Abstract
The leukocyte antigen related (LAR) family of receptor‐like protein tyrosine phosphatases has three members in humans – PTPRF, PTPRD and PTPRS – that have been implicated in diverse processes including embryonic development, inhibition of cell growth and axonal guidance. Mutations in the LAR family are associated with developmental defects such as cleft palate as well as various cancers including breast, neck, lung, colon and brain. Although this family of tyrosine phosphatases is important for many developmental processes, little is known of their substrates. This is partially due to functional redundancy within the LAR family, as deletion of a single gene in the LAR family does not have an appreciable phenotype, but a dual knockout is embryonically lethal in mouse models. To circumvent the inability to knockout multiple members of the LAR family in mouse models, we used a knockout of ptp‐3, which is the only known ortholog of the LAR family in Caenorhabditis elegans and allows for the study of the LAR family at the organismal level. Using SILAC‐based quantitative phosphoproteomics, we identified 255 putative substrates of ptp‐3, which included four of the nine known annotated substrates of the LAR family. A motif analysis of the identified phosphopeptides allowed for the determination of sequences that appear to be preferentially dephosphorylated. Finally, we discovered that kinases were overrepresented in the list of identified putative substrates and tyrosine residues whose phosphorylation is known to increase kinase activity were dephosphorylated by ptp‐3. These data are suggestive of ptp‐3 as a potential negative regulator of several kinase families, such as the mitogen activated kinases (MAPKs), and multiple tyrosine kinases including FER, MET, and NTRK2.
Keywords: Phosphoproteomics, Protein tyrosine phosphatases, Tyrosine phosphorylation
Highlights
Phosphoproteomics reveals 255 putative substrates of the tyrosine phosphatase ptp‐3.
Putative substrates of ptp‐3 are enriched for kinases.
Motif analysis reveals two motifs for the ptp‐3 phosphatase.
1. Introduction
Tyrosine phosphorylation is a reversible post‐translational modification involved in most cellular processes. Dysregulation of tyrosine phosphorylation has been implicated in a number of human cancers (Hanahan and Weinberg, 2000). The role of tyrosine kinases as potent oncogenes has been well described. For example, HER2 is found to be amplified in 20% of breast cancers (Slamon et al., 1987), the BCR‐ABL translocation is observed in nearly all chronic myelogenous leukemias (Buchdunger et al., 2001) and increased expression of EGFR is reported in 50% of primary lung cancers (Kolibaba and Druker, 1997). Protein tyrosine phosphatases, on the other hand, have been shown to act as tumor suppressors. Functional inactivation of protein tyrosine phosphatases through either mutation or decreased expression is associated with a multitude of cancers (Jacob and Motiwala, 2005; Li et al., 1997; The Cancer Genome Atlas Network, 2012; Veeriah et al., 2009). Recently, protein tyrosine phosphatases have also been found to play a role as oncogenes in addition to acting as tumor suppressors (Tonks, 2006). For example, deletion of the protein tyrosine phosphatase non‐receptor type 11 (PTPN11) gene results in increased development of hepatocellular carcinoma in mice (Bard‐Chapeau et al., 2011) whereas activating mutations or overexpression has been described in several forms of leukemia (Chan et al., 2008).
Though both protein tyrosine kinases and protein tyrosine phosphatases play integral roles in cancer biology, there is a marked discrepancy in our knowledge regarding phosphatases as compared to kinases. For instance, although there are approximately equal numbers of tyrosine phosphatases and tyrosine kinases in the human genome, the phosphorylation database PhosphoSitePlus lists over 1700 known tyrosine kinase substrates while the phosphatase database DEPOD contains ∼130 tyrosine phosphatase substrates (Duan et al., 2015; Hornbeck et al., 2015). This disparity in our knowledge is partly due to intrinsic differences in the experimental methodologies that are amenable to studying kinases versus phosphatases. Kinases result in a gain of signal that more easily lend themselves to experimental techniques such as radiolabeling ATP to identify substrates, or more recently, using mass spectrometry to identify phosphorylated sites. Phosphatases, on the other hand, lead to a loss of signal and cannot be studied in a straightforward fashion. One approach to identifying phosphatase substrates is the creation of substrate trapping mutants in which a catalytically inactive phosphatase is used to pull‐down interacting proteins and methods such as western blotting and mass spectrometry are used to identify substrates. While useful, this approach is an in vitro binding method and requires a functional, mutated fusion protein which may not be possible to generate easily.
To expand upon the known phosphatase substrates, we sought to apply a high‐throughput strategy that would lead to discovery of novel substrates of tyrosine phosphatases in vivo. We reasoned that a functionally inactive mutants or knockdowns of a phosphatase would result in an increase in the phosphorylation levels of its substrates. This, combined with mass spectrometry, could rapidly identify hyperphosphorylated peptides derived from putative substrates of a protein phosphatase. Moreover, because mass spectrometry is used to detect phosphopeptides, the actual sites that are differentially phosphorylated due to a mutation are revealed.
For this study, we chose to identify substrates of the LAR family of protein tyrosine phosphatases. These phosphatases are receptor‐like tyrosine phosphatase characterized by several extracellular fibronectin and immunoglobulin domains along with two intracellular tyrosine phosphatase domains. Within vertebrates, there are at least three related genes comprising the LAR family: the LAR receptor protein tyrosine phosphatase (PTPRF), protein tyrosine phosphatase receptor type delta (PTPRD), and protein tyrosine phosphatase receptor type sigma (PTPRS). PTPRD has been shown to be a tumor suppressor gene and has been found to be functionally inactivated in glioblastoma multiforme, head and neck cancer, and lung cancer (Veeriah et al., 2009). Loss of PTPRS, which is known to dephosphorylate EGFR, has been reported in ∼25% of head and neck cancers and its deletion in lung cancer results in increased resistance to the tyrosine kinase inhibitor erlotinib (Morris et al., 2011).
Given the association between mutations in the LAR family and a number of cancers, identification of substrates of the LAR family may help elucidate the molecular consequences of these mutations and how they might lead to oncogenesis. However, such studies in mouse have been complicated by the functional redundancy between the multiple members of the LAR family. This redundancy can be mitigated through the use of multiple gene knockouts; however, existing attempts to knockout multiple members of the LAR family have resulted in embryonic lethality (Uetani et al., 2006). To circumvent this issue, we used Caenorhabditis elegans as a model system to identify phosphatase substrates in an organism which has reduced redundancy but retains many important developmental effects of the loss of LAR. The single LAR family homolog in C. elegans, ptp‐3, is made up of the characteristic extracellular fibronectic and immunoglobulin domains and two intracellular tyrosine phosphatase domains that are observed in this family. Similar to its human homologs, only the membrane proximal tyrosine phosphatase domain is active and ptp‐3 has been implicated in gastrulation, epidermal development and embryonic morphogenesis. Further, deletion of ptp‐3 results in smaller brood size due to a diminished embryonic viability (Harrington et al., 2002). Here, through the use of quantitative phosphoproteomics by employing SILAC labeling of C. elegans, we identified 255 putative substrates of ptp‐3 and used computational methods to identify common motifs for this phosphatase. Further, we identified that kinases were overrepresented as substrates of ptp‐3, with their kinase activation loop being a common target of dephosphorylation by ptp‐3.
2. Materials and methods
2.1. Reagents
The Escherichia coli SLE1 strain and all C. elegans strains were purchased from the Caenorhabditis Genetics Center (CGC). Heavy labeled amino acids – arginine (13C6‐15N4) and lysine (13C6‐15N2) were purchased from Cambridge Isotopes Laboratories (Andover, MA). For phosphotyrosine enrichment, anti‐phosphotyrosine mouse monoclonal antibody (P‐Tyr‐1000) was purchased from Cell Signaling Technology (Danvers, MA). Nystatin (N1638‐100ML) was purchased from Sigma–Aldrich. Complete, EDTA‐free protease inhibitor cocktail tablets (10711400) were purchased from Roche. Sequencing grade trypsin and lys‐c enzymes were purchased from Promega and TPCK‐treated trypsin was purchased from Worthington Biochemical Corp. All other reagents were purchased from Sigma.
2.2. C. elegans metabolic labeling
For SILAC labeling of C. elegans, the bacterial food source was labeled and then fed to the C. elegans. For culturing bacteria, light cultures were grown in Lysogeny Broth (LB) and heavy cultures were grown in M9 Minimal Media supplemented with 40 μg/ml final concentrations of heavy arginine and heavy lysine. Each culture was grown overnight, and centrifuged at 8000 × g for 10 min at 4 °C. For each 250 ml of culture pelleted, 50 ml of S. Medium (Stiernagle, 2006) was added to resuspend the pellet. C. elegans were added to a 1 L flask containing 50 ml of resuspended E. coli supplemented with 10 μg/ml of Nystatin to prevent fungal growth and grown according to previously described methods (Stiernagle, 2006). The culture was monitored and when the bacteria was almost clear (approximately 3 days), the C. elegans were pelleted and resuspended in 0.1 M NaCl. To complete purification, an equal volume of 60% sucrose was added and centrifuged for 5 min at 900 × g at 4 °C. C. elegans at the surface were then pipetted off, and washed three times in 50 ml of 0.1 M NaCl to obtain a pure, live population of C. elegans.
2.3. C. elegans lysis and protein digestion
C. elegans were resuspended in 9 M urea containing a protease inhibitor cocktail (Roche 10711400), 50 mM beta‐galactosidase, 25 mM NaF, and 1 mM sodium orthovanadate. Next, the resuspension was sonicated and viewed under a microscope following sonication until the C. elegans were sufficiently broken apart. The lysate was cleared by centrifugation at 14,000 × g for 20 min at 4 °C. For whole proteome experiments, 250 μg from wild type and ptp‐3 lysates were combined, reduced with 5 mM of dithiothreitol, and alkylated with 5 mM of iodoacetamide. Then, the lysate was diluted to a final urea concentration of 3 M and digested with lys‐C with an enzyme to protein ratio of 1:100 for 4 h at room temperature. After digestion with lys‐C, the crude digests were diluted to a final urea concentration of 1.5 M and digested overnight with sequencing grade trypsin with an enzyme to protein ratio of 1:25. After confirming digestion efficiency, the sample was acidified with 1% trifluoroacetic acid (TFA) and centrifuged at 2000 × g for 5 min at room temperature. The supernatant was loaded onto a Sep‐Pak C18 cartridge (Waters, Cat#WAT051910) and equilibrated with 0.1% TFA. Desalted peptides were then lyophilized and subjected to basic reverse phase liquid chromatography. For phosphotyrosine peptide enrichment, 25 mg each from heavy and light lysates were combined, reduced with 5 mM of dithiothreitol, and alkylated with 5 mM of iodoacetamide. Then, lysates were diluted to a final urea concentration of 1.5 M and digested overnight with TPCK‐treated trypsin (Worthington) with an enzyme to protein ratio of 1:25. Peptide digests were desalted with a Sep‐Pak C18 cartridge (Waters, Cat#WAT051910) and then lyophilized for subsequent use in phosphotyrosine enrichment.
2.4. Immunoaffinity purification of tyrosine phosphopeptides
Immunoaffinity purification (IAP) of tyrosine phosphopeptides was carried out as previously described (Kim et al., 2014b; Rush et al., 2005). Briefly, after lyophilization, 50 mg of peptide mixture was dissolved in 1.4 ml of IAP buffer (50 mM MOPS pH 7.2, 10 mM sodium phosphate, 50 mM NaCl) and subjected to centrifugation at 2000 × g at room temperature for 5 min. Before IAP, P‐Tyr‐1000 beads (Cell Signaling Technology) were washed with IAP buffer twice at 4 °C and the pH of the supernatant containing peptides was adjusted to 7.2 by adding 1 M Tris Base. For IAP, the supernatant was incubated with P‐Tyr‐1000 beads (Cell Signaling Technology) at 4 °C for 30 min, and the beads were washed three times with IAP buffer followed by two washes with pure water. Enriched peptides were eluted twice from beads by incubating the beads with 0.15% TFA at room temperature.
2.5. LC‐MS/MS
Peptide samples were analyzed on an LTQ‐Orbitrap Elite mass spectrometer (Thermo Electron, Bremen, Germany) interfaced with Easy‐nLC II nanoflow liquid chromatography systems (Thermo Scientific) as previously described (Kim et al., 2014a). Peptides were reconstituted in solvent A (0.1% formic acid) and loaded onto a trap column (75 μm × 2 cm) packed in‐house with Magic C18 AQ (Michrom Bioresources, Inc., Auburn, CA, USA) (5 μm particle size, pore size 100 Å) at 280 bar with solvent A (0.1% formic acid in water). Peptides were resolved on an analytical column (75 μm × 15 cm) at a flow rate of 300 nl/min using a linear gradient of 5–35% solvent B (0.1% formic acid in 95% acetonitrile). Mass spectrometry analysis was carried out in a data dependent manner with full scans (300–1,700 m/z) acquired using an Orbitrap mass analyzer at a mass resolution of 120,000 at 400 m/z. The ten and fifteen most intense precursor ions from a survey scan were selected for MS/MS from each duty cycle for whole proteome and phosphopeptides, respectively, and detected at a mass resolution of 30,000. All the tandem mass spectra were produced using higher energy C‐trap dissociation method. Dynamic exclusion was set for 30 s with a 7 p.p.m. mass window. Internal calibration was carried out using lock‐mass from ambient air (m/z 445.1200025).
2.6. Data analysis
Tandem mass spectra were processed and searched using MASCOT (Version 2.2.0) (Perkins et al., 1999) and SEQUEST (Eng et al., 1994) search algorithms against a C. elegans WormBase (Harris et al., 2010) database (version 248) through the Proteome Discoverer platform (version 2.0, Thermo Scientific). For both algorithms, the search parameters included a maximum of two missed cleavages; carbamidomethylation at cysteine as a fixed modification; N‐terminal acetylation at protein N‐termini, deamidation at asparagine and glutamine, oxidation at methionine, phosphorylation at serine, threonine and tyrosine and SILAC labeling at 13C6 15N2 lysine and 13C6 15N4 arginine as variable modifications. The mass tolerance for peptide and fragment ions were set to 10 ppm and 0.1 Da, respectively. The false discovery rate was set to 1% at the peptide level. The probability of phosphorylation for each Ser/Thr/Tyr site on each peptide was calculated by the PhosphoRS node in Proteome Discoverer 2.0.
Following the database search, SILAC ratios were quantified with an in‐house python script and a 2‐fold cutoff was chosen to indicate a significant change in a given PSM between samples. For whole proteome measurements, the data was normalized to the median to control for loading bias. After normalization, peptide spectra matches (PSMs) were grouped at the peptide level and the median value was taken of each peptide group. Following this, the median value of a protein's peptide groups was taken to estimate the baseline protein abundance. Next, in‐house python scripts were used to provide protein level assignments of phosphotyrosine sites (e.g. VLPAATY(phos)LK becomes CE000001 Tyr582). To normalize loading bias during mixing of samples, the data were normalized against each enrichment's corresponding whole proteome experiment. Identified phosphotyrosine sites were then grouped together, and the median value of these groups was used to measure the phosphorylation level of each phosphotyrosine site. Finally, the abundance level of each site was normalized against the protein abundance measured from the whole proteome experiment. If possible, proteins were then mapped to human homologs and the corresponding human residues of phosphotyrosine sites were annotated.
For mapping C. elegans genes to human homologs, the Wormbase API was used. To perform Gene Ontology based clustering clustering and functional annotation, the R package clusterProfiler (Yu et al., 2012) and the PANTHER classification system (Mi et al., 2013) were used, respectively. For identification of motifs, the surrounding sequence (the 13 flanking residues) from each phosphotyrosine site was extracted from the WormBase protein database and analyzed using MEME (Bailey and Elkan, 1994) and GLAM2 (Frith et al., 2008) to generate motifs. For prediction of sequences containing these motifs, motifs generated with MEME were searched using FIMO and predictions with a q‐value below 10−4 were retained (Grant et al., 2011). For motifs generated using GLAM2, GLAM2SCAN was used to predict sequences containing the motif and predictions with a score above 15 were kept (Frith et al., 2008).
2.7. Data availability
Mass spectrometry derived proteomics data has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD003233.
3. Results
3.1. Deletion of the ptp‐3 phosphatase results in increased global tyrosine phosphorylation
A mutant C. elegans strain harboring a mutation in ptp‐3, designated RB1878, was previously generated by the C. elegans Gene Knockout Consortium. This mutant contains a premature stop codon leading to the truncation of ptp‐3 prior to its phosphatase domains. For quantitative measurements, wild type worms were labeled with heavy isotope‐labeled lysine and arginine amino acids, and mutant worms were labeled with light versions of these amino acids. Following a label check to confirm incorporation of the heavy isotope, samples were lysed and protein concentration was measured. From each sample, 25 mg of lysate was taken, mixed, and digested in‐solution with trypsin and subjected to an antibody‐based phosphotyrosine enrichment and LC/MS–MS analysis (Figure 1) as described previously by our group (Syed et al., 2015; Zahari et al., 2015; Zhong et al., 2012). Peptides were identified using Mascot and Sequest run through Proteome Discoverer 2.0 and quantified using PyQuant.
Figure 1.
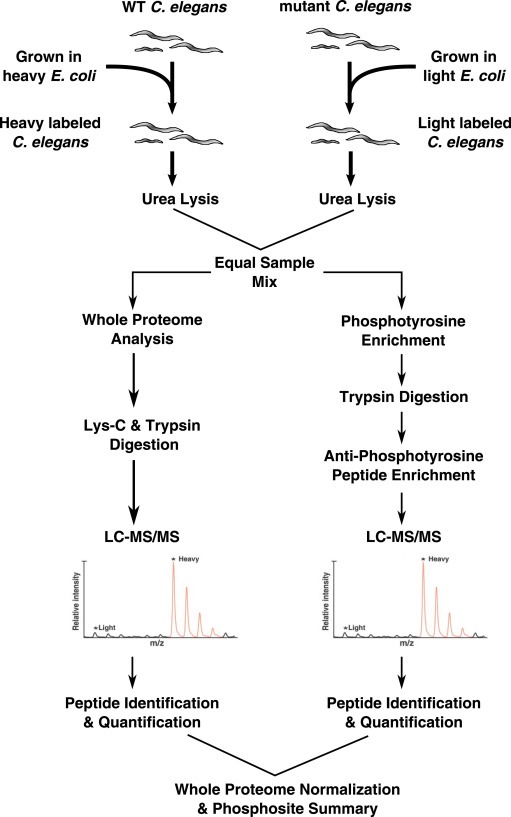
Overall strategy of phosphotyrosine peptide enrichment and LC‐MS/MS analysis. The experimental workflow for labeling of C. elegans, phosphotyrosine enrichment and LC‐MS/MS is shown. First, E. coli cultures were grown with SILAC isotopes and then fed to C. elegans over several generations until the worms were fully labeled. Following label incorporation, wild type and mutant samples were lysed and an equal protein amount of each labeled lysate was mixed together. Following mixing, one portion of the sample was subjected to whole proteome LC‐MS/MS analysis and the remaining portion was subjected to phosphotyrosine peptide enrichment and LC‐MS/MS analysis.
Two biological replicates of wild type and ptp‐3 mutants each were grown, lysed, and subjected to phosphotyrosine enrichment and whole proteome analysis. First, we assessed proteomic changes of the ptp‐3 knockout on C. elegans. From our analysis, we identified 2130 proteins encoded by 1266 genes, with 148 proteins encoded by 72 genes that were up‐regulated and 29 proteins encoded by 20 genes that were downregulated in the ptp‐3 knockout (Supplementary Figure 1A). To assess the possible impact of the knockout, we performed a Gene Ontology (GO) analysis (Ashburner et al., 2000) using the PANTHER classification system (Mi et al., 2013). GO analysis of genes revealed an enrichment for genes involved with developmental processes (Supplementary Figure 1B), which is consistent with the known phenotype of increased embryonic lethality and abnormal morphology in the ptp‐3 knockout.
Using antibody‐based phosphotyrosine peptide enrichment, we identified 3021 phosphotyrosine containing peptides. We grouped the identified peptides by their phosphotyrosine site (e.g. y543). This analysis identified 1463 different phosphotyrosine sites (Supplementary Table 1). Because global protein changes in ptp‐3 knockout worms may be falsely inferred as changes in phosphorylation levels, protein levels determined from whole proteome analysis were used to correct for protein fluctuations that may contribute to phosphotyrosine measurements. We considered a site hyper or hypophosphorylated if there was at least a 2‐fold change in the phosphorylation state between the wild type and mutant samples. Using these criteria, we identified 255 sites that were hyperphosphorylated and 264 that were hypophosphorylated in the ptp‐3 mutant (Figure 2). As a phosphatase, knockout of ptp‐3 predicts an increase in phosphorylation levels of its substrate(s); therefore, we consider these 255 hyperphosphorylated sites to be potential substrates of ptp‐3.
Figure 2.
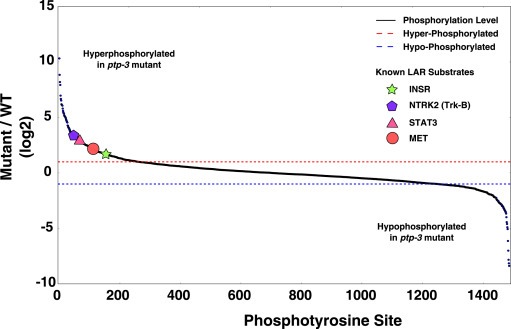
Distribution of phosphotyrosine sites identified. The relative change in phosphorylation levels of phosphotyrosine sites between the ptp‐3 mutant and wild type is shown. Higher values on the y‐axis correspond to increases in phosphotyrosine levels with a tyrosine phosphatase mutation. The red dashed line indicates a 2‐fold increase in tyrosine phosphorylation, and the blue dashed line indicates a 2‐fold decrease in tyrosine phosphorylation. Shapes with their corresponding gene are known substrates of human homologs of ptp‐3, and show that this study identifies known phosphotyrosine substrates as being hyperphosphorylated.
To evaluate the general impacts of the loss of ptp‐3 mediated dephosphorylation, we analyzed hyperphosphorylated tyrosine sites in the ptp‐3 knockout. A GO analysis of putative substrates was performed using clusterProfiler (Yu et al., 2012). Similar to the whole proteome, GO analysis revealed enrichment of genes related to reproduction, anatomical development and embryo development.
3.2. Conservation of putative substrates in humans
Due to evolutionary changes from C. elegans to Homo sapiens, putative substrates identified in C. elegans may not be conserved across species. Moreover, conservation is a strong predictor of functionality, and sites that are conserved across species are more likely to be true positives than those that are found only in C. elegans (Tan et al., 2009). Thus, for our subsequent analysis we focused only on putative substrates that were conserved in C. elegans and humans.
We performed a search using Wormbase (Harris et al., 2010) to locate homologous genes of the identified putative ptp‐3 substrates. This analysis revealed 145 putative substrates with clearly defined human homologs. Next, Clustal Omega (Sievers et al., 2011) was used to align each C. elegans protein sequence to its human homolog and to determine if the specific phosphotyrosine site was conserved in humans. Of the 145 putative substrates with homologs in humans, 45 contained a tyrosine residue in both species at the identified hyperphosphorylated tyrosine site. Interestingly, we also found that in 14 cases the tyrosine residue that was hyperphosphorylated in C. elegans was replaced by a phenylalanine in the orthologous sequence in humans. This is of note because mutational studies often mutate tyrosine residues to phenylalanine in order to prevent phosphorylation while retaining the structural importance of the aromatic side chain. Thus, these residues may not be crucial for signaling in humans, but could have important roles in structure‐based interactions.
3.3. Site level resolution of known substrates of the LAR family
To evaluate the success of our profiling approach, we compared the list of proteins that were hyperphosphorylated in the ptp‐3 mutant with C. elegans orthologs of previously described substrates of the LAR family. We chose to use the substrates of the LAR family members in the human dephosphorylation database DEPOD (Duan et al., 2015) because currently there are no databases listing dephosphorylation events in C. elegans, there is no entry for ptp‐3 in the Worm Interactome Database (Simonis et al., 2009), and we are unable to find known substrates of ptp‐3 in the literature through PubMed searches.
At this time, the DEPOD database contains nine known substrates of the LAR family, with three substrates belonging to the same gene family. Of the known substrates, only Tyr705 of STAT3 was annotated as the site that is dephosphorylated by PTPRD. Remarkably, in our data, we identified orthologs of four of the nine known substrates, NTRK2, MET, INSR, and STAT3 (sites indicated in Figure 2), to be hyperphosphorylated with a conserved tyrosine residue. Additionally, the C. elegans ortholog of STAT3, sta‐1, was found and the C. elegans site corresponding to Tyr705, Tyr588, exhibited a 5.6 fold increase in phosphorylation levels in the ptp‐3 knockout. This shows we are able to find known gene products which are dephosphorylated by the LAR family in addition to the site which has previously been shown to be dephosphorylated.
The DEPOD database listed only one substrate where the actual tyrosine residue that is dephosphorylated was confirmed. This is an important limitation to known enzyme–substrate relationships; that even when experimental evidence shows a protein is dephosphorylated by a phosphatase, the exact sites which are modified are often not known. Since mass spectrometry was used to identify substrates in this study, it inherently provides site level resolution as well as quantitative measures of phosphorylation. As discussed above, in our data three substrates, NTRK2, MET, and INSR, were known to be dephosphorylated in humans although their actual site or sites of dephosphorylation was unknown. We identified conserved hyperphosphorylated sites mapping to these three genes as well as a hyperphosphorylated site that has been previous described. This illustrates that our approach is able to provide highly confident sites for future functional studies that further dissect phosphorylation based signaling networks.
3.4. Putative substrates of ptp‐3 are enriched in kinases
Next, we performed a GO analysis for biological processes of the conserved putative substrates of ptp‐3, which revealed a significant enrichment for kinases (q‐value 5.9 * 10−27). Of these genes included in the GO analysis, we found that 15 conserved hyperphosphorylated sites were located in kinases. We then examined the location of the phosphosite in each kinase, and found that 6 sites were in the activation loop and have previously been shown to be critical for kinase activation (Figure 4). Additionally, several sites were identified that were not in the activation loop, but conferred important catalytic activity. For example, Tyr579 of PDGFRB and Tyr15 CDK5 were hyperphosphorylated, and phosphorylation of these sites has been shown to increase kinase activity (Baxter et al., 1998; Zukerberg et al., 2000). Tyr1356 of MET was hyperphosphorylated, which is not known to increase kinase activity, but has been shown to be crucial for transduction of signals involving cell motility, morphogenesis, and tubule formation (Dekel et al., 2003).
Figure 4.
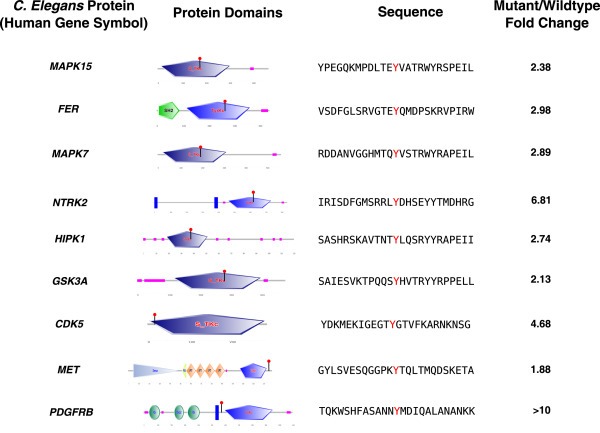
Activating residues of kinases that are hyperphosphorylated. Tyrosine residues with increased phosphorylation in the ptp‐3 knockout are depicted, with the identified residue indicated in red. The first six rows correspond to tyrosine residues that are located in the activation loop of the indicated gene, and the last three rows correspond to tyrosine residues that have been shown to alter kinase activity.
Next, we sought to determine whether a common motif exists within the putative substrates of ptp‐3. We selected the identified conserved hyperphosphorylated tyrosine sites and 13 amino acids surrounding each site as the input sequences for motif prediction. These sequences were scanned for common motifs using GLAM2 (Frith et al., 2008) and MEME (Bailey and Elkan, 1994), two motif identification programs that are part of the MEME suite (Bailey et al., 2009). This analysis revealed two motifs where the hyperphosphorylated tyrosine residue was located at the 0 position (Figure 3A; 3B). Following this, we compared the conservation of substrates containing each motif across C. elegans and humans. This revealed that for the motif in 3A, there is a 100% identity between the dominant residues comprising the motif (YxxxRxYRxPE) in humans. However, for the motif in 3B, we find there is a poor conservation of the dominant features of the motif ((A/Y/G)xxxxxYxxxxx(Y/N/F)xxP) in humans. This indicates that the motif in 3A may be common to both species. At this time, we do not know the significance of the motif identified in Figure 3B. Next, we used these motifs to predict additional substrates of ptp‐3 and the LAR family by searching against the human RefSeq protein database with GLAM2SCAN (Frith et al., 2008) and FIMO (Grant et al., 2011), two programs that take the respective output of GLAM2 and MEME and identify sequences containing a given motif. The motif identified by GLAM2 (Figure 3B), was present in 65 proteins encoded by 20 genes while the MEME motif (Figure 3A) was identified in 108 proteins encoded by 29 genes (Supplementary Table 2). Significantly, 12 of the 20 genes containing the GLAM2 motif (60%) and all 29 genes containing the MEME motif are kinases, implying an important role for ptp‐3 and the LAR family in the regulation of kinases.
Figure 3.
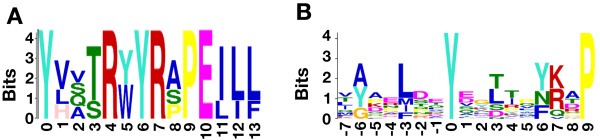
Motif analysis of hyperphosphorylated tyrosine sites. A) A motif identified by MEME for hyperphosphorylated tyrosine residues. The zero position for the motif indicates the hyperphosphorylated tyrosine residue. B) A motif identified by GLAM2 for hyperphosphorylated tyrosine residues. The zero position for the motif indicates the hyperphosphorylated tyrosine residue.
Given the abundance of kinases identified through our profiling approach, and that increased phosphorylation of many of these tyrosine residues is known to lead to increased kinase activity, functional inactivation of a LAR family member could lead to increased kinase activity. In the context of our study, this means that some of the identified sites underwent increased phosphorylation because of increased kinase activity and not because they were substrates of ptp‐3. To explore this possibility, we evaluated tyrosine residues that were phosphorylated >2.83 fold (1.5 log2) in the mutants to determine how many sites that were hyperphosphorylated were also known targets of tyrosine kinases (Table 1). This revealed 20 tyrosine residues to be highly phosphorylated of which eight were novel tyrosine phosphorylation sites. Of the 12 previously known sites, only 6 were annotated as interacting with tyrosine kinases and 3 of these sites possessed information on the tyrosine kinases that are known to phosphorylate them. One of these Tyr15 is located in CDK5, which is a substrate of ABL1. This is notable because an activating tyrosine site of ABL1 was hyperphosphorylated in our data suggesting that this site may be due to an active kinase as opposed to being a substrate of ptp‐3. For the remaining two sites, Tyr579 of PDGFRB and Tyr702 of NTRK2, both cases represented sites that are known to be autophosphorylated.
Table 1.
Manual annotations of sites with increase in tyrosine phosphorylation.
| Conserved human site | Protein | Human homolog | Previously reported site | Known tyrosine kinase interactions |
|---|---|---|---|---|
| y684 | CE35454 | FRMD4A | Yesb | None |
| y598 | CE02628 | RAD21 | Novel | None |
| y143 | CE06481 | TIMM50 | Novel | None |
| y285 | CE12272 | GNAQ | Novel | BTKa,d, CRKd |
| y579 | CE09274 | PDGFRB | Yesa,b | PDGFRBa,b,c,d,f |
| y162 | CE20412 | PABPC1 | Novel | None |
| y54 | CE01044 | RBM8A | Yesa,b,c | None |
| y136 | CE33442 | DYRK1A | Yesa,b | DYRK1Aa |
| y268 | CE15746 | LMNB1 | Yesb | None |
| y364 | CE02193 | PABPC4 | Yesb | None |
| y702 | CE40381 | NTRK2 | Yesb,c | NTRK2a,c,f, FYNa |
| y266 | CE04237 | RPS2 | Yesb | None |
| y4332 | CE43614 | MUC17 | Yesb | None |
| y131 | CE09710 | PSAT1 | Novel | None |
| y4126 | CE38607 | TTN | Novel | None |
| y705 | CE22342 | STAT5A | Yese | ETKa, c‐SRCa,b, EGFRa, ErbB4a, JAK1Aa, JAK3a,b, SYKa,b, TEKa, JAK2a,b, BTKa, LCKa,b, EK1b, ERK2b, LYNb |
| y15 | CE21213 | CDK5 | Yesa,b,c | ABL1a,b,f, FYNa,f, c‐SRCa, ErbB3a, MEK1a |
| y487 | CE18697 | PRDM2 | Novel | None |
| y250 | CE04237 | RPS2 | Yesb | None |
| y110 | CE30219 | DMGDH | Novel | None |
Annotated by Human Protein Reference Database (Keshava Prasad et al., 2009).
Annotated by PhosphoSitePlus (Hornbeck et al., 2015).
Annotated by PhosphoELM (Dinkel et al., 2010).
Annotated by NetworKIN (Horn et al., 2014).
Reported in literature (Chen et al., 2015; Zhang et al., 2007).
Kinase is reported to phosphorylate identified site.
Taking into account the known oncogenic potential of kinases, our data present a possible link between the mutation of various LAR family members and associated cancers (FUNATO et al., 2011; Giefing et al., 2011; Morris et al., 2011; Veeriah et al., 2009). Although our profiling approach was able to recapitulate many known substrates of the human homolog of ptp‐3 and provided broad implications to the regulatory roles of ptp‐3 and the LAR family in humans, it should be noted these are putative substrates and do not supplant directed mutational studies and in vitro assays for final confirmation. For example, while hyperphosphorylation of protein kinases has interesting biological consequences, increased tyrosine kinase activity may lead to hyperphosphorylation of its substrates that may be falsely be inferred as phosphatase substrates. However, given the ability of this approach to capture much of what is known, we feel the hyperphosphorylated peptides identified in this study are excellent candidates for further validation including functional studies to precisely map out the LAR family's dephosphorylation network.
4. Discussion
The C. elegans LAR‐like receptor tyrosine phosphatase, ptp‐3, is expressed in several tissues during C. elegans development and has been shown to play a role in gastrulation and neural morphogenesis (Chin‐Sang et al., 2002; Harrington et al., 2002). A human homolog of ptp‐3, PTPRD, has been found to be deleted or functionally inactivated in several cancers, with inactivation of PTPRD occurring in over 50% of GBM tumors (Veeriah et al., 2009). A recent study identified Tyr705 of STAT3 as a specific target of PTPRD. In our data, we found the C. elegans STAT orthologs in humans to be hyperphosphorylated at Tyr588, which upon alignment to the human STAT3 protein, corresponds to Tyr705 of STAT3. Interestingly, other receptor tyrosine phosphatases, PTPRK and PTPRT, have been shown to dephosphorylate Tyr705 of STAT3 as well (Chen et al., 2015; Zhang et al., 2007). Given that multiple homologs of ptp‐3 exist in humans, this may indicate that putative substrates found in our study may be common to multiple protein receptor type tyrosine phosphatases.
Many substrates of the LAR‐like receptor tyrosine phosphatases are known only at the protein level and lack site level resolution. PTPRF has been shown to dephosphorylate the insulin receptor (INSR), beta‐catenin, and hepatocyte growth factor receptor (MET) but there is no site level resolution (Hashimoto et al., 1992; Machide et al., 2006; Müller et al., 1999). In our data, we found hyperphosphorylated tyrosine residues corresponding to the C. elegans homologs of INSR and MET. In the C. elegans homolog of INSR, daf‐2, we find two hyperphosphorylated sites, Tyr1419 and Tyr1420, which are both conserved in humans. For MET, we find several hyperphosphorylated sites in its C. elegans homolog svh‐2 as well. While only one of these sites is conserved in humans, we note that there are nearby tyrosine residues which may be the functionally equivalent site in humans. PTPRS has been shown to dephosphorylate the epidermal growth factor receptor (EGFR) and members of the neutrotrophic tyrosine kinase receptors NTRK1, NTRK2, and NTRK3 (Faux et al., 2007; Vijayvargia et al., 2004). We find hyperphosphorylated tyrosine residues in both of the C. elegans homologs of these genes. For EGFR, like MET, the hyperphosphorylated residue is not conserved in the human homolog, but once again a conserved tyrosine residue is near the site identified in C. elegans. For the neutrotrophic tyrosine kinase receptors, we identified a hyperphosphorylated Tyr632 in the C. elegans homolog trk‐1, which is conserved in the human sequence as well.
When considering the known literature, our study was able to identify known proteins dephosphorylated by the LAR‐like receptor tyrosine phosphatases, and provided potential sites of substrates known to be dephosphorylated by PTPRF and PTPRS. Additionally, we provide an extensive catalog of potential substrates of ptp‐3 in C. elegans, many of which are related to the known developmental roles of ptp‐3 in C. elegans. In humans, many of the potential substrates were found to be kinase‐related, with 9 of the hyperphosphorylated tyrosine residues having known effects on kinase activity, which may explain why functional inactivation of PTPRD, PTPRF, and PTPRS have been found in multiple cancers. Given the numerous knockouts in C. elegans, our strategy can be applied to identify potential substrates of additional phosphatases as well as other post‐translational modifications to further characterize signaling networks in an organismal setting.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
The following are the supplementary data related to this article:
Supplementary Figure 1 Whole proteome analysis of the ptp‐3 mutant. A) The distribution of protein abundance between the ptp‐3 mutant and wild type strain measured at the global proteome level is shown. B) A GO analysis of genes up‐regulated in the whole proteome data shows an enrichment for genes involved in developmental processes, in agreement with the known phenotype of increased embryonic lethality of the ptp‐3 mutant.
Supplementary data
Supplementary data
Acknowledgments
This study was supported by NCI's Clinical Proteomic Tumor Analysis Consortium initiative (U24CA160036).
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2016.03.003.
Mitchell Christopher J., Kim Min-Sik, Zhong Jun, Raja Sekhar Nirujogi, Bose Anjun K., Pandey Akhilesh, (2016), Unbiased identification of substrates of protein tyrosine phosphatase ptp‐3 in C. elegans, Molecular Oncology, 10, doi: 10.1016/j.molonc.2016.03.003.
References
- Ashburner, M. , Ball, C.A. , Blake, J.A. , Botstein, D. , Butler, H. , Cherry, J.M. , Davis, A.P. , Dolinski, K. , Dwight, S.S. , Eppig, J.T. , Harris, M.A. , Hill, D.P. , Issel-Tarver, L. , Kasarskis, A. , Lewis, S. , Matese, J.C. , Richardson, J.E. , Ringwald, M. , Rubin, G.M. , Sherlock, G. , 2000. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L. , Boden, M. , Buske, F.A. , Frith, M. , Grant, C.E. , Clementi, L. , Ren, J. , Li, W.W. , Noble, W.S. , 2009. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L. , Elkan, C. , 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. ISMB Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36. [PubMed] [Google Scholar]
- Bard-Chapeau, E.A. , Li, S. , Ding, J. , Zhang, S.S. , Zhu, H.H. , Princen, F. , Fang, D.D. , Han, T. , Bailly-Maitre, B. , Poli, V. , Varki, N.M. , Wang, H. , Feng, G.-S. , 2011. Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular carcinogenesis. Cancer Cell. 19, 629–639. 10.1016/j.ccr.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter, R.M. , Secrist, J.P. , Vaillancourt, R.R. , Kazlauskas, A. , 1998. Full activation of the platelet-derived growth factor β-receptor kinase involves multiple events. J. Biol. Chem. 273, 17050–17055. 10.1074/jbc.273.27.17050 [DOI] [PubMed] [Google Scholar]
- Buchdunger, E. , Matter, A. , Druker, B.J. , 2001. Bcr-Abl inhibition as a modality of CML therapeutics. Biochim. Biophys. Acta BBA – Rev. Cancer. 1551, M11–M18. 10.1016/S0304-419X(01)00022-1 [DOI] [PubMed] [Google Scholar]
- Chan, G. , Kalaitzidis, D. , Neel, B.G. , 2008. The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer Metastasis Rev. 27, 179–192. 10.1007/s10555-008-9126-y [DOI] [PubMed] [Google Scholar]
- Chen, Y.-W. , Guo, T. , Shen, L. , Wong, K.-Y. , Tao, Q. , Choi, W.W.L. , Au-Yeung, R.K.H. , Chan, Y.-P. , Wong, M.L.Y. , Tang, J.C.O. , Liu, W.-P. , Li, G.-D. , Shimizu, N. , Loong, F. , Tse, E. , Kwong, Y.-L. , Srivastava, G. , 2015. Receptor-type tyrosine-protein phosphatase κ directly targets STAT3 activation for tumor suppression in nasal NK/T-cell lymphoma. Blood. 125, 1589–1600. 10.1182/blood-2014-07-588970 [DOI] [PubMed] [Google Scholar]
- Chin-Sang, I.D. , Moseley, S.L. , Ding, M. , Harrington, R.J. , George, S.E. , Chisholm, A.D. , 2002. The divergent C. elegans ephrin EFN-4 functions inembryonic morphogenesis in a pathway independent of the VAB-1 Eph receptor. Development. 129, 5499–5510. 10.1242/dev.00122 [DOI] [PubMed] [Google Scholar]
- Dekel, B. , Biton, S. , Yerushalmi, G.M. , Altstock, R.T. , Mittelman, L. , Faletto, D. , Smordinski, N.I. , Tsarfaty, I. , 2003. In situ activation pattern of Met docking site following renal injury and hypertrophy. Nephrol. Dial. Transpl. 18, 1493–1504. 10.1093/ndt/gfg215 [DOI] [PubMed] [Google Scholar]
- Dinkel, H. , Chica, C. , Via, A. , Gould, C.M. , Jensen, L.J. , Gibson, T.J. , Diella, F. , 2011 Jan. Phospho.ELM: a database of phosphorylation sites—update 2011. Nucleic Acids Res. 39, (Database issue) D261–D267. 10.1093/nar/gkq1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, G. , Li, X. , Köhn, M. , 2015. The human DEPhOsphorylation database DEPOD: a 2015 update. Nucleic Acids Res. 43, D531–D535. 10.1093/nar/gku1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, J.K. , McCormack, A.L. , Yates, J.R. , 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989. 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- Faux, C. , Hawadle, M. , Nixon, J. , Wallace, A. , Lee, S. , Murray, S. , Stoker, A. , 2007. PTPσ binds and dephosphorylates neurotrophin receptors and can suppress NGF-dependent neurite outgrowth from sensory neurons. Biochim. Biophys. Acta BBA – Mol. Cell Res. 1773, 1689–1700. 10.1016/j.bbamcr.2007.06.008 [DOI] [PubMed] [Google Scholar]
- Frith, M.C. , Saunders, N.F.W. , Kobe, B. , Bailey, T.L. , 2008. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 4, e1000071 10.1371/journal.pcbi.1000071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato, K. , Yamazumi, Y. , Oda, T. , Akiyama, T. , 2011. Tyrosine phosphatase PTPRD suppresses colon cancer cell migration in coordination with CD44. Exp. Ther. Med. 2, 457–463. 10.3892/etm.2011.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giefing, M. , Zemke, N. , Brauze, D. , Kostrzewska-Poczekaj, M. , Luczak, M. , Szaumkessel, M. , Pelinska, K. , Kiwerska, K. , Tönnies, H. , Grenman, R. , Figlerowicz, M. , Siebert, R. , Szyfter, K. , Jarmuz, M. , 2011. High resolution ArrayCGH and expression profiling identifies PTPRD and PCDH17/PCH68 as tumor suppressor gene candidates in laryngeal squamous cell carcinoma. Genes. Chromosomes Cancer. 50, 154–166. 10.1002/gcc.20840 [DOI] [PubMed] [Google Scholar]
- Grant, C.E. , Bailey, T.L. , Noble, W.S. , 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics. 27, 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. , Weinberg, R.A. , 2000. The hallmarks of Cancer. Cell. 100, 57–70. 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Harrington, R.J. , Gutch, M.J. , Hengartner, M.O. , Tonks, N.K. , Chisholm, A.D. , 2002. The C. elegans LAR-like receptor tyrosine phosphatase PTP-3 and the VAB-1 Eph receptor tyrosine kinase have partly redundant functions in morphogenesis. Development. 129, 2141–2153. [DOI] [PubMed] [Google Scholar]
- Harris, T.W. , Antoshechkin, I. , Bieri, T. , Blasiar, D. , Chan, J. , Chen, W.J. , Cruz, N.D.L. , Davis, P. , Duesbury, M. , Fang, R. , Fernandes, J. , Han, M. , Kishore, R. , Lee, R. , Müller, H.-M. , Nakamura, C. , Ozersky, P. , Petcherski, A. , Rangarajan, A. , Rogers, A. , Schindelman, G. , Schwarz, E.M. , Tuli, M.A. , Auken, K.V. , Wang, D. , Wang, X. , Williams, G. , Yook, K. , Durbin, R. , Stein, L.D. , Spieth, J. , Sternberg, P.W. , 2010. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 38, D463–D467. 10.1093/nar/gkp952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, N. , Feener, E.P. , Zhang, W.R. , Goldstein, B.J. , 1992. Insulin receptor protein-tyrosine phosphatases. Leukocyte common antigen-related phosphatase rapidly deactivates the insulin receptor kinase by preferential dephosphorylation of the receptor regulatory domain. J. Biol. Chem. 267, 13811–13814. [PubMed] [Google Scholar]
- Hornbeck, P.V. , Zhang, B. , Murray, B. , Kornhauser, J.M. , Latham, V. , Skrzypek, E. , 2015. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520. 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, H. , Schoof, E.M. , Kim, J. , Robin, X. , Miller, M.L. , Diella, F. , Palma, A. , Cesareni, G. , Jensen, L.J. , Linding, R. , 2014. KinomeXplorer: an integrated platform for kinome biology studies. Nat. Methods. 11, 603–604. 10.1038/nmeth.2968 [DOI] [PubMed] [Google Scholar]
- Jacob, S.T. , Motiwala, T. , 2005. Epigenetic regulation of protein tyrosine phosphatases: potential molecular targets for cancer therapy. Cancer Gene Ther. 12, 665–672. 10.1038/sj.cgt.7700828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad, T.S. , Goel, R. , Kandasamy, K. , Keerthikumar, S. , Kumar, S. , Mathivanan, S. , Telikicherla, D. , Raju, R. , Shafreen, B. , Venugopal, A. , Balakrishnan, L. , Marimuthu, A. , Banerjee, S. , Somanathan, D.S. , Sebastian, A. , Rani, S. , Ray, S. , Harrys Kishore, C.J. , Kanth, S. , Ahmed, M. , Kashyap, M.K. , Mohmood, R. , Ramachandra, Y.L. , Krishna, V. , Rahiman, B.A. , Mohan, S. , Ranganathan, P. , Ramabadran, S. , Chaerkady, R. , Pandey, A. , 2009. Human Protein Reference Database–2009 update. Nucleic Acids Res. 37, D767–D772. 10.1093/nar/gkn892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.-S. , Pinto, S.M. , Getnet, D. , Nirujogi, R.S. , Manda, S.S. , Chaerkady, R. , Madugundu, A.K. , Kelkar, D.S. , Isserlin, R. , Jain, S. , Thomas, J.K. , Muthusamy, B. , Leal-Rojas, P. , Kumar, P. , Sahasrabuddhe, N.A. , Balakrishnan, L. , Advani, J. , George, B. , Renuse, S. , Selvan, L.D.N. , Patil, A.H. , Nanjappa, V. , Radhakrishnan, A. , Prasad, S. , Subbannayya, T. , Raju, R. , Kumar, M. , Sreenivasamurthy, S.K. , Marimuthu, A. , Sathe, G.J. , Chavan, S. , Datta, K.K. , Subbannayya, Y. , Sahu, A. , Yelamanchi, S.D. , Jayaram, S. , Rajagopalan, P. , Sharma, J. , Murthy, K.R. , Syed, N. , Goel, R. , Khan, A.A. , Ahmad, S. , Dey, G. , Mudgal, K. , Chatterjee, A. , Huang, T.-C. , Zhong, J. , Wu, X. , Shaw, P.G. , Freed, D. , Zahari, M.S. , Mukherjee, K.K. , Shankar, S. , Mahadevan, A. , Lam, H. , Mitchell, C.J. , Shankar, S.K. , Satishchandra, P. , Schroeder, J.T. , Sirdeshmukh, R. , Maitra, A. , Leach, S.D. , Drake, C.G. , Halushka, M.K. , Prasad, T.S.K. , Hruban, R.H. , Kerr, C.L. , Bader, G.D. , Iacobuzio-Donahue, C.A. , Gowda, H. , Pandey, A. , 2014. A draft map of the human proteome. Nature. 509, 575–581. 10.1038/nature13302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.-S. , Zhong, Y. , Yachida, S. , Rajeshkumar, N.V. , Abel, M.L. , Marimuthu, A. , Mudgal, K. , Hruban, R.H. , Poling, J.S. , Tyner, J.W. , Maitra, A. , Iacobuzio-Donahue, C.A. , Pandey, A. , 2014. Heterogeneity of pancreatic cancer metastases in a single patient revealed by quantitative proteomics. Mol. Cell. Proteomics. 13, 2803–2811. 10.1074/mcp.M114.038547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolibaba, K.S. , Druker, B.J. , 1997. Protein tyrosine kinases and cancer. Biochim. Biophys. Acta BBA – Rev. Cancer. 1333, F217–F248. 10.1016/S0304-419X(97)00022-X [DOI] [PubMed] [Google Scholar]
- Li, J. , Yen, C. , Liaw, D. , Podsypanina, K. , Bose, S. , Wang, S.I. , Puc, J. , Miliaresis, C. , Rodgers, L. , McCombie, R. , Bigner, S.H. , Giovanella, B.C. , Ittmann, M. , Tycko, B. , Hibshoosh, H. , Wigler, M.H. , Parsons, R. , 1997. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 275, 1943–1947. [DOI] [PubMed] [Google Scholar]
- Machide, M. , Hashigasako, A. , Matsumoto, K. , Nakamura, T. , 2006. Contact inhibition of hepatocyte growth regulated by functional association of the c-Met/hepatocyte growth factor receptor and LAR protein-tyrosine phosphatase. J. Biol. Chem. 281, 8765–8772. 10.1074/jbc.M512298200 [DOI] [PubMed] [Google Scholar]
- Mi, H. , Muruganujan, A. , Casagrande, J.T. , Thomas, P.D. , 2013. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566. 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, L.G.T. , Taylor, B.S. , Bivona, T.G. , Gong, Y. , Eng, S. , Brennan, C.W. , Kaufman, A. , Kastenhuber, E.R. , Banuchi, V.E. , Singh, B. , Heguy, A. , Viale, A. , Mellinghoff, I.K. , Huse, J. , Ganly, I. , Chan, T.A. , 2011. Genomic dissection of the epidermal growth factor receptor (EGFR)/PI3K pathway reveals frequent deletion of the EGFR phosphatase PTPRS in head and neck cancers. Proc. Natl. Acad. Sci. U. S. A. 108, 19024–19029. 10.1073/pnas.1111963108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, T. , Choidas, A. , Reichmann, E. , Ullrich, A. , 1999. Phosphorylation and free pool of β-catenin are regulated by tyrosine kinases and tyrosine phosphatases during epithelial cell migration. J. Biol. Chem. 274, 10173–10183. 10.1074/jbc.274.15.10173 [DOI] [PubMed] [Google Scholar]
- Perkins, D.N. , Pappin, D.J.C. , Creasy, D.M. , Cottrell, J.S. , 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 20, 3551–3567. doi:10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2–2 [DOI] [PubMed] [Google Scholar]
- Rush, J. , Moritz, A. , Lee, K.A. , Guo, A. , Goss, V.L. , Spek, E.J. , Zhang, H. , Zha, X.-M. , Polakiewicz, R.D. , Comb, M.J. , 2005. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101. 10.1038/nbt1046 [DOI] [PubMed] [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. , Gibson, T.J. , Karplus, K. , Li, W. , Lopez, R. , McWilliam, H. , Remmert, M. , Söding, J. , Thompson, J.D. , Higgins, D.G. , 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis, N. , Rual, J.-F. , Carvunis, A.-R. , Tasan, M. , Lemmens, I. , Hirozane-Kishikawa, T. , Hao, T. , Sahalie, J.M. , Venkatesan, K. , Gebreab, F. , Cevik, S. , Klitgord, N. , Fan, C. , Braun, P. , Li, N. , Ayivi-Guedehoussou, N. , Dann, E. , Bertin, N. , Szeto, D. , Dricot, A. , Yildirim, M.A. , Lin, C. , de Smet, A.-S. , Kao, H.-L. , Simon, C. , Smolyar, A. , Ahn, J.S. , Tewari, M. , Boxem, M. , Milstein, S. , Yu, H. , Dreze, M. , Vandenhaute, J. , Gunsalus, K.C. , Cusick, M.E. , Hill, D.E. , Tavernier, J. , Roth, F.P. , Vidal, M. , 2009. Empirically-controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat. Methods. 6, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon, D.J. , Clark, G.M. , Wong, S.G. , Levin, W.J. , Ullrich, A. , McGuire, W.L. , 1987. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Sci. New Ser. 235, 177–182. [DOI] [PubMed] [Google Scholar]
- Stiernagle, T. , 2006. Maintenance of C. elegans Wormbook; 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed, N. , Barbhuiya, M.A. , Pinto, S.M. , Nirujogi, R.S. , Renuse, S. , Datta, K.K. , Khan, A.A. , Srikumar, K. , Prasad, T.S.K. , Kumar, M.V. , Kumar, R.V. , Chatterjee, A. , Pandey, A. , Gowda, H. , 2015. Phosphotyrosine profiling identifies ephrin receptor A2 as a potential therapeutic target in esophageal squamous-cell carcinoma. Proteomics. 15, 374–382. 10.1002/pmic.201400379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, C.S.H. , Bodenmiller, B. , Pasculescu, A. , Jovanovic, M. , Hengartner, M.O. , Jørgensen, C. , Bader, G.D. , Aebersold, R. , Pawson, T. , Linding, R. , 2009. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci. Signal. 2, ra39 10.1126/scisignal.2000316 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network, 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490, 61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks, N.K. , 2006. Protein tyrosine phosphatases: from genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 7, 833–846. 10.1038/nrm2039 [DOI] [PubMed] [Google Scholar]
- Uetani, N. , Chagnon, M.J. , Kennedy, T.E. , Iwakura, Y. , Tremblay, M.L. , 2006. Mammalian motoneuron axon targeting requires receptor protein tyrosine phosphatases σ and δ. J. Neurosci. 26, 5872–5880. 10.1523/JNEUROSCI.0386-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeriah, S. , Brennan, C. , Meng, S. , Singh, B. , Fagin, J.A. , Solit, D.B. , Paty, P.B. , Rohle, D. , Vivanco, I. , Chmielecki, J. , Pao, W. , Ladanyi, M. , Gerald, W.L. , Liau, L. , Cloughesy, T.C. , Mischel, P.S. , Sander, C. , Taylor, B. , Schultz, N. , Major, J. , Heguy, A. , Fang, F. , Mellinghoff, I.K. , Chan, T.A. , 2009. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc. Natl. Acad. Sci. 106, 9435–9440. 10.1073/pnas.0900571106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayvargia, R. , Kaur, S. , Krishnasastry, M.V. , 2004. α-Hemolysin-induced dephosphorylation of EGF receptor of A431 cells is carried out by rPTPσ. Biochem. Biophys. Res. Commun. 325, 344–352. 10.1016/j.bbrc.2004.10.038 [DOI] [PubMed] [Google Scholar]
- Yu, G. , Wang, L.-G. , Han, Y. , He, Q.-Y. , 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 16, 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahari, M.S. , Wu, X. , Blair, B.G. , Pinto, S.M. , Nirujogi, R.S. , Jelinek, C.A. , Malhotra, R. , Kim, M.-S. , Park, B.H. , Pandey, A. , 2015. Activating mutations in PIK3CA lead to widespread modulation of the tyrosine phosphoproteome. J. Proteome Res. 14, 3882–3891. 10.1021/acs.jproteome.5b00302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Guo, A. , Yu, J. , Possemato, A. , Chen, Y. , Zheng, W. , Polakiewicz, R.D. , Kinzler, K.W. , Vogelstein, B. , Velculescu, V.E. , Wang, Z.J. , 2007. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc. Natl. Acad. Sci. 104, 4060–4064. 10.1073/pnas.0611665104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, J. , Kim, M.-S. , Chaerkady, R. , Wu, X. , Huang, T.-C. , Getnet, D. , Mitchell, C.J. , Palapetta, S.M. , Sharma, J. , O'Meally, R.N. , Cole, R.N. , Yoda, A. , Moritz, A. , Loriaux, M.M. , Rush, J. , Weinstock, D.M. , Tyner, J.W. , Pandey, A. , 2012. TSLP signaling network revealed by SILAC-based phosphoproteomics. Mol. Cell. Proteomics. 11, 10.1074/mcp.M112.017764 M112.017764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukerberg, L.R. , Patrick, G.N. , Nikolic, M. , Humbert, S. , Wu, C.-L. , Lanier, L.M. , Gertler, F.B. , Vidal, M. , Van Etten, R.A. , Tsai, L.-H. , 2000. Cables links Cdk5 and c-Abl and facilitates Cdk5 tyrosine phosphorylation, kinase upregulation, and neurite outgrowth. Neuron. 26, 633–646. 10.1016/S0896-6273(00)81200-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary Figure 1 Whole proteome analysis of the ptp‐3 mutant. A) The distribution of protein abundance between the ptp‐3 mutant and wild type strain measured at the global proteome level is shown. B) A GO analysis of genes up‐regulated in the whole proteome data shows an enrichment for genes involved in developmental processes, in agreement with the known phenotype of increased embryonic lethality of the ptp‐3 mutant.
Supplementary data
Supplementary data
Data Availability Statement
Mass spectrometry derived proteomics data has been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD003233.


