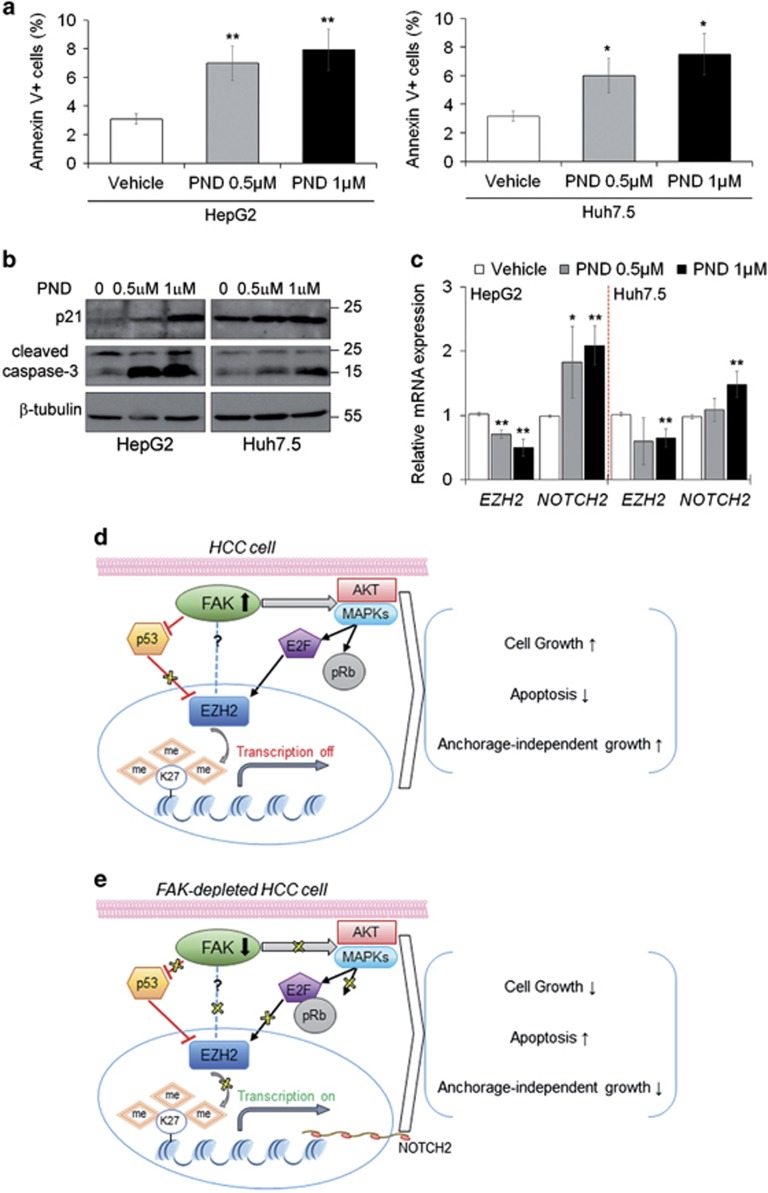
HCC cells were exposed to 0.5 μM and 1 μM of PND-1186. The treatment with PND-1186 decreased cell proliferation rate (Supplementary Figure S8a) by significantly inducing G0/G1 phase arrest and apoptosis at 48 h (Supplementary Figure S8b,Figure 8a). Accordingly, PND-1186 treatment caused downregulation of FAK Tyr-397 phosphorylation and cyclin D1 amount (Supplementary Figures S8c and d) and increased the expression of both p21 and cleaved caspase-3 at 48 h (Figure 8b). As expected, 1 μM PND-1186 significantly downregulated EZH2 mRNA and upregulated NOTCH2 expression (Figure 8c).
Figure 8.
Pharmacological inhibition of FAK mimics the effect of FAK silencing confirming FAK/EZH2 network in HCC cells. (a) Percentage of apoptotic HepG2 and Huh7.5 cells after 48 h of treatment with DMSO (Vehicle), 0.5 μM or 1 μM PND 1186 measured by Annexin V and flow cytometry. Values are plotted as mean±SD (*P<0.05; **P<0.01; versus Vehicle, n =3). (b) Representative WB for p21 and caspase-3 in HepG2 and Huh7.5 cells after 48 h of treatment with DMSO (0), 0.5 μM or 1 μM PND 1186. β-tubulin is reported as a loading control (n=2). (c) Relative mRNA expression of EZH2 and NOTCH2 genes as measured by qRT-PCR in HepG2 and Huh7.5 cells after 48 h of treatment with DMSO (Vehicle), 0.5 μM or 1 μM PND 1186 (*P<0.05; **P<0.01; versus Vehicle, n =3). (d) In HCC cells, upregulated FAK causes inhibition of p53 expression, which normally hampers EZH2 production, while in the meantime induces E2F transcriptional activity favoring EZH2 over-expression. The consequent transcriptional repression of EZH2 target genes supports the tumorigenic cell features. (e) Conversely, the depletion of FAK in the same cellular context results in the expression of p53 and the blockade of E2F functions, critically reducing the EZH2-dependent epigenetic silencing of target genes, thus impairing tumor growth. The dotted blue line indicates additional potential regulations of EZH2 functions by FAK physical interaction
Discussion
Here, we present data demonstrating that specific depletion of FAK by genetic silencing slows down/reduces HCC cell growth in vitro and in vivo. Thus, we unveil a new molecular crosstalk between FAK and the histone methyl transferase EZH2 (Figures 8d and e) that may explain the FAK knockdown-dependent biological effects in HCC.
The anti-tumorigenic in vitro effect of FAK depletion depended on the induction of apoptosis that reduced the proliferation rate by the activation of specific signaling pathways. In fact, the signaling of PTEN/AKT, and the activity of pro-mitogenic activated protein kinases were affected by FAK silencing in line with their already described role on HCC pathogenesis.39, 40, 41, 42 Furthermore, HCC cells with FAK depletion displayed an over-expression of p21, involved in the G2/M cell cycle arrest, which can precede apoptotic response.42 Accordingly, FAK silencing in human HCC cells was able to decrease their intrahepatic growth in immune-deficient mice.
Our data suggest that partial FAK knockdown may achieve these effects regulating the expression of several cancer-related genes. In fact, the analysis of common dysregulated genes, by either silencing or exogenous over-expression of FAK, led to the identification of different genes that can be flagged as new potential direct/indirect targets of FAK. Among these, we identified the gene encoding for EZH2, which was downregulated by FAK silencing in in vitro and in vivo HCC models. Moreover, the modulatory effect of FAK depletion is not only restricted to EZH2 transcriptional regulation but also encompasses its role on H3K27me3 activity. The parallel trend of FAK and EZH2 well correlated with their already-reported tumor promoting activity on HCC.20, 25 In fact, FAK silencing significantly reduced the well-characterized tumor-suppressor miR-200b, as previously reported by Au et al.25 in EZH2-knockdown cell lines and primary HCC samples. Furthermore, since both FAK and EZH2 downregulation may interfere with autophagy in cancer cells,43, 44 it is plausible that also this mechanism may contribute to cell cycle arrest and apoptosis in our system, but further studies are required to confirm or disprove this hypothesis.
To our knowledge, only few evidences of EZH2 and FAK connection have been reported so far. A recent study demonstrates that FAK activity is influenced by EZH2 in dendritic cells,45 while concomitant over-expression of these two proteins was also reported in endometrial carcinoma tissues.28 Accordingly, in our experiments, tumor area from human HCC samples exhibited concomitant over-expression of FAK, EZH2 and H3K27me3 compared to non-tumor zones. Further studies on large cohorts of pediatric and adult HCCs are necessary to explore the correlation of these three interactors with severity and progression of disease.
EZH2 function in transcriptional repression via H3K27me3has been extensively investigated.46 However, the mechanism by which EZH2 is regulated, in response to intrinsic cell-cycle signals or to extrinsic inputs, remains elusive. Although miR-101 has been shown to be a crucial repressor of EZH2 transcription,31 the FAK-dependent EZH2 down-modulation in HCC cells seems to be independent of this mRNA requiring alternative molecular factors.
A previous study showed that p53 over-expression resulted in the EZH2 transcriptional repression.32 Our results demonstrated that an inactive p53 reduced most of the FAK-dependent effects on HCC cell growth, apoptosis and EZH2 regulation suggesting a role of p53 as a crucial mediator of FAK/EZH2 crosstalk. Indeed, we found an increased binding of p53 to EZH2 promoter region upon FAK silencing, confirming the upstream role of p53 on EZH2 also in HCC cells. However, as FAK depletion preserves a quote of its reducing ability on EZH2 transcription and pro-apoptotic effects even in a condition of p53 loss of function, it is likely that additional factors may connect FAK to EZH2 and apoptosis. E2F might be one of these players since it is involved in the control of cell proliferation and may bind EZH2 gene promoter, positively regulating its transcription.33 In line with our hypothesis and literature data,33, 34, 35 we found that E2F2/3 binding to EZH2 promoter is impaired upon FAK silencing in HCC cells and probably as a consequence of decrease of pRb phosphorylation and increase of E2F2/3 retention. In addition, our preliminary results provide evidence that FAK may physically associate with EZH2 at least in p53-functional HCC cells, suggesting an additional mechanism of interaction between these two proteins, which deserves further consideration.
The inhibition of FAK in HCC cells might upregulate NOTCH2 gene expression by reducing the enrichment of EZH2 and H3K27me3 at NOTCH2 promoter. Gao et al.37 previously reported the repression activity of EZH2 on NOTCH2 in HCC. Accordingly, we found that the expression of some genes upregulated by FAK silencing, such as NOTCH2, was downregulated again under EZH2 and PTK2 rescue in FAK-depleted HCC cells, which exhibited apoptosis reduction and unlocking of G2/M-phase arrest.
Finally, the use of the specific FAK inhibitor PND-118647 demonstrated that the impair of FAK autophosphorylation at tyrosine 397 was able to reduce HCC cell growth via induction of G0/G1 arrest and apoptosis, and downregulation of EZH2 gene expression with consequent upregulation of NOTCH2 gene expression. However, the effect of FAK activity inhibition on HCC growth appears to be not as strong as that observed by FAK silencing, thus suggesting that the G2/M arrest and consequent apoptosis might be mainly associated to the scaffold function of FAK.
Our results also highlight that disruption of FAK/EZH2 network may induce apoptosis by both a p53-mediated and p53-independent mechanism, though further studies are required to investigate the performance of FAK/EZH2 network on the other modulated genes.
In conclusion, our results complete the picture from previous studies on the key role of FAK as tumor promoting gene in HCC,20, 21, 22 but more importantly, provide the first evidence of a connection between FAK and EZH2, introducing a new protein network that is active in the control of HCC cell proliferation. In this FAK/EZH2 nexus, p53 and E2F may act as mediators while NOTCH2, and other unexplored genes, would act as potential target genes.
All our findings may have strong therapeutic implications. In fact, the effects, resulting from FAK-dependent deregulation of EZH2 transcriptional repression of specific targets, may converge to reinforce HCC sensitivity to selective FAK inhibition possibly reducing both local tumor growth and distant metastasis. Furthermore, since Azzariti et al. have recently reported48 a novel mechanism by which Sorafenib may induce HCC cell resistance through Laminin-332/α3 integrin-dependent escape of FAK from ubiquitination and consequent degradation, it is plausible that pharmacological inhibition of FAK may also enhance Sorafenib therapeutic performance. This information provides a proof-of-concept that drugs acting against FAK and EZH2, either those already approved by Food and Drug Administration or new ones, could represent a therapeutic option for HCC.11, 49
Materials and Methods
Cell lines and cultures
The human HepG2 cell line was purchased from ATCC (Manassas, VA, USA) that provided certificated authentication, and the Huh6 and HuH7.5 cell lines were provided by a collaboration with Dr. Balsano that performed authentication in 2014. Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 1% L-glutamine, 1% penicillin/streptomycin at 37 °C in 5% CO2 in a 95% humidified atmosphere. All cell lines were intermittently tested for the presence of mycoplasma.
To follow tumor growth in vivo, HepG2 cells were transfected with an inducible plasmid encoding firefly luciferase and selected with G418 to produce HepG2-Luc cells.
shRNA lentiviral transduction
MISSION shRNA Lentiviral Transduction Particles (Sigma Aldrich, Poole, UK) were used to knockdown PTK2 in HepG2 and Huh7.5 cell lines. TRC1-pLKO.1-puro vector containing a short hairpin insert with gene-specific sequence was used for cell transduction according to the manufacturers' protocol in presence of hexadimethrine bromide (8 μg/ml), to enhance transduction efficiency. Sequences of inserts in shRNA constructs targeting the PTK2 gene (3UTR and CDS) are 5′-CCGGGAGAGCATGAAGCAAAGAATTCTCGAGAATTCTTTGCTTCATGCTCTCTTTTT-3′ and 5′-CCGGGTCTAGAAATACGGCGATCATCTCGAGATGATCGCCGTATTTCTAGACTTTTTG-3′ respectively. Pilot studies were performed to define the optimal multiplicity of infection without cell toxicity (MOI=4). In addition, MISSION pLKO.1-puro Non-Target shRNA Control Transduction Particles (Sigma Aldrich) were used as negative control and reference for interpretation of knockdown. Stable gene knockdown was established by cellular resistance to puromycin (5 μg/ml). Obtained polyclones were crio-preserved, tested for FAK mRNA and protein expression and used for the experiments.
Plasmids and siRNA transfection
Details on cell transfection, plasmids and siRNAs were provided in Supplementary Experimental Procedures.
Animals
All procedures involving mice and care were performed in compliance with our institutional animal care guidelines and following national and international directives (D.L. 4 March 2014, no. 26; directive 2010/63/EU of the European parliament and of the council).
CD-1 nude (12 animals) and NOD/SCID (14 animals) male mice 6–8 weeks old were used (Charles River laboratories, Calco, MI, Italy). HepG2-Luc silenced or not for FAK cells were injected intramuscularly in nude mice at 3 × 106 cells/mouse or in the liver of NOD/SCID mice at 106 cells/mice. Intramuscular tumors were measured twice per week, and tumor sizes were calculated using the following formula: V=(a/b2)/2, where a is the largest dimension and b the smallest. Mice bearing intramuscularly or intrahepatic tumors were imaged using the IVIS imaging system 200 series (Caliper Life Sciences, Hopkinton, MA, USA). Briefly, mice were anesthetized and then were injected intraperitoneally with 150 mg/kg D-luciferin and then imaged. At the end of the experiments, mice were killed; livers were collected and imaged. Data were acquired and analyzed using the living image software version 3.0 (Caliper Life Sciences).
Cell growth
Cell proliferation was assessed by trypan blue exclusion assay. Briefly, for each experiment Control and shFAK HCC cells were seeded in T25 flasks in triplicate and cultured. At 24, 48, 72, 96 and 120 h cells were trypsinized and the number of viable cells was counted under a light microscope. At least four counts were performed per flask on three different experiments.
Bromodeoxyuridine-based cell proliferation assay
Control or shFAK HCC cells were plated in a 96 microplate well and allowed to grow. The DELFIA (Dissociation-Enhanced Lanthanide Fluorescent Immunoassay) Cell Proliferation Assay was performed following the manufacturer's instructions (PerkinElmer, Waltham, MA, USA) as previously described.50
Cell cycle analysis
Cell cycle phase distribution was analyzed at 24 h from post-starvation serum replacement by flow cytometry using PI staining (Sigma Aldrich). Briefly, Control or shFAK HCC cells were collected by trypsinization, washed with PBS, then fixed in a solution of a cold 4 : 1 methanol/acetone solution. Cells were first incubated with RNase A at +37 °C then stained with a solution containing 100 μg/ml PI, at +37 °C for 20 min. Stained nuclei were analyzed for DNA-PI fluorescence using a Becton Dickinson FACSCanto II flow cytometer (Becton-Dickinson, Milan, Italy). Resulting DNA distributions were analyzed for the proportions of cells in G0/G1, S phase, and G2/M phases of the cell cycle by DiVa Software, version 6.3 (Becton-Dickinson).
Apoptosis
Apoptosis was assessed by Annexin V staining at 24 h from post-starvation serum replacement. Briefly, cells were washed in PBS and re-suspended in Annexin Binding Buffer (10 mmol/L HEPES pH 7.4, 140 mmol/L NaCl, and 2.5 mmol/L CaCl). Cells were then stained with 0.5 mg/mL Annexin V-FITC (Becton-Dickinson, Milan, Italy) for 15 min before analyzing. Acquisition and analysis were carried out on a Becton Dickinson FACSCanto II flow cytometer, using DiVa Software, version 6.3.
Analysis of mRNAs and protein expression
Details for the analysis of mRNA and protein expression are provided in the Supplementary Experimental Procedures.
Soft agar assay
Anchorage-independent growth was determined by soft agar assay. A base layer of complete culture medium in 1% agar was established in 60-mm culture dishes. HCC cells were plated at 1 × 105 cells per dish in the top layer composed of the same culture medium and 0.7% agar mixture. Growth medium was refreshed every 2–3 days. After 28 days, colonies were stained with Crystal Violet (Sigma Aldrich) and counted under a light microscope.
Immunohistochemistry and immunofluorescence
Immunohistochemical and immunoflorescent staining on human and murine livers were performed as previously reported.51 Major details are available in the Supplementary Experimental Procedures.
Open array
A pre-designed TaqMan OpenArray Human Cancer Panel (Life Technologies, Thermo Fisher Scientific Corporation, Foster City, CA, USA) was used to assess the effect of FAK depletion or over-expression in HepG2 cells on a signature panel of 624 well-defined genes validated for the characterization of cancers, plus 24 endogenous control genes. cDNAs were loaded onto the Open Array platform and run as recommended by the manufacturer on the QuantStudio 12K Flex Real-Time PCR system (Life Technologies, Thermo Fisher Scientific corporation). Details for the analysis of gene profile are provided in the Supplementary Experimental Procedures.
Heatmaps generation and pathway enrichment
Relative quantity (RQ) expression values above 20 were considered unreliable and thus discarded. For visualization purposes, heatmaps were generated by the function heatmap.2 in the R library gplots starting from log2(RQ+e), where e=0.01 was added to avoid infinite values when RQ=0.
For Venn diagrams, genes were considered ‘upregulated' (resp. ‘downregulated') if RQ>1 (resp. RQ<1). Pathway analysis was conducted by querying Reactome annotations using the R/Bioconductor library reactome.db.52, 53
ChIP assay
ChIP assay protocol is described in Supplementary Experimental Procedures.
Antibody list
All used antibodies were validate for the appropriate application (Supplementary Table S5).
Drug treatment and cell proliferation assay
PND-1186 (Selleck Chemicals, Houston, TX, USA) was dissolved in dimethyl sulfoxide and stored at −80 °C until time of use. Cell proliferation was then assessed using Cell Proliferation Kit II (XTT) (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer's recommendations.
Statistical analysis
The data are presented as mean±SD. Comparisons were made between means from at least three independent experiments repeated in duplicate. Statistical differences were analyzed using the Student t-test. P-values <0.05 were considered to be statistically significant.
Acknowledgments
This work was mainly supported by MFAG12936 Grant of AIRC (Associazione Italiana per la Ricerca sul Cancro) to A. Alisi. Contributors were supported by AIRC (R. Rota), AIRC Special Grant ‘5xmille'-9962 (F. Locatelli). We thank SIBBM (Società Italiana di Biofisica e Biologia Molecolare) for the fellowship to Dr. Gnani for visiting Dr. Pasini Laboratory. We also thank Nicolò Valenti Onlus for donation of 2100 EnvisionTM Multilabel Reader.
Author contributions
Conception and design: A Alisi. Development of methodology: D Gnani, I Romito, S Artuso, N Panera, A Crudele, S Ceccarelli, E Carcarino, K Ferrari. Execution of experiments: D Gnani, I Romito, S. Artuso, C De Stefanis, N Panera, A Crudele, S Ceccarelli, E Carcarino. Acquisition of data: C De Stefanis, E Giorda, V D'Oria, M Porru. Analysis and interpretation of data: A Alisi, D Gnani, I Romito, S Artuso, M Chierici, L Miele, E Villa, C Balsano, C Furlanello, V Nobili, R. Rota, C Leonetti Writing, review, and/or revision of the manuscript: A Alisi, D Gnani, D Pasini, F Locatelli, R Rota, C Leonetti. Study supervision: A Alisi
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by S Fulda
The authors declare no conflict of interest.
Supplementary Material
References
- El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011; 365: 1118–1127. [DOI] [PubMed] [Google Scholar]
- Njei B, Rotman Y, Ditah I, Lim JK. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology 2015; 61: 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Schwartz M, Mazzaferro V. Resection and liver transplantation for hepatocellular carcinoma. Semin Liver Dis 2005; 25: 181–200. [DOI] [PubMed] [Google Scholar]
- Xu Q, Kobayashi S, Ye X, Meng X. Comparison of hepatic resection and radiofrequency ablation for small hepatocellular carcinoma: a meta-analysis of 16,103 patients. Sci Rep 2014; 4: 7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozen C, Yildiz G, Dagcan AT, Cevik D, Ors A, Keles U et al. Genetics and epigenetics of liver cancer. Nat Biotechnol 2013; 30: 381–384. [DOI] [PubMed] [Google Scholar]
- Li S, Mao M. Next generation sequencing reveals genetic landscape of hepatocellular carcinomas. Cancer Lett 2013; 340: 247–253. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Yasui K, Zhao C, Arii S, Inazawa J. PTK2 and EIF3S3 genes may be amplification targets at 8q23-q24 and are associated with large hepatocellular carcinomas. Hepatology 2003; 38: 1242–1249. [DOI] [PubMed] [Google Scholar]
- Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res 2013; 23: 1422–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JT. Focal adhesion kinase: the first ten years. J Cell Sci 2003; 116: 1409–1416. [DOI] [PubMed] [Google Scholar]
- Schaller MD. Cellular functions of FAK kinases: insight into molecular mechanisms and novel functions. J Cell Sci 2010; 123: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer 2014; 14: 598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev 2009; 28: 35–49. [DOI] [PubMed] [Google Scholar]
- Lim ST. Nuclear FAK: a new mode of gene regulation from cellular adhesions. Mol Cells 2013; 36: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrels A, Lund T, Serrels B, Byron A, McPherson RC, von Kriegsheim A et al. Nuclear FAK controls chemokine transcription, Tregs, and evasion of anti-tumor immunity. Cell 2015; 163: 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh S, Maeda T, Shimada M, Aishima S, Shirabe K, Tanaka S et al. Role of expression of focal adhesion kinase in progression of hepatocellular carcinoma. Clin Cancer Res 2004; 10: 2812–2817. [DOI] [PubMed] [Google Scholar]
- Fujii T, Koshikawa K, Nomoto S, Okochi O, Kaneko T, Inoue S et al. Focal adhesion kinase is overexpressed in hepatocellular carcinoma and can be served as an independent prognostic factor. J Hepatol 2004; 41: 104–111. [DOI] [PubMed] [Google Scholar]
- von Sengbusch A, Gassmann P, Fisch KM, Enns A, Nicolson GL, Haier J. Focal adhesion kinase regulates metastatic adhesion of carcinoma cells within liver sinusoids. Am J Pathol 2005; 166: 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Huang XH, Wang Q, Chen XL, Fu XH, Tan HX et al. FAK is involved in invasion and metastasis of hepatocellular carcinoma. Clin Exp Metastasis 2010; 27: 71–82. [DOI] [PubMed] [Google Scholar]
- Shang N, Arteaga M, Zaidi A, Stauffer J, Cotler SJ, Zeleznik-Le NJ et al. FAK is required for c-Met/β-catenin-driven hepatocarcinogenesis. Hepatology 2015; 61: 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung IH, Chen CY, Lin YH, Chi HC, Huang YH, Tai PJ et al. Thyroid hormone-mediated regulation of lipocalin 2 through the Met/FAK pathway in liver cancer. Oncotarget 2015; 6: 15050–15064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WX, Chen LP, Sun MY, Li JT, Liu HZ, Zhu W. 3'3-Diindolylmethane inhibits migration, invasion and metastasis of hepatocellular carcinoma by suppressing FAK signaling. Oncotarget 2015; 6: 23776–23792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillory LA, Stewart JE, Megison ML, Nabers HC, Mroczek-Musulman E, Beierle EA. FAK inhibition decreases hepatoblastoma survival both in vitro and in vivo. Transl Oncol 2013; 6: 206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lin MC, Yao H, Wang H, Zhang AQ, Yu J et al. Lentivirus-mediated RNA interference targeting enhancer of zeste homolog 2 inhibits hepatocellular carcinoma growth through down-regulation of stathmin. Hepatology 2007; 46: 200–208. [DOI] [PubMed] [Google Scholar]
- Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology 2011; 54: 1679–1689. [DOI] [PubMed] [Google Scholar]
- Au SL, Wong CC, Lee JM, Wong CM, Ng IO. Enhancer of zeste homolog 2 (EZH2) epigenetically silences multiple tumor suppressor miRNAs to promote liver cancer metastasis. Hepatology 2012; 56: 622–631. [DOI] [PubMed] [Google Scholar]
- Salvati E, Scarsella M, Porru M, Rizzo A, Iachettini S, Tentori L et al. PARP1 is activated at telomeres upon G4 stabilization: possible target for telomere-based therapy. Oncogene 2010; 29: 6280–6293. [DOI] [PubMed] [Google Scholar]
- Shen SQ, Li K, Zhu N, Nakao A. Expression and clinical significance of NET-1 and PCNA in hepatocellular carcinoma. Med Oncol 2008; 25: 341–345. [DOI] [PubMed] [Google Scholar]
- Zhou J, Roh JW, Bandyopadhyay S, Chen Z, Munkarah AR, Hussein Y et al. Overexpression of enhancer of zeste homolog 2 (EZH2) and focal adhesion kinase (FAK) in high grade endometrial carcinoma. Gynecol Oncol 2013; 128: 344–348. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298: 1039–1043. [DOI] [PubMed] [Google Scholar]
- Cao Q, Mani RS, Ateeq B, Dhanasekaran SM, Asangani IA, Prensner JR et al. Coordinated regulation of polycomb group complexes through microRNAs in cancer. Cancer Cell 2011; 20: 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008; 322: 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Milyavsky M, Shats I, Erez N, Goldfinger N, Rotter V. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene 2004; 23: 5759–5769. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. Embo J 2003; 22: 5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. The retinoblastoma protein and cell cycle control. Cell 1995; 81: 323–330. [DOI] [PubMed] [Google Scholar]
- Zhao J, Pestell R, Guan JL. Transcriptional activation of cyclin D1 promoter by FAK contributes to cell cycle progression. Mol Biol Cell 2001; 12: 4066–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloche S, Pouysségur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene 2007; 26: 3227–3239. [DOI] [PubMed] [Google Scholar]
- Gao SB, Zheng QF, Xu B, Pan CB, Li KL, Zhao Y et al. EZH2 represses target genes through H3K27-dependent and H3K27-independent mechanisms in hepatocellular carcinoma. Mol Cancer Res 2014; 12: 1388–1397. [DOI] [PubMed] [Google Scholar]
- Golubovskaya VM. Targeting FAK in human cancer: from finding to first clinical trials. Front Biosci (Landmark Ed) 2014; 19: 687–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YH, Wu TT, Huang CY, Hsieh YS, Hwang JM, Liu JY. p38 mitogen-activated protein kinase pathway is involved in protein kinase C alpha-regulated invasion in human hepatocellular carcinoma cells. Cancer Res 2007; 67: 4320–4327. [DOI] [PubMed] [Google Scholar]
- Sze KM, Wong KL, Chu GK, Lee JM, Yau TO, Ng IO. Loss of phosphatase and tensin homolog enhances cell invasion and migration through AKT/Sp-1 transcription factor/matrix metalloproteinase 2 activation in hepatocellular carcinoma and has clinicopathologic significance. Hepatology 2011; 53: 1558–1569. [DOI] [PubMed] [Google Scholar]
- Wang S, Huang X, Li Y, Lao H, Zhang Y, Dong H et al. RN181 suppresses hepatocellular carcinoma growth by inhibition of the ERK/MAPK pathway. Hepatology 2011; 53: 1932–1942. [DOI] [PubMed] [Google Scholar]
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009; 9: 400–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilands E, Serrels B, McEwan DG, Morton JP, Macagno JP, McLeod K et al. Autophagic targeting of Src promotes cancer cell survival following reduced FAK signalling. Nat Cell Biol 2011; 14: 51–60. [DOI] [PubMed] [Google Scholar]
- Yao Y, Hu H, Yang Y, Zhou G, Shang Z, Yang X et al. Downregulation of enhancer of Zeste Homolog 2 (EZH2) is essential for the induction of autophagy and apoptosis in colorectal cancer cells. Genes (Basel) 2016; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawan M, Venkatesan N, Loh JT, Wong JF, Berger H, Neo WH et al. The methyltransferase Ezh2 controls cell adhesion and migration through direct methylation of the extranuclear regulatory protein talin. Nat Immunol 2015; 16: 505–516. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 2009; 10: 697–708. [DOI] [PubMed] [Google Scholar]
- Panera N, Crudele A, Romito I, Gnani D, Alisi A. Focal adhesion kinase: insight into molecular roles and functions in hepatocellular carcinoma. Int J Mol Sci 2017; 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzariti A, Mancarella S, Porcelli L, Quatrale AE, Caligiuri A, Lupo L et al. Hepatic stellate cells induce HCC cell resistance to sorafenib through the laminin-332/α3 integrin axis recovery of FAK ubiquitination. Hepatology 2016; 64: 2103–2117. [DOI] [PubMed] [Google Scholar]
- Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med 2016; 22: 128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli S, Panera N, Mina M, Gnani D, De Stefanis C, Crudele A et al. LPS-induced TNF-α factor mediates pro-inflammatory and pro-fibrogenic pattern in non-alcoholic fatty liver disease. Oncotarget 2015; 6: 41434–41452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alisi A, Ceccarelli S, Panera N, Prono F, Petrini S, De Stefanis C et al. Association between serum atypical fibroblast growth factors 21 and 19 and pediatric nonalcoholic fatty liver disease. PLoS One 2013; 8: 67160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milacic M, Haw R, Rothfels K, Wu G, Croft D, Hermjakob H et al. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers (Basel) 2012; 4: 1180–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R et al. The Reactome pathway knowledgebase. Nucleic Acids Res 2016; 44: D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.