Abstract
Metastasis is the primary cause of death in prostate cancer (PCa) patients. Small nucleolar RNAs (snoRNAs) have long been considered “housekeeping” genes with no relevance for cancer biology. Emerging evidence has challenged this assumption, suggesting that snoRNA expression is frequently modulated during cancer progression. Despite this, no study has systematically addressed the prognostic and functional significance of snoRNAs in PCa.
We performed RNA Sequencing on paired metastatic/non‐metastatic PCa xenografts derived from clinical specimens. The clinical significance of differentially expressed snoRNAs was further investigated in two independent primary PCa cohorts (131 and 43 patients, respectively). The snoRNA demonstrating the strongest association with clinical outcome was quantified in PCa patient‐derived serum samples and its functional relevance was investigated in PCa cells via gene expression profiling, pathway analysis and gene silencing.
Our comparison revealed 21 differentially expressed snoRNAs in the metastatic vs. non‐metastatic xenografts. Of those, 12 were represented in clinical databases and were further analyzed. SNORA55 emerged as a predictor of shorter relapse‐free survival (results confirmed in two independent databases). SNORA55 was reproducibly detectable in serum samples from PCa patients. SNORA55 silencing in PCa cell lines significantly inhibited cell proliferation and migration. Pathway analysis revealed that SNORA55 expression is significantly associated with growth factor signaling and pro‐inflammatory cytokine expression in PCa.
Our results demonstrate that SNORA55 up‐regulation predicts PCa progression and that silencing this non‐coding gene affects PCa cell proliferation and metastatic potential, thus positioning it as both a novel biomarker and therapeutic target.
Keywords: Prostate cancer, SNORA55, Non‐coding RNAs, Patient‐derived xenograft, Next generation sequencing, Antisense oligonucleotide
Highlights
The role of small nucleolar RNAs (SnoRNAs) in prostate cancer progression has been scarcely investigated.
We analyzed the transcriptome of patient‐derived prostate cancer xenografts.
Our analysis revealed that SNORA55 is up‐regulated in prostate cancer and associated with worse prognosis.
SNORA55 inhibition impaired prostate cancer cell growth and metastatic potential.
Pathway analysis revealed that SNORA55 interacts with pro‐oncogenic and inflammatory pathways.
1. Introduction
Prostate cancer (PCa) is a heterogeneous disease that constitutes the most frequently diagnosed neoplasm in North American males (Siegel et al., 2012). Patients with localized PCa can be treated by radical prostatectomy or radiotherapy, often resulting in complete disease remission. Conversely, the vast majority of PCa‐related deaths result from progression of localized disease to metastatic castration‐resistant PCa (mCRPC) (Kirby et al., 2011). Despite recent therapeutic advancements (Bishr and Saad, 2013), mCRPC remains an incurable disease. Novel insights into PCa progression might therefore identify more effective therapeutic targets. In addition, clinico‐pathological factors such as T (tumor) stage, Gleason score and prostate‐specific antigen (PSA) are still the gold standard for PCa prognostication. However, it has been shown that their discriminatory value for identifying potentially lethal disease is limited (D'Elia et al., 2014). As a result, the discovery of more effective and potentially non‐invasive prognostic biomarkers is of paramount importance.
Recent analyses have revealed that most RNA molecules produced in human cells are not translated into proteins (Kapranov et al., 2007). Included among these non‐coding transcripts are microRNAs, which have emerged as critical mediators of cancer progression (Di Leva et al., 2014) and potentially useful biomarkers in oncology (Watahiki et al., 2013, 2011), as well as long non‐coding RNAs (lncRNAs) and small nucleolar RNAs (snoRNAs). Many research groups, including ours, have identified numerous PCa‐associated lncRNAs (Chakravarty et al., 2014; Crea et al., 2014b; Iyer et al., 2015; Prensner et al., 2014). Indeed it has been shown that the PCA3 lncRNA is detectable in biological fluids and discriminates PCa patients vs. healthy subjects (Wei et al., 2014), suggesting that the latter two classes of non‐coding RNAs may also be useful as biomarkers.
On the contrary, snoRNAs are a poorly investigated class of non‐coding RNAs that are encoded by at least 400 distinct genomic regions. They have traditionally been considered “housekeeping” genes, mainly involved in alternative splicing and ribosomal RNA (rRNA) modifications (Martens‐Uzunova et al., 2013). In light of their presumptive stable expression, some snoRNAs have been selected as reference genes in studies investigating the expression of microRNAs in human neoplasm (Gee et al., 2011). However, recent findings have challenged these assumptions, revealing that snoRNAs can migrate to the cytoplasm and modulate a wide range of cellular functions (Taulli and Pandolfi, 2012). At the same time, it has become evident that snoRNA expression patterns are deregulated in cancer cells and in serum samples from cancer patients (Baraniskin et al., 2013). In addition, some snoRNAs are emerging as critical mediators of cancer progression. For example SNORA42 is often amplified in lung cancer, and its silencing leads to cancer cell apoptosis, thereby impairing in vivo cancer growth (Mei et al., 2012). SNORD50A is frequently deleted and acts as a tumor suppressor in breast cancer and PCa (Dong et al., 2009, 2008). This gene also represents, to date, the only functionally characterized snoRNA in PCa.
We have utilized novel patient tumor tissue‐derived xenograft models to identify snoRNAs that were differentially expressed in non‐metastatic versus metastatic xenografts derived from the primary tumor of one patient. Differentially expressed snoRNAs were then queried in a test cohort of PCa patients to identify their correlation with disease progression and prognosis. These analyses revealed that SNORA55 was reproducibly associated with worse PCa outcome. In addition, we found that SNORA55 is required for PCa proliferation and migration. Taken together our studies support the hypothesis that SNORA55 may be a novel prognostic biomarker, as well as a novel therapeutic target, in PCa.
2. Patients and methods
2.1. Patient‐derived prostate cancer xenografts
The generation and propagation of the LTL313B and LTL313H tumor tissue xenograft models, as well as their characterization and their use in identification of differentially expressed transcripts, has been described previously (Watahiki et al., 2011) (Wang et al., 2005) (Crea et al., 2014b). Tumor tissue was accrued under UBC/BCCA REB protocol #: H04‐60131 and engraftment was performed according to the University of British Columbia Animal Care Committee protocol: A10‐0100.
2.2. RNA sequencing
Total RNA extraction, RNA Sequencing and transcript annotation have been described previously, and have been performed on one sample per model (Crea et al., 2014b). Importantly for this project, total RNA was extracted using Trizol, a method that preserves small RNAs (Rio et al., 2010). Reverse transcription was performed using random hexamers (Otogenetics, Oto‐Rseq2‐20N). In addition, transcript annotation (DNA Nexus) was performed using 36 nt alignment sequences, which are much shorter than the typical snoRNA length (70–120 nt) (Stepanov et al., 2015). Gene expression was quantified through the RPKM value reads per kilobase of transcript per million mapped reads (Mortazavi et al., 2008). RPKM values were normalized to the root mean square (RMS) for each sample. All genes annotated as “non‐coding” were further filtered to exclude microRNAs, ribosomal RNAs, transfer RNAs, piwi‐interacting RNAs and long non‐coding RNAs. The remaining transcripts contained 372 typical small nucleolar RNAs and 24 small Cajal body RNAs (scaRNAs), which were selected for further characterization. Collectively, classical small nucleolar RNAs and scaRNAs are denoted as “snoRNAs” in this manuscript. Differential expression of SNORA55 was confirmed on an independent microarray experiment (LTL‐313H vs. LTL‐313B), which is publically available: http://www.livingtumorcentre.com/GeneExpression/searchform.php.
2.3. Database analysis
Selected snoRNAs were analyzed through the cBio cancer genomic portal (Cerami et al., 2012). This database includes clinico‐pathological and gene expression information from 29 normal prostate, 131 primary and 19 metastatic PCa samples (Taylor et al., 2010). Samples containing more than 70% neoplastic cells based on histological assessment were used for RNA extraction. Metastatic cases were defined as those having clinical or pathological evidence of cancer dissemination to any of the following: lymph nodes, bones or soft tissues (lung, brain, spine, testis). Annotated clinical data from patients who underwent radical prostatectomy was available including PSA every three months for the first year post‐prostatectomy, semi‐annually for the second year and annually thereafter. Recurrence‐free survival (RFS) was calculated as the time between prostatectomy and the occurrence of two, consecutive rising plasma PSA values ≥ 0.2 ng/ml (Mottet et al., 2011).
Gene expression data were downloaded from the portal as log2 whole transcript normalized RNA expression values (Affymetrix Human Exon 1.0 ST arrays). For survival analysis (log‐rank test), Z score vs. normal values were employed, since this is the only option available in the portal. The optimal Z score threshold was determined as the one giving the lowest p value (Z score range: 2 ± 0.25). Based on these criteria, a Z score > 2.25 was indicated as “high” SNORA55 expression.
Significance Analysis of Microarray data (SAM) was performed in R using the MSKCC database and clinical and gene expression information available through the cBio portal. We identified 663 positively associated and 220 negatively associated genes (Q < 0.001%, fold change higher than 2). Genes positively associated with SNORA55 were then uploaded in Ingenuity Pathway Analysis (Ingenuity® Systems, www.ingenuity.com).
2.4. Clinical samples
2.4.1. Validation dataset
Samples from 43 PCa patients treated with primary radical prostate‐vesiculectomy with pelvic lymph node sampling were collected at the Stephanshorn Clinic in St. Gallen Switzerland, following study protocol approval by the local ethics committee (Protocol number: ZeTuP19/04). Resected specimens were immediately transferred on ice to the Institute for Pathology of the Kantons Hospital, St. Gallen for examination. Small tissue samples from macroscopically visible tumor and non‐tumor prostate tissue were dissected, snap frozen in liquid nitrogen and cryo‐preserved at −80 °C. Samples were cut in a cryo‐microtome and a slide of each probe was stained with hematoxylin‐eosin for histological verification. Localized PCa cases were defined as those with no clinical or pathological evidence of metastatic diffusion. PSA measurements were performed pre‐operation, and after operation every 6 ± 2 months. Recurrence‐free survival was calculated as the time between prostatectomy and the occurrence of two, consecutive rising plasma PSA values ≥ 0.2 ng/ml. RNA was isolated from frozen materials using the TRI‐reagent (Ambion) method according to the manufacturer's guidelines. cDNA was synthesized from 1 μg of total RNA using Superscript II RNase H‐reverse transcriptase and DNAse digestion (Invitrogen).
2.4.2. Serum samples from PCa patients
Serum samples from patients with newly diagnosed prostate cancer were collected at the Royal Surrey and Frimley Park Hospital, Guildford, UK. All samples were collected prior to any prostate cancer treatment. Patients donated samples following a process of informed consent and an ethical approval granted by the NHS Ethics Committee number 08/H1306/115. Healthy controls (males 45 years old or younger) were recruited and assessed for absence of clinical signs of prostatic neoplasms, including normal PSA levels, as described (Crea et al., 2014b). Control subjects' age was chosen to minimize the possibility of occult PCa, which is very infrequent in males younger than 45 (Aubert et al., 1991).
2.5. Quantitative PCR (qPCR)
qPCR was performed on RNA extracted and reverse‐transcribed from the validation and serum sample datasets. For the validation dataset, we employed TaqMan gene expression assays and performed the PCR reaction as previously described (Crea et al., 2011). SNORA55, TNF and GHRHR expression (Applied Biosystems assays: Hs03298696_s1, Hs01113624_g1, Hs00173457_m1, respectively) was normalized to HPRT1 (Applied Biosystems assay: Hs01003267_m1) and quantified through the 2−ΔΔCt method. HPRT1 was chosen because its expression was identified as the most stable among a panel of putative reference genes tested in normal and neoplastic tissues (including PCa samples) (de Kok et al., 2005). Since we intended to validate gene expression data obtained in a different platform and expressed in different unit of measure (Z scores from MSKCC database) we dichotomized SNORA55 expression based on the expression level that most efficiently predicted relapse‐free survival (lowest p value, log rank‐test; at least 5 patients in the smaller group). RNA derived from the serum samples was reverse transcribed using the NCode VILO cDNA Synthesis kit (Applied Biosystems). For quantification, we employed the SNORA55 TaqMan assay as described above. Mir‐30e (assay number: 000602) was used as the internal reference, since we have previously described this gene as the most stably expressed circulating small RNA in PCa patients (Watahiki et al., 2013).
2.6. In vitro experiments
PCa cell lines were grown in appropriate cell culture media and screened for SNORA55 expression, as previously described (Crea et al., 2014b). Second‐generation (2′OMe/PS gapmers) antisense oligonucleotides (ASOs) specifically targeting SNORA55 were designed and produced by Integrated DNA Technologies, based on the mature sequence of SNORA55 (Gene ID: 677834). The negative control ASO was designed to avoid silencing of any known human transcript. ASO transfection was performed as previously described (Narita et al., 2008). Cell proliferation and percentage of cell death were measured on Trypan Blue stained cells using a TC20 Automated Cell Counter (Bio‐Rad). Cell migration was measured using a wound healing assay, as previously described by our laboratory (Chiang et al., 2014). The following modifications were introduced to the protocol: cells were transfected with ASO1 or NC; pictures were taken 0, 12, 18, 30, 36 and 48 h post transfections. For the highly migrating DU145 cell line, pictures were taken till full “healing” of the NC‐treated cells (20 h). Cell fractionation and dihydrotestosterone (DHT) treatment of LNCaP cells have been previously described (Parolia et al., 2015).
2.7. Statistical analysis
Unless otherwise specified, all statistical analyses were performed using Graph Pad Prism 6 software (La Jolla, CA). MSKCC database clinical annotation and gene expression levels (available through Cbio portal) were used to dichotomize all patients in 2 groups: high SNORA55 expression (threshold as described in “Database analysis”) vs. low SNORA55 expression (all other patients).
3. Results
3.1. Small nucleolar RNA transcriptome analysis of matched non‐metastatic and metastatic PCa xenografts
Identification of metastasis‐associated genes in primary PCa using patient samples is a major challenge due to the heterogeneity of such specimens. They typically consist of subpopulations with different metastatic abilities, making it difficult to identify genes with critical roles in the development of metastasis (Fidler, 2002; Lin et al., 2010). For these reasons, molecular “signatures” for metastatic PCa have been extremely difficult to identify. To overcome these hurdles, we developed a multi‐step profiling strategy to identify snoRNAs associated with PCa progression (Figure 1A). First, we profiled snoRNAs in a paired localized/metastatic tumor tissue PCa xenograft model (Watahiki et al., 2011) that has already been exploited to successfully identify miRNAs (Watahiki et al., 2011) and long‐non‐coding RNAs (Crea et al., 2014b) associated with PCa progression. Our analysis showed that the global expression profiles of protein‐coding and non‐coding RNAs were very similar in Metastatic vs. Non‐Metastatic PCa xenografts, indicating that no general alteration in the synthesis of these two kinds of RNAs was associated with PCa progression (Suppl. Figure 1). Notably, snoRNAs and ribonuclear‐associated RNAs were the most abundantly expressed transcripts in both tumor tissue lines. We identified a total of 396 snoRNAs expressed in at least one PCa xenograft. Of those, 15 were up‐regulated and 6 down‐regulated in Metastatic vs. Non‐Metastatic PCa xenografts (Table 1). These transcripts were further analyzed in clinical samples.
Figure 1.
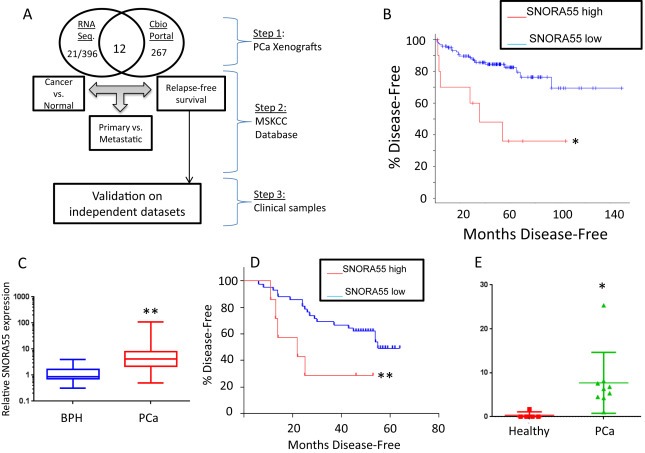
A, Analytical strategy employed for this study. 21 out of 396 differentially expressed snoRNAs (Met. vs. Non‐met. xenografts) were searched in the MSKCC database containing 267 snoRNAs. 12/21 genes were also represented in this platform and were chosen for further analyses. The gene(s) associated with PCa prognosis were validated in an independent clinical dataset. B, Relapse‐free survival according to SNORA55 expression in primary tumor at diagnosis, (MSKCC database, dichotomization based on Z score, as described in “Methods”) *p < 0.05 (log rank test, Bonferroni correction). C, SNORA55 expression measured by qPCR in 9 BPH and 43 primary PCa samples. **p < 0.01 (unpaired T test). D, Relapse‐free survival according to SNORA55 expression in primary tumor at diagnosis (qPCR on 43 samples). **p < 0.01 (log rank test). E, Expression of SNORA55 in serum samples from PCa patients and healthy controls. *p < 0.05 (unpaired, two‐sided T test).
Table 1.
Differentially expressed snoRNAs (Metastatic vs. Non‐Metastatic PCa xenograft). RMS, root mean square. RPKM, reads per kilobase of transcript per million mapped reads. We report transcripts with RMS‐normalized RPKM ratio>2 or <0.5 (Metastatic vs. Non‐Metastatic xenograft, RNA Seq. data). Transcripts up‐regulated in the metastatic xenograft are in red, transcripts down‐regulated in the metastatic xenograft are in blue. Transcripts up‐regulated in the metastatic xenograft are ranked based on absolute expression level in LTL313H cells. Transcripts down‐regulated in the non‐metastatic xenograft are ranked based on absolute expression in LTL313B cells.
| Gene name | Met. | Non‐met. | RMS‐normalized RPKM ratio | Represented in MSKCC database? |
|---|---|---|---|---|
| SNORA6 | 52.24 | 17.85 | 3.383431953 | Yes |
| SNORA42 | 28.86 | 10.59 | 3.153692615 | Yes |
| SNORA62 | 25.9 | 12.75 | 2.349627175 | Yes |
| SNORD88A | 13.78 | 6.397 | 2.491743725 | No |
| SNORA55 | 10.8 | 3.559 | 3.511427723 | Yes |
| SCARNA20 | 8.448 | 3.751 | 2.605070423 | No |
| SNORA11B | 8.337 | 3.166 | 3.045045045 | No |
| SNORA18 | 5.064 | 0.6716 | 8.721094856 | Yes |
| SNORA45 | 4.374 | 1.692 | 2.990630856 | Yes |
| SNORA72 | 1.809 | 0.3358 | 6.228373702 | Yes |
| SNORA26 | 1.174 | 0.3633 | 3.736551323 | No |
| SNORD33 | 1.151 | 0.534 | 2.490603363 | Yes |
| SNORA22 | 1.077 | 0.3333 | 3.736925515 | Yes |
| SNORD88B | 0.9845 | 0.457 | 2.492485549 | No |
| SCARNA18 | 0.7235 | 0.3358 | 2.491349481 | No |
| SNORA64 | 233.5 | 70.91 | 0.351062867 | Yes |
| SNORA63 | 32.83 | 13.09 | 0.461068211 | No |
| SNORA81 | 13.94 | 4.829 | 0.400454545 | No |
| SNORA31 | 4.432 | 1.102 | 0.287416587 | Yes |
| SNORA51 | 4.365 | 1.447 | 0.383349468 | Yes |
| SNORA76 | 2.015 | 0.7235 | 0.415312008 | No |
3.2. Expression profile of snoRNAs in clinical samples
In order to validate and establish the clinical relevance of our RNA Sequencing data, we analyzed differentially expressed snoRNAs though the cBio MSKCC database, which holds gene expression and clinical data from a collection of 131 localized and 19 metastatic PCa samples, as well as 29 benign prostatic tissues (Cerami et al., 2012; Taylor et al., 2010). We found that 12 out of 21 differentially expressed snoRNAs were represented in this database (Table 1). We hypothesized that snoRNAs up‐regulated in the metastatic xenograft could contribute to PCa progression, and should be therefore up‐regulated in neoplastic vs. normal and/or in metastatic vs. localized PCa samples. Similarly, the down‐regulated snoRNAs should also be down‐regulated in the clinical database samples studied. Those transcripts were therefore investigated for correlations with cancer progression and for their prognostic value. In order to reduce our false discovery ratio, we applied the Bonferroni correction to all analyses performed on this dataset. We identified 6 snoRNAs with statistically significant differential expression in normal prostate vs. localized PCa and/or in metastatic vs. localized PCa (Suppl. Figure 2; corrected p < 0.05, ANOVA and Tukey's post‐test). Notably, all differentially expressed transcripts were modulated in accordance to our pre‐clinical model's predictions. For example SNORA6, which was the most highly up‐regulated snoRNA in our RNA Sequencing database (Table 1), was progressively up‐regulated in normal prostate, localized PCa and metastatic PCa samples (Suppl. Figure 2A). Similarly, SNORA55 expression was higher in metastatic PCa xenografts, and progressively increased in normal prostate, primary PCa and metastatic PCa samples (Suppl. Figure 2C). The only snoRNA significantly repressed in metastatic vs. normal PCa samples was snoRNA31 (SNORA31), which was also down‐regulated in Metastatic vs. Non‐Metastatic xenografts (Table 1). These results corroborate the clinical relevance of the transcription profiles derived from our pre‐clinical model.
3.3. Diagnostic and prognostic value of SNORA55
In light of our initial findings, we set out to test whether snoRNAs were able to predict recurrence‐free survival (RFS) when measured in primary PCa samples at the time of surgery. Among the 12 selected snoRNAs (Table 1), only SNORA55 was associated with clinical outcome. Univariate analysis (Figure 1B) indicated that PCa samples with SNORA55 up‐regulation displayed a significantly shorter RFS after surgery (corrected log‐rank p = 0.015, median survival = 35.35 vs. >140 months, 6/11 vs. 21/120 events). Subgroup analysis revealed no correlation between SNORA55 expression and ERG fusion status, Gleason grade, T stage or circulating PSA levels (Suppl. Figure 3A–D).
To further strengthen our analysis, we explored the diagnostic and prognostic value of SNORA55 in an independent dataset including benign prostatic hyperplasia (BPH) and localized PCa samples with long‐term follow‐up (patient characteristics summarized in Suppl. Tab. 1). In this case, SNORA55 expression was measured by qPCR, using RNA extracted from primary PCa biopsies at the time of diagnosis. Our analysis confirmed significant up‐regulation of SNORA55 in PCa vs. BPH samples (Figure 1C; mean expression: 8.345 ± 2.555 vs. 1.342 ± 0.3729, p = 0.0095). We also confirmed that SNORA55 up‐regulation was associated with shorter RFS after prostatectomy (Figure 1D; log‐rank p value = 0.0054, Hazard Ratio 95% Confidence interval: 1.134–8.230).
Recent reports indicate that snoRNAs can be detected in serum samples from cancer patients and that those findings might have a diagnostic value (Baraniskin et al., 2013). In light of the prognostic role of SNORA55, we investigated its expression in a small but homogenous cohort of 9 PCa patients treated with brachytherapy vs. 5 healthy subjects (Suppl. Table 2). Our results indicated that circulating SNORA55 is significantly up‐regulated in PCa patients vs. healthy controls (Figure 1E).
3.4. Functional characterization of SNORA55
Our results to this point indicate that SNORA55 is increasingly up‐regulated during PCa progression, and that higher SNORA55 expression is associated with poorer prognosis and worse response to therapy. SNORA55 locus maps on chromosome 1p34 and co‐localizes with an intron of the PABPC4 protein‐coding gene (Figure 2A). Notably, clinical data indicate that SNORA55 and PABC4 expression profiles are not correlated, and that PABPC4 has no prognostic value in PCa (Suppl. Figure 4). To investigate the specific role of SNORA55 in PCa biology, we conducted functional experiments in PCa cell lines. First we screened the expression of this gene in a panel of human prostatic cell lines (Figure 2B). SNORA55 was easily detectable (Ct value < 30) in all the investigated cell lines, with highest expression in LNCaP, an androgen‐receptor (AR) positive cell line derived from a metastatic PCa lesion (Horoszewicz et al., 1983). RNA fractionation analyses revealed that SNORA55 is predominantly nuclear (Figure 2C). Since the expression of some PCa‐associated non‐coding RNAs is regulated by AR activation (Crea et al., 2014b), we tested this possibility for SNORA55. Data from LNCaP cells exposed to different DHT concentrations demonstrated that SNORA55 expression is not controlled by AR activity (Figure 2D).
Figure 2.
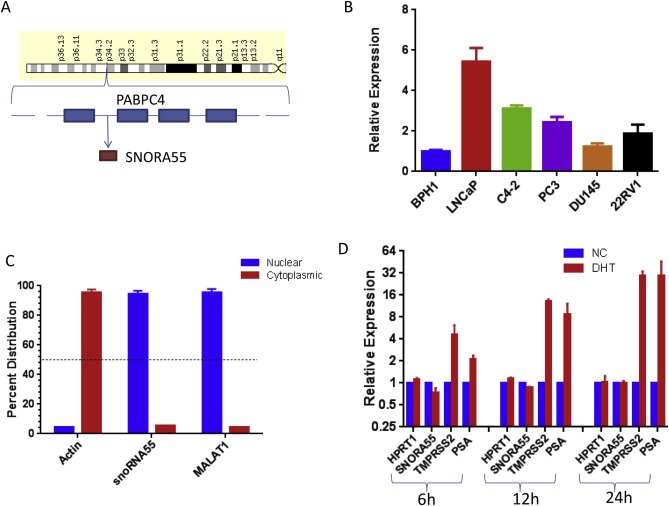
A, Chromosomal localization of SNORA55 (data from Gene Cards). B, SNORA55 expression in prostate cell lines. C, Subcellular localization of SNORA55 in LNCaP cells (MALAT1 is a nuclear non‐coding RNA, actin mRNA is prevalently cytoplasmic). D, Effects of DHT treatment on SNORA55 expression (LNCaP cells). HPRT1 is an AR‐independent gene (negative control), PSA and TMPRSS2 are AR‐dependent genes (positive controls). The experiment was performed as previously described (Parolia et al., 2015).
Subsequently, we designed three ASOs specifically targeting SNORA55 and measured their gene silencing efficacy in LNCaP cells. ASO1 induced a dose‐dependent SNORA55 silencing, when compared with control ASO at the same concentration (Figure 3A). ASO1 was therefore selected for functional studies. SNORA55 silencing induced a dose‐dependent inhibition of cell proliferation (Figure 3B). Since our findings were limited to one AR‐positive cell line expressing high levels of SNORA55, we tested the effects of ASO1 on DU145 cells (AR‐negative cells expressing relatively lower levels of SNORA55). Our results indicate that SNORA55 silencing in this cell line induced significant, even if slightly less emphatic, cell growth inhibition (Figure 3C). In order to elucidate the mechanism underlying this growth inhibitory effect, we measured the percentage of cell death in DU145 and LNCaP cells exposed to ASO1 vs. NC. Our results indicate that SNORA55 silencing significantly increases cell death in both cell lines (Figure 3D). Notably, this effect is more evident in LNCaP cells. Similarly, we found that SNORA55 silencing hinders the migration of both LNCaP and DU145 cells (Figure 4A,B). We cannot rule out the possibility that reduced migration is at least in part a consequence of ASO1‐induced cell death. However, the anti‐migratory effect is equally emphatic in LNCaP as in DU145 cells, whereas cell death is more marked in LNCAP cells. It is therefore conceivable that effect of SNORA55 on cell migration is at least in part cell death‐independent.
Figure 3.
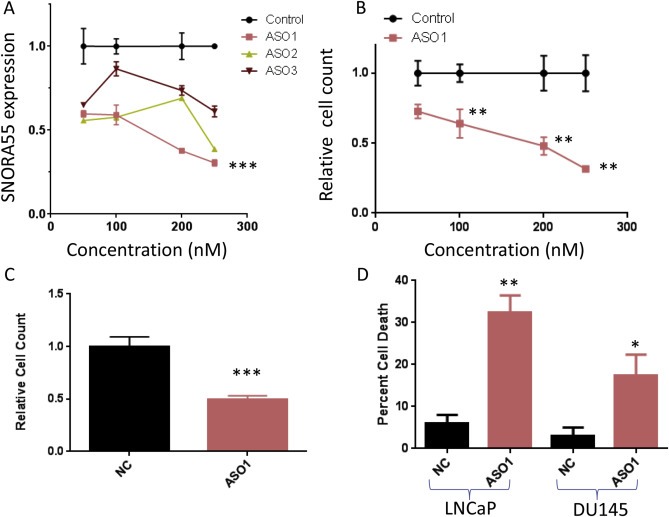
A, SNORA55 expression in LNCaP cells exposed to different ASO concentrations (48 h). ***p < 0.001 vs. control (1 way ANOVA and Tukey multiple comparison post‐test, highest concentration of each treatment). B, Relative LNCaP cell count after treatment with different ASO concentrations (48 h). **p < 0.01 (2 way ANOVA and Sidak post‐test). C, Relative DU145 cell count after treatment with ASOs (250 nM, 48 h). ***p < 0.001 (unpaired T test). D, Percentage of cell death in LNCaP and DU145 cells exposed to NC or ASO1 (250 nM, 48 h). *p < 0.05; **p < 0.01 (unpaired T test).
Figure 4.
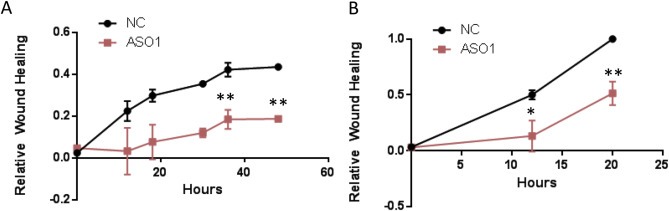
LNCaP (A) and Du145 (B) cell migration in cells exposed to control or ASO1 (250 nM). *p < 0.05; **p < 0.01 (2 way ANOVA and Sidak post‐test).
3.5. Pathway analysis
Our results indicate that SNORA55 is up‐regulated during PCa progression and that it is functionally relevant for PCa cell proliferation and migration. Since this gene had never been associated with cancer before, we investigated the molecular pathways associated with its expression. To this end, we performed SAM analysis and identified 663 genes significantly correlated with SNORA55 expression. We designated those genes as SNORA55‐Related Signature (55‐RS) and analyzed them through Ingenuity Pathway Analysis. The 55‐RS was significantly associated with several pathways, including “Cell‐to‐Cell Signaling and interaction” and “Cell death and Survival” (Table 2). The top molecular network associated with 55‐RS was “Protein Synthesis, Cell‐to‐Cell Signaling and Interaction, Lipid Metabolism” (Suppl. Figure 5). Notably, tumor necrosis factor (TNF) and growth hormone releasing hormone receptor (GHRHR) occupied a central role in this molecular network and were highly correlated with SNORA55 expression. In keeping with these predictions, SNORA55 silencing reduced the expression of TNF and, to a lesser extent, of GHRHR (Suppl. Figure 6).
Table 2.
Top 5 Molecular and Cellular Functions associated with SNORA55 expression. Ingenuity Pathway Analysis was used for this analysis. Genes significantly associated with SNORA55 (in red) were identified through SAM analysis (see “Methods” for details).
| Function name | P value | Number of Molecules |
|---|---|---|
| Cell‐to‐Cell Signaling and interaction | 9.04E−18–3.91E−2 | 130 |
| Cell death and survival | 1.46E−4–3.91E−2 | 35 |
| Cell Signaling | 2.52E−4–2.07E−2 | 36 |
| Drug metabolism | 3.89E−4–3.91E−2 | 4 |
| Molecular transport | 3.89E−4–3.91E−2 | 50 |
4. Discussion
In the present study, we identified SNORA55 as a previously uncharacterized gene that is potentially associated with PCa progression. Our results were generated through a multi‐step strategy that takes advantage of unique pre‐clinical models developed at the Living Tumor Lab (www.livingtumorlab.com). These patient‐derived, PCa xenografts accurately reproduce clinico‐pathological and molecular features of the original neoplasm (Lin et al., 2014). SnoRNAs identified as differentially expressed in this pre‐clinical model were further investigated through 2 independent clinical databases (193 total samples). Our experimental strategy is based on the observation that PCa samples are generally composed of multi‐clonal subpopulations, each with a peculiar transcriptional landscape and metastatic potential (Lin et al., 2010; Liu et al., 2009). PCa heterogeneity often masks the expression profile of truly metastatic cells, thereby hampering the identification of an effective prognostic signature. As a consequence, several multi‐genic PCa prognostic algorithms have been described (Cooperberg et al., 2013; Glinsky et al., 2004; Irshad et al., 2013; Varambally et al., 2005) but none of them have been currently integrated in the clinical practice. The only risk stratification method that is commonly employed is still based on plasma PSA levels, T stage and Gleason grade (Heidenreich et al., 2014). The lack of more accurate and easy‐to‐detect prognostic markers hinders an optimal exploitation of the therapeutic armamentarium against lethal PCa (Cersosimo, 2012).
Our results from two independent clinical datasets demonstrated that SNORA55 is: 1) easily detectable in PCa specimens, including serum samples; 2) significantly up‐regulated in neoplastic vs. normal prostatic tissue; and 3) a predictor of post‐prostatectomy outcome. While the first two features are shared with other investigated snoRNAs, the third is a distinctive feature of SNORA55. A previous study mainly focusing on miRNAs also reported that some snoRNAs are differentially expressed in primary vs. lymph node‐derived lesions from PCa patients (Martens‐Uzunova et al., 2012). In accordance with our findings, SNORA6 was listed as up‐regulated in metastatic lesions. However, this study did not address the prognostic and functional value of snoRNAs in PCa. Another very recent study showed that snoRNA expression profiles are deregulated during PCa initiation and progression, and suggested that they can be employed as prognostic markers (Martens‐Uzunova et al., 2015). However, our report is the first one showing that the expression levels of a snoRNA (measured in primary PCa specimens) efficiently predict post‐operative PCa prognosis. Notably, the prognostic value of SNORA55 was confirmed in two clinical datasets with different characteristics: the MSKCC dataset comprises relatively younger men (median age = 58.3) and lower‐grade neoplasms (percentage of Gleason grade>7 = 9%) (Taylor et al., 2010), compared to our validation cohort (median age = 68, percentage of Gleason Grade >7 = 23%).
SnoRNAs have been long considered “housekeeping” genes, whose only function was to regulate basic cellular processes in the nucleus. Recent evidence indicated that snoRNAs also contribute to cancer progression through nucleolar and extra‐nucleolar functions (reviewed in Crea et al. (2014a)). For example, some snoRNAs are able to translocate into the cytoplasm, where they control reactive oxygen species production, thereby fueling chemoresistance and metastatic progression (Chu et al., 2012; Crea et al., 2014a). Other snoRNAs generate miRNA‐like transcripts, which in turn regulate the expression of several cancer‐related genes (Ono et al., 2011). Our study shows that SNORA55 is specifically expressed and functionally relevant in cancer cells. Our gene expression analysis revealed that, unlike other disease‐associated snoRNAs, SNORA55 is mainly retained in the nucleus. Due to its sub‐cellular localization, we could not employ small interfering RNAs (siRNAs) to target this gene. For this reason, we silenced its expression via ASOs, which proved effective in silencing nuclear RNAs (Gutschner et al., 2013). Our results indicate that SNORA55 is required for PCa cell proliferation and migration. These findings may explain the strong correlation between SNORA55 expression and PCa prognosis.
In keeping with those findings, our Pathway Analysis sheds new light on the molecular mechanisms by which snoRNAs may contribute to cancer progression. We found that SNORA55 expression is significantly associated with known oncogenic pathways and particularly with the expression of TNF‐ and GHRH‐dependent signaling (Suppl. Figure 5). The pro‐inflammatory cytokine TNFα is over‐expressed in PCa specimens (Rodriguez‐Berriguete et al., 2013), and it may contribute to prostate carcinogenesis and neoplastic progression (De Marzo et al., 2007). Notably, high TNFα expression has been associated with poor PCa prognosis (Rodriguez‐Berriguete et al., 2013). GHRH was first discovered as a hypothalamic neuropeptide (Bowers, 2012). The GHRH‐FSH/LH axis is primarily responsible for triggering testosterone production by the testis. Recent evidence showed that GHRH receptors are also expressed by PCa cells, and that their inhibition is an effective therapeutic strategy in PCa pre‐clinical models (Rick et al., 2012). Therefore, the GHRH‐dependent signal transduction pathway is functionally relevant for PCa cells and might mediate the functional role of SNORA55.
5. Conclusions
The non‐coding transcriptome represents a vast and mainly uncharted region of the human genome whose relevance in neoplasms has been overlooked for many years. SnoRNAs are one of the most abundant non‐coding RNA classes and some of them show deregulated expression and/or epigenetic alterations in human neoplasms (Esteller, 2011; Martens‐Uzunova et al., 2013). Notably, snoRNAs are also detectable in biological fluids (Baraniskin et al., 2013), a feature that makes them particularly attractive cancer biomarkers. For these reasons, some authors have predicted that snoRNAs will emerge as novel diagnostic and prognostic tools in the near future (Mannoor et al., 2012). We anticipate that our findings, along with similar studies performed on other neoplasms (Gee et al., 2011; Valleron et al., 2012), will pave the way to the discovery of novel clinically relevant biomarkers and therapeutic targets in oncology.
Funding
Canadian Institutes of Health Research (YW, MG), Terry Fox Research Institute (YW, MG), BC Cancer Foundation (YW), Canadian Cancer Society Research Institute (CDH, YW). FC holds a Michael Smith Foundation for Health Research postdoctoral fellowship award and a Prostate Cancer Foundation‐BC award. AAA is supported by a CJ Martin Biomedical Overseas Fellowship from the National Health & Medical Research Council.
Conflict of interest
The authors declare that they have no competing interests.
Supporting information
The following are the supplementary data related to this article:
Suppl. Figure 1: Global RPKM plot for protein coding and non‐coding genes in LTL313H (Metastatic) and LTL313B (Non‐metastatic) cells. SnoRNAs and ribonuclease‐associated RNAs (RMRP and RPPH1) are indicated by arrows.
Suppl. Figure 2: Differentially expressed snoRNAs in the MSKCC database *p < 0.05, **p < 0.01 (ANOVA and Tukey's post‐test, Bonferroni correction for multiple comparisons). We only show results with corrected p < 0.05. Horizontal bars represent median value, vertical bars pan from lowest to highest value in the group. Data are from the MSKCC database (cBio portal).
Suppl. Figure 3: SNORA55 expression based on Gleason Score (A), T stage (B), ERG fusion status (C) and circulating PSA levels (D). P values (>0.05 for all the analysis) were calculated using ANOVA and Tukey's post‐test (A), unpaired two‐tailed T test (B–C) and linear regression (D). Horizontal bars represent median value, vertical bars pan from lowest to highest value in the group Data are from the MSKCC database (Cbio portal).
Suppl. Figure 4: A, SNORA55 and PABPC4 mRNA expression in 131 primary PCa samples from the MSKCC database. B, Relapse‐free survival according to PABPC4 expression in primary tumor at diagnosis, (MSKCC database, dichotomization based on Z score, as for SNORA55). P value refers to log‐rank test.
Suppl. Figure 5: Top‐molecular networks associated with SNORA55 expression (Ingenuity Pathway Analysis was used for analysis and visualization). Genes significantly associated with SNORA55 (in red) were identified through SAM analysis (see “Methods” for details).
Suppl. Figure 6: qPCR analysis of TNF and GHRHR expression in LNCaP cells exposed to NC or SNORA55‐targeting ASO1.
Suppl. Table 1: Clinical characteristics of the validation dataset (primary PCa samples).
Suppl. Table 2: Clinical characteristics of the serum sample PCa dataset.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2015.12.010.
Crea Francesco, Quagliata Luca, Michael Agnieszka, Liu Hui Hsuan, Frumento Paolo, Azad Arun A., Xue Hui, Pikor Larissa, Watahiki Akira, Morant Rudolf, Eppenberger-Castori Serenella, Wang Yuwei, Parolia Abhijit, Lennox Kim A., Lam Wan L., Gleave Martin, Chi Kim N., Pandha Hardev, Wang Yuzhuo, Helgason Cheryl D., (2016), Integrated analysis of the prostate cancer small‐nucleolar transcriptome reveals SNORA55 as a driver of prostate cancer progression, Molecular Oncology, 10, doi: 10.1016/j.molonc.2015.12.010.
Contributor Information
Yuzhuo Wang, Email: ywang@bccrc.ca.
Cheryl D. Helgason, Email: chelgaso@bccrc.ca
References
- Aubert, J. , Dore, B. , Irani, J. , Aubert, M.N. , 1991. Prostatic cancer before the age of 50–report of 7 cases. Progres en urologie : journal de l'Association francaise d'urologie et de la Societe francaise d'urologie. 1, 432–439. [PubMed] [Google Scholar]
- Baraniskin, A. , Nopel-Dunnebacke, S. , Ahrens, M. , Jensen, S.G. , Zollner, H. , Maghnouj, A. , Wos, A. , Mayerle, J. , Munding, J. , Kost, D. , Reinacher-Schick, A. , Liffers, S. , Schroers, R. , Chromik, A.M. , Meyer, H.E. , Uhl, W. , Klein-Scory, S. , Weiss, F.U. , Stephan, C. , Schwarte-Waldhoff, I. , Lerch, M.M. , Tannapfel, A. , Schmiegel, W. , Andersen, C.L. , Hahn, S.A. , 2013. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for pancreatic and colorectal adenocarcinoma. Int. Cancer J. Int. Cancer. 132, E48–E57. [DOI] [PubMed] [Google Scholar]
- Bishr, M. , Saad, F. , 2013 Sep. Overview of the latest treatments for castration-resistant prostate cancer. Nat. Rev. Urol. 10, (9) 522–528. [DOI] [PubMed] [Google Scholar]
- Bowers, C.Y. , 2012. History to the discovery of ghrelin. Methods Enzymol. 514, 3–32. [DOI] [PubMed] [Google Scholar]
- Cerami, E. , Gao, J. , Dogrusoz, U. , Gross, B.E. , Sumer, S.O. , Aksoy, B.A. , Jacobsen, A. , Byrne, C.J. , Heuer, M.L. , Larsson, E. , Antipin, Y. , Reva, B. , Goldberg, A.P. , Sander, C. , Schultz, N. , 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cersosimo, R.J. , 2012. New agents for the management of castration-resistant prostate cancer. Ann. Pharmacother. 46, 1518–1528. [DOI] [PubMed] [Google Scholar]
- Chakravarty, D. , Sboner, A. , Nair, S.S. , Giannopoulou, E. , Li, R. , Hennig, S. , Mosquera, J.M. , Pauwels, J. , Park, K. , Kossai, M. , MacDonald, T.Y. , Fontugne, J. , Erho, N. , Vergara, I.A. , Ghadessi, M. , Davicioni, E. , Jenkins, R.B. , Palanisamy, N. , Chen, Z. , Nakagawa, S. , Hirose, T. , Bander, N.H. , Beltran, H. , Fox, A.H. , Elemento, O. , Rubin, M.A. , 2014. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 5, 5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, Y.T. , Wang, K. , Fazli, L. , Qi, R.Z. , Gleave, M.E. , Collins, C.C. , Gout, P.W. , Wang, Y. , 2014. GATA2 as a potential metastasis-driving gene in prostate cancer. Oncotarget. 5, 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, L. , Su, M.Y. , Maggi, L.B. , Lu, L. , Mullins, C. , Crosby, S. , Huang, G. , Chng, W.J. , Vij, R. , Tomasson, M.H. , 2012. Multiple myeloma-associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J. Clin. Invest. 122, 2793–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperberg, M.R. , Simko, J.P. , Cowan, J.E. , Reid, J.E. , Djalilvand, A. , Bhatnagar, S. , Gutin, A. , Lanchbury, J.S. , Swanson, G.P. , Stone, S. , Carroll, P.R. , 2013. Validation of a cell-cycle progression gene panel to improve risk stratification in a contemporary prostatectomy cohort. J. Clin. Oncol. Off J. Am. Soc. Clin. Oncol. 31, 1428–1434. [DOI] [PubMed] [Google Scholar]
- Crea, F. , Clermont, P.L. , Parolia, A. , Wang, Y. , Helgason, C.D. , 2014 Mar. The non-coding transcriptome as a dynamic regulator of cancer metastasis. Cancer Metastasis Rev. 33, (1) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea, F. , Duhagon Serrat, M.A. , Hurt, E.M. , Thomas, S.B. , Danesi, R. , Farrar, W.L. , 2011. BMI1 silencing enhances docetaxel activity and impairs antioxidant response in prostate cancer. Int. J. Cancer J. Int. Cancer. 128, 1946–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crea, F. , Watahiki, A. , Quagliata, L. , Xue, H. , Pikor, L. , Parolia, A. , Wang, Y. , Lin, D. , Lam, W.L. , Farrar, W.L. , Isogai, T. , Morant, R. , Castori-Eppenberger, S. , Chi, K.N. , Wang, Y. , Helgason, C.D. , 2014 Feb 15. Identification of a long non-coding RNA as a novel biomarker and potential therapeutic target for metastatic prostate cancer. Oncotarget. 5, (3) 764–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Elia, C. , Cerruto, M.A. , Cioffi, A. , Novella, G. , Cavalleri, S. , Artibani, W. , 2014. Upgrading and upstaging in prostate cancer: from prostate biopsy to radical prostatectomy. Mol. Clin. Oncol. 2, 1145–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok, J.B. , Roelofs, R.W. , Giesendorf, B.A. , Pennings, J.L. , Waas, E.T. , Feuth, T. , Swinkels, D.W. , Span, P.N. , 2005. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab. Invest. J. Tech. Methods Pathol. 85, 154–159. [DOI] [PubMed] [Google Scholar]
- De Marzo, A.M. , Platz, E.A. , Sutcliffe, S. , Xu, J. , Gronberg, H. , Drake, C.G. , Nakai, Y. , Isaacs, W.B. , Nelson, W.G. , 2007. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer. 7, 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva, G. , Garofalo, M. , Croce, C.M. , 2014. MicroRNAs in Cancer. Annu. Rev. Pathol. 9, 287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X.Y. , Guo, P. , Boyd, J. , Sun, X. , Li, Q. , Zhou, W. , Dong, J.T. , 2009. Implication of snoRNA U50 in human breast cancer. J. Genet. Genom. = Yi Chuan Xue Bao. 36, 447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X.Y. , Rodriguez, C. , Guo, P. , Sun, X. , Talbot, J.T. , Zhou, W. , Petros, J. , Li, Q. , Vessella, R.L. , Kibel, A.S. , Stevens, V.L. , Calle, E.E. , Dong, J.T. , 2008. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum. Mol. Genet. 17, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller, M. , 2011. Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. [DOI] [PubMed] [Google Scholar]
- Fidler, I.J. , 2002. Critical determinants of metastasis. Semin. Cancer Biol. 12, 89–96. [DOI] [PubMed] [Google Scholar]
- Gee, H.E. , Buffa, F.M. , Camps, C. , Ramachandran, A. , Leek, R. , Taylor, M. , Patil, M. , Sheldon, H. , Betts, G. , Homer, J. , West, C. , Ragoussis, J. , Harris, A.L. , 2011. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br. J. Cancer. 104, 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky, G.V. , Glinskii, A.B. , Stephenson, A.J. , Hoffman, R.M. , Gerald, W.L. , 2004. Gene expression profiling predicts clinical outcome of prostate cancer. J. Clin. Invest. 113, 913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner, T. , Hammerle, M. , Eissmann, M. , Hsu, J. , Kim, Y. , Hung, G. , Revenko, A. , Arun, G. , Stentrup, M. , Gross, M. , Zornig, M. , MacLeod, A.R. , Spector, D.L. , Diederichs, S. , 2013. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 73, 1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich, A. , Bastian, P.J. , Bellmunt, J. , Bolla, M. , Joniau, S. , van der Kwast, T. , Mason, M. , Matveev, V. , Wiegel, T. , Zattoni, F. , Mottet, N. , 2014. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent-update 2013. Eur. Urol. 65, 124–137. [DOI] [PubMed] [Google Scholar]
- Horoszewicz, J.S. , Leong, S.S. , Kawinski, E. , Karr, J.P. , Rosenthal, H. , Chu, T.M. , Mirand, E.A. , Murphy, G.P. , 1983. LNCaP model of human prostatic carcinoma. Cancer Res. 43, 1809–1818. [PubMed] [Google Scholar]
- Irshad, S. , Bansal, M. , Castillo-Martin, M. , Zheng, T. , Aytes, A. , Wenske, S. , Le Magnen, C. , Guarnieri, P. , Sumazin, P. , Benson, M.C. , Shen, M.M. , Califano, A. , Abate-Shen, C. , 2013. A molecular signature predictive of indolent prostate cancer. Sci. Transl. Med. 5, 202ra122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, M.K. , Niknafs, Y.S. , Malik, R. , Singhal, U. , Sahu, A. , Hosono, Y. , Barrette, T.R. , Prensner, J.R. , Evans, J.R. , Zhao, S. , Poliakov, A. , Cao, X. , Dhanasekaran, S.M. , Wu, Y.M. , Robinson, D.R. , Beer, D.G. , Feng, F.Y. , Iyer, H.K. , Chinnaiyan, A.M. , 2015. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov, P. , Cheng, J. , Dike, S. , Nix, D.A. , Duttagupta, R. , Willingham, A.T. , Stadler, P.F. , Hertel, J. , Hackermuller, J. , Hofacker, I.L. , Bell, I. , Cheung, E. , Drenkow, J. , Dumais, E. , Patel, S. , Helt, G. , Ganesh, M. , Ghosh, S. , Piccolboni, A. , Sementchenko, V. , Tammana, H. , Gingeras, T.R. , 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 316, 1484–1488. [DOI] [PubMed] [Google Scholar]
- Kirby, M. , Hirst, C. , Crawford, E.D. , 2011. Characterising the castration-resistant prostate cancer population: a systematic review. Int. J. Clin. Pract. 65, 1180–1192. [DOI] [PubMed] [Google Scholar]
- Lin, D. , Bayani, J. , Wang, Y. , Sadar, M.D. , Yoshimoto, M. , Gout, P.W. , Squire, J.A. , 2010. Development of metastatic and non-metastatic tumor lines from a patient's prostate cancer specimen-identification of a small subpopulation with metastatic potential in the primary tumor. The Prostate. 70, 1636–1644. [DOI] [PubMed] [Google Scholar]
- Lin, D. , Wyatt, A.W. , Xue, H. , Wang, Y. , Dong, X. , Haegert, A. , Wu, R. , Brahmbhatt, S. , Mo, F. , Jong, L. , Bell, R.H. , Anderson, S. , Hurtado-Cull, A. , Fazli, L. , Sharma, M. , Beltran, H. , Rubin, M.A. , Cox, M.E. , Gout, P.W. , Morris, J. , Goldenberg, L. , Volik, S.V. , Gleave, M.E. , Collins, C.C. , Wang, Y. , 2014 Feb 15. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res. 74, (4) 1272–1283. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Laitinen, S. , Khan, S. , Vihinen, M. , Kowalski, J. , Yu, G. , Chen, L. , Ewing, C.M. , Eisenberger, M.A. , Carducci, M.A. , Nelson, W.G. , Yegnasubramanian, S. , Luo, J. , Wang, Y. , Xu, J. , Isaacs, W.B. , Visakorpi, T. , Bova, G.S. , 2009. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat. Med. 15, 559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannoor, K. , Liao, J. , Jiang, F. , 2012. Small nucleolar RNAs in cancer. Biochim. Biophys. Acta. 1826, 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Uzunova, E.S. , Hoogstrate, Y. , Kalsbeek, A. , Pigmans, B. , Vredenbregt-van den Berg, M. , Dits, N. , Nielsen, S.J. , Baker, A. , Visakorpi, T. , Bangma, C. , Jenster, G. , 2015. C/D-box snoRNA-derived RNA production is associated with malignant transformation and metastatic progression in prostate cancer. Oncotarget. 6, 17430–17444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Uzunova, E.S. , Jalava, S.E. , Dits, N.F. , van Leenders, G.J. , Moller, S. , Trapman, J. , Bangma, C.H. , Litman, T. , Visakorpi, T. , Jenster, G. , 2012. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 31, 978–991. [DOI] [PubMed] [Google Scholar]
- Martens-Uzunova, E.S. , Olvedy, M. , Jenster, G. , 2013 Nov 1. Beyond microRNA – novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 340, (2) 201–211. [DOI] [PubMed] [Google Scholar]
- Mei, Y.P. , Liao, J.P. , Shen, J. , Yu, L. , Liu, B.L. , Liu, L. , Li, R.Y. , Ji, L. , Dorsey, S.G. , Jiang, Z.R. , Katz, R.L. , Wang, J.Y. , Jiang, F. , 2012. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 31, 2794–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi, A. , Williams, B.A. , McCue, K. , Schaeffer, L. , Wold, B. , 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Mottet, N. , Bellmunt, J. , Bolla, M. , Joniau, S. , Mason, M. , Matveev, V. , Schmid, H.P. , Van der Kwast, T. , Wiegel, T. , Zattoni, F. , Heidenreich, A. , 2011. EAU guidelines on prostate cancer. Part II: treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur. Urol. 59, 572–583. [DOI] [PubMed] [Google Scholar]
- Narita, S. , So, A. , Ettinger, S. , Hayashi, N. , Muramaki, M. , Fazli, L. , Kim, Y. , Gleave, M.E. , 2008. GLI2 knockdown using an antisense oligonucleotide induces apoptosis and chemosensitizes cells to paclitaxel in androgen-independent prostate cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 14, 5769–5777. [DOI] [PubMed] [Google Scholar]
- Ono, M. , Scott, M.S. , Yamada, K. , Avolio, F. , Barton, G.J. , Lamond, A.I. , 2011. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucl. Acids Res. 39, 3879–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolia, A. , Crea, F. , Xue, H. , Wang, Y. , Mo, F. , Ramnarine, V. , Liu, H. , Lin, D. , Saidy, N. , Clermont, P.L. , Cheng, H. , Collins, C. , Wang, Y. , Helgason, C.D. , 2015. The long non-coding RNA PCGEM1 is regulated by androgen receptor activity in vivo. Mol. Cancer. 14, 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner, J.R. , Chen, W. , Han, S. , Iyer, M.K. , Cao, Q. , Kothari, V. , Evans, J.R. , Knudsen, K.E. , Paulsen, M.T. , Ljungman, M. , Lawrence, T.S. , Chinnaiyan, A.M. , Feng, F.Y. , 2014. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 16, 900–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick, F.G. , Schally, A.V. , Szalontay, L. , Block, N.L. , Szepeshazi, K. , Nadji, M. , Zarandi, M. , Hohla, F. , Buchholz, S. , Seitz, S. , 2012. Antagonists of growth hormone-releasing hormone inhibit growth of androgen-independent prostate cancer through inactivation of ERK and Akt kinases. Proc. Natl. Acad. Sci. United States America. 109, 1655–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio, D.C. , Ares, M. , Hannon, G.J. , Nilsen, T.W. , 2010 Jun. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harbor Protoc. 2010, (6) 10.1101/pdb.prot5439 pdb.prot5439 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Berriguete, G. , Sanchez-Espiridion, B. , Cansino, J.R. , Olmedilla, G. , Martinez-Onsurbe, P. , Sanchez-Chapado, M. , Paniagua, R. , Fraile, B. , Royuela, M. , 2013. Clinical significance of both tumor and stromal expression of components of the IL-1 and TNF-alpha signaling pathways in prostate cancer. Cytokine. 64, 555–563. [DOI] [PubMed] [Google Scholar]
- Siegel, R. , Naishadham, D. , Jemal, A. , 2012. Cancer statistics, 2012. CA Cancer J. Clin. 62, 10–29. [DOI] [PubMed] [Google Scholar]
- Stepanov, G.A. , Filippova, J.A. , Komissarov, A.B. , Kuligina, E.V. , Richter, V.A. , Semenov, D.V. , 2015. Regulatory role of small nucleolar RNAs in human diseases. Biomed. Res. Int. 206849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulli, R. , Pandolfi, P.P. , 2012. “Snorkeling” for missing players in cancer. J. Clin. Invest. 122, 2765–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, B.S. , Schultz, N. , Hieronymus, H. , Gopalan, A. , Xiao, Y. , Carver, B.S. , Arora, V.K. , Kaushik, P. , Cerami, E. , Reva, B. , Antipin, Y. , Mitsiades, N. , Landers, T. , Dolgalev, I. , Major, J.E. , Wilson, M. , Socci, N.D. , Lash, A.E. , Heguy, A. , Eastham, J.A. , Scher, H.I. , Reuter, V.E. , Scardino, P.T. , Sander, C. , Sawyers, C.L. , Gerald, W.L. , 2010. Integrative genomic profiling of human prostate cancer. Cancer Cell. 18, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valleron, W. , Ysebaert, L. , Berquet, L. , Fataccioli, V. , Quelen, C. , Martin, A. , Parrens, M. , Lamant, L. , de Leval, L. , Gisselbrecht, C. , Gaulard, P. , Brousset, P. , 2012. Small nucleolar RNA expression profiling identifies potential prognostic markers in peripheral T-cell lymphoma. Blood. 120, 3997–4005. [DOI] [PubMed] [Google Scholar]
- Varambally, S. , Yu, J. , Laxman, B. , Rhodes, D.R. , Mehra, R. , Tomlins, S.A. , Shah, R.B. , Chandran, U. , Monzon, F.A. , Becich, M.J. , Wei, J.T. , Pienta, K.J. , Ghosh, D. , Rubin, M.A. , Chinnaiyan, A.M. , 2005. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 8, 393–406. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Xue, H. , Cutz, J.C. , Bayani, J. , Mawji, N.R. , Chen, W.G. , Goetz, L.J. , Hayward, S.W. , Sadar, M.D. , Gilks, C.B. , Gout, P.W. , Squire, J.A. , Cunha, G.R. , Wang, Y.Z. , 2005. An orthotopic metastatic prostate cancer model in SCID mice via grafting of a transplantable human prostate tumor line. Lab. Invest. J. Tech. Methods Pathol. 85, 1392–1404. [DOI] [PubMed] [Google Scholar]
- Watahiki, A. , Macfarlane, R.J. , Gleave, M.E. , Crea, F. , Wang, Y. , Helgason, C.D. , Chi, K.N. , 2013. Plasma miRNAs as biomarkers to identify patients with castration-resistant metastatic prostate cancer. Int. J. Mol. Sci. 14, 7757–7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watahiki, A. , Wang, Y. , Morris, J. , Dennis, K. , O'Dwyer, H.M. , Gleave, M. , Gout, P.W. , Wang, Y. , 2011. MicroRNAs associated with metastatic prostate cancer. PloS One. 6, e24950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J.T. , Feng, Z. , Partin, A.W. , Brown, E. , Thompson, I. , Sokoll, L. , Chan, D.W. , Lotan, Y. , Kibel, A.S. , Busby, J.E. , Bidair, M. , Lin, D.W. , Taneja, S.S. , Viterbo, R. , Joon, A.Y. , Dahlgren, J. , Kagan, J. , Srivastava, S. , Sanda, M.G. , 2014. Can urinary PCA3 supplement PSA in the early detection of prostate cancer?. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 32, 4066–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Suppl. Figure 1: Global RPKM plot for protein coding and non‐coding genes in LTL313H (Metastatic) and LTL313B (Non‐metastatic) cells. SnoRNAs and ribonuclease‐associated RNAs (RMRP and RPPH1) are indicated by arrows.
Suppl. Figure 2: Differentially expressed snoRNAs in the MSKCC database *p < 0.05, **p < 0.01 (ANOVA and Tukey's post‐test, Bonferroni correction for multiple comparisons). We only show results with corrected p < 0.05. Horizontal bars represent median value, vertical bars pan from lowest to highest value in the group. Data are from the MSKCC database (cBio portal).
Suppl. Figure 3: SNORA55 expression based on Gleason Score (A), T stage (B), ERG fusion status (C) and circulating PSA levels (D). P values (>0.05 for all the analysis) were calculated using ANOVA and Tukey's post‐test (A), unpaired two‐tailed T test (B–C) and linear regression (D). Horizontal bars represent median value, vertical bars pan from lowest to highest value in the group Data are from the MSKCC database (Cbio portal).
Suppl. Figure 4: A, SNORA55 and PABPC4 mRNA expression in 131 primary PCa samples from the MSKCC database. B, Relapse‐free survival according to PABPC4 expression in primary tumor at diagnosis, (MSKCC database, dichotomization based on Z score, as for SNORA55). P value refers to log‐rank test.
Suppl. Figure 5: Top‐molecular networks associated with SNORA55 expression (Ingenuity Pathway Analysis was used for analysis and visualization). Genes significantly associated with SNORA55 (in red) were identified through SAM analysis (see “Methods” for details).
Suppl. Figure 6: qPCR analysis of TNF and GHRHR expression in LNCaP cells exposed to NC or SNORA55‐targeting ASO1.
Suppl. Table 1: Clinical characteristics of the validation dataset (primary PCa samples).
Suppl. Table 2: Clinical characteristics of the serum sample PCa dataset.


