Abstract
MicroRNAs (miRNAs) are single‐stranded, small non‐coding RNA molecules that participate in important biological processes. Although the functions of many miRNAs in breast cancer metastasis have been established, the role of others remains to be characterized. To identify additional miRNAs involved in metastasis, we performed a genetic screen by transducing a Lenti‐miR™ virus library into MCF‐7 cells. Using transwell invasion assays we identified human miR‐548j as an invasion‐inducing miRNA. The endogenous levels of miR‐548j expression in breast cancer cell lines were shown to correlate with invasiveness. Moreover, miR‐548j was shown to stimulate breast cancer cell invasion and metastasis in vitro and in vivo, but had no effect on proliferation. Next, using a series of in vitro and in vivo experiments, we found that Tensin1 served as a direct and functional target of miR‐548j. Both miR‐548j and Tensin1 modulated the activation of Cdc42 to regulate cell invasion and siCdc42 or the selective Cdc42 inhibitor ML141 suppressed the pathway of miR‐548j‐mediated cell invasion. Furthermore, a strong correlation between miR‐548j, Tensin1, metastasis and survival was observed using two sets of clinical breast cancer samples. Our findings demonstrate that miR‐548j functions as a metastasis‐promoting miRNA to regulate breast cancer cell invasion and metastasis by targeting Tensin1 and activating Cdc42, suggesting a potential therapeutic application in breast cancer.
Keywords: miR‐548j, Tensin1, Cdc42, Breast cancer, Metastasis
Highlights
New microRNAs involved in metastasis can be identified using a Lenti‐miR™ virus library.
miR‐548j promotes breast cancer cell invasion in vitro and metastasis in vivo.
Tensin1 is the direct target of miR‐548j.
Cdc42 is activated in miR‐548j‐induced metastasis.
miR‐548j has a correlation with clinical breast cancer metastasis and survival.
1. Introduction
Metastasis, a fundamental hallmark of cancer, is a complex multistep process whereby cancer cells spread from a primary site and form tumors at distant sites (Fidler, 2003; Hanahan and Weinberg, 2011). Metastasis is responsible for almost 90% of cancer‐associated mortality (Chaffer and Weinberg, 2011). Breast cancer is one of the most common malignant tumors in women. The world‐wide incidence of breast cancer keeps rising dramatically, especially in China. Despite improvements in diagnosis and treatment methods, fatality associated with metastasis is still the leading cause of death in patients with breast cancer. Therefore, a thorough understanding of the molecular mechanisms of breast cancer metastasis is critical to the diagnosis and development of therapy for advanced breast cancer patients.
MicroRNAs (miRNAs) are highly conserved, single‐strand, small non‐coding RNA molecules that are known to play critical roles in multiple species. Through post‐transcriptional regulation, they participate in almost all the physiological and pathological processes, such as cell cycle control, cell proliferation, apoptosis and metastasis (Di Leva et al., 2014; Huntzinger and Izaurralde, 2011). For breast cancer, multiple miRNAs have been implicated in regulating the metastasis process. For example, miR‐10b, miR‐9, miR‐373 and miR‐520c have been reported to promote metastasis (Huang et al., 2008, 2007, 2010). Conversely, the miR‐200 family, the miR‐34 family and miR‐205 have been shown to suppress metastasis (Gregory et al., 2008; Yang et al., 2013). Due to the large number of miRNAs with functions that have yet to be characterized, further understanding of the function and mechanism of additional miRNAs that regulate breast cancer metastasis may provide more insights into better treatment for advanced breast cancer patients.
The miR‐548 family is a large, primate‐specific gene family that is derived from repetitive elements in the genome (Yuan et al., 2011). More than 100 members of the miR‐548 family have been identified in primates, including Homo sapiens (hsa, 72), Pongopygmaeus (ppy, 5), Pan troglodytes (ptr, 19), Gorilla gorilla (ggo, 6) and Macacamulatta (mml, 10) (http://www.mirbase.org/). Non‐human primate members have primarily been predicted by software based on their sequence similarity to hsa‐miR‐548. Many microRNA families we known, such as let‐7 family, are highly conserved, even among various animal species. They share the same ‘seed sequence’, suggesting their targets and function may be similar (Lee et al., 2016). However, the miR‐548 family is a poorly conserved miRNA family. It is originated from the transposable elements (TEs), which is probably derived from the evolutionary origin, leading to the diversity of miRNA sequences. The family members share similar, instead of the same ‘seed sequence’ (2–8 nucleotide), nucleotide substitution, insertion/deletion in this region or 5′ end differences will make their ‘seed sequence’ diversity, and further impact their biological functions via targeting on various genes, including some key proteins and signaling pathways in cancer (Liang et al., 2012). Recently, miR‐548k has been suggested to promote esophageal squamous cell carcinoma (ESCC) cell growth and mobility (Song et al., 2014), while a novel human miR‐548 family member has been shown to reside within the FHIT tumor‐suppressor gene and to harbor a tumor‐suppressor function in a mouse xenograft model (Hu et al., 2014). Furthermore, miR‐548d‐3p is reported to directly regulate the expression of Her2, a common oncogene in breast cancer (Chen et al., 2009). However, the function and mechanism of the miR‐548 family in human breast cancer development is not entirely clear.
In this study, we used a forward genetic screen to explore new miRNAs that positively regulate the invasion process in vitro, which may also positively regulate the metastasis process in vivo, and we identified several members of the miR‐548 family. We demonstrate for the first time that miR‐548j promotes breast cancer metastasis in vitro and in vivo. Furthermore, we identify Tensin1, a component of focal adhesion, as a direct and functional target of miR‐548j that impacts the Cdc42 pathway and the cytoskeleton. Our findings suggest that miR‐548j may serve as a candidate target for advanced breast cancer treatment.
2. Materials and methods
2.1. Cell culture
All cell lines except SKBR3 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained according to manufacturer's recommendations. SKBR3 was maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 10 μg/ml insulin and antibiotics.
2.2. Lenti‐miR™ library infection
MCF‐7 cells were plated at a density of 1 × 105 cells per well in a 6‐well plate format the day before viral infection. Dilute the virus to infect at an MOI of 3 into 0.5 ml of complete medium, and added polybrene to a final concentration of 5 μg/ml. Infect MCF‐7 cells by adding the viral stock dilutions to the wells. Seventy two hours later, GFP expression in infected cells was detected with fluorescence microscope.
2.3. Plasmid construction
The short‐length 3′UTR of Tensin1 containing two miR‐548j binding sites was cloned from MCF‐10A complementary DNA and then inserted into pIS0 luciferase plasmid. The mutant Tensin1 3′UTR sites (site 1 and site 2) were generated using a KOD‐plus‐Mutagenesis kit (Toyobo Co., LTD., Osaka, Japan). Pri‐miR‐548j was amplified from MCF‐10A genomic DNA and inserted into pLVX‐EGFP‐C1. For stable knockdown of Tensin1 (shTNS1‐1, shTNS1‐2) and the shControl, short hairpin RNA (shRNA) oligos were annealed and cloned into pSIH1‐H1‐Puro. The target sequences of these three shRNAs are detailed in Supplementary Materials and Methods. For stable knockdown of miR‐548j, miRZip‐548j was designed as described previously (Chou et al., 2013), synthesized by Invitrogen (Invitrogen, Camarillo, CA, USA) and then cloned into pSIHI‐H1‐Puro. The sequence of this shRNA is provided in Supplementary Materials and Methods.
2.4. Oligonucleotide and plasmid transfection
For miR‐548j overexpression, double‐strand small RNA (miR‐548j mimics) containing the sequence of mature miR‐548j was used. For miR‐548j knock down, a single‐strand small RNA (miR‐548j inhibitor) containing the antisense sequence of mature miR‐548j was designed. miR‐548j mimics, inhibitor and relevant negative controls (NC for miR‐548j mimics, NC inhibitor for miR‐548j inhibitor) were synthesized by Invitrogen (Invitrogen, Camarillo, CA, USA). siRNAs for Tensin1 and Cdc42 and relevant negative control siNC were ordered from Qiagen (Qiagen, Germantown, MD, USA). Oligonucleotides and plasmids transfection were done by using HiperFect (Qiagen) or Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. The sequences of oligonucleotides are shown in Supplementary Table S1.
2.5. Invasion assay
Invasion assays were performed using 24‐well Boyden chambers (Corning Incorporated, Corning, NY, USA) coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Cells were plated in the upper chambers (3 × 104 cells/chamber for MDA‐MB‐231, 5 × 104 cells/chamber for SKBR3, 2 × 105 cells/chamber for MCF‐7) and then cultured in 37 °C (20 h for MDA‐MB‐231, 48 h for SKBR3 and MCF‐7). Then cells that remained on the upper side of the filter were removed. The remaining cells were fixed and stained with 0.5% crystal violet. Cells from at least four randomly selected microscopic fields were counted.
2.6. Animal studies
All research involving animals complied with protocols approved by the Beijing Medical Experimental Animal Care Commission. For experimental metastasis assays, six‐week‐old female NOD/SCID mice were injected with 1 × 106 231‐Luc cells infected with control, miR‐548j, miRZip‐548j or shTNS1‐1, shTNS1‐2 via tail vein. Fourteen weeks later, the mice were anesthetized and injected with D‐luciferin (150 mg/kg, i.p. PerkinElmer, Waltham, MA, USA) 15 min before imaging. Metastasis was determined using a Xenogen optical in vivo imaging system (IVIS Lumina, Caliper Life Sciences, Waltham, MA, USA). For tumor growth, cell suspensions (1 × 107 231‐Luc cells infected with control or miR‐548j per mouse) were mixed 1:1 with Matrigel (BD Biosciences) and injected into the mammary fat pads of six‐week‐old female mice. Ten weeks later, the tumor sizes were determined in the same way as the metastasis assays. All values of bioluminescence signal were measured, quantified and expressed as photon counts per area.
2.7. Clinical specimens
A total of 117 infiltrating ductal carcinoma samples were collected from November 2011 to November 2013 in our hospital at the time of surgery and immediately stored in liquid nitrogen until use. None of the patients had received chemotherapy or radiotherapy before surgery. Clinicopathological characteristics are shown in Supplementary Table S2. This study was approved by the ethical committee of the Cancer Institute & Hospital, Chinese Academy of Medical Sciences, and informed consent was obtained from each patient. For survival analysis, human breast cancer tissue microarrays were purchased from Outdo Biotech (Outdo Biotech Co. Ltd., Shanghai, China). All the cases covered in tissue microarrays were followed up for 9–12.5 years.
2.8. Immunohistochemistry and in situ hybridization
Immunohistochemical staining was performed as described previously (He et al., 2015). Tensin1 antibody (Abcam) was used at a 1:100 dilution at 4 °C overnight. miR‐548j LNA™ probe was purchased from Exiqon (Vedbaek, Denmark), and in situ hybridization was performed according to manufacturer's recommendations. Images were visualized and annotated with Aperio ImageScope software (Aperio Technologies, Inc., CA, USA), and the number of positive cells at ×200 magnification was quantified.
2.9. Statistical analyses
Statistical analysis was performed using Prism 6 (GraphPad Software, La Jolla, USA) or SPSS 20.0 (IBM SPSS software, NY, USA). All data are presented as mean ± SEM, unless otherwise stated. The Student's t‐test was used, unless otherwise stated. For analysis of tumor growth and metastasis in mice, the Mann–Whitney U‐test was used to determine significance by comparing photo counts per area. We considered P < 0.05 as significant.
3. Results
3.1. Identification of miR‐548j as an inducer of cell invasion using a forward genetic screen
To identify miRNAs that promote tumor invasion, we used a Lenti‐miR™ virus library to screen the candidate miRNAs by transwell invasion assay (Figure 1A). We transduced MCF‐7, a non‐migratory breast cancer cell line, with a Lenti‐miR™ virus library (MOI = 3) purchased from System Biosciences (SBI, Mountain View, CA, USA), which contains approximately 1000 pri‐miRNAs that are tagged with GFP. We subjected the transduced MCF‐7 cells to a transwell invasion assay to collect subsets expressing GFP that had gained the capacity of invasion (Figure 1B), and then we sequenced the miRNA inserts to identify the miRNAs that had stimulated cell invasion. The frequency of each miRNA in the invasive population was compared with the abundance of the total invasive population (Figure 1C). Interestingly, we observed that about 30 percent of the overexpressed miRNAs were derived from the same family: the human miR‐548 family. The members of the miR‐548 family we identified owned homologous miRNA sequences, with only several inconsistent nucleotides insertion or deletion, and some of them share the same seed sequence (Supplementary Figure S1). We first synthesized several of the selected miRNAs and transfected them into MDA‐MB‐231 cells to verify the efficiency of this screen, and we observed that the invasive potential had been promoted more or less (Supplementary Figure S2). Because miR‐548j appeared to make the cells gain the strongest ability of invasion, we selected it for further validation as an inducer of tumor invasion.
Figure 1.
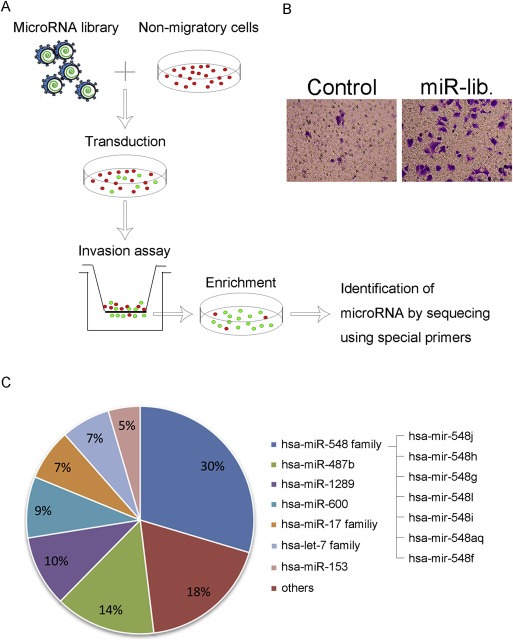
Identification of miR‐548j as an inducer of cell invasion using a forward genetic screen. (A) General scheme of a genetic screen for miRNAs that promote breast cancer cell invasion using a Lenti‐miR™ virus library tagged with GFP. (B) Verification of invading capability of subsets selected by the forward genetic screen using a transwell invasion assay. (C) Pie distribution of miRNAs and their proportions obtained by sequencing subsets.
3.2. miR‐548j promotes breast cancer cell invasion in vitro
To verify that miR‐548j is capable of promoting MCF‐7 cell invasion, we first transfected MCF‐7 cells with either miR‐548j mimics or NC, and then used a transwell invasion assay to test the invasiveness. The results showed that enforced expression of miR‐548j produced an invasive phenotype, compared with the NC group (Figure 2A). To rule out the influence of proliferation, we measured cell proliferation of these two groups simultaneously, and did not observe any significant differences (Supplementary Figure 3A), suggesting that miR‐548j has no effect on MCF‐7 cell proliferation.
Figure 2.
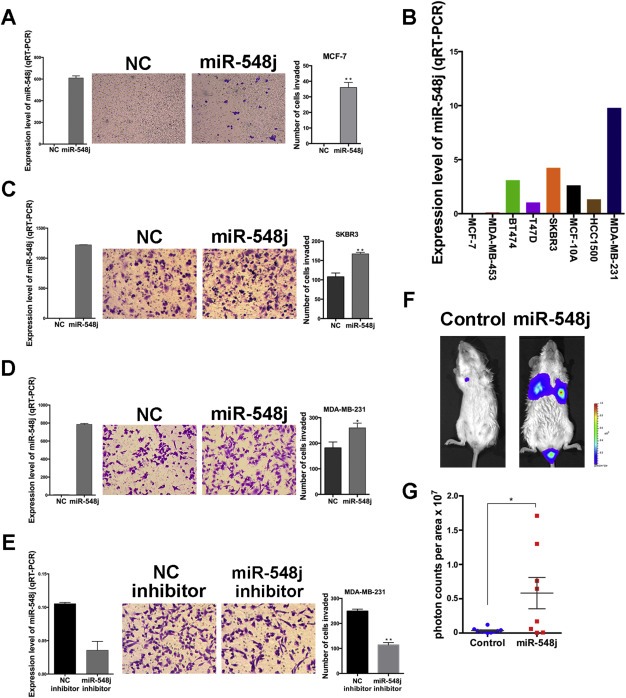
miR‐548j promotes breast cancer cell invasion in vitro and in vivo. (A) Expression level of miR‐548j (left), representative micrographs (middle) and quantification of invading cells (right) of MCF‐7 cells transfected with NC or miR‐548j mimics. (B) The endogenous expression level of miR‐548j in some breast cancer cell lines as determined by quantitative RT‐PCR. The values were normalized to U6. (C–D) Expression level of miR‐548j (left), representative micrographs (middle) and quantification of invading cells (right) of SKBR3 (C) and MDA‐MB‐231 (D) transfected with NC or miR‐548j mimics. (E) Expression level of miR‐548j (left), representative micrographs (middle) and quantification of invading cells (right) in MDA‐MB‐231 cells transfected with NC inhibitor or miR‐548j inhibitor. (F) Representative photos of bioluminescence of mice from each group injected with the indicated cells. The metastasis animal model was carried out as described in the Materials and Methods. (G) Quantitative analysis of metastasis. The values of bioluminescence signals from each group were quantified and expressed as photon counts per area. P‐values were obtained using the Mann–Whitney U‐test. Error bars in this figure represent mean ± SEM. *P < 0.05, **P < 0.01.
To further assess the contribution of miR‐548j on cell invasion, we screened several breast cancer cell lines for their endogenous expression of miR‐548j, the invasive potential of which increases gradually (Neve et al., 2006). We observed an increase in endogenous expression of miR‐548j in these breast cancer cell lines, accompanied by increasing invasive potential from left to right (Figure 2B). This pattern of expression is consistent with the possibility that miR‐548j regulates invasion. To directly determine whether miR‐548j can promote invasion, we chose other two breast cancer cell lines, SKBR3 and MDA‐MB‐231, with median or high endogenous expression level of miR‐548j. We overexpressed miR‐548j in these two cell lines, and observed that miR‐548j presented an enhanced invasiveness (Figure 2C and 2D), which was not explained by effects on cell proliferation (Supplementary Figure 3B and 3C) in both of these two cell lines. As MDA‐MB‐231 had the highest endogenous expression of miR‐548j among all the cell lines, we transfected a single‐stranded, modified, synthesized RNA (miR‐548j inhibitor) into MDA‐MB‐231 cells to block endogenous miR‐548j, and significant reduction of invasiveness was observed as expected (Figure 2E). These results clearly show that miR‐548j promotes invasiveness of breast cancer cells in vitro, without any influence on cell proliferation.
3.3. miR‐548j promotes breast cancer cell metastasis in vivo
To examine the in vivo effect of miR‐548j on breast cancer metastasis, we injected luciferase‐tagged MDA‐MB‐231 (231‐Luc) cells stably overexpressing miR‐548j or a control miRNA into NOD/SCID female mice via tail‐vein injection. Twelve weeks after injection, metastatic nodules developed in the lungs. Using a Xenogen optical in vivo imaging system, we demonstrated that metastasis of the miR‐548j‐overexpressing group was significantly increased compared with that of the control group (Figure 2F). The results were further confirmed by quantification of in vivo luciferase activity (Figure 2G). We also injected the same two 231‐Luc subsets into breast fat pads of NOD/SCID mice to detect the effects of miR‐548j on tumor growth by measurement of in vivo luciferase activity, but did not find any significant difference between the two groups (Supplementary Figure S3D), indicating that miR‐548j promotes metastasis in vivo without affecting tumor growth, which is consistent with the in vitro study. Thus, overexpression of miR‐548j increases the metastatic potential but does not affect tumor growth of breast cancer cells in vitro or in vivo.
3.4. Tensin1 is a direct target of miR‐548j
To further explore the mechanism by which miR‐548j regulates breast cancer cell invasion and metastasis, we adopted two widely used bioinformatics algorithms (TargetScan, Miranda) to predict targets, and then we functionally classified the predicted targets using the DAVID program (http://david.abcc.ncifcrf.gov) (Supplementary Figure S4). One of the predicted targets, Tensin1 (TNS1), is localized to focal adhesions (Lo et al., 1994) and acts as a link between the extracellular matrix and the cytoskeleton, thereby mediating signaling for cell shape and motility (Lo, 2004). Tensin1 is expressed in normal tissues (Chen et al., 2000), and is greatly reduced in cancer, especially in metastatic cancer (Rhodes et al., 2004). Therefore, we selected Tensin1 for further assessment as a miR‐548j‐regulated target with a potential role in regulating metastasis in breast cancer.
To assess the potential role of Tensin1, we first measured endogenous Tensin1 expression levels in the same breast cancer cell lines detected in Figure 2B. The expression level of Tensin1 decreased with the increased in levels of invasiveness in these cell lines (Figure 3A), which was the converse of the expression pattern for miR‐548j (Figure 2B). These findings raise the possibility that Tensin1 could negatively regulate invasiveness in breast cancer metastasis and that miR‐548j may induce metastasis by targeting Tensin1. To verify our prediction, we introduced each of the two putative miR‐548j binding sites of the Tensin1 3′UTR (site 1 or site 2) into a luciferase reporter plasmid (Figure 3B and Supplementary Figure S5A). Luciferase reporter assays showed that miR‐548j markedly suppressed the expression of a luciferase gene containing site 1 in a dose dependent manner (Figure 3C), but not mutant site 1 (Figure 3D) or site 2 (Supplementary Figure S5B and S5C). These results indicate that miR‐548j directly targets site 1 of the Tensin1 3′UTR. Consistently, miR‐548j overexpression in each of the three breast cancer cell lines led to a clear decrease in endogenous Tensin1 levels (Figure 3E). Furthermore, transfection of SKBR3 and MDA‐MB‐231 cells with miR‐548j inhibitor led to an upregulation of Tensin1 (Figure 3F), as the endogenous expression of miR‐548j in MCF‐7 cells was too low to be blocked by miR‐548j inhibitor. Taken together, these results suggest that miR‐548j directly targets Tensin1 through its 3′UTR.
Figure 3.
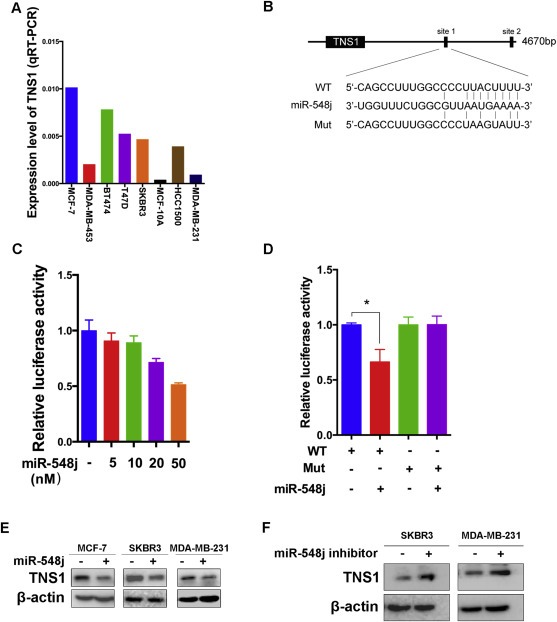
Tensin1 is a direct target of miR‐548j. (A) The endogenous expression of Tensin1 was examined in breast cancer cell lines using quantitative RT‐PCR. The values were normalized to GAPDH. (B) A description of two predicted miR‐548j binding sites within the Tensin1 3′UTR, as well as sequence alignment of the miR‐548j binding site 1 (WT) and its mutation (Mut) with the miR‐548j targeting sequence. (C) Luciferase reporter assay of HEK293T cells co‐transfected with miR‐548j mimics in a dose dependent manner. (D) Luciferase reporter assay of HEK293T cells co‐transfected with miR‐548j mimics and either WT or Mut luciferase plasmid. *P < 0.05. (E) Western blot analysis of the protein levels of Tensin1 (TNS1) in the indicated cells transfected with NC or miR‐548j mimics. (F) Western blot analysis of Tensin1 protein levels in cells transfected with NC inhibitor or miR‐548j inhibitor.
3.5. Tensin1 is involved in the regulation of cell invasion and metastasis by miR‐548j
To examine the role of Tensin1 in miR‐548j‐mediated breast cancer cell invasion and metastasis, we transfected a Tensin1 siRNA pool into MCF‐7, SKBR3 and MDA‐MB231 cells to knock down Tensin1 expression (Figure 4A), and then we used a transwell invasion assay to examine the effects of Tensin1 knock down on invasiveness. Consistent with previous reports (Rhodes et al., 2004), knock down of Tensin1 increased the invasiveness in all three cell lines (Figure 4B). To determine whether Tensin1 is a direct functional mediator of miR‐548j‐induced breast cancer cell invasion in vitro, we performed a ‘Rescue’ experiments in 231‐Luc cells with stable knockdown of endogenous miR‐548j using miRZip‐548j as described in Materials and Methods. Tensin1 was blocked by two different siRNAs from the siRNA pool (Figure 4C). Our results demonstrate that miRZip‐548j inhibits invasion by 231‐Luc cells, but that shTNS1‐1 and shTNS1‐2 can each reverse this inhibition (Figure 4D).
Figure 4.
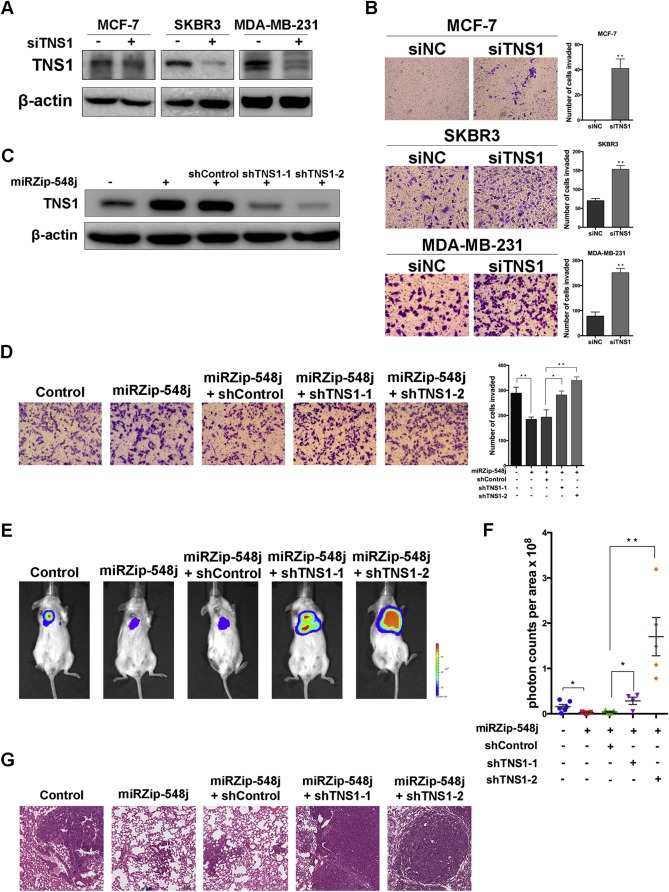
Tensin1 is involved in the regulation of cell invasion and metastasis by miR‐548j. (A) Tensin1 protein levels in MCF‐7, SKBR3 and MDA‐MB‐231 cells transfected with control siRNA (siNC) or Tensin1 siRNA (siTNS1) were analyzed by Western blotting. β‐actin is shown as a loading control. (B) Transwell invasion assays were performed to examine the invasiveness of the indicated cells after transfection with control siRNA (siNC) or Tensin1 siRNA (siTNS1). Results are quantified (right panel). (C) Western blot of Tensin1 protein levels in 231‐Luc cells infected with a control or miRZip‐548j together with a control shRNA or one of two different shRNAs targeting Tensin1. (D) Representative micrographs and quantification of ‘Rescue’ experiments performed by transwell invasion assays. (E) Representative photos of bioluminescence signals of mice from each group in ‘Rescue’ experiments. In vivo metastasis was assayed as described in Materials and Methods. (F) Quantitative analysis of metastasis. The values of the bioluminescence signals from each group were quantified and analyzed using the Mann–Whitney U‐test. (G) Lung metastasis in mice in which the indicated cells were implanted was confirmed by H&E staining. Error bars in B, D and F represent mean ± SEM. *P < 0.05, **P < 0.01.
To further verify that Tensin1 plays a similar role in miR‐548j‐mediated cell metastasis in vivo, we introduced the same five subsets of 231‐Luc cells into female NOD/SCID mice via tail vein injection. Twelve weeks after injection, we examined the metastasis of the five groups. Similar to the in vitro results, the inhibition of metastasis by miRZip‐548j was rescued by Tensin1 knockdown by either shTNS1‐1 or shTNS1‐2 (Figure 4E). These in vivo results were confirmed by quantification of luciferase activity (Figure 4F) and histological analysis (Figure 4G). Taken together, these findings indicate that Tensin1 is a functional target of miR‐548j in breast cancer metastasis.
3.6. miR‐548j and Tensin1 regulate cell invasion via activation of Cdc42
Tensin1 is known to localize to focal adhesion sites upon ligation with integrin (Miyamoto et al., 1995), where it mediates focal adhesion signal transduction (Lo, 2004). To define the pathway of miR‐548j and Tensin1‐mediated regulation of breast cancer metastasis, we evaluated the potential role of other molecules that localize to focal adhesion assembly sites and participate in integrin‐mediated focal adhesion signal transduction. First, we examined the expression and phosphorylation of FAK, an integral component of the focal adhesion complex that plays a key role in the focal adhesion signaling pathway. Transfection of MCF‐7, SKBR3 and MDA‐MB‐231 cells with miR‐548j mimics did not elicit any obvious changes in the levels of FAK or p‐FAK (Supplementary Figure S6). Because Tensin1 has been reported to interact with DLC1, a RhoGAP family member, to inhibit cell migration by reducing Rho‐GTP (Qian et al., 2007), we evaluated the possibility that miR‐548j may have an effect on Tensin1‐mediated reduction of Rho‐GTP. To verify this hypothesis, we first examined the expression of Cdc42, a prominent member of the Rho‐GTPase family, and its active form, Cdc42‐GTP. Interestingly, an obvious accumulation of Cdc42‐GTP was apparent in all three cell lines upon miR‐548j overexpression (Figure 5A). In addition, we observed similar increase in active Cdc42‐GTP in these cell lines with Tensin1 knock down (Supplementary Figure S7A) and conversely a decrease in Cdc42‐GTP in cells expressing miRZip‐548j, with reversal of this effect upon rescue with Tensin1 knock down (Supplementary Figure S7B). These results suggest that miR‐548j may promote cell invasion by regulating Tensin1‐mediated inactivation of Cdc42.
Figure 5.
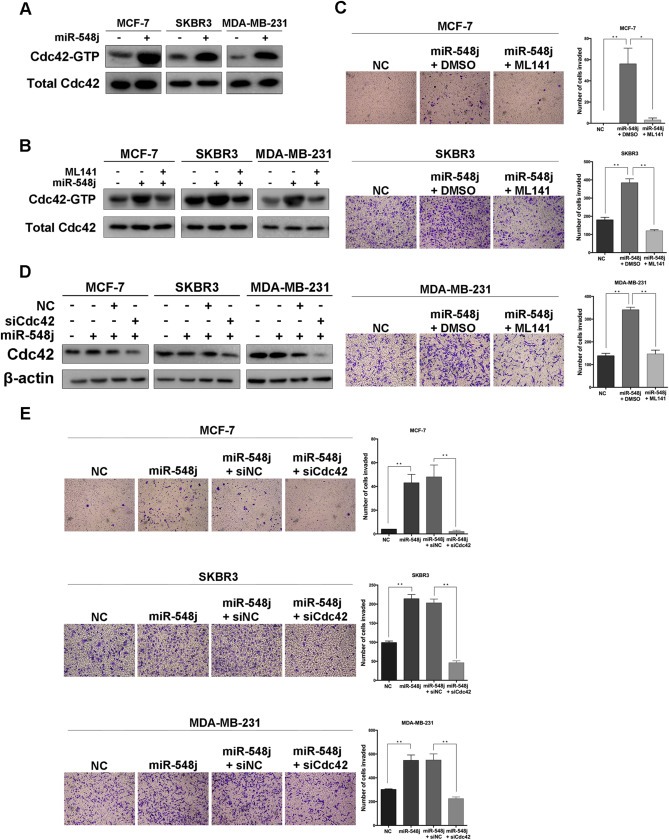
miR‐548j and Tensin1 regulate cell invasion via activation of Cdc42. (A) The active form of Cdc42 (Cdc42‐GTP) and total Cdc42 were measured by Western blotting of the indicated cells upon transfection of NC or miR‐548j mimics. (B) The efficiency of the Rho inhibitor ML141 on breast cancer cells with miR‐548j overexpression, represented by the expression level of Cdc42‐GTP. (C) Representative micrographs and quantification of invading cells transfected with a control miRNA or miR‐548j mimics and without or with treatment with either ML141. (D) To confirm the efficiency of siCdc42, total Cdc42 protein levels in the three breast cancer cells overexpressing miR‐548j and transfected with control siRNA (siNC) or Tensin1 siRNA (siTNS1) were analyzed by Western blotting. β‐actin is shown as a loading control. (E) Representative micrographs and quantification of invading cells transfected with a control miRNA or miR‐548j mimics and transfected with a siNC or siCdc42 RNA. Error bars in C and E represent mean ± SEM. *P < 0.05, **P < 0.01.
To further confirm the role of Cdc42 activation in miR‐548j‐promoted cell invasion, we used ML141, a selective inhibitor of Cdc42 (Hong et al., 2013; Surviladze et al., 2010), to reduce Cdc42 activity (Figure 5B). The level of invasiveness induced by miR‐548j was diminished sharply after ML141 treatment in MCF‐7, SKBR3 and MDA‐MB‐231 cells (Figure 5C). In addition, after the transfection of a Cdc42 siRNA pool (Figure 5D), the miR‐548j‐promoted invasiveness was dramatically reduced (Figure 5E). Moreover, we added either ML141 or Cdc42 siRNA into these three breast cancer cell lines with Tensin1 knock down, and a similar suppression of invasiveness was obtained as that of miR‐548j overexpression (Supplementary Figure S7C–F). These results clearly show that miR‐548j promotes cell invasion through activation of Cdc42, and further demonstrate that miR‐548j targets Tensin1 to activate Cdc42.
3.7. Clinical association of miR‐548j with metastasis and survival in human breast cancer
To further understand the clinical relevance of our findings to human breast cancer, we examined the expression levels of miR‐548j and Tensin1 in 117 human breast cancer tissue specimens, and found that miR‐548j was significantly increased in human breast tumors with lymph node metastasis (Figure 6A). Conversely, as a direct and functional target of miR‐548j, Tensin1 was markedly reduced in metastatic breast cancer tissue specimens, compared with the non‐metastatic group (Figure 6B). These data clearly demonstrate that both miR‐548j and Tensin1 correlate with metastasis in breast cancer patients. Additionally, we tested both miR‐548j and Tensin1 expression histologically using a human breast cancer tissue microarray containing an additional 104 human breast cancer tissue specimens, with mean follow‐up time of 9–12.5 years for all the cases (Figure 6C). Kaplan–Meier curve analysis of the histological findings and survival data showed that high miR‐548j expression was significantly associated with shorter metastasis‐free survival (Figure 6D), whereas high Tensin1 expression was associated with longer metastasis‐free survival (Figure 6E). However, given the high similarity of the miR‐548 family (Supplementary Figure S1), the probe used to detected miR‐548j in tissue microarray could not distinguish miR‐548j with miR‐548 family members completely. These data demonstrate that miR‐548j, maybe together with other miR‐548 family members, correlate with breast cancer patient survival, as well as Tensin1.
Figure 6.
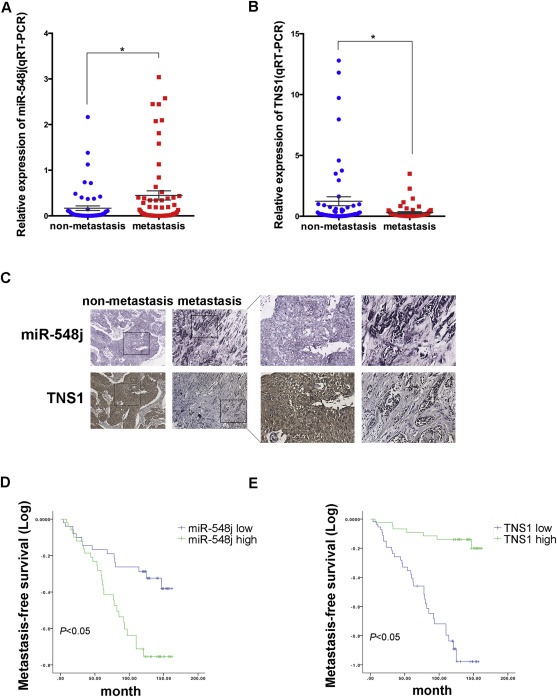
Clinical association of miR‐548j with metastasis and survival in human breast cancer. (A–B) Quantitative RT‐PCR analysis of the expression levels of miR‐548j (A) and Tensin1 (B) in 117 human breast tumors with or without lymph node metastasis. (C) Representative cases of histological analysis of miR‐548j and Tensin1 in human breast cancer tissue microarrays. Original magnification, ×400. (D–E) Kaplan–Meier metastasis‐free survival curves comparing high and low miR‐548j expression groups (D) and high or low Tensin1 expression groups (E) in breast cancer tissue microarrays containing 104 breast cancer cases; P value was based on a log‐rank test.
4. Discussion
Metastasis has always been the main cause of poor prognosis and death in breast cancer patients. Therefore, clarification of the molecular mechanism of metastasis is critical for new insights into better therapy for advanced breast cancer. miRNAs, as a class of molecules involved in metastasis, are attractive candidates for clinical breast cancer therapy (Kaboli et al., 2015). Using a forward genetic screen, we identified miR‐548j and several other microRNAs promoting breast cancer cell migration (data not shown) and invasion in vitro. We chose miR‐548j, which gave the most aggressive phenotype in selected subsets, for further study, and we confirmed that miR‐548j acted as an inducer of breast cancer cell metastasis in vivo. Notably, high expression of miR‐548j in breast cancer patients correlates with lymph node metastasis and poor outcome, implicating miR‐548j as a new potential target of breast cancer diagnosis and therapy.
miR‐548j belongs to a large primate‐specific miRNA family. There are 72 human miR‐548 family members located on almost all of the human chromosomes, especially chromosome 6, 8 and X. This family has been predicted to play important roles in physiological and pathological processes (Liang et al., 2012); however, there have been a limited number of studies that directly address the function of this family. Recently, miR‐548d‐3p has been reported to targeting Her2, an oncogene that is highly expressed in a subset of breast cancer, through directly binding to its 3′UTR, whereas the effects of miR‐548d‐3p on Her2 protein level and biological function in breast cancer needs further exploration (Chen et al., 2009). However, miR‐548d‐3p was not identified in our screen. miR‐548k has also been recently determined to promote ESCC cell growth and mobility (Song et al., 2014). Furthermore, the chromosome region 11q13.3–13.4 in which miR‐548k located is amplified in ESCC (Gao et al., 2014; Song et al., 2014), suggesting that miR‐548k may function as an oncomiR in ESCC. We did not identify miR‐548k in our screen, but we identified several other miR‐548 family members, including miR‐548j, which had the most obvious effect on the transduced cells. Functional analyses showed that miR‐548j promotes increased invasiveness in three breast cancer cell lines and increases the amount of metastasis in NOD/SCID mice; however, miR‐548j does not appear to have an effect on proliferation nor obvious amplification of the region 22q12.1 in which miR‐548j is located (data not shown). These results indicate that miR‐548 family members may assume different functions in different cancers. Unlike other conserved miRNA families, such as let‐7, of which the highly consensus sequence leads to similar targets and functions (Lee et al., 2016). The 7 miR‐548 family members selected by our screening have the same function, some of them share the same seed sequence, such as miR‐548h and miR‐548i. However, the nucleotide substitution or insertion/deletion in their sequences make others of them (e.g. miR‐548l and miR‐548f), to be predicted to act on different targets (Supplementary Figure S1), such as Tensin1.
Tensin1 is a cytoplasmic phosphoprotein. It shares a highly conserved N‐terminus and C‐terminus with three other Tensin family members. The N‐terminus contains an actin‐binding domain that interacts with actin filaments, and a focal adhesion‐binding (FAB) domain, whereas the C‐terminus contains a SH2 domain, PTB domain and another FAB site (Lo, 2004). Based on the structure, Tensin1 is reported to be important in focal adhesion and maintenance of cell morphology (Chen et al., 2000). Interestingly, Tensin1 is critical for normal cell migration processes (Chen and Lo, 2003). It acts as a tumor suppressor and inhibits cell migration in many cancers (Martuszewska et al., 2009; Qian et al., 2007). We have confirmed that Tensin1 plays a similar role in breast cancer cell metastasis in vitro and in vivo as a direct and functional target of miR‐548j.
As a signaling effector between the extracellular matrix and the cytoskeleton, Tensin1 is rapidly translocated to assembling focal adhesion sites upon integrin ligation to promote focal adhesion and mediate signal transduction (Miyamoto et al., 1995). Therefore, we first examined the expression and activation of FAK, a key protein in the focal adhesion complex and signaling pathway, yet no obvious change of FAK expression or activation was observed. It means that simply change of miR‐548j expression levels in cells had little effect on translocating Tensin1 to assemble focal adhesion sites and then activate FAK. On the other hand, it is reported that Tensin1 can interact with DLC‐1 or other proteins to inhibit cell migration via reduction of Rho‐GTP (Hall et al., 2009; Qian et al., 2007). In this study, we verified the activation of the Rho family member Cdc42 in promoting cell invasion induced by either miR‐548j overexpression or Tensin1 knock down. Interestingly, a more remarkable suppression of invasiveness was observed in cells treated with the Rho‐GTPase inhibitor ML141 than those transfected with Cdc42 siRNA. This difference could be explained by a contribution of other members of the Rho‐GTPase family that may also be activated by miR‐548j and inhibited by ML141. The possible role of additional Rho‐GTPase awaits further exploration.
Given the vast heterogeneity of breast cancer, we verified miR‐548j function on invasion promotion in three different breast cancer cell lines with different levels of invasiveness, representing different subtypes of breast cancer (MCF‐7: ER positive; SKBR3: ER negative, Her2 positive; MDA‐MB‐231: triple negative) (Neve et al., 2006). Our results demonstrate an association between the expression of miR‐548j and invasiveness, which is also conserved in clinical breast cancer samples. Although, we could not completely distinguish miR‐548j with other miR‐548 family members, the analysis of breast cancer tissues indicate that miR‐548j expression correlates inversely with Tensin1 expression and patient survival; however, no significant difference was observed between the expression levels of miR‐548j and ER, PR or Her2 (Supplementary Table S2). These results suggest that promotion of metastasis by miR‐548j is not limited to certain subtypes but is widespread in breast cancer.
In summary, we determined that elevated expression of miR‐548j is directly related to cell invasion and metastasis, as well as poor prognosis for breast cancer patients. The function of miR‐548j is mediated by targeting Tensin1 via activation of Cdc42. These findings uncover a novel miRNA and its mechanism in breast cancer metastasis and provide a candidate for diagnosis and therapeutic targeting of breast cancer.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ZL and QZ planned and guided all the experiments. YZ, XL and YL conceived and carried out experiments. YZ and LL carried out animal models. FD cultured all the cell lines. BW and XW collected clinical breast cancer tissues specimens. YZ analyzed the data and wrote the manuscript. All authors had reviewed and had final approval of the submitted version.
Supporting information
The following is the supplementary data related to this article:
Supplementary data
Acknowledgments
We thank Mr. Yongquan Wang and Shenghua Zhang for providing technical support. This work was supported by Natural Science Foundation of China (81461148025, 81130043, 81472660) and National Basic Research Program of China (2011CB504205, 2013CB911004).
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2016.02.002.
Zhan Yun, Liang Xiaoshuan, Li Lin, Wang Baona, Ding Fang, Li Yi, Wang Xiang, Zhan Qimin, Liu Zhihua, (2016), MicroRNA‐548j functions as a metastasis promoter in human breast cancer by targeting Tensin1, Molecular Oncology, 10, doi: 10.1016/j.molonc.2016.02.002.
References
- Chaffer, C.L. , Weinberg, R.A. , 2011. A perspective on cancer cell metastasis. Science. 331, 1559–1564. [DOI] [PubMed] [Google Scholar]
- Chen, H. , 2000. Molecular characterization of human tensin. Biochem. J. 351, (Pt 2) 403–411. [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Lo, S.H. , 2003. Regulation of tensin-promoted cell migration by its focal adhesion binding and Src homology domain 2. Biochem. J. 370, 1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , 2009. Preliminary validation of ERBB2 expression regulated by miR-548d-3p and miR-559. Biochem. Biophys. Res. Commun. 385, 596–600. [DOI] [PubMed] [Google Scholar]
- Chou, J. , 2013. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat. Cell Biol. 15, 201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva, G. , 2014. MicroRNAs in cancer. Annu. Rev. Pathol. 9, 287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler, I.J. , 2003. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 3, 453–458. [DOI] [PubMed] [Google Scholar]
- Gao, Y.B. , 2014. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 46, 1097–1102. [DOI] [PubMed] [Google Scholar]
- Gregory, P.A. , 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601. [DOI] [PubMed] [Google Scholar]
- Hall, E.H. , 2009. Tensin1 requires protein phosphatase-1alpha in addition to RhoGAP DLC-1 to control cell polarization, migration, and invasion. J. Biol. Chem. 284, 34713–34722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. , Weinberg, R.A. , 2011. Hallmarks of cancer: the next generation. Cell. 144, 646–674. [DOI] [PubMed] [Google Scholar]
- He, H. , 2015. Kruppel-like factor 4 promotes esophageal squamous cell carcinoma differentiation by up-regulating keratin 13 expression. J. Biol. Chem. 290, 13567–13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, L. , 2013. Characterization of a Cdc42 protein inhibitor and its use as a molecular probe. J. Biol. Chem. 288, 8531–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , 2014. Identification of a tumor-suppressive human-specific microRNA within the FHIT tumor-suppressor gene. Cancer Res. 74, 2283–2294. [DOI] [PubMed] [Google Scholar]
- Huang, Q. , 2008. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol. 10, 202–210. [DOI] [PubMed] [Google Scholar]
- Huntzinger, E. , Izaurralde, E. , 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 12, 99–110. [DOI] [PubMed] [Google Scholar]
- Kaboli, P.J. , 2015. MicroRNA-based therapy and breast cancer: a comprehensive review of novel therapeutic strategies from diagnosis to treatment. Pharmacol. Res. 97, 104–121. [DOI] [PubMed] [Google Scholar]
- Lee, H. , 2016. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell. 7, 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, T. , 2012. Genome-wide analysis of mir-548 gene family reveals evolutionary and functional implications. J. Biomed. Biotechnol. 2012, 679563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, S.H. , 2004. Tensin. Int. J. Biochem. Cell Biol. 36, 31–34. [DOI] [PubMed] [Google Scholar]
- Lo, S.H. , 1994. Molecular cloning of chick cardiac muscle tensin. Full-length cDNA sequence, expression, and characterization. J. Biol. Chem. 269, 22310–22319. [PubMed] [Google Scholar]
- Ma, L. , 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 449, 682–688. [DOI] [PubMed] [Google Scholar]
- Ma, L. , 2010. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 12, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martuszewska, D. , 2009. Tensin3 is a negative regulator of cell migration and all four Tensin family members are downregulated in human kidney cancer. PLoS One. 4, e4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, S. , 1995. Synergistic roles for receptor occupancy and aggregation in integrin transmembrane function. Science. 267, 883–885. [DOI] [PubMed] [Google Scholar]
- Neve, R.M. , 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 10, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, X. , 2007. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc. Natl. Acad. Sci. U. S. A. 104, 9012–9017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes, D.R. , 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 6, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y. , 2014. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 509, 91–95. [DOI] [PubMed] [Google Scholar]
- Surviladze, Z. , 2010. A Potent and Selective Inhibitor of Cdc42 GTPase. [PubMed] [Google Scholar]
- Yang, S. , 2013. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 32, 4294–4303. [DOI] [PubMed] [Google Scholar]
- Yuan, Z. , 2011. MicroRNA genes derived from repetitive elements and expanded by segmental duplication events in mammalian genomes. PLoS One. 6, e17666 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following is the supplementary data related to this article:
Supplementary data


