Abstract
The presence of circulating tumor cells (CTCs) in the blood of ovarian cancer patients was shown to correlate with decreased overall survival, whereby CTCs with epithelial–mesenchymal‐transition (EMT) or stem‐like traits are supposed to be involved in metastatic progression and recurrence. Thus, investigating the transcriptional profiles of CTCs might help to identify therapy resistant tumor cells and to overcome treatment failure. For this purpose, we established a multi‐marker panel for the molecular characterization of single CTCs, detecting epithelial (EpCAM, Muc‐1, CK5/7), EMT (N‐cadherin, Vimentin, Snai1/2, CD117, CD146, CD49f) and stem cell (CD44, ALDH1A1, Nanog, SOX2, Notch1/4, Oct4, Lin28) associated transcripts.
First primer specificity and PCR‐performance of the multiplex‐RT‐PCRs were successfully validated on genomic DNA and cDNA isolated from OvCar3 cells. The assay sensitivity of the epithelial panel was evaluated by adding defined numbers of tumor cells into the blood of healthy donors and performing a subsequent immunomagnetic tumor cell enrichment (AdnaTest OvarianCancerSelect), resulting in a 100% concordance for the epithelial markers EpCAM and Muc‐1 to the AdnaTest OvarianCancerDetect. Additionally, by processing blood from ovarian cancer patients, high assay sensitivity could be verified. In blood of healthy donors no signals for epithelial markers were detected, for EMT and stem cell markers, however, signals were obtained mainly originating from leukocytes which calls for single cell analysis.
To that aim by using the ovarian cancer cell line OvCar3, we successfully established a workflow enabling the characterization of single CTCs. It consists of a density gradient‐dependent enrichment for nucleated cells, a depletion of CD45‐positive cells of hematopoietic origin followed by immunofluorescent labeling of CTCs by EpCAM and Muc‐1. Single CTCs are then isolated by micromanipulation and processed for panel gene expression profiling. Finally, fifteen single CTCs from three ovarian cancer patients were analyzed and found to be positive for stem cell (CD44, ALDH1A1, Nanog, Oct4) and EMT markers (N‐cadherin, Vimentin, Snai2, CD117, CD146). Albeit, inter‐cellular and intra/inter‐patient heterogeneity and co‐expression of epithelial, mesenchymal and stem cell transcripts on the same CTC was observed.
We have established a robust workflow to perform sensitive single cell panel gene expression analysis without the need of pre‐amplification steps. Our data point towards a heterogeneous expression of stem cell and EMT associated transcripts in ovarian cancer CTCs.
Keywords: Single cell expression analysis, Circulating tumor cells, Ovarian cancer, Multiplex‐RT‐PCR
Highlights
Workflow for detection and gene expression profiling of single ovarian cancer CTCs.
Multiplex‐PCR method detects 19 transcripts on single CTCs at once.
Single ovarian CTCs express stem cell and EMT markers.
Abbreviations
- CTC(s)
circulating tumor cell(s)
- DAPI
4,6-diamidino-2-phenylindole
- DTC(s)
disseminated tumor cell(s)
- DPO
dual priming nucleotides
- 6-FAM
6-carboxyfluorescein
- JOE
6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein
- FITC
fluorescein-isothiocyanate
- TRITC
tetramethyl-rhodamine-isothiocyanate
- PDH
pyruvate dehydrogenase
- CK
cytokeratin
- Muc-1
mucin-1
- EpCAM
epithelial cell adhesion molecule
- CD117
cluster of differentiation 117 also known as proto-oncogene c-Kit or tyrosine-protein kinase Kit
- ALDH1A1
aldehyde dehydrogenase 1 family, member A1
- SOX2
SRY (sex determining region Y)-box 2
- Oct4
octamer-binding transcription factor 4
- FIGO
Fédération Internationale de Gynécologie et d'Obstétrique
- MPC
magnetic particle concentrator
- Pt
patient
- EMT
epithelial–mesenchymal-transition
1. Introduction
Ovarian cancer is a highly aggressive tumor entity, due to the lack of specific symptoms and screening methods most patients are diagnosed in an advanced stage, which correlates with poor prognosis. Initial debulking surgery aiming at macroscopic complete tumor resection combined with subsequent platinum‐ and paclitaxel‐ based chemotherapy is highly effective at inducing remission in patients with advanced ovarian cancer (du Bois et al., 2009). However, more than half of the patients will relapse shortly after an initial response to chemotherapy (Aktas et al., 2011; Fehm et al., 2013; Martin and Schilder, 2009; Rubin et al., 1999). So far, residual postoperative tumor load is one of the most important prognostic factors for the outcome of ovarian cancer (Goodman et al., 2003).
Resistance to platinum‐based chemotherapy constitutes a major clinical challenge for ovarian cancer treatment. Several cellular processes can cause platinum resistance, including increased tolerance towards DNA‐platinum adducts or enhanced DNA repair capacity of tumor cells (Galluzzi et al., 2012). Moreover, intra‐tumor heterogeneity can contribute to chemo‐resistance in different ways, which take place on the genomic, transcriptomic, epigenetic and clonal level: i) chemotherapy leads to clonal expansion of intrinsically resistant and pre‐existing resistant tumor cells. ii) Chemosensitive tumor cells increasingly convert to a chemo‐resistant state and acquire “de novo” therapy resistance. iii) Both mechanisms co‐exist (Kuhlmann et al., 2015). Though the link between drug resistance and cellular heterogeneity was initially explored in the context of cancer stem cells, which are present as a small subgroup within the primary tumor (Pribluda et al., 2015; Shah and Landen, 2014). Due to their intrinsic ability to self‐renew these CSCs are regarded as the source of metastatic tumor spread and to enhance tumorigenesis and drug‐resistance (Dyall et al., 2010; Reya et al., 2001). CSCs have been identified in ovarian cancer cell lines and tissues and their presence has been associated with aggressive tumor behavior (Bapat et al., 2005; Boesch et al., 2014; Hosonuma et al., 2011). CSC heterogeneity may also be increased by the process of epithelial–mesenchymal transition (EMT), which is capable of generating cells with stem cell‐like properties from differentiated epithelial cells (Brabletz, 2012; Mani et al., 2008). EMT is a process essential for embryonic development, but also plays a role in tumor progression and metastasis (Thiery, 2002). During EMT epithelial cells of the primary tumor upregulate mesenchymal genes causing them to lose their cell‐to‐cell adhesions and apico‐basal cell polarity, leading to an increase in the cells mobility and invasiveness (Guarino, 2007). It is assumed, that in some cases the combination of EMT and stem cell traits allows tumor cells to escape from the primary tumor, to enter the blood stream and may act as potential metastasis initiating cell.
Several studies have confirmed the prognostic impact of CTCs in ovarian cancer (Abu‐Rustum et al., 1999; Aktas et al., 2011; Kuhlmann et al., 2014; Poveda et al., 2011; Zeng et al., 2015; Zhou et al., 2015). Beyond their quantification, a further molecular characterization of CTCs is of high interest to develop CTC‐based therapy regimen. Additionally, since CTCs supposedly consist of heterogeneous cell populations with different potentials to survive chemotherapy (Aktas et al., 2011) and to initiate secondary tumors or metastases the use of single cell analysis is required. Only single cell analysis of CTCs allows us to distinguish cells with different expression profiles which give a hint towards the evolution of CTCs during treatment. It will dissect cellular heterogeneity since only a small subset of CTCs from one patient may exhibit the genotype or phenotype responsible for development of therapy resistance. Thereby, single CTC analysis represents a ‘liquid biopsy’ for the selection of an appropriate therapy and for real time monitoring of its effectiveness (Aktas et al., 2009; Barriere et al., 2012; Giordano et al., 2013; Kasimir‐Bauer et al., 2012).
To date, there is no data available showing the expression of stem cell‐ and EMT associated transcripts expressed in ovarian cancer CTC. 5 . However, multiple studies in breast cancer have documented that the expression of stem cell/EMT markers in CTCs is associated with poor prognosis and resistance against chemotherapy (Aktas et al., 2009; Mego et al., 2012) suggesting that their presence should also be investigated in single CTCs from ovarian cancer patients. In order to determine expression of such transcripts in single CTCs, we developed a multiplex PCR approach for genes associated with epithelial [mucin‐1 (Muc‐1), epithelial cell adhesion molecule (EpCAM), cytokeratin 5&7 (CK5&7)], EMT [N‐cadherin, Vimentin, Snai2, cluster of differentiation 117, 146 & 49f (CD117, CD146 & CD49f), Snai1] and stem cell features [cluster of differentiation 44 (CD44), aldehyde dehydrogenase 1 family, member A1 (ALDH1A1), Nanog, SRY (sex determining region Y)‐box 2 (SOX2), Notch1, Notch4, Octamer‐binding transcription factor 4 (Oct4) and Lin28]. For verification we benchmarked our method to the well‐known AdnaTest OvarianCancer.
2. Materials and methods
2.1. Cell line and cell culture
The human ovarian cancer cell line OvCar3 was purchased from the American Type Culture Collection (ATCC, Manassas, VA, US) and cultured in RPMI 1640 containing 10% fetal calf serum and 1% (100 U/ml) Penicillin–Streptomycin (Gibco™ by Thermo Fisher Scientific, Waltham MA, US). Cells were grown at 37 °C in a humidified atmosphere with 5% CO2. The ovarian carcinoma cell line OvCar3 was used because all markers studied (except for Snai2) are expressed in that cell line.
2.2. Patient samples
The present work is a joint project of the Departments of Gynecology and Obstetrics of the University Hospitals of Duesseldorf and Dresden, Germany. Only patients with histologically confirmed epithelial ovarian cancer were enrolled. Written informed consent was obtained from all participating patients and the study was approved by the local research ethics committees in Duesseldorf (3768) and Dresden (EK 236082012). Clinical patient data are summarized in Table 2 Supplementary Material. Tumors were classified according to the World Health Organization (WHO) classification, grading was done according to Silverberg (Silverberg, 2000) and tumor staging was classified according to FIGO (Fédération Internationale de Gynécologie et d'Obstétrique) (FIGO Committee on Gynecologic Oncology, 2009). The whole study population received primary radical surgery aiming at macroscopic complete tumor resection. Total abdominal hysterectomy, bilateral salpingo‐oophorectomy, infragastric omentectomy, peritoneal stripping, and pelvic as well as paraaortic lymphadenectomy were performed, where feasible. For bulk analysis a total of 5 ml of peripheral blood was collected in K2 EDTA tubes (Becton Dickinson, Plymouth, UK) and processed within 4 h after phlebotomy. For single cell analysis 7.5 ml of peripheral blood was processed within 8 h after blood collection. To avoid contamination with epithelial skin cells the first blood sample was always discarded.
2.3. Blood sample processing
2.3.1. Tumor cell enrichment and isolation for bulk analysis
Cells from ovarian cancer cell lines were dissociated with Accutase (Gibco™ by Thermo Fisher Scientific, Waltham MA, US) and separated with a cellstrainer (Greiner Bio‐one, Kremsmünster, Austria) into a single cell suspension. Defined cell numbers (5, 10 and 25) were transferred into 5 ml blood of healthy donors by the MoFlo™ XDP cell sorter (Beckman Coulter, Brea, CA, US). Cells were distinguished based on size and granularity using defined Forward Scatter (FSC) and Side Scatter (SSC) parameters. Cell clumps were excluded by means of SSC‐W. All blood samples of patients and healthy donors were subjected to AdnaTest OvarianCancerSelect and/or to the AdnaTest EMT‐1/StemCell Select (QIAGEN GmbH, Hilden, Germany) which enable immunomagnetic enrichment of tumor cells positive for epithelial antigens EpCAM and Muc‐1. Both tests are using the same CTC enrichment strategy, with a special washing procedure (AdnaWash buffer) included in the AdnaTest EMT‐1/StemCell Select to reduce contaminating leukocytes. All experimental steps were performed according to the manufacturer's protocol. In brief, EpCAM‐ and Muc‐1‐positive cells were captured and extracted by the Dynal MPC®‐S and L magnetic particle concentrators (Invitrogen™ Dynal® by Thermo Fisher Scientific, Waltham, MA). Enriched cells were lysed and RNA was transcribed into cDNA using Sensiscript® Reverse Transcription Kit (QIAGEN GmbH). All spiking experiments were performed at least three times. Blood of healthy donors was examined additionally.
2.3.2. Tumor cell enrichment, identification and isolation for single cell analysis
For spiking experiments 10 dissociated OvCar3 cells were sorted into 7.5 ml of blood from a healthy volunteer as described above. Spiking experiments were performed eight times. These samples and ‘control’ blood from both ovarian cancer patients and healthy donors was processed as follows. Peripheral blood mononuclear cells (PBMCs) and neutrophils were extracted by Biocoll Separation Solution (Biochrom by Merck, Darmstadt, Germany) gradient centrifugation (density 1.077 g/ml), according to the manufacturer's recommendations. To reduce the number of contaminating leukocytes the PBMC phase was washed for 5 min with 10 ml PBS (Gibco™ by Thermo Fisher Scientific, Waltham MA, US) and CD45 depletion was performed. In brief, 1.5 ml Dynabeads® CD45 (Invitrogen™ by Thermo Fisher Scientific, Waltham, MA, US) were washed two times for 5 min and incubated with the PBMC phase for 30 min whilst rotating. Leukocytes bound to the Dynabeads® CD45 were removed using Dynal MPC®‐S (Invitrogen™ Dynal® by Thermo Fisher Scientific, Waltham, MA). Cells of interest were washed twice in 1 ml PBS for 5 min.
To identify CTCs, unfixed and unpermeabilized cells were immunofluorescently stained in suspension with a FITC conjugated mouse monoclonal antibody to EpCAM (clone VU‐1D9; 1:50; Cell Signaling Technology by Merck, Darmstadt, Germany), a FITC conjugated mouse monoclonal antibody to pan‐cytokeratin (C11, 1:400, GeneTex, Irvine, CA, US) and 4′,6‐Diamidine‐2′‐phenylindole dihydrochloride ((DAPI) 1 μg/ml Roche, Basel, Switzerland). Leukocytes were identified using a PE‐Cy5 conjugated CD45‐specific mouse antibody (1:25, Santa Cruz Biotechnology, Dallas, TX, US). Following 2 h incubation in the dark at 4 °C and centrifugation at 450 rpm for 2 min the cell pellet was washed three times with 1 ml PBS for 5 min and processed further.
Single cells were isolated using CellCelector (ALS GmbH, Jena, Germany). This system combines microscopic detection of labeled cells and their automated micromanipulation with a vertical glass capillary fixed to a robotic arm. For microscopy the following set‐up was used: Olympus CKX 41; camera system: CCD camera XM10‐IR (Olympus, Tokyo, Japan); for analysis ALS CellCelector‐Software 3.0, (ALS, Jena, Germany). In detail labeled cell solutions were transferred to a glass slide and cells were allowed to settle. Then CK‐ and/or EpCAM‐positive cells were detected in the FITC channel at a 40× magnification. CK and/or EpCAM‐positive cells were selected by the software and additionally recorded in the remaining channels (brightfield (BF), DAPI, and Cy5) at 40× magnification to verify morphology and CD45‐negativity of isolated cells. Single cells fullfilling the ‘CTC‐criteria’ i.e. DAPI and CK/EpCAM positivity and CD45 negativity were micromanipulated in DAPI at 40× magnification. Accordingly, selected cells were aspirated with a volume of 20–100 nl using a 30 μm glass capillary. To achieve optimal cell deposition 2–9 μl PBS buffer were taken up into the capillary prior to the picking process. Single cells were deposited into PCR tubes containing 100 μl of lysis buffer of the Dynabeads® mRNA DIRECT Micro Kit (Invitrogen™ Dynal® by Thermo Fisher Scientific, Waltham, MA, US) and stored for up to two weeks at −20 °C until further processing.
Subsequently, oligo (dT) based mRNA isolation was performed for all single cell lysates, according to the manufacturer's protocol with the Dynabeads® mRNA DIRECT Micro Kit (Invitrogen™ Dynal® by Thermo Fisher Scientific, Waltham, MA, US), which is part of the AdnaTest OvarianCancerDetect. For reverse transcription the Sensiscript® Reverse Transcription Kit (QIAGEN GmbH) was used. The resulting/obtained cDNA served as template for tumor cell detection and characterization by multiplex‐PCR.
2.4. Tumor cell detection/analysis of CTCs
2.4.1. AdnaTest OvarianCancerDetect
The AdnaTest OvarianCancerDetect was used to amplify transcripts of the epithelial markers EpCAM, Muc‐1 and CA125, whereas the AdnaTest EMT‐1/StemCell Detect was employed to analyze ALDH1 expression in a singleplex PCR assay and EMT markers Akt‐2, TWIST, PI3Kα in a multiplex‐RT‐PCR assay. Resulting PCR products were separated using the DNA 1000 LabChips and visualized with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) using the Expert Software Package (version B.02.07.SI532). Samples analyzed with the AdnaTest OvarianCancerDetect were considered as positive if signals for at least one epithelial marker and the housekeeping transcript β‐actin were detected. For the AdnaTest EMT‐1/StemCell Detect PCR signals for at least ALDH1 or one EMT product plus a fragment of β‐actin had to be present in a positive sample.
2.5. Multiplex‐RT‐PCR panels
2.5.1. Primer design
Dual priming oligonucleotide primers (DPO) for three independent multiplex‐RT‐PCR panels were designed using Primer3 Plus software (Rozen and Skaletsky, 2000). In contrast to conventional primers, DPO primers contain two priming sites, which improve binding specificity (Chun et al., 2007). All primers were designed to the 3′ end of each target mRNA to ensure amplification of mRNA degradation products, as well. Forward primers were alternately labeled either with the fluorescent dye 6‐Carboxyfluorescein (6‐FAM) or JOE (6‐Carboxy‐4′,5′‐Dichloro‐2′,7′‐Dimethoxyfluorescein), respectively. Additionally, in between one multiplex panel transcript‐specific amplicons were designed with at least 20 bp size differences to distinguish them by gel or capillary electrophoresis. Primer sequences, their annealing temperatures and labeling are given in Table1 Supplemented Material. Primers were purchased from Biomers (biomers.net GmbH, Ulm, Germany).
2.5.2. Multiplex‐RT‐PCR conditions
Multiplex‐RT‐PCR for all three panels was performed using the KAPA2G Fast Multiplex ReadyMix (Peqlab, Erlangen, Germany) in a final reaction volume of 25 μl. The reaction mixture contained 1 × KAPA2G Fast HotStart DNA Polymerase (1 U/25 μl) KAPA2G Fast HotStart buffer, dNTPs (0.2 mM of each dNTP), MgCl2 (3 mM) and stabilizers. Primers were used in a final concentration of 0.08 μM. For bulk analysis the following amounts of cDNA were used: 4 μl for the epithelial marker panel and 7 μl for stem cell and EMT marker panels. Single cell analysis for all three marker panels (epithelial, EMT and stem cell) was performed with 12.5 μl cDNA. Genomic DNA and cDNA from OvCar3 cells were used as positive control.
The thermal profile used for all multiplex‐RT‐PCRs was as follows: after an initial denaturation step at 95 °C for 15 min 10 PCR cycles were carried out consisting of denaturation at 95 °C for 45 s, primer annealing/extension at 57 °C for 45 s and elongation for 45 s at 72 °C. Subsequently, annealing/extension temperature was increased for 1 °C per 10 cycles up to 61 °C. Finally, leading to 50 cycles in total. Samples were stored at 4 °C over night or at −20 °C for long‐term storage. PCR products were visualized by capillary electrophoresis.
2.6. Capillary electrophoresis
Forward primers were alternately labeled with either 6‐FAM or JOE to distinguished PCR products from each other in the same panel. For each sample 15.5 μl of Hi‐Di™ Formamide and 1.5 μl of an internal size standard (GeneScan500 ROX, both Applied Biosystems by Thermo Fisher Scientific, Waltham, MA, US) were mixed with 3 μl PCR product (1 μl for controls) and transferred to an ABgene Thermo‐Fast 96 PCR Detection Plate (Thermo Fisher Scientific, Waltham, MA, US). For separation of the PCR products an ABI PRISM 3130XL Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA) with a capillary of 36 cm length and POP‐7 polymer was used. Analysis was performed with the Peak Scanner™ Software Version: 1.0 (Applied Biosystems by Thermo Fisher Scientific, Waltham, MA). A peak was considered as positive if its height excelled 750 rfu (relative fluorescence units). Samples analyzed for epithelial markers by multiplex‐RT‐PCR were considered as positive for CTCs if signals for at least one epithelial marker and the housekeeping transcript for PDH were detected.
2.7. Detection and characterization of single leukocytes
CD45‐positive cells were micromanipulated, RNA was isolated and cDNA was synthesized. Then cDNA was analyzed by multiplex‐RT‐PCR for expression of epithelial, mesenchymal and stem cell transcripts. The remaining 2.5 μl of cDNA were used for CD45‐specific PCR using the same thermal profile as applied to all multiplex‐RT‐PCRs. As positive control reverse transcribed cDNA from 1000 leukocytes was employed. CD45‐specific RT‐PCR products were visualized with a 2100 Bioanalyzer as described above.
3. Results
3.1. Establishing three independent multiplex‐RT‐PCR panels for the detection and characterization of CTCs
Primer specificity and PCR performance of all three multiplex panels, epithelial (Muc‐1, EpCAM, CK5 and CK7), EMT (N‐cadherin, Vimentin, Snai2, CD117, CD146, CD49f and Snai1) and stem cell markers (CD44, ALDH1A1, Nanog, SOX2, Notch1, Notch4, Oct4 and Lin28), were successfully established with genomic DNA and cDNA from the ovarian cancer cell line OvCar3. An intron‐spanning amplicon for the reference gene pyruvate dehydrogenase (PDH) was included into the epithelial panel to distinguish signals between ‘contaminating’ genomic DNA (183 bp) and cDNA (100 bp) (Figure 1). With adapted PCR conditions all amplicons were optimized in single PCR and in multiplex reactions (Supplementary Material Figure 1) ensuring an optimal PCR performance without unspecific priming.
Figure 1.
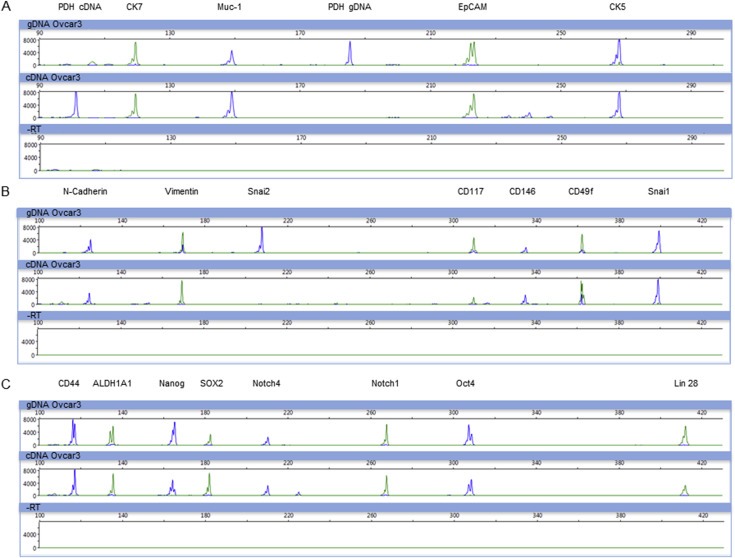
Multiplex‐RT‐PCR for the detection of epithelial, EMT and stem cell markers. Depicted are electropherograms of three multiplex‐RT‐PCR panels after analysis by capillary electrophoresis. For all panels, upper row: gDNA, middle row: cDNA, lower row: ‐RT. Primer specificity and PCR‐performance was successfully validated on genomic DNA and cDNA of OvCar3 cells. A) Epithelial marker panel: CK7 (124 bp), Muc‐1 (149 bp), EpCAM (222 bp) and CK5. PDH (100/183 bp) was used as control to distinguish between gDNA and cDNA. B) EMT marker panel: N‐cadherin (127 bp), Vimentin (170 bp), Snai2 (208 bp), CD117 (309 bp), CD146 (335 bp), CD49f (363 bp) and Snai1 (402 bp). C) Stem cell marker panel: CD44 (120 bp), ALDH1A1 (139 bp), Nanog (162 bp), SOX2 (185 bp), Notch4 (210 bp), Notch1 (268 bp), Oct4 (310 bp) and Lin28 (413 bp).
3.2. Validation of multiplex‐RT‐PCR panels by spiking experiments into blood of healthy donors and blood from ovarian cancer patients
3.2.1. Analysis of spiked blood samples
OvCar3 cells express the epithelial cell surface proteins Muc‐1 and EpCAM and are thereby suitable for spiking experiments and subsequent enrichment with the AdnaTest OvarianCancer Kit to validate the analytic sensitivity of the epithelial multiplex‐PCR panel. Low numbers of OvCar3 cells (5, 10 and 25) were spiked into 5 ml blood of healthy donors and processed with the AdnaTest OvarianCancerSelect. Resulting cDNA of the same sample was used to compare the presence of the epithelial markers EpCAM and Muc‐1 by the AdnaTest OvarianCancerDetect and multiplex‐RT‐PCR. Both tests were 100% concordant (Figure 2).
Figure 2.
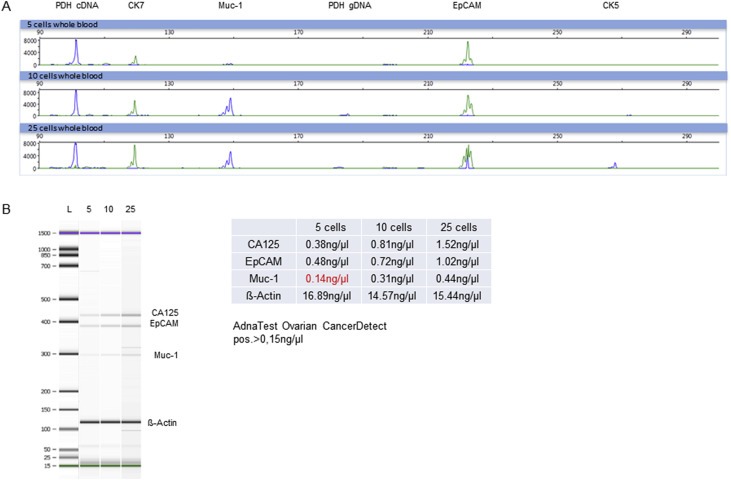
Detection of epithelial markers in spiked blood samples after immunomagnetic enrichment with the AdnaTest OvarianCancerSelect. Signals derived from defined numbers of OvCar3 cells (5, 10 and 25) spiked into blood. Positivity for EpCAM and Muc‐1 of samples spiked with 5, 10 and 25 OvCar3 cells was 100% concordant to EpCAM and Muc‐1 positivity detected by the AdnaTest OvarianCancerDetect A) Visualized are electropherograms of the epithelial multiplex‐RT‐PCR panel after analysis by capillary electrophoresis. Amplified fragments of the epithelial transcripts CK7 (124 bp), Muc‐1 (149 bp), EpCAM (222 bp) and CK5 (265 bp) are shown. PDH is used as control to distinguish between gDNA (183 pb) and cDNA (100 bp). B) Shown are the amplified transcripts Muc‐1 (299 bp), EpCAM (395 bp), CA125 (432 bp) and β‐Actin (120 bp) by AdnaTest OvarianCancerDetect after electrophoretic separation. Corresponding values (ng/μl) of each transcript are calculated out of the tape thickness and are represented in the table. A signal was considered as positive in case of >15 ng/μl.
3.2.2. Analysis of blood from ovarian cancer patients
Next, the sensitivity/performance of the epithelial transcript panel was determined with patient blood samples. In total, 10 blood samples from ovarian cancer patients were examined for epithelial markers by the AdnaTest OvarianCancerDetect and the multiplex‐RT‐PCR. The multiplex‐RT‐PCR not only confirmed both patients tested positive for CTCs with the AdnaTest OvarianCancerDetect (Table 1 Pt.3 and 4) but also identified a further patient as ‘CTC‐positive’ (Table 1 Pt.1).
Table 1.
Detection of epithelial markers in 10 ovarian cancer patients, analyzed with multiplex‐RT‐PCR and AdnaTest OvarianCancerDetect. Samples analyzed by the AdnaTest OvarianCancerDetect were considered as positive for CTCs, if signals for at least one epithelial marker and the housekeeper β‐actin were detected. Samples analyzed for epithelial markers by multiplex‐RT‐PCR were considered as positive for CTCs, if signals for at least one epithelial marker and the housekeeper PDH were detected. Pt. = patient.
| Patients | Multiplex RT‐PCR epithelial markers | AdnaTest OvarianCancerDetect | Muc‐1 | EpCAM | CA125 | Actin |
|---|---|---|---|---|---|---|
| Pt.1 | pos | neg | 0 ng/μl | 0 ng/μl | 0 ng/μl | 13.37 ng/μl |
| Pt.2 | neg | neg | 0 ng/μl | 0 ng/μl | 0 ng/μl | 15.43 ng/μl |
| Pt.3 | pos | pos | 0.22 ng/μl | 0 ng/μl | 0 ng/μl | 17.2 ng/μl |
| Pt.4 | pos | pos | 0.38 ng/μl | 0 ng/μl | 0 ng/μl | 14.69 ng/μl |
| Pt.5 | neg | neg | 0 ng/μl | 0 ng/μl | 0 ng/μl | 15.06 ng/μl |
| Pt.6 | neg | neg | 0 ng/μl | 0 ng/μl | 0 ng/μl | 13.47 ng/μl |
| Pt.7 | neg | neg | 0 ng/μl | 0 ng/μl | 0 ng/μl | 14.31 ng/μl |
| Pt.8 | neg | neg | 0 ng/μl | 0 ng/μl | 0 ng/μl | 10.95 ng/μl |
| Pt.9 | neg | neg | 0 ng/μl | 0 ng/μl | 0 ng/μl | 7.38 ng/μl |
| Pt.10 | neg | neg | 0 ng/μl | 0 ng/μl | 0 ng/μl | 10.35 ng/μl |
AdnaTest OvarianCancerDetect pos. > 0.15 ng/μl.
3.2.3. Analysis of blood from healthy donors
In a next step, blood from healthy donors was assessed as described in 3.2.2. No epithelial markers could be detected with neither AdnaTest OvarianCancerDetect nor multiplex‐RT‐PCR (Figure 3). However, stem cell‐ and EMT associated marker signals were obtained by both tests, probably deriving from leukocytes (Supplementary Material Figures 2 and 3). Despite using a washing buffer recommended to reduce co‐isolation of leukocytes during the CTC capture phase false positive results were detected.
Figure 3.
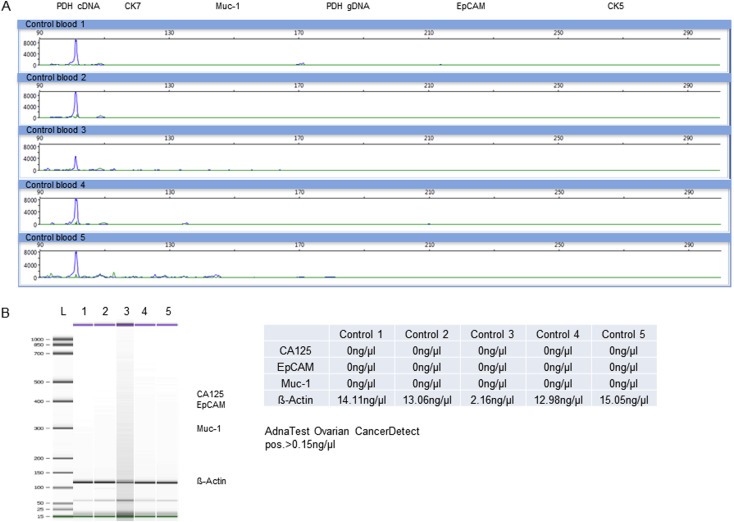
Detection of epithelial markers in blood from healthy donors after immunomagnetic enrichment with the AdnaTest OvarianCancerSelect. For both tests no epithelial markers, only housekeepers (PDH (100 bp) or β‐Actin (120 bp)) were detected. A) Visualized are electropherograms of 5 healthy donor samples for the epithelial multiplex‐RT‐PCR panel after analysis by capillary electrophoresis. B) Displayed are the transcripts of 5 healthy donor samples amplified by the AdnaTest OvarianCancerDetect and separated by capillary electrophoresis. Corresponding values (ng/μl) of each transcript are calculated out of the tape thickness and are represented in the table. A signal was considered as positive in case of >15 ng/μl.
3.3. Panel gene expression profiling of single ovarian cancer cells
In order to avoid false positive results from non‐malignant cells such as leukocytes, a workflow based on spiking experiments of OvCar3 cells into blood of healthy volunteers was established to isolate and characterize single CTCs. It consists of density gradient centrifugation of the blood, depletion of CD45‐positive cells, immunofluorescent labeling of CTCs, their micromanipulation, RNA isolation, reverse transcription and analysis by multiplex‐RT‐PCR (Figure 4A). A tumor cell was classified as CTC and selected for micromanipulation when it was positive for DAPI and CK/EpCAM and CD45‐negative (Figure 4B).
Figure 4.
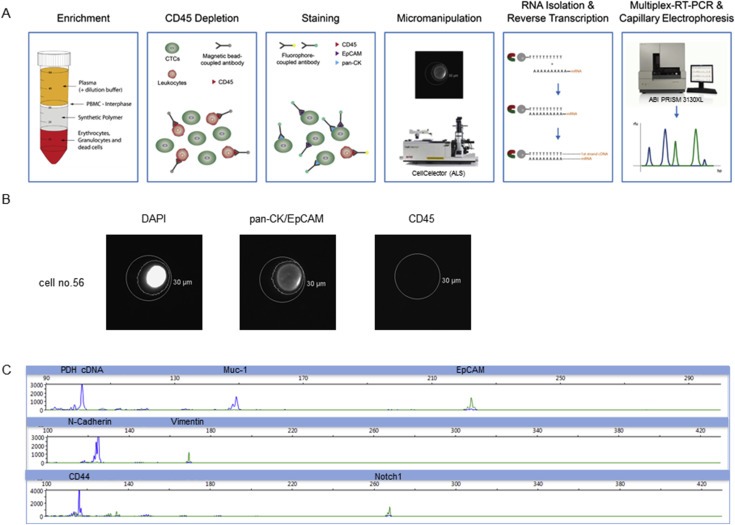
CTC/tumor cell isolation and analysis on the single cell level. A) Workflow: CTCs were enriched by density gradient centrifugation and CD45‐positive cells were depleted. The remaining cells were labeled for EpCAM, pan‐CK and CD45 and isolated by micromanipulation. RNA of each single cell was isolated and reverse transcribed into cDNA. Finally, multiplex‐RT‐PCRs for all 3 panels were performed and PCR products were visualized by capillary electrophoresis. B) Illustration of a single cell (no. 56). A cell was characterized as a CTC in case of DAPI, pan‐CK/EpCAM positivity and CD45 negativity as evaluated by microscopy. The outer circle constitutes the capillary, sized with 30 μm. The inner circle is created by the analyzing software indicating a recognized fluorescence signal. B) Electropherograms of epithelial (Muc‐1, EpCAM), EMT (N‐cadherin, Vimentin) and stem cell markers (CD44) exemplified for one single cell (no. 56). Cells were denoted/designated as CTC‐positive in case of positivity for at least one epithelial marker and the housekeeper PDH (cDNA).
Regarding multiplex PCR analysis samples were classified as CTC‐positive when PCR signals in capillary electrophoresis were present for the housekeeper PDH (cDNA) and at least one epithelial marker and (Figure 4C).
Panel expression profiling of 77 single OvCar3 cells, which were spiked into blood from healthy donors isolated and then processed using selection criteria revealed distinct heterogeneity between single OvCar3 cells especially for mesenchymal and stem cell markers (Figure 5):Twenty‐one out of 77 cells simultaneously expressed Muc‐1 and EpCAM. Fifteen out of 77 co‐expressed CK7, Muc‐1 and EpCAM. Only one cell was positive for all four tested epithelial markers (CK7, Muc‐1, EpCAM and CK5) (Figure 5 cell no. 7). Only 3 out of 77 cells expressed CK5. N‐cadherin was the most prevalent EMT marker (35/77 cells); in 12/35 cells it was co‐expressed with vimentin. In 43 of the 77 analyzed cells no stem cell markers were detected. Expression of only CD44 (16/77) or co‐expression of Oct4 and Lin28 (4/77) or Notch1 and Notch4 (3/77) was registered. Distribution of all other stem cell markers was non‐uniform. Co‐expression of epithelial and mesenchymal markers (57/77) as well as mesenchymal and stem cell markers (28/77) or epithelial and stem cell markers (34/77) could be observed frequently. The expression of epithelial, mesenchymal and stem cell markers within one cell was found in 22 of 77 analyzed cells (Figure 5).
Figure 5.
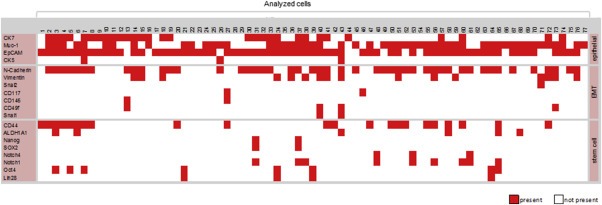
Expression profiles of 77 single OvCar3 cells spiked into blood of healthy volunteers. Depicted is the expression of epithelial, EMT and stem cell markers (different rows) in 77 single OvCar‐3 cells (different columns). A signal was counted as positive reaching a signal intensity of >750 rfu in capillary electrophoresis and is color coded as present (red square). In general a cell was characterized as a tumor cell in case of positivity for PDH (cDNA) and at least one epithelial marker (constituted as present). A distinct intercellular heterogeneity between the 77 analyzed OvCar3 cells originating from one cell line was determined. Not all cells were found to be positive for mesenchymal (EMT) and/or stem cell markers. Furthermore cells with a co‐expression of epithelial–mesenchymal (EMT), stem cell‐mesenchymal (EMT) and epithelial–mesenchymal (EMT)‐stem cell markers could be observed.
Underlining their contaminating potential in bulk analysis single leukocytes were analyzed for all three marker panels and yielded no signals for epithelial markers, while signals for stem cell‐ and EMT markers were identified (Supplementary Material Figure 4A and B). Their hematopoietic origin was confirmed by detection of CD45 transcript (Supplementary Material Figure 4C Supplemented Data).
3.3.1. Panel gene expression profiling of single CTCs from ovarian cancer patients
For single CTC gene expression analysis blood samples from 3 ovarian cancer patients were processed as described in 3.2.2. CTC counts ranged from 4 (Pt. 1 and 3) to 7 (Pt. 2). They were isolated via micromanipulation (Figure 6A) and further profiled for their gene expression for epithelial, EMT and stem cell markers (Figure 6B). All CTCs were positive for Muc‐1, whereas the presence of other epithelial markers varied. Four of the 15 analyzed CTCs expressed stem cell markers (CD44, ALDH1A1, Nanog and Oct4) and 13 out of 15 were positive for EMT markers (N‐cadherin, Vimentin, Snai2, CD117 and CD146) (Figure 6B). Expression profiles differed in CTCs obtained from different patients (inter‐patient heterogeneity) and in CTCs isolated from the same patient (intra‐patient and inter‐cellular heterogeneity). Furthermore, co‐expression of EMT and stem cell markers could be observed.
Figure 6.
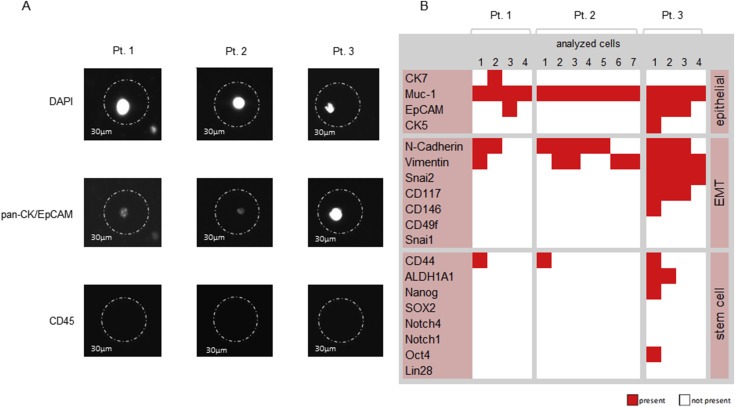
Single cell expression profiling of CTCs from 3 ovarian cancer patients analyzed by multiplex‐RT‐PCR for epithelial, EMT and stem cell markers. A) Representative images of single CTCs for each patient after fluorescence microscopy. A cell was characterized as a CTC in case of DAPI, pan‐CK/EpCAM positivity and CD45 negativity, as evaluated by microscopy. The circle constitutes the capillary, sized with 30 μm. B) Expression profile of CTCs. A signal was counted as positive reaching a signal intensity of >750 rfu in capillary electrophoresis and is color coded as present (red square). A cell was characterized as a CTC in case of positivity for PDH (cDNA) and at least on epithelial marker after capillary electrophoresis (constituted as present). Expression patterns of all patient samples revealed a ubiquitous expression of Muc‐1 in all analyzed CTCs, whereas the expression of additional epithelial markers varied between the cells, even within one patient. In every patient CTCs expressing epithelial, mesenchymal and stem cell markers could be detected even if the amount of CTCs harboring stem cell‐ and EMT markers was various. Pt. = patient.
4. Discussion
Even though huge efforts have been undertaken to elucidate the biological nature of primary tumors and to identify new biomarkers in order to improve the treatment (Serio and Billack, 2011), many patients who were considered to be cured relapse even several years after resection/treatment of the primary tumor (Aktas et al., 2011; Martin and Schilder, 2009). It has already been shown that the presence of CTCs in ovarian cancer patients has predictive and prognostic relevance for the patient's OS and/or PFS (Abu‐Rustum et al., 1999; Aktas et al., 2011; Kuhlmann et al., 2014; Poveda et al., 2011; Zeng et al., 2015; Zhou et al., 2015). The pheno‐ and genotype of disseminated cancer cells have been found to differ from the bulk cell mass of the primary tumor, and CTCs are thought to initiate and establish a secondary tumor at distant sites, thereby worsening the clinical outcome (Polzer et al., 2014). Up to now, therapy strategies for patients diagnosed with ovarian cancer do not consider the phenotype of CTCs. However, revealing unique features of these cells can help to understand the mechanisms underlying the development of resistances and/or therapy failure. CTCs circulate through the bloodstream, they are therefore easily accessible and can be regarded as a ‘liquid biopsy’ for real time monitoring. Accordingly, characterizing CTCs may support the selection of an appropriate therapy in the near future, thus leading to a more personalized treatment.
Our main goal was to develop a method that combines both the detection and the molecular characterization of single ovarian cancer CTCs. It consists of a CTC enrichment by density gradient centrifugation, CD45 depletion, immunofluorescent labeling, single cell isolation via micromanipulation, reverse transcription and multiplex‐RT‐PCR analysis.
At first, we successfully validated primer specificity and PCR‐performance of all three multiplex panels on genomic‐DNA and cDNA of the ovarian cancer cell line OvCar3 (Figure 1). Consequently, we benchmarked the multiplex‐RT‐PCR approach for the detection of epithelial markers to the commercially available AdnaTest OvarianCancerDetect. Positivity rates for EpCAM and Muc‐1 signals for blood samples spiked with different numbers of OvCar3 were 100% concordant with AdnaTest OvarianCancerDetect. Next we confirmed the applicability of our technique for blood samples derived from ovarian cancer patients. As a result the multiplex‐RT‐PCR not only confirmed both patients tested positive for CTCs by the AdnaTest OvarianCancerDetect but also identified an additional patient as ‘CTC‐positive’, indicating a higher sensitivity/performance of our PCR‐based method. Another advantage of our method is, that it only requires half the amount of cDNA as used in the AdnaTest Ovarian Cancer Detect. The reason for this advanced performance most likely lies in the use of DPO primers, which bind with a higher specificity to their target sequences than conventional primers.
So far, analysis of enriched bulk CTC populations is the most common approach to characterize the expression of transcripts (Aktas et al., 2009; Barriere et al., 2012; Kasimir‐Bauer et al., 2012) However, this approach may lead to false observations and interpretations due to contamination with non‐CTC cells such as leukocytes (Aktas et al., 2009; Sieuwerts et al., 2009). Although they are depleted beforehand, a number of leukocytes usually remain after different CTC enrichment and CD45‐based depletion strategies (Sieuwerts et al., 2009). Especially due to their mesenchymal nature, leukocytes interfere with the detection of EMT markers, while incompletely differentiated leukocytes express stem cell markers due to their hematopoietic origin (Bryder et al., 2006). These expression patterns can lead to false positive results. In our experiments this issue was also encountered as blood samples of healthy donors were tested positive for EMT and stem cell transcripts by using both, the AdnaTest OvarianCancerDetect and two multiplex‐RT‐PCR panels (Supplementary Material Figures 2 and 3). Therefore, we believe that molecular characterization of CTCs is more accurate on the single cell level.
In this study, we describe the development and establishment of a workflow to isolate CTCs from ovarian cancer patients and to perform subsequent gene expression profiling on single cells. This consists of a density gradient‐dependent enrichment for nucleated cells, a depletion of CD45‐positive cells of hematopoietic origin followed by immunofluorescent labeling of CTCs. Potential CTCs are then isolated by micromanipulation and processed for panel gene expression profiling. We were able to successfully detect and characterize single OvCar3 cells and CTCs isolated from ovarian cancer patients.
We found CTCs expressing both epithelial and mesenchymal genes, which has also been described for CTCs of breast cancer patients on protein level (Kallergi et al., 2011; Yu et al., 2013). An explanation for this phenomenon is that malignant cells escape from the primary tumor and develop a migratory phenotype, which allows them to enter the circulatory system. In addition, disseminated tumor cells persisting in the bone marrow also undergo EMT in order to re‐enter the bloodstream. Dynamic changes in the epithelial and mesenchymal proportion of CTCs derived from metastatic breast cancer patients were monitored and an association between treatment resistance and the presence of CTCs with mesenchymal features was detected (Yu et al., 2013). Interestingly, we found both, stem cell and EMT markers co‐expressed on single OvCar3 and patient‐derived CTCs, supporting the theory that EMT generates a cell population with stem cell‐like properties (Mani et al., 2008; Morel et al., 2008).
Similarly to the cellular heterogeneity of the primary tumor, CTCs themselves are likely to consist of a heterogeneous cell population. Our analysis of 77 single OvCar3 cells revealed heterogeneous – gene expression patterns for epithelial, stem cell and EMT markers. We additionally observed this heterogeneity between the blood samples (inter‐patient heterogeneity) obtained from our 3 analyzed patients as well as in CTCs of the same patient (intra‐patient and inter‐cellular heterogeneity).
Our results are in line with previous observations, which has already proposed a heterogeneous cell population with regard to their morphology, molecular characteristics and their metastatic potential (Fehm et al., 2010; Krawczyk et al., 2013; Lianidou et al., 2013) Supporting the hypothesis of different CTC subpopulations, which were selected during platinum‐based therapy and may be responsible for drug resistance (Bapat et al., 2005; Dyall et al., 2010). However, it has to be noted that all analyzed CTCs expressed Muc‐1, in contrast to EpCAM or cytokeratins. This could be due to the small number of patients investigated.
To this day there is no data available on EMT or stem cells markers in ovarian cancer CTCs. Therefore, the aim of this study was to establish a multimarker panel to detect and characterize CTCs. In spite of a small patient cohort, our study offers a unique workflow for the isolation, detection and characterization of single CTCs from ovarian cancer patients.
However, defining which cell is ‘THE’ CTC with metastasis initiating capacity and the ability to induce recurrence, remains one of the most essential questions in the CTC research field. We believe that sensitive and more accurate assays for molecular characterization of single cells, as described herein, are the first steps to correlate molecular “snapshots” of single ovarian cancer CTCs with clinically relevant phenotypes. For instance, it was already shown that EpCAM‐negative CTCs with a defined signature (HER2+/EGFR+/HPSE+/Notch1+) are highly invasive and specifically competent for generating brain and lung metastases in a breast cancer model (Zhang et al., 2013). Therefore, our next goal is to perform a long‐time study investigating single CTCs from ovarian cancer patients regarding their expression of epithelial, stem cell and EMT markers before, during and after chemotherapy. By doing this we hope to gain deeper insights into potential changes of their expression profile during treatment, which in the future could help physicians to improve therapeutic intervention.
Limitation of the study: There are multiple techniques available for the quantitative transcriptomic profiling of CTCs. So far, pre‐amplification of the mRNA is needed to quantify its quality and to analyze multiple transcripts (Powell et al., 2012; Ting et al., 2015). This approach is prone for technical errors due to an amplification bias.
The multiplex‐RT‐PCR described herein was designed as a “non‐quantitative” approach to simultaneously detect 19 transcripts without prior pre‐amplification in order to provide a time‐ and cost‐efficient screening tool for longitudinal molecular characterization of CTCs e.g. during chemotherapies. For subsequent quantification of identified therapy‐relevant candidate transcripts RT‐qPCR can be applied by using the same primers.
So far our workflow exclusively focuses on epithelial‐associated markers to enrich for CTCs, which only allows characterization of a restricted set of CTCs. However, we would like to refrain from naming those cells of interest “epithelial CTCs”, since we believe that CTCs captured with EpCAM/CK antibodies do not necessarily exhibit a completely epithelial phenotype. In this context, we and others have already observed co‐expression of mesenchymal/epithelial markers in CTCs enriched by epithelial epitopes, which may represent an “intermediate state” (Aktas et al., 2009; Kasimir‐Bauer et al., 2012; Schneck et al., 2015). This CTC subpopulation expresses epithelial‐associated surface antigens, which makes them accessible for enrichment by EpCAM as well as CK. Nevertheless they may already express EMT or stem cell associated genes which indicate the beginning of EMT. This is in accordance with the hypothesis that EMT stem or epithelial associated CTC traits represent rather a “continuum”, with a high degree of plasticity than a sharply defined phenotypes (Scheel and Weinberg, 2012; Yu et al., 2013).
However, the enrichment strategy used herein is exemplarily and our multiplex assay for CTC detection is designed to allow combinations with any other enrichment strategy (such as selection marker independent filtration or microfluidic separation). The only prerequisite is that cells remain intact during isolation, which excludes e.g. cell permeabilization. In future studies we plan to apply our multiplex panel also downstream of other enrichment strategies, e.g. for the molecular characterization of EpCAM‐negative CTC. We recently published a detection and isolation strategy for such kind of CTCs derived from breast cancer patients (Schneck et al., 2015) and we plan to adapt this method to ovarian cancer.
5. Conclusion
Taken together, we developed a workflow for the detection and gene expression profiling of single CTCs from ovarian cancer patients. Our multiplex‐RT‐PCR is inexpensive, versatile and applicable to upstream CTC enrichment strategies, working in a non‐invasive manner to the cells. This multiplex assay allows the detection of 19 epithelial‐, EMT‐ or tumor stem cell‐associated markers of single cells without pre‐amplification of transcripts or co‐isolation of contaminating non‐malignant cells. Amplicons of all 3 panels can be combined and panels can be extended underscoring the usability and versatility of our technique.
Conflict of interest
All authors declare that they have no conflicts of interest regarding the contents of this manuscript.
Supporting information
The following are the supplementary material related to this article:
Supplementary Material Table 1 Primer sequences, amplicon sizes and labeling for multiplex PCR products.
Supplementary Material Table 2 Patient characteristics Pt. = patient.
Supplementary Material Figure 1 Establishment of a multiplex‐RT‐PCR exemplified for the detection of stem cell markers. All amplicons were tested in single RT‐PCR reactions and multiplex‐RT‐PCRs for up to 8 markers. PCR products were visualized by gel electrophoresis on an agarose gel (3%).
Supplementary Material Figure 2 Detection of stem cell markers in blood from healthy donors after immunomagnetic enrichment with the AdnaTest EMT‐1/StemCell Select. A) Visualized are electropherograms of the stem cell multiplex‐RT‐PCR panel after analysis by capillary electrophoresis. Amplified fragments of the stem cell transcripts CD44 (120 bp), ALDH1A1 (139 bp) and Notch1 (268 bp) are shown. In all control blood samples CD44 and Notch1 could be detected, whereas ALDH1A1 was identified in 3 out of 7. B) Noted is the amount of the amplified transcript ALDH1 in ng/μl after AdnaTest EMT‐1/StemCell Select, which was detected in 2/7 analyzed blood samples from healthy donors. The CTC enriched fraction still contained leucocytes, which interfered with our stem cell panel, as well as with those of the AdnaTest EMT‐1/Stem Cell Detect in blood of healthy donors.
Supplementary Material Figure 3 Detection of EMT markers in blood from healthy donors after immunomagnetic enrichment with the AdnaTest EMT‐1/StemCell Select. A) Visualized are electropherograms of the EMT multiplex‐PCR panel after analysis by capillary electrophoresis. The amplified fragment of Vimentin (170 bp) was detected in all blood samples. B) Noted is the amount of the amplified transcripts PIK3CA, Akt2, TWIST1 and β‐Actin in ng/μl after AdnaTest EMT‐1/StemCell Select. In 3 out of 7 analyzed blood samples Akt2 was detected as positive. The CTC enriched fraction still contained leucocytes, which interfered with our EMT‐panel, as well as with those of the AdnaTest EMT‐1/Stem Cell Detect in blood of healthy donors.
Supplementary Material Figure 4 Expression profiling of single leukocytes analyzed by multiplex‐RT‐PCR for epithelial, EMT and stem cell markers A) Electropherograms of epithelial, EMT and stem cell‐markers exemplified for a single leukocyte. No epithelial markers could be observed, whereas the stem cell marker CD44 and the EMT markers N‐cadherin, Vimentin and Snai2 were detected. B) Expression profile of 10 single leukocytes analyzed by multiplex‐RT‐PCR for epithelial, EMT and stem cell markers. In none of the analyzed leukocytes epithelial markers could be observed, whereas EMT markers were detected in all cases, and stem cell markers in 6 out of 10 cells. C) Detection of CD45 on single leukocytes. CD45 PCR fragments from single leukocytes were visualized with the Bioanalyzer 2100 (Agilent Technologies) and cells could be identified as leukocytes.
Acknowledgments
We would like to thank Katharina Raba (Core Flow Cytometry Facility, University of Duesseldorf) for excellent assistance in flow cytometry/sorting.
This project was funded by Deutsche Forschungsgemeinschaft (DFG, NE805/4‐1).
Supplementary material 1.
1.1.
Supplementary material related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2016.04.002.
Blassl Christina, Kuhlmann Jan Dominik, Webers Alessandra, Wimberger Pauline, Fehm Tanja, Neubauer Hans, (2016), Gene expression profiling of single circulating tumor cells in ovarian cancer – Establishment of a multi‐marker gene panel, Molecular Oncology, 10, doi: 10.1016/j.molonc.2016.04.002.
Footnotes
Pubmed search from 15.01.2016 (January fifteenth two thousand and sixteen), Keywords: ovarian cancer; circulating tumor cells; mesenchymal; EMT; stem cell.
Contributor Information
Christina Blassl, Email: Christina.Blassl@med.uni-duesseldorf.de.
Jan Dominik Kuhlmann, Email: Jan.Kuhlmann@uniklinikum-dresden.de.
Alessandra Webers, Email: al.c.webers@gmail.com.
Pauline Wimberger, Email: Pauline.Wimberger@uniklinikum-dresden.de.
Tanja Fehm, Email: Tanja.Fehm@med.uni-duesseldorf.de.
Hans Neubauer, Email: Hans.Neubauer@med.uni-duesseldorf.de.
References
- Abu-Rustum, N.R. , Chi, D.S. , Curtin, J.P. , 1999. Prognostic analysis of invasive circulating tumor cells (iCTCs) in epithelial ovarian cancer. Curr. Probl. Surg. 36, 1–53. 10.1016/j.ygyno.2014.06.013 [DOI] [PubMed] [Google Scholar]
- Aktas, B. , Kasimir-Bauer, S. , Heubner, M. , Kimmig, R. , Wimberger, P. , 2011. Molecular profiling and prognostic relevance of circulating tumor cells in the blood of ovarian cancer patients at primary diagnosis and after platinum-based chemotherapy. Int. J. Gynecol. Cancer. 21, 822–830. 10.1097/IGC.0b013e318216cb91 [DOI] [PubMed] [Google Scholar]
- Aktas, B. , Tewes, M. , Fehm, T. , Hauch, S. , Kimmig, R. , Kasimir-Bauer, S. , 2009. Stem cell and epithelial–mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 11, R46 10.1186/bcr2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapat, S. , Koppikar, C.B. , Kurrey, N.K. , 2005. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 3025–3029. [DOI] [PubMed] [Google Scholar]
- Barriere, G. , Riouallon, A. , Renaudie, J. , Tartary, M. , Michel, P.R. , 2012. Mesenchymal and stemness circulating tumor cells in early breast cancer diagnosis. BMC Cancer. 12, 114 10.1186/1471-2407-12-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesch, M. , Zeimet, A.G. , Reimer, D. , Schmidt, S. , Parson, W. , Spoeck, F. , Hatina, J. , Wolf, D. , Sopper, S. , 2014. The side population of ovarian cancer cells defines a heterogeneous compartment exhibiting stem cell characteristics. Oncotarget. 5, 7027–7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabletz, T. , 2012. EMT and MET in metastasis: where are the cancer stem cells?. Cancer Cell. 22, 699–701. 10.1016/j.ccr.2012.11.009 [DOI] [PubMed] [Google Scholar]
- Bryder, D. , Rossi, D.J. , Weissman, I.L. , 2006. Hematopoietic stem cells: the paradigmatic tissue-specific stem cell. Am. J. Pathol. 169, 338–346. 10.2353/ajpath.2006.060312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun, J.Y. , Kim, K.J. , Hwang, I.T. , Kim, Y.J. , Lee, D.H. , Lee, I.K. , Kim, J.K. , 2007. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 35, e40 10.1093/nar/gkm051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Bois, A. , Reuss, A. , Pujade-Lauraine, E. , Harter, P. , Ray-Coquard, I. , Pfisterer, J. , 2009. Role of surgical outcome as prognostic factor in advanced epithelial ovarian cancer: a combined exploratory analysis of 3 prospectively randomized phase 3 multicenter trials. Cancer. 115, 1234–1244. 10.1002/cncr.24149 [DOI] [PubMed] [Google Scholar]
- Dyall, S. , Gayther, S.A. , Dafou, D. , 2010. Cancer stem cells and epithelial ovarian cancer. J. Oncol. 2010, 1–9. http://dx.doi.org/10.1155/2010/105269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehm, T. , Banys, M. , Rack, B. , Janni, W. , Marth, C. , Blassl, C. , Hartkopf, A. , Trope, C. , Kimmig, R. , Krawczyk, N. , Wallwiener, D. , Wimberger, P. , Kasimir-Bauer, S. , 2013. Pooled analysis of the prognostic relevance of disseminated tumor cells in the bone marrow of patients with ovarian cancer. Int. J. Gynecol. Cancer. 23, 839–845. 10.1097/IGC.0b013e3182907109 [DOI] [PubMed] [Google Scholar]
- Fehm, T. , Müller, V. , Aktas, B. , Janni, W. , Schneeweiss, A. , Stickeler, E. , Lattrich, C. , Löhberg, C.R. , Solomayer, E. , Rack, B. , Riethdorf, S. , Klein, C. , Schindlbeck, C. , Brocker, K. , Kasimir-Bauer, S. , Wallwiener, D. , Pantel, K. , 2010. HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial. Breast Cancer Res. Treat. 124, 403–412. 10.1007/s10549-010-1163-x [DOI] [PubMed] [Google Scholar]
- FIGO Committee on Gynecologic Oncology, 2009. Current FIGO staging for cancer of the vagina, fallopian tube, ovary, and gestational trophoblastic neoplasia. Int. J. Gynaecol. Obstet. 105, (1) 3–4. 10.1016/j.ijgo.2008.12.015 [DOI] [PubMed] [Google Scholar]
- Galluzzi, L. , Senovilla, L. , Vitale, I. , Michels, J. , Martins, I. , Kepp, O. , Castedo, M. , Kroemer, G. , 2012. Molecular mechanisms of cisplatin resistance. Oncogene. 31, 1869–1883. 10.1038/onc.2011.384 [DOI] [PubMed] [Google Scholar]
- Giordano, A. , Gao, H. , Anfossi, S. , Cohen, E. , Mego, M. , Tin, S. , De, Laurentiis M. , Parker, C.A. , Alvarez, R.H. , Ueno, N.T. , De, Placido S. , Mani, S.A. , Esteva, F.J. , 2013. Epithelial-mesenchymal transition and stem cell markers in patients with HER2-positive metastatic breast cancer. Mol. Cancer Ther. 11, 2526–2534. 10.1158/1535-7163.MCT-12-0460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, M.T. , Howe, H.L. , Tung, K.H. , Hotes, J. , Miller, B.A. , Coughlin, S.S. , Chen, V.W. , 2003. Incidence of ovarian cancer by race and ethnicity in the United States, 1992–1997. Cancer. 97, 2676–2685. 10.1002/cncr.11349 [DOI] [PubMed] [Google Scholar]
- Guarino, M. , 2007. Epithelial-mesenchymal transition and tumour invasion. Int. J. Biochem. Cell Biol. 39, 2153–2160. 10.1016/j.biocel.2007.07.011 [DOI] [PubMed] [Google Scholar]
- Hosonuma, S. , Kobayashi, Y. , Kojo, S. , Wada, H. , Seino, K.I. , Kiguchi, K. , Ishizuka, B. , 2011. Clinical significance of side population in ovarian cancer cells. Hum. Cell. 24, 9–12. 10.1007/s13577-010-0002-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallergi, G. , Papadaki, M.A. , Politaki, E. , Mavroudis, D. , Georgoulias, V. , Agelaki, S. , 2011. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 13, R59 10.1186/bcr2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimir-Bauer, S. , Hoffmann, O. , Wallwiener, D. , Kimmig, R. , Fehm, T. , 2012. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 14, R15 10.1186/bcr3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk, N. , Banys, M. , Hartkopf, A. , Hagenbeck, C. , Melcher, C. , Fehm, T. , 2013. Circulating tumour cells in breast cancer. Ecancermedicalscience. 7, 352 10.3332/ecancer.2013.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann, J.D. , Hein, L. , Kurth, I. , Wimberger, P. , Dubrovska, A. , 2015. Targeting cancer stem cells: promises and challenges. Anticancer Agents Med. Chem. 16, 38–58. [DOI] [PubMed] [Google Scholar]
- Kuhlmann, J.D. , Wimberger, P. , Bankfalvi, A. , Keller, T. , Schöler, S. , Aktas, B. , Buderath, P. , Hauch, S. , Otterbach, F. , Kimmig, R. , Kasimir-Bauer, S. , 2014. ERCC1-positive circulating tumor cells in the blood of ovarian cancer patients as a predictive biomarker for platinum resistance. Clin. Chem. 1–8. 10.1373/clinchem.2014.224808 [DOI] [PubMed] [Google Scholar]
- Lianidou, E.S. , Mavroudis, D. , Georgoulias, V. , 2013. Clinical challenges in the molecular characterization of circulating tumour cells in breast cancer. Br. J. Cancer. 108, 2426–2432. 10.1038/bjc.2013.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani, S.A. , Guo, W. , Liao, M. , Eaton, E.N. , Zhou, A.Y. , Brooks, M. , Reinhard, F. , Zhang, C.C. , Campbell, L.L. , Polyak, K. , Brisken, C. , Yang, J. , Weinberg, R.A. , 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133, 704–715. 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, L.P. , Schilder, R.J. , 2009. Management of recurrent ovarian carcinoma: current status and future directions. Semin. Oncol. 36, 112–125. 10.1053/j.seminoncol.2008.12.003 [DOI] [PubMed] [Google Scholar]
- Mego, M. , Gao, H. , Lee, B.N. , Cohen, E.N. , Tin, S. , Giordano, A. , Wu, Q. , Liu, P. , Nieto, Y. , Champlin, R.E. , Hortobagyi, G.N. , Cristofanilli, M. , Ueno, N.T. , Reuben, J.M. , 2012. Prognostic value of EMT-circulating tumor cells in metastatic breast cancer patients undergoing high-dose chemotherapy with autologous hematopoietic stem cell transplantation. J. Cancer. 3, 369–380. 10.7150/jca.5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, A.P. , Lièvre, M. , Thomas, C. , Hinkal, G. , Ansieau, S. , Puisieux, A. , 2008. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 3, e2888 10.1371/journal.pone.0002888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzer, B. , Medoro, G. , Pasch, S. , Fontana, F. , Zorzino, L. , Pestka, A. , Andergassen, U. , Meier-Stiegen, F. , Czyz, Z.T. , Alberter, B. , Treitschke, S. , Schamberger, T. , Sergio, M. , Bregola, G. , Doffini, A. , Gianni, S. , Calanca, A. , Signorini, G. , Bolognesi, C. , Hartmann, A. , Fasching, P.A. , Sandri, M.T. , Rack, B. , Fehm, T. , Giorgini, G. , Manaresi, N. , Klein, C.A. , 2014. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol. Med. 6, 1371–1386. 10.15252/emmm.201404033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poveda, A. , Kaye, S.B. , McCormack, R. , Wang, S. , Parekh, T. , Ricci, D. , Lebedinsky, C.A. , Tercero, J.C. , Zintl, P. , Monk, B.J. , 2011. Circulating tumor cells predict progression free survival and overall survival in patients with relapsed/recurrent advanced ovarian cancer. Gynecol. Oncol. 122, 567–572. 10.1016/j.ygyno.2011.05.028 [DOI] [PubMed] [Google Scholar]
- Powell, A.A. , Talasaz, A.H. , Zhang, H. , Coram, M.A. , Reddy, A. , Deng, G. , Telli, M.L. , Advani, R.H. , Carlson, R.W. , Mollick, J.A. , Sheth, S. , Kurian, A.W. , Ford, J.M. , Stockdale, F.E. , Quake, S.R. , Pease, R.F. , Mindrinos, M.N. , Bhanot, G. , Dairkee, S.H. , Davis, R.W. , Jeffrey, S.S. , 2012. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 7, e33788 10.1371/journal.pone.0033788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribluda, A. , de la Cruz, C.C. , Jackson, E.L. , 2015. Intratumoral heterogeneity: from diversity comes resistance. Clin. Cancer Res. 21, 2916–2923. 10.1158/1078-0432.CCR-14-1213 [DOI] [PubMed] [Google Scholar]
- Reya, T. , Morrison, S.J. , Clarke, M.F. , Weissman, I.L. , 2001. Stem cells, cancer, and cancer stem cells. Nature. 414, 105–111. 10.1007/978-1-60327-933-8 [DOI] [PubMed] [Google Scholar]
- Rozen, S. , Skaletsky, H. , 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. 10.1385/1-59259-192-2:365 [DOI] [PubMed] [Google Scholar]
- Rubin, S.C. , Randall, T.C. , Armstrong, K.A. , Chi, D.S. , Hoskins, W.J. , 1999. Ten-year follow-up of ovarian cancer patients after second-look laparotomy with negative findings. Obstet. Gynecol. 93, 21–24. 10.1016/S0029-7844(98)00334-2 [DOI] [PubMed] [Google Scholar]
- Scheel, Weinberg, R.A. , 2012. Phenotypic plasticity and epithelial–mesenchymal transitions in cancer and normal stem cells?. Int. J. Cancer. 129, 2310–2314. 10.1002/ijc.26311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneck, H. , Gierke, B. , Uppenkamp, F. , Behrens, B. , Niederacher, D. , Stoecklein, N.H. , Templin, M.F. , Pawlak, M. , Fehm, T. , Neubauer, H. , 2015. EpCAM-independent enrichment of circulating tumor cells in metastatic breast cancer. PLoS One. 10, e0144535 10.1371/journal.pone.0144535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio, R. , Billack, B. , 2011. Potential Tumor Biomarkers for Ovarian Cancer. Researchgate.Net 125 [Google Scholar]
- Shah, M.M. , Landen, C.N. , 2014. Ovarian cancer stem cells: are they real and why are they important?. Gynecol. Oncol. 132, 483–489. 10.1016/j.ygyno.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieuwerts, A.M. , Kraan, J. , Bolt-De Vries, J. , Van Der Spoel, P. , Mostert, B. , Martens, J.W.M. , Gratama, J.W. , Sleijfer, S. , Foekens, J.A. , 2009. Molecular characterization of circulating tumor cells in large quantities of contaminating leukocytes by a multiplex real-time PCR. Breast Cancer Res. Treat. 118, 455–468. 10.1007/s10549-008-0290-0 [DOI] [PubMed] [Google Scholar]
- Silverberg, S.G. , 2000. Histopathologic grading of ovarian carcinoma: a review and proposal. Int. J. Gynecol. Pathol. 19, 7–15. 10.1097/00004347-200001000-00003 [DOI] [PubMed] [Google Scholar]
- Thiery, J.P. , 2002. Epithelial–mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2, 442–454. 10.1038/nrc822 [DOI] [PubMed] [Google Scholar]
- Ting, D.T. , Wittner, B.S. , Ligorio, M. , Jordan, N.V. , Ajay, M. , Miyamoto, D.T. , Aceto, N. , Bersani, F. , Brian, W. , Xega, K. , Ciciliano, J.C. , Zhu, H. , Mackenzie, O.C. , Trautwein, J. , Arora, K.S. , Shahid, M. , Ellis, H.L. , Qu, N. , Bardeesy, N. , Rivera, M.N. , Deshpande, V. , Ferrone, C.R. , Ramaswamy, S. , Shioda, T. , Toner, M. , 2014. Single-cell rna sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 8, 1905–1918. http://dx.doi.org/10.1016/j.celrep.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M. , Bardia, A. , Wittner, B.S. , Stott, S.L. , Smas, M.E. , Ting, D.T. , Isakoff, S.J. , Ciciliano, J.C. , Wells, M.N. , Shah, A.M. , Concannon, K.F. , Donaldson, M.C. , Sequist, L.V. , Brachtel, E. , Baselga, J. , Ramaswamy, S. , Toner, M. , Haber, D.A. , 2013. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 339, (80) 580–584. 10.1126/science.1228522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L. , Liang, X. , Liu, Q. , Yang, Z. , 2015. The predictive value of circulating tumor cells in ovarian cancer. Int. J. Gynecol. Cancer. 1 10.1097/IGC.0000000000000459 [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Ridgway, L.D. , Wetzel, M.A. , Ngo, J. , Yin, W. , Kumar, D. , Goodman, J.C. , Groves, M.D. , Marchetti, D. , 2013. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 5, 10.1126/scitranslmed.3005109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y. , Bian, B. , Yuan, X. , Xie, G. , Ma, Y. , Shen, L. , 2015. Prognostic value of circulating tumor cells in ovarian cancer: a meta-analysis. PLoS One. 10, e0130873 10.1371/journal.pone.0130873 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary material related to this article:
Supplementary Material Table 1 Primer sequences, amplicon sizes and labeling for multiplex PCR products.
Supplementary Material Table 2 Patient characteristics Pt. = patient.
Supplementary Material Figure 1 Establishment of a multiplex‐RT‐PCR exemplified for the detection of stem cell markers. All amplicons were tested in single RT‐PCR reactions and multiplex‐RT‐PCRs for up to 8 markers. PCR products were visualized by gel electrophoresis on an agarose gel (3%).
Supplementary Material Figure 2 Detection of stem cell markers in blood from healthy donors after immunomagnetic enrichment with the AdnaTest EMT‐1/StemCell Select. A) Visualized are electropherograms of the stem cell multiplex‐RT‐PCR panel after analysis by capillary electrophoresis. Amplified fragments of the stem cell transcripts CD44 (120 bp), ALDH1A1 (139 bp) and Notch1 (268 bp) are shown. In all control blood samples CD44 and Notch1 could be detected, whereas ALDH1A1 was identified in 3 out of 7. B) Noted is the amount of the amplified transcript ALDH1 in ng/μl after AdnaTest EMT‐1/StemCell Select, which was detected in 2/7 analyzed blood samples from healthy donors. The CTC enriched fraction still contained leucocytes, which interfered with our stem cell panel, as well as with those of the AdnaTest EMT‐1/Stem Cell Detect in blood of healthy donors.
Supplementary Material Figure 3 Detection of EMT markers in blood from healthy donors after immunomagnetic enrichment with the AdnaTest EMT‐1/StemCell Select. A) Visualized are electropherograms of the EMT multiplex‐PCR panel after analysis by capillary electrophoresis. The amplified fragment of Vimentin (170 bp) was detected in all blood samples. B) Noted is the amount of the amplified transcripts PIK3CA, Akt2, TWIST1 and β‐Actin in ng/μl after AdnaTest EMT‐1/StemCell Select. In 3 out of 7 analyzed blood samples Akt2 was detected as positive. The CTC enriched fraction still contained leucocytes, which interfered with our EMT‐panel, as well as with those of the AdnaTest EMT‐1/Stem Cell Detect in blood of healthy donors.
Supplementary Material Figure 4 Expression profiling of single leukocytes analyzed by multiplex‐RT‐PCR for epithelial, EMT and stem cell markers A) Electropherograms of epithelial, EMT and stem cell‐markers exemplified for a single leukocyte. No epithelial markers could be observed, whereas the stem cell marker CD44 and the EMT markers N‐cadherin, Vimentin and Snai2 were detected. B) Expression profile of 10 single leukocytes analyzed by multiplex‐RT‐PCR for epithelial, EMT and stem cell markers. In none of the analyzed leukocytes epithelial markers could be observed, whereas EMT markers were detected in all cases, and stem cell markers in 6 out of 10 cells. C) Detection of CD45 on single leukocytes. CD45 PCR fragments from single leukocytes were visualized with the Bioanalyzer 2100 (Agilent Technologies) and cells could be identified as leukocytes.


