Abstract
It is well established that lncRNAs are aberrantly expressed in cancer where they have been shown to act as oncogenes or tumor suppressors. RNA profiling of 314 colorectal adenomas/adenocarcinomas and 292 adjacent normal colon mucosa samples using RNA‐sequencing demonstrated that the snoRNA host gene 16 (SNHG16) is significantly up‐regulated in adenomas and all stages of CRC. SNHG16 expression was positively correlated to the expression of Wnt‐regulated transcription factors, including ASCL2, ETS2, and c‐Myc. In vitro abrogation of Wnt signaling in CRC cells reduced the expression of SNHG16 indicating that SNHG16 is regulated by the Wnt pathway. Silencing of SNHG16 resulted in reduced viability, increased apoptotic cell death and impaired cell migration. The SNHG16 silencing particularly affected expression of genes involved in lipid metabolism. A connection between SNHG16 and genes involved in lipid metabolism was also observed in clinical tumors. Argonaute CrossLinking and ImmunoPrecipitation (AGO‐CLIP) demonstrated that SNHG16 heavily binds AGO and has 27 AGO/miRNA target sites along its length, indicating that SNHG16 may act as a competing endogenous RNA (ceRNA) “sponging” miRNAs off their cognate targets. Most interestingly, half of the miRNA families with high confidence targets on SNHG16 also target the 3′UTR of Stearoyl‐CoA Desaturase (SCD). SCD is involved in lipid metabolism and is down‐regulated upon SNHG16 silencing. In conclusion, up‐regulation of SNHG16 is a frequent event in CRC, likely caused by deregulated Wnt signaling. In vitro analyses demonstrate that SNHG16 may play an oncogenic role in CRC and that it affects genes involved in lipid metabolism, possible through ceRNA related mechanisms.
Keywords: Colorectal cancer, Wnt pathway, Long non-coding RNAs, SNHG16, AGO-CLIP, Functional analyses
Highlights
SNHG16 is highly up‐regulated in adenomas and all stages of colorectal cancer.
SNHG16 is regulated by the Wnt signaling pathway in colorectal cancer.
SNHG16 is localized in the cytoplasm and associated with polysomes.
Silencing of SNHG16 increases apoptotic death and impairs cell migration.
SNHG16 heavily binds AGO and contains 27 AGO/miRNA target sites.
Abbreviations
- ACLY
ATP citrate lyase
- AGO
Argonaute
- AGO-CLIP
Argonaute Crosslinking and ImmunoPrecipitation
- AGO-PAR-CLIP
Argonaute PhotoActivatable-Ribonucleoside-enhanced CrossLinking and ImmunoPrecipitation
- ASCL2
Achaete-Scute Complex-Like 2
- ceRNA
competing endogenous RNA
- ChIP
Chromatin Immunoprecipitation
- CLIP
CrossLinking and ImmunoPrecipitation
- c-Myc
v-myc avian myelocytomatosis viral oncogene
- CRC
colorectal cancer
- EDTA
EthyleneDiamineTetraacetic Acid
- ELAVL1
Embryonic Lethal, Abnormal Vision, Drosophila-like 1
- ETS2
V-Ets Erythroblastosis Virus E26 Oncogene
- FC(s)
Fold Change(s)
- FPKM
fragment per kilobase of exon per million mapped sequence reads
- GAPDH
Glyceraldehyde 3-Phosphate Dehydrogenase
- HNRNPA1
Heterogeneous Nuclear RiboNucleoProtein A1
- HuR
Hu antigen R
- IP
immunoprecipitation
- IPA
Ingenuity Pathway Analysis
- IPKB
Ingenuity Pathways Knowledge Base
- LDH
lactate dehydrogenase
- lncRNA
long non-coding RNA
- MeV
Multi Experiment Viewer
- miRNA
microRNA
- MSI
microsatellite instable
- MSS
microsatellite stable
- MTT
3-[4,5-dimethylthiazol-2-yl]-2.5-diphenyltetrazolium bromide
- ncRNA
non-coding RNA
- Nek2
NIMA-related kinase 2
- NMD
nonsense-mediated decay
- NR3C1
Nuclear Receptor Subfamily 3, Group C, Member 1
- ORF(s)
open reading frame(s)
- PCSK9
Proprotein Convertase Subtilisin/Kexin Type 9
- RIN
RNA Integrity Number
- PBS
Phosphate Buffered Saline
- Seq
sequencing
- RNAseq
RNA sequencing
- RXRA
Retinoic X Receptor, Alpha
- SCD
Stearoyl-CoA Desaturase
- SNHG16
snoRNA host gene 16
- snoRNA
small nucleolar RNA
- scr
scrambled
- sd
standard deviation
- TF
transcription factor
- TNM
Tumor, Node, Metastasis
- TSS
transcription start site
- UBC
Ubiquitin
- URMs
U-rich motifs
- vs
versus
1. Introduction
Colorectal cancer (CRC) is the third most common malignant disease and the fourth most common cause of cancer death worldwide (Haggar and Boushey, 2009). The molecular alterations in CRC have been intensively studied in order to discover diagnostic and prognostic markers (reviewed in (Grady and Pritchard, 2014)). Among others, expression profiling has been widely used to identify differentially expressed genes with prognostic and diagnostic implications. However, at present none of these have been translated into clinical practice and consequently, there is still a need for further molecular characterization and classification of CRC.
It has recently become clear that most of the genome is transcribed into RNA, although the classical protein‐coding mRNAs only account for approximately 2% of the genome (Djebali et al., 2012). Hence non‐coding RNAs (ncRNA) including the classical house‐keeping ncRNAs (i.e. ribosomal RNA (rRNA), transfer RNAs (tRNA), small nuclear RNA (snRNA) and small nucleolar RNAs (snoRNAs)), microRNAs (miRNAs) and the more recently discovered long non‐coding RNAs (lncRNAs) make up a large fraction of the encoded transcripts. Apart from the house‐keeping ncRNAs, the vast majority of ncRNAs are lncRNAs (>200 bases). Many lncRNAs share common characteristics with protein‐coding transcripts i.e. they are transcribed by RNA polymerase II, spliced using canonical splice site motifs and frequently poly‐adenylated at the 3′‐end (Derrien et al., 2012; Prensner and Chinnaiyan, 2011). Functional characterization of lncRNAs has shown that they carry out various biological functions such as transcriptional regulation (cis/trans), titration of miRNAs (competing endogenous RNA (ceRNA)) or proteins (molecular decoys) and bridging of proteins or chromatin regions (scaffolds) (reviewed in (Ulitsky and Bartel, 2013; Wang and Chang, 2011)). Through the above different mechanisms of action, lncRNAs are involved in the regulation of numerous biological processes, including cell cycle, apoptosis, histone modifications, chromosome imprinting and cell differentiation (Wang and Chang, 2011). Accordingly, profiling of lncRNAs has revealed that they are deregulated in various types of cancer, suggesting their potential as cancer biomarkers or therapeutic targets (Gibb et al., 2011). Few studies have dealt with the overall expression of lncRNAs in CRC (Gibb et al., 2015; Hu et al., 2014). Nevertheless several lncRNAs such as CRNDE, HOTAIR, MALAT‐1, PCAT1 and PTENP1, have been shown to be dysregulated in CRC (Ge et al., 2013; Graham et al., 2011; Johnsson et al., 2013; Kogo et al., 2011; Xu et al., 2011) (reviewed by (Ragusa et al., 2015)). SNHG16 (also named non‐coding RNA expressed in aggressive neuroblastoma (ncRAN)) (Entrez gene ID: 100507246) was originally identified as an oncogene in neuroblastoma. Increased levels of SNHG16 expression has been reported to associated with poor patient outcome in neuroblastoma (Yu et al., 2009) and with invasiveness of bladder cancer (Zhu et al., 2011). On the contrary, reduced SNHG16 expression was recently demonstrated to be associated with poor prognosis in colorectal cancer (Qi et al., 2015). In line with the analysis in clinical samples SNHG16 exhibited oncogenic phenotypes in bladder and neuroblastoma cell lines and tumor suppressor like phenotypes in CRC cell lines in vitro (Qi et al., 2015; Yu et al., 2009; Zhu et al., 2011). Finally, silencing of SNHG16 improved chemotherapy sensitivity of bladder cells lines suggesting that targeting of SNHG16 may improve chemoresponse in bladder cancer patients (Zhu et al., 2011).
The main objectives of the current study were to investigate the expression of SHNG16 in a large cohort of colorectal adenomas and CRCs (n > 300), to elucidate the mechanism behind SNHG16 deregulation, and to investigate the functional role of SNHG16 deregulation. We found SNHG16 up‐regulated in the vast majority of CRCs, similarly to what has been reported for neuroblastoma and bladder cancer but contrary to the previous finding in CRC. Our in vitro analyses indicate that SNHG16 is positively regulated by the Wnt pathway and plays an oncogenic role in CRC among others through de‐regulation of genes involved in lipid metabolism. Notably, Argonaute CrossLinking and ImmunoPrecipitation (AGO‐CLIP) data suggest that SNHG16 may act as a ceRNA targeting up to 26 miRNA families.
2. Materials and methods
Details are provided in Supplemental Materials and Methods.
2.1. Ethics statement
The use of the human tissue samples for research purpose was approved by the Central Denmark Region Committees on Biomedical Research Ethics (DK; 1999/4678). Informed written consent was given by all participants.
2.2. Clinical samples and cell lines
A total of 281 fresh frozen microsatellite stable (MSS) or microsatellite instable (MSI), primary stage I–IV (T2‐4, N0‐3, M0/1) CRCs, 33 adenomas and 292 adjacent normal colon mucosa samples (of which 290 were matched to an included tumor i.e. collected from the same resected specimen) selected from the colorectal cancer biobank at the Department of Molecular Medicine, Aarhus University Hospital, Skejby, Denmark were used for largeRNA sequencing (largeRNAseq) (largeRNAseq cohort) (patients and sample characteristics are summarized in Supplemental Table 1). Among this cohort a subpopulation of 102 MSI and MSS primary stage I–IV (T2‐4, N0‐3, M0/1), 5 adenomas and 151 normal colon mucosa samples were used for smallRNA sequencing (smallRNAseq) (smallRNAseq cohort) (patients and sample characteristics in Supplemental Table 2). Finally, a cohort consisting of 44 MSS and MSI primary stage I–IV(T2‐4, N0‐3, M0/1) CRCs, 39 adenomas and 20 normal colon mucosa samples were used for validation (patients and sample characteristics in Supplemental Table 3). Patients who had received preoperative chemotherapy and/or radiation of rectal cancers were excluded. Postoperatively the tumors were histologically classified and staged according to the pTNM system. Cases with hereditary colorectal cancer syndromes were not included in the study.
Cell lines, growth conditions and cell line authentication can be found in Supplemental Material and Methods.
2.3. Isolation of RNA from clinical samples and cell lines
RNA from clinical samples was isolated as follows. Large RNAs (>200 bases) were isolated from fresh frozen tissue sample using RNeasy Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The small RNAs (<200 bases) were recovered from the flow‐through fraction using RNeasy Micro Kit together with the RNeasy MinElute spin columns (Qiagen). Total RNA from cell lines harvested with Qiazol (Qiagen) was purified using miRNeasy mini kit (Qiagen) according to the manufacturer's instructions.
2.4. mRNA/ncRNA profiling in clinical samples using RNAseq
The mRNA/ncRNA expression profiling was performed using RNA sequencing (RNAseq). In brief, 500 ng of fragmented large RNA (fragment size 200 bases) was used for largeRNAseq (>200 bases). Subsequently, the TruSeq RNA sample preparation Kit v2 (Illumina, San Diego, CA, USA) was used to generate paired‐end and indexed libraries. The small RNAseq libraries were generated using small RNAs (<200 bases) and the TruSeq small RNA sample preparation Kit (Illumina). The RNAseq libraries were loaded on a TruSeq PE v3 flowcells (Illumina) and amplified with TrueSeq PE Cluster Kit v3 on a cBot (automated cluster generation system) (Illumina). Finally indexed paired‐end sequencing was carried out on a HiSeq 2000 using TruSeq SBS Kit v3 chemistry (Illumina). Fastq files were generated using Illuminas CASAVA software (v1.7) stripped from adapters using AdapterRemoval (v1.2) (Lindgreen, 2012) and overlapping read pairs were joined. The reads were processed using the Tuxedo Suite, consisting of Tophat (v2.0.10), Bowtie (v2.1.0.0) and Cufflinks (v2.0.2) using default settings without passing gene annotations to Tophat (Trapnell et al., 2012). RNA statistics were generated using SAMtools (v0.1.19.0) and Picard (v1.96). Cufflinks (v2.0.2) was used to assemble transcripts and call their relative expression valued based on GENCODE (v15) annotations stripped for pseudogenes. Expression was quantified as fragments per kilobase of exon per million mapped sequence reads (FPKM). Isoform specific expression analysis using Tophat was based on the Ensemble human transcriptome annotation (v15). A gene or isoform was considered expressed in a given sample if log2(FPKM+1) was above 2.
2.5. mRNA/ncRNA profiling of clinical samples using microarrays
The NimbleGen HD2‐12 platform (135K 60mer probes) (Roche NimbleGen, Madison, WI, USA) was used to design custom made microarrays containing probes against 29,291 non‐coding transcripts and 6856 protein‐coding transcripts. Further details about the microarray design, the sample preparation, array normalization and expression analysis have been described previously (Nielsen et al., 2014).
2.6. mRNA/ncRNA profiling of HCT116 cells transfected with SNHG16 siRNAs using RNAseq
HCT116 cells (5 × 105) were reverse transfected with 20 nM of either SNHG16 siRNA_1, siRNA_2 or the negative control siRNA (Scrambled (Scr)) in triplicates for 48 h. The sequences of the SNHG16 siRNAs and Scr (Gene Pharma, Shanghai, China) are listed in the Supplemental Table 4 and their position in SNHG16 is shown in Supplemental Figure 1A. Subsequently, the cells were harvested and RNA (>200 bases) was isolated from cell pellets using the RNeasy Mini Kit (Qiagen). The RNA concentration was determined using RiboGreen quantification (Quant‐iT™ RiboGreen® RNA Assay Kit (Invitrogen)). Prior to RNA library construction ribosomal RNA was removed from total‐RNA preparations using the Ribo‐Zero Magnetic Gold Kit (Human/Mouse/Rat, Epicentre, Madison, WI, USA) in combination with the Ribo‐Zero Magnetic Meta‐Bacteria Kit (Epicentre). Subsequently, paired‐end and indexed RNASeq libraries were synthesized using the ScriptSeq v2 kit (Epicentre) and enriched using the FailSafe PCR Enzyme, according to the manufactures recommendations. The RNASeq libraries were loaded into TruSeq PE v3 flowcells (Illumina) on an Illumina cBot followed by indexed paired‐end sequencing on an Illumina HiSeq 2000 using TruSeq SBS Kit v3 chemistry.
2.7. mRNA/ncRNA and snoRNA RT‐qPCR
The TaqMan Assay ID's (Applied Biosystems, Life Technologies, Foster City, CA, USA) and the custom made primers/probes used for RT‐qPCR are listed in Supplemental Table 5.
2.8. c‐Myc knockdown and Wnt pathway model systems
HCT116 cells (5 × 105) were reverse transfected with 50 nM of c‐Myc siRNAs and Scr (Supplemental Table 4) for 48 h. Inactivation of the Wnt signaling pathway in DLD1 cells by knockdown of β‐catenin or over‐expression of dominant negative TCF1 (dnTCF1) or dominant negative TCF4 (dnTCF4) in DLD1 and subsequent isolation of RNA were carried out as described previously (Thorsen et al., 2011). The DLD1 dnTCF1 and dnTCF4 cell line models were a kind gift from Dr Hans Clevers The Hubrect Laboratory, Utrecht, The Netherlands. The over‐expression of c‐Myc in human immortalized fibroblasts (BJ/hTERT) and the subsequent RNA expression profiling using custom‐made microarrays have been described previously (Gingold et al., 2014).
2.9. Polysome analysis
HCT116 cells were grown to 70% confluence followed by incubation with cycloheximide (100 μg/ml) at 37 °C for 10 min to inhibit protein synthesis. Subsequently, the cells were lysed and loaded onto a sucrose gradient (10–56%) and centrifuged at 40,000 rpm for 2.5 h to separate actively translated RNA (polysome bound) and untranslated RNA (free RNA). Finally, the sucrose gradient was fractionated and total RNA was isolated from each fraction. The RNA was used to determine the expression of SNHG16 and c‐Myc using RT‐qPCR (Supplemental Table 5). Upon RNA isolation the 28s/18s ratios (agarose gel analysis) were used to determine the fractions containing free RNA (no ribosomes and) (28s/18s ≠ 2) and fractions with polysome bound RNA (28s/18s = 2). As a control, an EDTA (EthyleneDiamineTetraacetic Acid) release experiment was carried out in which the lysis buffer was supplemented with 25 mM EDTA (pH 8.0).
2.10. Cell fractionation
The Protein And RNA Isolation System (PARIS) (Ambion, Life Technologies, Foster City, CA, USA) was used to partition HCT116 and SW480 cells into cytoplasmic and nuclear fractions prior to isolation of RNA according to the manufacturer's instructions. Briefly, 5 × 106 HCT116 and SW480 cells were harvested and re‐suspended in cell fractionation buffer followed by low speed centrifugation (500 × g) at 4 °C for 5 min. Finally, RNA was isolated from the supernatant (cytoplasmic fraction) and the pellet (whole nuclei) as described by the manufacturer. In parallel RNA was isolated from 5 × 106 unfractionated cells (total cell).
2.11. Cell viability/death, apoptosis and real‐time analysis
Cell viability/proliferation was measured using 3‐[4,5‐dimethylthiazol‐2‐yl]‐2.5‐diphenyltetrazolium bromide (MTT) assay (Roche Applied Science, Penzberg, Germany). Cellular death (Lactate Dehydrogenase (LDH) activity) was measured using the Cytotoxicity Detection KitPLUS(LDH) (Roche Applied Science). The Caspase 3/7 activity assay was used to measure apoptotic death and performed mainly as described previously (Ostenfeld et al., 2005). The xCELLigence system (Roche Applied Science) was used for real‐time monitoring of cell proliferation and migration (Atienza et al., 2006). The sequences of the SNHG16 siRNAs and the negative control siRNA (Scr) (GenePharma) are listed in the Supplemental Table 4.
2.12. Ingenuity Pathway Analysis
Ingenuity Pathway Analysis (IPA) software (IPA, QIAGEN, Redwood city, CA, USA) was used to gain insight into the overall biological changes introduced upon ectopic knockdown of SNHG16 using siRNAs. Filtered RNAseq data were uploaded to IPA. Using the Ingenuity Pathways Knowledge Base (IPKB) each gene was linked to specific functions, pathways and diseases and an enrichment analysis was performed examining whether the data were enriched for genes associated with a particular function.
2.13. HuR immunoprecipitation (HuR‐IP)
HCT116 cells were hypertonically lysed and centrifuged. A fraction of the supernatant was collected as input control (RNA‐Input). The remaining part was subjected to immunoprecipitation by incubation with monoclonal Hu antigen R (HuR) antibody (Sc5261, Santa Cruz)‐bound Protein G‐coupled Magnetic Dynabeads slurry (Life technologies) following the manufacturer's recommendation. Anti‐FLAG immunoprecipitation was done in parallel as a negative control (antibody F1804, Sigma). Total RNA was isolated from the RNA‐Input fractions and the immunoprecipitated fractions (RNA‐IP) (HuR‐IP or FLAG‐IP) using QIAZol (Qiagen).
2.14. Motif enrichment analysis
A motif enrichment analysis tool, Regmex, capable of using regular expression, was used to analyze whether the differentially expressed genes in the siRNA analysis contained motifs (words) with perfect match(es) to the seed regions of siRNA_1 and siRNA_2 (Nielsen et al., 2016). In brief, Regmex evaluates enrichment of motifs in ranked lists of sequences by calculating a per sequence p‐value for finding the observed number of motifs or more in a sequence, using a Markov chain embedding approach. Sequences were ranked by fold change (FC) of expression, and occurrences of motifs (siRNA_1 and siRNA_2 seed regions) were correlated with gene ranks. To identify additional common motifs in the genes affected by SNHG16 silencing, the above analysis was repeated for all 16384 possible 7‐mers. The 7‐mers occurring in the TOP 100 of correlating 7‐mers in both knockdown experiments were selected (n = 40) (Supplemental Table 6). Out of these, five 7‐mers overlapped with 6 bases (CCTCAGC, CTCAGCC, TCAGCCT, CAGCCTG and AGCCTGG) and two 7‐mers (AGGCTGG and CAGGCTG) overlapped with 6 bases and had one mismatch (mismatch underlined), thus defining a 7‐mer consensus sequence (CAG(C/G)CTG). All the 7‐mers associated with the motif correlate with expression in a manner that down‐regulated genes are enriched for the presence of the motif in their 3′UTR sequences. Furthermore, the 7‐mer consensus sequence was identified at two positions in the reverse complement sequence of SNHG16.
2.15. AGO‐CLIP target analysis
AGO‐CLIP coupled to high throughput sequencing (Chi et al., 2009; Hafner et al., 2010) (AGO‐CLIP‐seq) defines global miRNA binding activity in the cell. PhotoActivatable‐Ribonucleoside‐enhanced CrossLinking and ImmunoPrecipitation of Argonaute (AGO‐PAR‐CLIP) is a subset of AGO‐CLIP methods that allows improved target identification through induction of T → C transitions in bound RNA reads. CrossLinking and ImmunoPrecipitation (CLIP) data was mined in a manner similar to that described previously (Hamilton et al., 2013) with minor modifications. To determine putative miRNAs bound to SNHG16, high confidence miRNA clusters (atlas occurrences ≥3, conserved miRNA families) (Hamilton et al., 2013) on SNHG16 were mined from the atlas. Cognate targets of these miRNAs were subsequently mined to predict the potential SNHG16 ceRNA target spectrum. CLIP binding data was mapped using integrative genomics viewer (Robinson et al., 2011) and Circos (Krzywinski et al., 2009).
2.16. Statistical analysis
The significance of mRNA/ncRNA expression changes were analyzed using the Mann Whitney U test in the Multi Experiment Viewer (MeV) array analyzer software (v4.9.0) (Saeed et al., 2006). Student's unpaired t‐test (two‐tailed) was applied to compare siRNA induced changes with respective controls in the MTT assay, LDH assay, Caspase 3/7 assay and RT‐qPCR analysis. The Student's paired t‐test was used for paired analysis of the expression of SNHG16, SnoRD1A and snoRD1C. Whereas the Student's unpaired t‐test was used to compare expression of SNHG16 in normal mucosa to adenomas and adenocarcinomas. The significance of the enrichment analyses was evaluated using Fisher's exact test. Spearman's correlation was used to measure the strength and direction of associations between SNHG16, c‐Myc and other selected transcription factors in clinical samples. A p‐value <0.05 was considered statistically significant.
3. Results
3.1. SNHG16 up‐regulation is an early event in colorectal cancer
RNA profiling of colorectal adenomas/adenocarcinomas and matched adjacent normal colon mucosa, using RNAseq (characteristics largeRNAseq cohort in Supplemental Table 1) demonstrated that SNHG16 was significantly up‐regulated in 70% (208/290) of the adenomas and adenocarcinomas pairs (FClog2 > 1, p < 0.001) (Table 1). The mean expression of SNHG16 in adenomas and all stages of CRC (n = 314) was also significantly higher than the mean expression in adjacent normal mucosa (n = 292) ((FC)log2 >1, p < 0.001) (Figure 1A). SNHG16 was among the TOP25 up‐regulated lncRNAs (Supplemental File 1 – which shows the TOP25 up‐ and down regulated lncRNAs). Furthermore, SNHG16 was up‐regulated at comparable levels in MSS and MSI CRCs (Figure 1B). According to the Ensemble human transcriptome annotation (v15) the SNHG16 locus potentially encodes 8 different transcript isoforms (Supplemental Figure 1B). To investigate which isoforms are expressed in CRC we applied TopHat to the largeRNAseq data. The analysis revealed that only a single transcript (ENST00000493536) was expressed in more than 10% of the samples, indicating that this is the primary SNHG16 transcript in CRC (Supplemental Figure 1B). Intronically the SNHG16 locus encode three snoRNAs (snoRD1A, snoRD1B and snoRD1C) (Supplemental Figure 1A) and their expression levels were estimated by small RNAseq (characteristics smallRNAseq cohort in Supplemental Table 2). In contrast to SNHG16 which was up‐regulated in 70% of the tumors and unchanged in the rest, the expression of the snoRNAs was much more variable. Paired analysis of matched tumor and normal mucosa samples showed the snoRNAs to be up‐regulated in some tumors (ranging from 14 to 55%) and down regulated (ranging from 7 to 28%) and unchanged in others (ranging from 38 to 57%) (Table 1). A recent study has shown that snoRNAs may be regulated independently of their host gene via alternative splicing and nonsense‐mediated decay (NMD), which may explain the differential expression of SNHG16 and the snoRNAs (Lykke‐Andersen et al., 2014). Given that the snoRNAs were only differentially expressed in a small subset of the CRC samples, all subsequent analyses were focused on SNHG16. The SNHG16 up‐regulation was validated in an independent cohort (20 normal mucosa, 39 adenomas and 44 adenocarcinomas) profiled using a custom‐made expression‐array platform (Supplemental Table 3 and Supplemental Figure 2A) and in a subset of 6 normal colon mucosa and 8 CRCs from the validation cohort using RT‐qPCR (Supplemental Figure 2B). All together the SNHG16 expression analysis demonstrates that SNHG16 up‐regulation is a frequent and early occurring event in CRC.
Table 1.
Expression analysis of SNHG16 and snoRNAs in paired samples.a
| Gene name | Gene id | Pairs with FC(log2) >1.0 (%) | Pairs with FC(log2) < −1.0 (%) | Pairs with −1.0 < FC(log2) < 1.0 (%) | Total number of pairs | FC(log2) all samples | Corrected p‐valueb |
|---|---|---|---|---|---|---|---|
| SNHG16 | ENSG00000163597.9 | 208 (72) | 0 (0) | 82 (28) | 290 | 1.24 | <0.001 |
| snoRD1A | ENST00000364968.1 | 40 (55) | 5 (7) | 28 (38) | 73 | 1.13 | <0.001 |
| snoRD1B | ENSG00000199961.1 | 14 (14) | 28 (28) | 57 (58) | 99 | −0.34 | 0.006 |
| snoRD1C | ENSG00000200185.1 | 25 (30) | 10 (12) | 47 (57) | 82 | 0.57 | <0.001 |
SNHG16 was measured in the largeRNAseq cohort, and the snoRNAs in the smallRNAseq cohort. Only pairs with expression of the respective genes in both the normal and the paired tumor sample were included in the analysis.
Student's paired t‐test. Bonferroni corrected p‐values.
Figure 1.
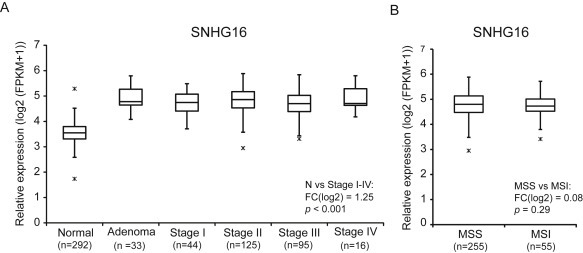
SNHG16 expression in CRC (largeRNAseq cohort). Expression was up‐regulated in adenomas and all stages of adenocarcinomas when compared to normal colorectal mucosa (A). Equal expression in microsatellite stable (MSS) and unstable (MSI) colorectal cancers (B).
3.2. SNHG16 is regulated by the Wnt pathway and c‐Myc in CRC
The up‐regulation of SNHG16 has previously been linked to the amplification of the SNHG16 (17q25) and the MYCN (2p24) locus in neuroblastoma (Yu et al., 2009). In house copy number analyses on a subset of our CRC samples showed no correlation between 17q25 gain and up‐regulation of SNHG16 (data not shown). Additionally, if amplification of the SNHG16 locus was a driving mechanism one would also expect neighboring genes to be frequently up‐regulated. However, the flanking genes were all significantly down‐regulated (Supplemental Figure 3). Taken together this indicates that amplifications are not driving the SNHG16 up‐regulation observed in CRC. Furthermore, the expression of N‐Myc, encoded by the MYCN locus, was generally very low and did not correlate with expression of SNHG16 (data not shown). Hence, we speculated that other transcription factors must be responsible for the up‐regulation of SNHG16 in CRC. A recent study identified global binding patterns of transcription factors (TFs) in the human CRC cell line LoVo using high‐throughput chromatin immunoprecipitation (ChIP) combined with DNA sequencing (ChIP‐seq) (Yan et al., 2013). Interestingly, a cluster of 55 TFs binding close to the SNHG16 transcription start site (TSS) (Chr17:72,064,795–72,066,401, Hg18) was identified. Moreover, 18 of these TFs were significantly dysregulated in the CRC largeRNAseq cohort including several targets of the Wnt pathway (Table 2). Most strikingly, SNHG16 was positively correlated to the expression of the Wnt targets c‐Myc (v‐myc avian myelocytomatosis viral oncogene), ASCL2 (Achaete‐Scute Complex‐Like 2) and ETS2 (V‐Ets Erythroblastosis Virus E26 Oncogene) (Table 2). The strongest correlation was found to c‐Myc (Spearman's ρ = 0.4) (Figure 2A). Like N‐Myc, c‐Myc belongs to the MYC family of transcription factors (Zimmerman and Alt, 1990). Using the ENCODE open chromatin TFBS by ChIP‐seq data, we also found strong and common binding of c‐Myc to the SNHG16 promoter in cell lines from various tissues (Supplemental Figure 4). Interestingly, SNHG16 was also negatively correlated to RXRA (Retinoic X Receptor, Alpha), a negative regulator of the Wnt pathway and to NR3C1 (Nuclear Receptor Subfamily 3, Group C, Member 1), which is known to be silenced by hypermethylation in CRC (Table 2) (Dillard and Lane, 2007; Li et al., 2015; Lind et al., 2006). To investigate if SNHG16 expression is regulated by the Wnt pathway we abrogated the pathway in two CRC model systems and correlated this to the expression of SNHG16. In one system the Wnt‐pathway is abrogated by knockdown of β‐catenin in the other by over‐expression of dominant negative forms of TCF1 (dnTCF1) or TCF4 (dnTCF4) (Thorsen et al., 2011). Indeed, siRNA knockdown of β‐catenin in the colon cancer cell line DLD1 resulted in reduced expression of SNHG16 as well as c‐Myc (Figure 2B). Likewise, SNHG16 was down‐regulated in DLD1 cells upon induction of dnTCF1 or dnTCF4 (Figure 2C). To specifically analyze the role of c‐Myc in the regulation of SNHG16 expression, c‐Myc was knocked‐down in HCT116 cells using two different siRNAs. Both siRNAs resulted in 85% knockdown of c‐Myc and a simultaneous 60% knockdown of SNHG16 (Figure 2D). Finally, up‐regulation of SNHG16 was demonstrated in immortalized human fibroblasts (BJ/hTERT) upon c‐Myc over‐expression (Supplemental Figure 5) mimicking the c‐Myc up‐regulation seen in colorectal cells after Wnt‐activation. All together the above results strongly indicate that the transcriptional activity of the SNHG16 locus in CRC is controlled by Wnt pathway regulated transcription factors, including c‐Myc.
Table 2.
Differential expression of SNHG16 promotor binding TFs and their correlation to the expression of SNHG16 in CRC clinical samples.
| Gene name | Gene id | Wnt targeta | FC(log2) | rs SNHG16 N + A + C | rs SNHG16 A + C | rs SNHG16 N |
|---|---|---|---|---|---|---|
| MYC | ENSG00000136997.10 | + | 2.1 | 0.8 | 0.4 | 0.4 |
| ASCL2 | ENSG00000183734.4 | + | 3.3 | 0.7 | 0.2 | 0.4 |
| ETS2 | ENSG00000157557.7 | + | 1.2 | 0.6 | 0.2 | 0.2 |
| MYBL2 | ENSG00000101057.11 | − | 1.6 | 0.6 | 0.03 | 0.4 |
| E2F7 | ENSG00000165891.11 | − | 0.8 | 0.6 | 0.1 | 0.3 |
| CEBPB | ENSG00000172216.4 | − | 1.4 | 0.6 | 0.02 | 0.2 |
| RARG | ENSG00000172819.12 | + | 0.9 | 0.5 | 0.05 | 0.4 |
| GMEB2 | ENSG00000101216.6 | − | 0.5 | 0.5 | 0.03 | 0.3 |
| ETV7 | ENSG00000010030.9 | − | 1.1 | 0.5 | 0.07 | 0.1 |
| RFX5 | ENSG00000143390.13 | − | 0.6 | 0.5 | 0.1 | 0.2 |
| SMC1A | ENSG00000072501.12 | − | 0.7 | 0.5 | 0.1 | 0.2 |
| TBX3 | ENSG00000135111.10 | + | 1.0 | 0.5 | 0.1 | 0.002 |
| TP73 | ENSG00000078900.9 | − | 0.6 | 0.5 | −0.05 | 0.2 |
| HOXA10 | ENSG00000253293.3 | + | 0.5 | 0.1 | −0.2 | 0.3 |
| E2F2 | ENSG00000007968.6 | − | −0.6 | −0.4 | −0.02 | 0.1 |
| FOXD2 | ENSG00000186564.5 | − | −0.8 | −0.4 | −0.01 | 0.1 |
| RXRA b | ENSG00000186350.8 | − | −0.6 | −0.5 | −0.2 | −0.4 |
| NR3C1 | ENSG00000113580.10 | − | −1.1 | −0.6 | −0.2 | 0.01 |
18/55 TFs binding to SNHG16 promotor on chromosome 17 position 72064795‐72066401 (Hg18) were significantly dysregulated in CRC clinical samples (FC(log2) >0.5 or < −0.5 with a Bonferroni corrected p‐value <0.001 (Student's unpaired t‐test)).
Genes that are significantly correlated to the expression of SNHG16 in adenomas/adenocarcinomas (A + C) are in bold (Spearman's ρ (rs) ≥ 0.2 or ≤ −0.2 and p < 0.05).
FC: fold change.
N: normal colon mucosa (n = 292).
A + C: adenoma + adenocarcinoma (n = 314).
Known Wnt target (direct/indirect).
Negative regulator of Wnt.
Figure 2.
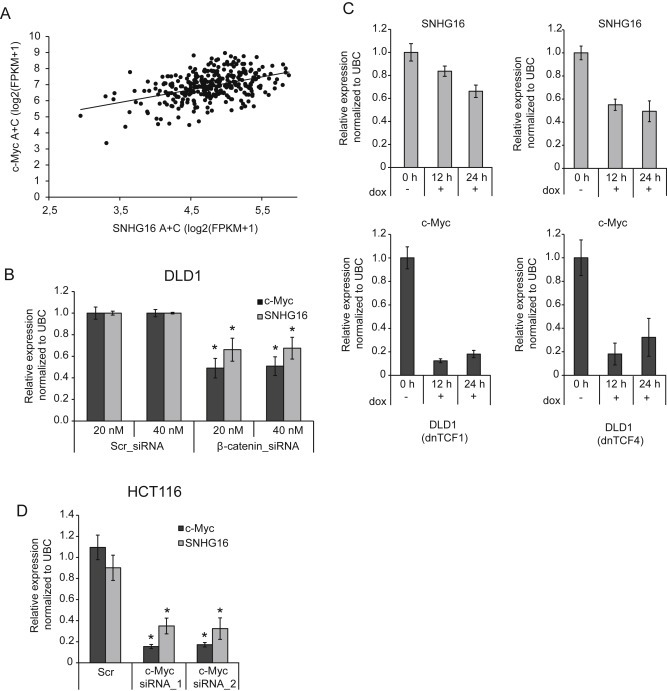
SNHG16 expression is regulated by the Wnt signaling pathway. Correlation of c‐Myc and SNHG16 expression in CRC (largeRNAseq cohort) (n = 314). Spearman's ρ = 0.4, p < 0.05 (A). SNHG16 expression in DLD1 cells transfected with 20 or 40 nm Scr or β‐catenin siRNA, quantified by RT‐qPCR. As a positive control of Wnt‐inactivation the well‐known Wnt target c‐Myc was also quantified (B). The expression of dnTCF1 or dnTCF4 was induced by doxycycline (dox) in stably transfected DLD1 cells. The SNHG16 expression was measured at different time points using RT‐qPCR. c‐Myc was included as a positive control (C). Expression analysis of SNHG16 and c‐Myc in HCT116 cells transfected with 50 nM of c‐Myc siRNAs or Scr (RT‐qPCR) (D). Data are presented as ±sd. of three biological replicates, *p < 0.05 (B–D).
3.3. SNHG16 is primarily expressed in the cytoplasm of CRC cell lines
To further characterize SNHG16, we analyzed its expression in nine different CRC cell lines. As shown in Figure 3A SNHG16 was present in all the analyzed cell lines at varying levels. By profiling RNA isolated from cytoplasmic and nuclear fractions from HCT116 and SW480 cells, selected as representatives of cell lines with high to medium expression, we found that SNHG16 primarily localizes to the cytoplasm (Figure 3B), indicating that it is likely to exert its functional role at the post‐transcriptional level (Ulitsky and Bartel, 2013). As expected the snoRNAs hosted by SNHG16 were expressed almost exclusively in the nucleus (Supplemental Figure 6A). Expression of HNRNPA1 (non‐coding isoform) and GAPDH confirmed the successful isolation of pure nuclear and cytoplasmic fractions (Supplemental Figure 6B).
Figure 3.
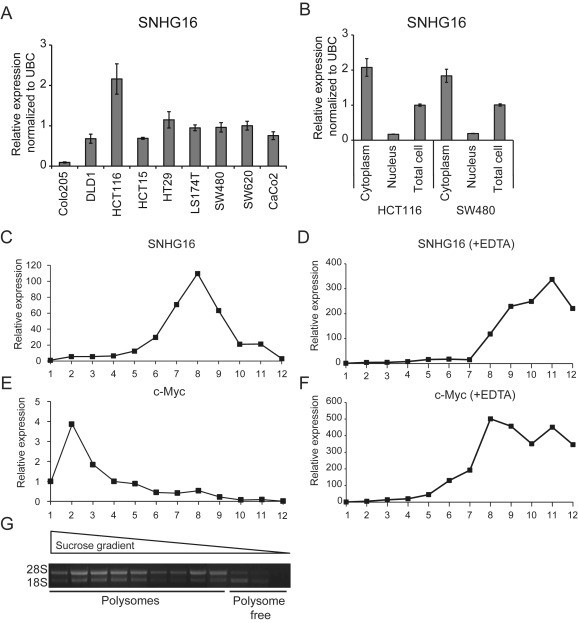
Expression analysis of SNHG16 in CRC cell lines and in polysome fractions. Expression of SNHG16 in nine CRC cell lines (RT‐qPCR). The results are presented as ±sd. of three biological replicates (A). Expression of SNHG16 in fractionated HCT116 and SW480 cells. The cells were fractionated into cytoplasmic and nuclear fractions followed by RT‐qPCR analyses. The data are presented as the relative expression in the nuclear/cytoplasmic fractions normalized to the expression in unfractionated cells (total cells). The experiment was repeated twice and the result of one representative experiment ±sd. is shown (B). The polysome profile of SNHG16 (C) and c‐Myc (positive control of polysome association) (D) was determined by isolation of RNA from each fraction collected from a 10–56% sucrose gradient. An EDTA release experiment, abrogating binding between RNAs and polysomes, was also performed (E–F). The relative expression of SNHG16 and c‐Myc was determined by RT‐qPCR. The result from one representative experiment is shown (C–F) Polysome fractions were identified by running RNA isolated from each fraction on an agarose gel to determine the 28S/18S rRNA ratios (G).
3.4. SNHG16 is associated with light polysomes
Cytoplasmic lncRNAs that contain small open reading frames (ORFs) are often associated with ribosomes (Bazzini et al., 2014; Carlevaro‐Fita et al., 2016; Chew et al., 2013). However, only in rare cases have these lncRNAs been shown to give rise to functional peptides (Pauli et al., 2014). The SNHG16 locus encodes transcripts with small ORFs (<60 aa) (Yu et al., 2009). The results of previous bioinformatic and comparative genomic analyses and in vitro [35S]‐methionine labeling in cell lines from neuroblastoma have all indicated that SNHG16 is a non‐coding RNA (Yu et al., 2009). In order to analyze the ribosomal association of the SNHG16 transcripts in colorectal cells, polysome analysis was carried out. This showed that SNHG16 is associated with the light polysome/monosome fractions (Figure 3C and G) compared to the known protein‐coding gene c‐Myc which is present in the heavy polysome containing fractions (Figure 3E and G). To ascertain whether SNHG16 was bound to polysomes an EDTA release experiment was carried out. Upon EDTA treatment SNHG16 redistributed toward less‐dense fractions, similar to the control c‐Myc (Figure 3D and F), indicating that while SNHG16 is not heavily bound by polysomes, it is nevertheless recruited to the ribosomes in CRC cells.
3.5. Knockdown of SNHG16 reduces cell viability, induces apoptotic death and decreases migration
To elucidate the functional role of SNHG16 in CRC, in vitro siRNA mediated loss‐of function analyses were carried out. Knockdown efficiencies of ∼90% were obtained with two independent siRNAs (Figure 4A and Supplemental Figure 7A). We hypothesized that the snoRNAs hosted by SNHG16 are most likely spliced out prior to siRNA‐mediated degradation of the host transcript. In agreement with this hypothesis snoRNA expression levels were not affected by SNHG16 knockdown (Supplemental Figure 7B). Accordingly, any phenotypic change observed following SNHG16 knockdown are most likely driven by the knockdown of the host transcript. Initial phenotypic analyses using an MTT assay demonstrated that loss of SNHG16 resulted in reduced viability of HCT116 (Figure 4B). This finding was corroborated by real‐time growth monitoring of the HCT116 cells, using xCELLigence (Figure 4C and D). To elucidate whether the growth suppression was related to cellular death we performed LDH (cellular death) and Caspase 3/7 activity (apoptosis) assays. Indeed knockdown of SNHG16 increased cellular death and apoptosis in HCT116 cells (Figure 4E and F). Furthermore, the apoptotic death could be inhibited with Z‐DEVD‐fmk (Caspase 3/7 inhibitor) (Figure 4F), demonstrating that the induced apoptosis is dependent on Caspase 3/7 activity. These results indicate that induction of apoptosis is part of the explanation for the reduced growth rate observed after SNHG16 knockdown, which is in line with previous analyses of neuroblastoma and bladder cancer. To further assess the functional role of elevated SNHG16, a migration assay was carried out. SNHG16 knockdown significantly inhibited HCT116 cell migration as early as 25 h after siRNA transfection (Figure 4G and H). Since SNHG16 silencing also affected proliferation we cannot rule out that the reduced number of migrating cells could also be due to increased apoptotic death. However, the effect on proliferation/apoptosis was most pronounced at later time‐points (60–80 h and 48 h, respectively) (Figure 4C, D and F). Accordingly, the increased migration at the early time points (25–35 h) are most likely due to an actual decreased migration ability of siRNA treated cells. Overall, the results of the migration assays add further evidence to an oncogenic role of SNHG16 in CRC (Figure 4G and H). In conclusion, knockdown of SNHG16 induces apoptotic death and has an inhibitory effect on cell migration.
Figure 4.
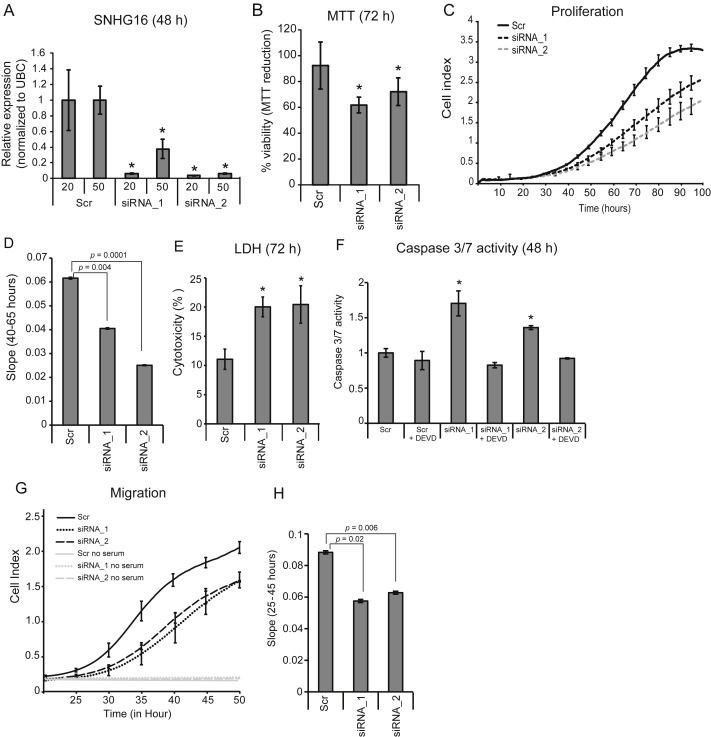
Knockdown of SNHG16 suppresses growth, increases cellular death and decreases migration in vitro. The relative expression of SNHG16 RNA in HCT116 cells transfected with SNHG16 siRNA_1 or siRNA_2. The siRNAs were used in two concentrations (20 and 50 nM) and the cells were harvested after 48 h. Shown are the mean of 3 biological replicates ± sd. *p < 0.05 when compared to Scr (A). The effect of SNHG16 knockdown on the viability HCT116 cells (MTT assay). Data are presented as the mean of at least 3 independent experiments ±sd., each with three biological replicates and normalized to Scr. *p < 0.05 (B). Real‐time monitoring of cell proliferation following SNHG16 knockdown using an xCELLigence instrument. The cell index from time 0–100 h is shown (C). Following the real‐time monitoring in C, the slope (rate of changes in cell index) was calculated from 40 to 65 h (i.e. when changes in proliferation were apparent) and presented graphically (D). The effect of SNHG16 knockdown on cellular death (LDH release assay) in HCT116 cells. The cellular death is expressed as percentage of released LDH out of total cellular LDH. At least two independent experiments were carried out and performed in triplicates. The result of one representative experiment ±sd. is shown. *p < 0.05 (E). Induction of apoptosis (Caspase 3/7 activity) in the lysate of siRNA transfected cells was examined by fluorometric kinetic analysis and expressed relative to the Caspase 3/7 activity in “Scr” transfected cells. The Caspase inhibitor Z‐DEVD‐fmk (DEVD) was added to the cells six hours post‐transfection. Data are presented as ±sd. of at least 2 independent experiments each with three biological replicates. *p < 0.05 (F). Real‐time monitoring of cell migration of HCT116 cells transfected with siRNA was performed using an xCELLigence instrument. The cell index from time 20–50 h is shown (G). Following the real‐time monitoring in G, the slope (rate of changes in cell index) was calculated from 25 to 45 h (i.e. when changes in migration were apparent) and presented graphically (H).
3.6. Knockdown of SNHG16 affects genes involved in lipid metabolism
In order to shed light on the molecular mechanism underlying the knockdown phenotypes we performed genome wide transcriptional profiling of HCT116 cells treated individually with SNHG16 siRNA_1 or siRNA_2. It is well‐known that siRNAs may exhibit off target effects (Birmingham et al., 2006; Jackson et al., 2006; Jackson and Linsley, 2010). Therefore, after having identified the transcripts deregulated by the siRNA treatments these were searched for 3′UTR motifs with perfect match(es) to the seed region of siRNA_1 and siRNAS_2, indicating that they may be deregulated due to a direct interaction with the siRNAs rather than through SNHG16. These transcripts were excluded from subsequent analyses and left a total of 124 transcripts that were significantly dysregulated by both siRNA_1 and siRNA_2 (p < 0.05, FC(log2) <−0.5 or >0.5). To gain insight into the over‐all biological changes introduced by SNHG16 knockdown the 124 transcripts (99 down‐ and 25 up‐regulated) were analyzed using Ingenuity Pathway analysis (IPA). IPA demonstrated that SNHG16 knockdown predominantly affected the expression of genes associated with lipid metabolism, gastrointestinal diseases and cancer (Figure 5A). To investigate whether SNHG16 potentially also regulate these transcripts in clinical samples, we set out to explore their expression pattern in the largeRNAseq cohort. Twenty‐four of 124 genes were significantly dysregulated between normal and tumor samples (p < 0.05, FC(log2) <−0.5 or >0.5) (Figure 5B and Supplemental Table 7). Accordingly, we defined them as clinically relevant SNHG16 regulated candidate genes. Most interestingly, 12 of the 24 genes, have functions associated with lipid metabolism and/or have previously been associated with gastrointestinal cancer (Supplemental Table 7). To further analyze the relation between the expression of SNHG16 and the genes related to lipid metabolism and gastrointestinal cancer, the clinical samples were ranked according to their SNHG16 FCs (290 adenomas/adenocarcinomas with paired normal colon mucosa). Subsequently, the median FCs of the lipid and cancer genes were calculated for the upper 25% quartile and the lower 25% quartile of the ranked samples (73 normal colon mucosa vs. 73 adenomas/adenocarcinomas). Strikingly, the FCs of 9/12 genes related to lipid metabolism and/or gastrointestinal cancer were significantly higher in the pairs belonging to the 25% upper quartile of SNHG16 FCs compared to the FCs of the pairs in the lower 25% quartile (Figure 5C). Using a “lipid gene list” containing 685 genes derived from GO term GO:006629 (lipid metabolic process) and Reactome:Metabolism_of_lipids_and_ lipoproteins we found that genes involved in lipid metabolism were generally enriched among the significantly dysregulated genes in the clinical samples (Enrichment score 2.87 (95% CI 2.3–3.46 and p = 2.2 × 10−16)) indicating that changes in lipid metabolism are a common trait of CRC. In summary, it is likely that SNHG16 is involved in the regulation of lipid metabolism in CRC.
Figure 5.
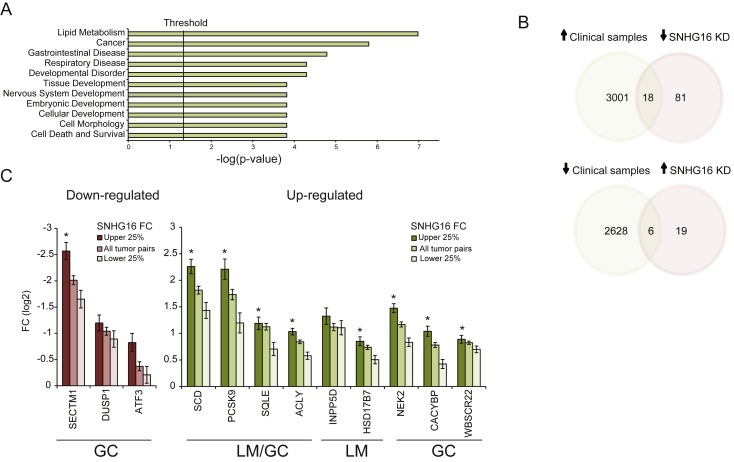
Ingenuity Pathway Analysis and identification of clinical SNHG16 targets. Ingenuity Pathway Analysis (IPA) was performed to investigate if particular diseases and biological functions were associated with the 124 genes significantly dysregulated upon SNHG16 siRNA knockdown. The top three enriched features were lipid metabolism, cancer, and gastrointestinal diseases (A). Venn diagrams showing the number of overlapping differentially expressed genes between clinical samples and siRNA transfected cells (p < 0.05, FC(log2) < −0.5 or >0.5). Upper diagram: significantly up‐regulated in the clinical samples and down‐regulated as a result of SNHG16 knockdown. Lower diagram: significantly down‐regulated in the clinical samples and up‐regulated as a result of SNHG16 knockdown. KD: knockdown (B). Overall FC of lipid and gastrointestinal cancer genes in all tumor pairs (n = 290) (adenomas/adenocarcinomas compared to matched normal colon mucosa) and in the pairs with high (25% upper quartile, n = 73) or low (25% lower quartile, n = 73) SNHG16 FCs. Left diagram (red bars): down‐regulated genes and right diagram (green bars): up‐regulated genes. LM: lipid metabolism, GC: gastrointestinal cancer. Data are presented as mean FC ± standard error of the mean. *Significant difference between the FCs in the 25% upper quartile compared to the FCs in the 25% lower quartile (p < 0.05) (C).
3.7. Genes affected by SNHG16 knockdown contain common sequence motifs
Like other classes of non‐coding RNA, cytoplasmic lncRNAs have been shown to bind to specific mRNA targets through sequence complementarity (Carrieri et al., 2012; Kretz et al., 2013; Yoon et al., 2012). In some cases specific motifs have been shown to be enriched in mRNAs targeted by a specific lncRNA (Kretz et al., 2013). To further investigate the RNA interacting potential of SNHG16, we asked if common sequence motifs, with complementarity to SNHG16 were found in the mRNAs affected by SNHG16 silencing. Initially we searched for 7‐mers correlating with the differential expression observed after SNHG16 knockdown. Five 7‐mers were identified which overlapped with 6 bases and two motifs had only a single mismatch to the 7‐mers (p < 8.4e‐07), thus defining a 7‐mer consensus sequence (CAG(C/G)CTG) (Supplemental Table 6 and Supplemental Figure 8A; details in Materials and Methods). The 7‐mer consensus sequence was present twice in the antisense sequence of SNHG16 (Supplemental Figure 8B). Finally, we analyzed whether the 12 clinically relevant genes involved in lipid metabolism and gastrointestinal cancer contained the above consensus sequence. Indeed, 9/12 genes contained 1–8 copies of the consensus sequence in their mRNA sequences. Interestingly, the PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) mRNA involved in lipid metabolism contained 8 copies of the motif. Observing 8 copies of the motif, as inPCSK9 is unlikely a coincidental finding (p = 1.3e10−6). Thus, identification of a 7‐mer consensus motif enriched in mRNAs affected by SNHG16 silencing suggests that SNHG16 may exert its function through pairing with specific mRNAs.
3.8. SNHG16 binds AGO and HuR and may function as a ceRNA
Apart from the consensus sequence described above, U‐rich motifs (URMs) were enriched in the genes that were dysregulated upon SNHG16 silencing (Supplemental Table 6). URMs are known from previous work to be enriched in transcripts that bind and interact with miRNAs (Jacobsen et al., 2010). AGO‐CLIP coupled to high throughput sequencing is a genomic technology that provides experimental evidence of global miRNA binding in the cell (Chi et al., 2009; Hafner et al., 2010). In order to determine if SNHG16 is actively bound by AGO we queried an updated atlas of AGO‐CLIP miRNA targets identified in multiple cell types (Hamilton et al., 2013). We found that SNHG16 contains 27 high confidence AGO/miRNA target sites along its length, corresponding to binding of 26 unique miRNA families (Figure 6A and Supplemental File 2). This data strongly suggests that the Argonaute protein heavily binds SNHG16 across multiple cell types. To determine if any specific mRNAs are targeted by multiple SNHG16 bound miRNAs (“co‐targeting”) we again used the AGO‐CLIP atlas to define the target spectrum of the miRNAs in question. SNHG16 bound miRNAs co‐target the 3′UTR of multiple important mRNA transcripts (Figure 6B, Supplemental File 2). Most interestingly, SCD involved in lipid synthesis and identified as a clinically interesting SNHG16 target in the knockdown analysis, had the strongest co‐targeting spectrum of 3305 3′UTRs in our CLIP analysis (Figure 6C and D), 13 out of 26 unique miRNA families with high confidence targets on SNHG16 also target the SCD 3′UTR. We next were curious if other mRNAs down‐regulated upon SNHG16 silencing could also be linked to SNHG16 through the AGO‐CLIP data. Indeed, 35/99 of these mRNAs were also bound by SNHG16 binding miRNAs and of these, 18 were up‐regulated in the clinical samples (p < 0.05). Alternatively, association of SNHG16 with AGO could also indicate that SNHG16 is being targeted for miRNA mediated degradation. However, out of 49 miRNAs (expressed in >80% of the CRCs in the small RNAseq cohort) belonging to the 26 miRNA families none where significantly negatively correlated to the expression of SNHG16, either individually or when using the geometric mean of the expression of all 49 miRNAs. This indicates that SNHG16 is generally not regulated by these miRNAs in CRC.
Figure 6.
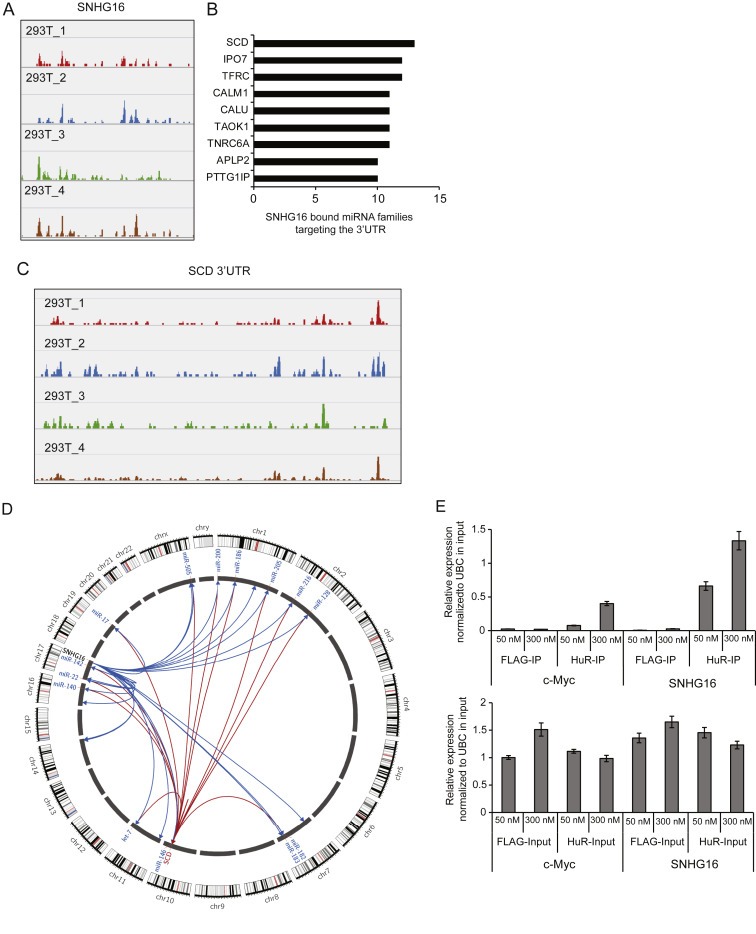
AGO‐CLIP binding analysis and HuR‐IP. SNHG16 AGO loading in 293T cells (shown are representative data from four individual AGO‐CLIP analyses) (A). The 3′UTR of multiple mRNA transcripts are heavily co‐targeted by miRNAs predicted as binding to SNHG16 in AGO‐CLIP data from 293T cells (B). The SCD mRNA has multiple CLIP defined miRNA binding sites along its 3′UTR (C). Circos plot depicting SNHG16 and SCD interaction with its CLIP predicted miRNA targets. SNHG16‐miRNA interactions are represented as blue links and miRNA‐SCD interactions are represented as red links. Representative members of miRNA families targeting SCD are labeled. Half of the miRNA families predicted to bind SNHG16 co‐target the SCD 3′UTR (D). HuR‐IP from HCT116 cell lysates (IP) followed by SNHG16 or c‐Myc (positive control) expression analysis using RT‐qPCR (upper panel). As a control SNHG16 and c‐Myc expression was also measured in the cell lysates used as input (lower panel). Immunoprecipitation with a FLAG antibody was used as negative control. The experiment was repeated twice and the result of one representative experiment ± sd. is shown (E).
A recent study has shown that the RNA binding protein HuR (encoded by ELAVL1) is required in the cytoplasm to facilitate the miRNA recruitment of linc‐MD1 (Legnini et al., 2014). RNA immunoprecipitation (RNA‐IP) analysis performed by the ENCODE project consortium demonstrated that HuR binds the SNHG16 transcript in K562 and GM12878 cells (Supplemental Figure 9A) (ENCODE_Project_Consortium, 2012). The RNA‐Protein Interaction Prediction program (RPIseq) also predicted high probability of HuR/SNHG16 interaction (Supplemental Figure 9B) (Muppirala et al., 2011). Previous analyses have shown that HuR is predominantly expressed in the nucleus in normal colon mucosa whereas in colon cancer HuR is up‐regulated and exhibits cytoplasmic localization (reviewed in (Ignatenko and Gerner, 2008)). The above findings and the cytoplasmic localization of both SNHG16 and HuR encouraged us to investigate the potential interaction/binding between SNHG16 and HuR in colon cancer cells. HuR‐IP followed by RT‐qPCR analysis of the RNA isolated from HuR‐IP fractions clearly demonstrated that SNHG16 is enriched in immunopreciptates from HCT116 cells in a HuR dependent manner (Figure 6E) indicating that the SNHG16 transcript also binds HuR in CRC cells.
In conclusion, SNHG16 binds AGO and HuR and may act as a ceRNA “sponging” miRNAs off their cognate targets, thus relieving miRNA mediated target repression.
4. Discussion
It is well established that lncRNAs are important for normal cellular development and that abnormal expression of lncRNAs play a key role in tumorigenesis. SNHG16 has been reported to be up‐regulated and act as a potential oncogene in neuroblastoma and bladder cancer (Yu et al., 2009; Zhu et al., 2011). Accordingly, these previous findings are in line with the results of the present paper. However, in contrast to our results, a recent study has reported that SNHG16 is down‐regulated in CRC (Qi et al., 2015). We speculated whether the use of different methodologies for SNHG16 quantification, detection of different isoforms, differences in RNA quality, differences in cancer cell percentage, and differences in the use of preoperative chemotherapy could possibly explain the discrepancy? To address this we investigated the two primer sets used by Qi et al. using the in silico PCR tool at the UCSC genome browser and found that these detect the same SNHG16 isoform detected in the present study using RNAseq, RT‐qPCR, and custom‐made microarrays (Supplemental Figure 1C). Accordingly the used methodologies and detected isoform are unlikely explanations of the discrepancy. The same goes for differences in prior therapy as the patients in the present study did not receive therapy prior to surgery nor did the patients in the Qi et al. paper. Data on RNA quality and cancer cell percentage was not included in the study by Qi et al. In the present study all RNAs were of high quality, with median RIN scores above 8.9 (Table 1). The median cancer cell percentage of the samples in the present study was 85% for the adenomas and 75% for the colorectal cancers indicating only minor contamination of non‐tumorous cells. In summary, the discrepancy between the two studies cannot be explained by differences in methodology/detection of different isoforms or preoperative therapy. Differences in RNA quality and/or cancer cell percentage in tissue samples may explain the differences but, these issues cannot be addressed since such data are not provided by Qi et al. Nevertheless, the results of the present study was based on data obtained using three different methods and two independent cohorts. Additionally, up‐regulation of SNHG16 was found in 7/9 additional cancer types from eight different tissue using data from the TCGA research network (http://cancergenome.nih.gov/) (Supplemental Figure 10). All together our data and the data from TCGA clearly indicate an oncogenic role of SNHG16 in colorectal cancer as well as in other cancers. In line with the up‐regulation of SNHG16 in multiple cancers, functional analyses carried out in the present study and in bladder and neuroblastoma cell lines also suggest oncogenic properties of SNHG16 (Yu et al., 2009; Zhu et al., 2011). On the contrary, previous phenotypic analysis in CRC suggested tumor suppressor like functions of SNHG16 (Qi et al., 2015). The siRNAs used in the present study as well as the RNAi used by Qi et al. were designed to target the same SNHG16 isoforms. Hence silencing of different isoforms does not explain the differences. Alternatively, variations in the used HCT116 cells may explain the divergent results in the phenotype assays. The HCT116 cell lines used in the present paper was authenticated according to the recommendation of ATCC using short tandem repeat (STR) profiling. The obtained STR profiles all matched those published by ATCC (data not shown). Such data are not provided for the SNHG16 cell lines in the study by Qi et al.
Although it is well established that lncRNAs play important roles in a large variety of cellular processes the function and mechanisms of action of most lncRNAs are unknown. Knowledge about the cellular localization of lncRNAs is an important factor in the understanding of their function and means of action. We show that SNHG16 is enriched in the cytoplasm of CRC cell lines and associated with ribosomes. Recent studies have shown that cytoplasmic enriched lncRNAs are indeed associated with ribosomes, although they are not actively translated and suggest that the main function of lncRNAs may be related to translational regulation (Carlevaro‐Fita et al., 2016; Guttman et al., 2013; Ingolia et al., 2011; Slavoff et al., 2013; van Heesch et al., 2014). Alternatively, ribosome associated lncRNA may be regulated by the non‐sense mediated RNA degradation pathway as previously shown for the lncRNA GAS5 (Chew et al., 2013; Tani et al., 2013). In support of this notion, some SNHG16 isoforms contain an in frame stop codon, >50 bases upstream of a splice‐junction, which is known to stimulate NMD (Schweingruber et al., 2013). Publicly available sequencing data globally assessing start codon usage and susceptibility to NMD also suggest that some SNHG16 isoforms can undergo NMD (Lee et al., 2012; Lykke‐Andersen et al., 2014).
Pathway analysis of the RNA profiles of HCT116 cells after silencing of SNHG16 showed that SNHG16 knockdown predominantly affected transcripts associated with lipid metabolism and gastrointestinal cancer, a finding which we corroborated by showing that many of these genes were also significantly dysregulated in clinical CRC tumors. Aberrant de novo lipid biosynthesis has frequently been observed in cancerous tissue (reviewed in (Baenke et al., 2013)). First of all, enhanced lipid synthesis is required for the metabolic reprogramming of cancer cells. However, there is compelling evidence that lipids also play a more active role in cell transformation and cancer development. Moreover, activation of hepatic β‐catenin has been found to increase the expression of genes involved in lipid metabolism, indicating that the Wnt pathway may also play a role in aberrant lipid synthesis in CRC (Liu et al., 2011). The inhibitory effect on pathways related to lipid metabolism/cancer as a result of SNHG16 knockdown is also in line with the observed phenotypic changes. As examples, knockdown of SCD, ACLY (ATP citrate lyase) and Nek2 (NIMA‐related kinase 2) has previously been shown to inhibit growth and induce apoptotic death in CRC cells (Mason et al., 2012; Suzuki et al., 2010; Zaidi et al., 2012).
It is well documented that lncRNAs may act as ceRNAs by sponging miRNAs off their cognate targets, and thus relieving miRNA mediated target repression (Salmena et al., 2011; Tay et al., 2014). Given that the majority of the mRNAs affected by SNHG16 silencing were down‐regulated and that SNHG16 contain high confidence AGO/miRNA target sites the results of the present paper suggest that SNHG16 may function as a ceRNA. Previous studies have primarily described the selective sponging of single miRNAs by specific lncRNA. However, it is well known that multiple miRNAs may bind and suppress the same mRNA 3′UTR (Tsang et al., 2010) suggesting that to obtain an effect at the expression level of target mRNAs several miRNAs must be repressed at the same time. Most interestingly our results indicate that a broad spectrum of lncRNA miRNA targets may converge on a few heavily co‐targeted genes suggesting that lncRNAs may also function by modulating the activity of a broad range of miRNAs.
5. Conclusions
We have shown that SNHG16 is regulated by the Wnt pathway and up‐regulated as an early event in CRC. In vitro functional analysis demonstrated that SNHG16 is present in the cytoplasm and associated with polysomes. Furthermore, knockdown of SNHG16 induced apoptotic death and increased cellular migration and showed that reduced expression of SNHG16 among others affects genes involved in lipid metabolism. Finally, AGO‐CLIP analysis and HuR‐IP led to the hypothesis that SNHG16 may act as ceRNA for miRNAs in CRC. Generally, our data suggest that lncRNAs may target multiple miRNAs rather than selectively sponging single miRNAs.
Financial support
This work was supported by the Danish National Advanced Technology Foundation, the John and Birthe Meyer Foundation, the Danish Council for Independent Research (Medical Sciences), the Danish Council for Strategic Research, Lundbeckfonden, The Danish Cancer Society and The European Union's Seventh Framework Program (SYSCOL HEALTH‐F5‐2010‐258236).
Conflict of interest
The authors declare that there are no conflicts of interest.
Supporting information
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
We are especially grateful to Susanne Bruun, Pamela Celis, Birgitte Trolle, Margaret Gellett and Louise Nielsen for skillful technical assistance. The Danish Cancer Biobank (DCB) is acknowledged for biological material and for information regarding handling and storage.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2016.06.003.
Christensen Lise Lotte, True Kirsten, Hamilton Mark P., Nielsen Morten M., Damas Nkerorema D., Damgaard Christian K., Ongen Halit, Dermitzakis Emmanouil, Bramsen Jesper B., Pedersen Jakob S., Lund Anders H., Vang Søren, Stribolt Katrine, Madsen Mogens R., Laurberg Søren, McGuire Sean E., Ørntoft Torben F., Andersen Claus L., (2016), SNHG16 is regulated by the Wnt pathway in colorectal cancer and affects genes involved in lipid metabolism, Molecular Oncology, 10, doi: 10.1016/j.molonc.2016.06.003.
Contributor Information
Lise Lotte Christensen, Email: liselotte.christensen@clin.au.dk.
Kirsten True, Email: Kirsten_true@hotmail.com.
Mark P. Hamilton, Email: mphamilt@bcm.edu
Morten M. Nielsen, Email: morten.muhlig@clin.au.dk
Nkerorema D. Damas, Email: Nkerorema.Damas@bric.ku.dk
Christian K. Damgaard, Email: ckd@mbg.au.dk
Halit Ongen, Email: Halit.Ongen@unige.ch.
Emmanouil Dermitzakis, Email: Emmanouil.Dermitzakis@unige.ch.
Jesper B. Bramsen, Email: bramsen@clin.au.dk
Jakob S. Pedersen, Email: jakob.skou@clin.au.dk
Anders H. Lund, Email: anders.lund@bric.ku.dk
Søren Vang, Email: vang@clin.au.dk.
Katrine Stribolt, Email: katrine.stribolt@rm.dk.
Mogens R. Madsen, Email: mogmad@rm.dk
Søren Laurberg, Email: soerlaur@rm.dk.
Sean E. McGuire, Email: Sean.McGuire@bcm.edu
Torben F. Ørntoft, Email: orntoft@clin.au.dk
Claus L. Andersen, Email: cla@clin.au.dk
References
- Atienza, J.M. , Yu, N. , Kirstein, S.L. , Xi, B. , Wang, X. , Xu, X. , Abassi, Y.A. , 2006. Dynamic and label-free cell-based assays using the real-time cell electronic sensing system. Assay. Drug Dev. Technol. 4, 597–607. [DOI] [PubMed] [Google Scholar]
- Baenke, F. , Peck, B. , Miess, H. , Schulze, A. , 2013. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 6, 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini, A.A. , Johnstone, T.G. , Christiano, R. , Mackowiak, S.D. , Obermayer, B. , Fleming, E.S. , Vejnar, C.E. , Lee, M.T. , Rajewsky, N. , Walther, T.C. , Giraldez, A.J. , 2014. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 33, 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham, A. , Anderson, E.M. , Reynolds, A. , Ilsley-Tyree, D. , Leake, D. , Fedorov, Y. , Baskerville, S. , Maksimova, E. , Robinson, K. , Karpilow, J. , Marshall, W.S. , Khvorova, A. , 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204. [DOI] [PubMed] [Google Scholar]
- Carlevaro-Fita, J. , Rahim, A. , Guigo, R. , Vardy, L.A. , Johnson, R. , June 2016. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 22, 867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri, C. , Cimatti, L. , Biagioli, M. , Beugnet, A. , Zucchelli, S. , Fedele, S. , Pesce, E. , Ferrer, I. , Collavin, L. , Santoro, C. , Forrest, A.R. , Carninci, P. , Biffo, S. , Stupka, E. , Gustincich, S. , 2012. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457. [DOI] [PubMed] [Google Scholar]
- Chew, G.L. , Pauli, A. , Rinn, J.L. , Regev, A. , Schier, A.F. , Valen, E. , 2013. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development 140, 2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, S.W. , Zang, J.B. , Mele, A. , Darnell, R.B. , 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien, T. , Johnson, R. , Bussotti, G. , Tanzer, A. , Djebali, S. , Tilgner, H. , Guernec, G. , Martin, D. , Merkel, A. , Knowles, D.G. , Lagarde, J. , Veeravalli, L. , Ruan, X. , Ruan, Y. , Lassmann, T. , Carninci, P. , Brown, J.B. , Lipovich, L. , Gonzalez, J.M. , Thomas, M. , Davis, C.A. , Shiekhattar, R. , Gingeras, T.R. , Hubbard, T.J. , Notredame, C. , Harrow, J. , Guigo, R. , 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard, A.C. , Lane, M.A. , 2007. Retinol decreases beta-catenin protein levels in retinoic acid-resistant colon cancer cell lines. Mol. Carcinog 46, 315–329. [DOI] [PubMed] [Google Scholar]
- Djebali, S. , Davis, C.A. , Merkel, A. , Dobin, A. , Lassmann, T. , Mortazavi, A. , Tanzer, A. , Lagarde, J. , Lin, W. , Schlesinger, F. , Xue, C. , Marinov, G.K. , Khatun, J. , Williams, B.A. , Zaleski, C. , Rozowsky, J. , Roder, M. , Kokocinski, F. , Abdelhamid, R.F. , Alioto, T. , Antoshechkin, I. , Baer, M.T. , Bar, N.S. , Batut, P. , Bell, K. , Bell, I. , Chakrabortty, S. , Chen, X. , Chrast, J. , Curado, J. , Derrien, T. , Drenkow, J. , Dumais, E. , Dumais, J. , Duttagupta, R. , Falconnet, E. , Fastuca, M. , Fejes-Toth, K. , Ferreira, P. , Foissac, S. , Fullwood, M.J. , Gao, H. , Gonzalez, D. , Gordon, A. , Gunawardena, H. , Howald, C. , Jha, S. , Johnson, R. , Kapranov, P. , King, B. , Kingswood, C. , Luo, O.J. , Park, E. , Persaud, K. , Preall, J.B. , Ribeca, P. , Risk, B. , Robyr, D. , Sammeth, M. , Schaffer, L. , See, L.H. , Shahab, A. , Skancke, J. , Suzuki, A.M. , Takahashi, H. , Tilgner, H. , Trout, D. , Walters, N. , Wang, H. , Wrobel, J. , Yu, Y. , Ruan, X. , Hayashizaki, Y. , Harrow, J. , Gerstein, M. , Hubbard, T. , Reymond, A. , Antonarakis, S.E. , Hannon, G. , Giddings, M.C. , Ruan, Y. , Wold, B. , Carninci, P. , Guigo, R. , Gingeras, T.R. , 2012. Landscape of transcription in human cells. Nature 489, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE_Project_Consortium, 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, X. , Chen, Y. , Liao, X. , Liu, D. , Li, F. , Ruan, H. , Jia, W. , 2013. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med. Oncol. 30, 588 [DOI] [PubMed] [Google Scholar]
- Gibb, E.A. , Brown, C.J. , Lam, W.L. , 2011. The functional role of long non-coding RNA in human carcinomas. Mol. Cancer 10, 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb, E.A. , Warren, R.L. , Wilson, G.W. , Brown, S.D. , Robertson, G.A. , Morin, G.B. , Holt, R.A. , 2015. Activation of an endogenous retrovirus-associated long non-coding RNA in human adenocarcinoma. Genome Med. 7, 22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold, H. , Tehler, D. , Christoffersen, N.R. , Nielsen, M.M. , Asmar, F. , Kooistra, S.M. , Christophersen, N.S. , Christensen, L.L. , Borre, M. , Sorensen, K.D. , Andersen, L.D. , Andersen, C.L. , Hulleman, E. , Wurdinger, T. , Ralfkiaer, E. , Helin, K. , Gronbaek, K. , Orntoft, T. , Waszak, S.M. , Dahan, O. , Pedersen, J.S. , Lund, A.H. , Pilpel, Y. , 2014. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292. [DOI] [PubMed] [Google Scholar]
- Grady, W.M. , Pritchard, C.C. , 2014. Molecular alterations and biomarkers in colorectal cancer. Toxicol. Pathol. 42, 124–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, L.D. , Pedersen, S.K. , Brown, G.S. , Ho, T. , Kassir, Z. , Moynihan, A.T. , Vizgoft, E.K. , Dunne, R. , Pimlott, L. , Young, G.P. , Lapointe, L.C. , Molloy, P.L. , 2011. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer 2, 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman, M. , Russell, P. , Ingolia, N.T. , Weissman, J.S. , Lander, E.S. , 2013. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner, M. , Landthaler, M. , Burger, L. , Khorshid, M. , Hausser, J. , Berninger, P. , Rothballer, A. , Ascano, M. , Jungkamp, A.C. , Munschauer, M. , Ulrich, A. , Wardle, G.S. , Dewell, S. , Zavolan, M. , Tuschl, T. , 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggar, F.A. , Boushey, R.P. , 2009. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin. Colon Rectal. Surg. 22, 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, M.P. , Rajapakshe, K. , Hartig, S.M. , Reva, B. , McLellan, M.D. , Kandoth, C. , Ding, L. , Zack, T.I. , Gunaratne, P.H. , Wheeler, D.A. , Coarfa, C. , McGuire, S.E. , 2013. Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat. Commun. 4, 2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Chen, H.Y. , Yu, C.Y. , Xu, J. , Wang, J.L. , Qian, J. , Zhang, X. , Fang, J.Y. , 2014. A long non-coding RNA signature to improve prognosis prediction of colorectal cancer. Oncotarget 5, 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatenko, N.A. , Gerner, E.W. , 2008. HuRrying colon cancer progression. Cancer Biol. Ther. 7, 428–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia, N.T. , Lareau, L.F. , Weissman, J.S. , 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, A.L. , Burchard, J. , Schelter, J. , Chau, B.N. , Cleary, M. , Lim, L. , Linsley, P.S. , 2006. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12, 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, A.L. , Linsley, P.S. , 2010. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nature reviews. Drug Discov. 9, 57–67. [DOI] [PubMed] [Google Scholar]
- Jacobsen, A. , Wen, J. , Marks, D.S. , Krogh, A. , 2010. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 20, 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson, P. , Ackley, A. , Vidarsdottir, L. , Lui, W.O. , Corcoran, M. , Grander, D. , Morris, K.V. , 2013. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 20, 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo, R. , Shimamura, T. , Mimori, K. , Kawahara, K. , Imoto, S. , Sudo, T. , Tanaka, F. , Shibata, K. , Suzuki, A. , Komune, S. , Miyano, S. , Mori, M. , 2011. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 71, 6320–6326. [DOI] [PubMed] [Google Scholar]
- Kretz, M. , Siprashvili, Z. , Chu, C. , Webster, D.E. , Zehnder, A. , Qu, K. , Lee, C.S. , Flockhart, R.J. , Groff, A.F. , Chow, J. , Johnston, D. , Kim, G.E. , Spitale, R.C. , Flynn, R.A. , Zheng, G.X. , Aiyer, S. , Raj, A. , Rinn, J.L. , Chang, H.Y. , Khavari, P.A. , 2013. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski, M. , Schein, J. , Birol, I. , Connors, J. , Gascoyne, R. , Horsman, D. , Jones, S.J. , Marra, M.A. , 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Liu, B. , Lee, S. , Huang, S.X. , Shen, B. , Qian, S.B. , 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. USA 109, E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini, I. , Morlando, M. , Mangiavacchi, A. , Fatica, A. , Bozzoni, I. , 2014. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell 53, 506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Chanrion, M. , Sawey, E. , Wang, T. , Chow, E. , Tward, A. , Su, Y. , Xue, W. , Lucito, R. , Zender, L. , Lowe, S.W. , Bishop, J.M. , Powers, S. , 2015. Reciprocal interaction of Wnt and RXR-alpha pathways in hepatocyte development and hepatocellular carcinoma. PLoS One 10, e0118480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind, G.E. , Kleivi, K. , Meling, G.I. , Teixeira, M.R. , Thiis-Evensen, E. , Rognum, T.O. , Lothe, R.A. , 2006. ADAMTS1, CRABP1, and NR3C1 identified as epigenetically deregulated genes in colorectal tumorigenesis. Cell. Oncol. 28, 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgreen, S. , 2012. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res. Notes 5, 337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Fergusson, M.M. , Wu, J.J. , Rovira, Liu, J. , Gavrilova, O. , Lu, T. , Bao, J. , Han, D. , Sack, M.N. , Finkel, T. , 2011. Wnt signaling regulates hepatic metabolism. Sci. Signal 4, ra6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, S. , Chen, Y. , Ardal, B.R. , Lilje, B. , Waage, J. , Sandelin, A. , Jensen, T.H. , 2014. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 28, 2498–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, P. , Liang, B. , Li, L. , Fremgen, T. , Murphy, E. , Quinn, A. , Madden, S.L. , Biemann, H.P. , Wang, B. , Cohen, A. , Komarnitsky, S. , Jancsics, K. , Hirth, B. , Cooper, C.G. , Lee, E. , Wilson, S. , Krumbholz, R. , Schmid, S. , Xiang, Y. , Booker, M. , Lillie, J. , Carter, K. , 2012. SCD1 inhibition causes cancer cell death by depleting mono-unsaturated fatty acids. PLoS One 7, e33823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muppirala, U.K. , Honavar, V.G. , Dobbs, D. , 2011. Predicting RNA-protein interactions using only sequence information. BMC Bioinformatics 12, 489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, M.M. , Tehler, D. , Vang, S. , Sudzina, F. , Hedegaard, J. , Nordentoft, I. , Orntoft, T.F. , Lund, A.H. , Pedersen, J.S. , 2014. Identification of expressed and conserved human noncoding RNAs. RNA 20, 236–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, M.N. , Tataru, P. , Madsen, T. , Hobolth, A. , Pedersen, J.S. , 2016. RegMex, motif analysis in ranked lists of sequences. BioRxiv 10.1101/035956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostenfeld, M.S. , Fehrenbacher, N. , Hoyer-Hansen, M. , Thomsen, C. , Farkas, T. , Jaattela, M. , 2005. Effective tumor cell death by sigma-2 receptor ligand siramesine involves lysosomal leakage and oxidative stress. Cancer Res. 65, 8975–8983. [DOI] [PubMed] [Google Scholar]
- Pauli, A. , Norris, M.L. , Valen, E. , Chew, G.L. , Gagnon, J.A. , Zimmerman, S. , Mitchell, A. , Ma, J. , Dubrulle, J. , Reyon, D. , Tsai, S.Q. , Joung, J.K. , Saghatelian, A. , Schier, A.F. , 2014. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 343, 1248636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner, J.R. , Chinnaiyan, A.M. , 2011. The emergence of lncRNAs in cancer biology. Cancer Discov. 1, 391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, P. , Xu, M.D. , Ni, S.J. , Shen, X.H. , Wei, P. , Huang, D. , Tan, C. , Sheng, W.Q. , Zhou, X.Y. , Du, X. , 2015. Down-regulation of ncRAN, a long non-coding RNA, contributes to colorectal cancer cell migration and invasion and predicts poor overall survival for colorectal cancer patients. Mol. Carcinog. 54, 742–750. [DOI] [PubMed] [Google Scholar]
- Ragusa, M. , Barbagallo, C. , Statello, L. , Condorelli, A.G. , Battaglia, R. , Tamburello, L. , Barbagallo, D. , Di Pietro, C. , Purrello, M. , 2015. Non-coding landscapes of colorectal cancer. World J. Gastroenterol. 21, 11709–11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. , Thorvaldsdottir, H. , Winckler, W. , Guttman, M. , Lander, E.S. , Getz, G. , Mesirov, J. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed, A.I. , Bhagabati, N.K. , Braisted, J.C. , Liang, W. , Sharov, V. , Howe, E.A. , Li, J. , Thiagarajan, M. , White, J.A. , Quackenbush, J. , 2006. TM4 microarray software suite. Methods Enzymol. 411, 134–193. [DOI] [PubMed] [Google Scholar]
- Salmena, L. , Poliseno, L. , Tay, Y. , Kats, L. , Pandolfi, P.P. , 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language?. Cell 146, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber, C. , Rufener, S.C. , Zund, D. , Yamashita, A. , Muhlemann, O. , 2013. Nonsense-mediated mRNA decay – mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta 1829, 612–623. [DOI] [PubMed] [Google Scholar]
- Slavoff, S.A. , Mitchell, A.J. , Schwaid, A.G. , Cabili, M.N. , Ma, J. , Levin, J.Z. , Karger, A.D. , Budnik, B.A. , Rinn, J.L. , Saghatelian, A. , 2013. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 9, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K. , Kokuryo, T. , Senga, T. , Yokoyama, Y. , Nagino, M. , Hamaguchi, M. , 2010. Novel combination treatment for colorectal cancer using Nek2 siRNA and cisplatin. Cancer Sci. 101, 1163–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani, H. , Torimura, M. , Akimitsu, N. , 2013. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS One 8, e55684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay, Y. , Rinn, J. , Pandolfi, P.P. , 2014. The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen, K. , Mansilla, F. , Schepeler, T. , Oster, B. , Rasmussen, M.H. , Dyrskjot, L. , Karni, R. , Akerman, M. , Krainer, A.R. , Laurberg, S. , Andersen, C.L. , Orntoft, T.F. , 2011. Alternative splicing of SLC39A14 in colorectal cancer is regulated by the Wnt pathway. Mol. Cell Proteomics 10, M110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts, A. , Goff, L. , Pertea, G. , Kim, D. , Kelley, D.R. , Pimentel, H. , Salzberg, S.L. , Rinn, J.L. , Pachter, L. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang, J.S. , Ebert, M.S. , van Oudenaarden, A. , 2010. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol. Cell 38, 140–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky, I. , Bartel, D.P. , 2013. lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesch, S. , van Iterson, M. , Jacobi, J. , Boymans, S. , Essers, P.B. , de Bruijn, E. , Hao, W. , MacInnes, A.W. , Cuppen, E. , Simonis, M. , 2014. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 15, R6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K.C. , Chang, H.Y. , 2011. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. , Yang, M. , Tian, J. , Wang, X. , Li, Z. , 2011. MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int. J. Oncol. 39, 169–175. [DOI] [PubMed] [Google Scholar]
- Yan, J. , Enge, M. , Whitington, T. , Dave, K. , Liu, J. , Sur, I. , Schmierer, B. , Jolma, A. , Kivioja, T. , Taipale, M. , Taipale, J. , 2013. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154, 801–813. [DOI] [PubMed] [Google Scholar]
- Yoon, J.H. , Abdelmohsen, K. , Srikantan, S. , Yang, X. , Martindale, J.L. , De, S. , Huarte, M. , Zhan, M. , Becker, K.G. , Gorospe, M. , 2012. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 47, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M. , Ohira, M. , Li, Y. , Niizuma, H. , Oo, M.L. , Zhu, Y. , Ozaki, T. , Isogai, E. , Nakamura, Y. , Koda, T. , Oba, S. , Yu, B. , Nakagawara, A. , 2009. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int. J. Oncol. 34, 931–938. [DOI] [PubMed] [Google Scholar]
- Zaidi, N. , Swinnen, J.V. , Smans, K. , 2012. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res. 72, 3709–3714. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Yu, M. , Li, Z. , Kong, C. , Bi, J. , Li, J. , Gao, Z. , Li, Z. , 2011. ncRAN, a newly identified long noncoding RNA, enhances human bladder tumor growth, invasion, and survival. Urology 77, 510–515. [DOI] [PubMed] [Google Scholar]
- Zimmerman, K. , Alt, F.W. , 1990. Expression and function of myc family genes. Crit. Rev. Oncog 2, 75–95. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data


