Abstract
1. Background
Pancreatic adenocarcinoma patients have low survival rates due to late‐stage diagnosis and high rates of cancer recurrence even after surgical resection. It is important to understand the molecular characteristics associated with survival differences in pancreatic adenocarcinoma tumors that may inform patient care.
2. Results
RNA sequencing was performed for 51 patient tumor tissues extracted from patients undergoing surgical resection, and expression was associated with overall survival time from diagnosis. Our analysis uncovered 323 transcripts whose expression correlates with survival time in our pancreatic patient cohort. This genomic signature was validated in an independent RNA‐seq dataset of 68 additional patients from the International Cancer Genome Consortium. We demonstrate that this transcriptional profile is largely independent of markers of cellular division and present a 19‐transcript predictive model built from a subset of the 323 transcripts that can distinguish patients with differing survival times across both the training and validation patient cohorts. We present evidence that a subset of the survival‐associated transcripts is associated with resistance to gemcitabine treatment in vitro, and reveal that reduced expression of one of the survival‐associated transcripts, Angiopoietin‐like 4, impairs growth of a gemcitabine‐resistant pancreatic cancer cell line.
3. Conclusions
Gene expression patterns in pancreatic adenocarcinoma tumors can distinguish patients with differing survival outcomes after undergoing surgical resection, and the survival difference could be associated with the intrinsic gemcitabine sensitivity of primary patient tumors. Thus, these transcriptional differences may impact patient care by distinguishing patients who would benefit from a non‐gemcitabine based therapy.
Keywords: Pancreatic adenocarcinoma, ANGPTL4, Gemcitabine, RNA-seq
Highlights
RNA expression patterns are associated with pancreatic cancer patient survival time.
A 19‐transcript model can predict patient survival time in multiple cohorts.
A subset of survival transcripts is associated with gemcitabine response in vitro.
Reduced expression of ANGPTL4 impairs growth of a gemcitabine resistant cell line.
Abbreviations
- ANGPTL4
Angiopoietin‐like 4
- B3GNT5
UDP‐GlcNAc: betaGal beta‐1,3‐N‐acetylglucosaminyltransferase 5
- CTSH
Cathepsin H
- FDR
False Discovery Rate
- ICGC
International Cancer Genome Consortium
- KLF6
Kruppel‐Like Factor 6
- LASSO
L1 penalized log partial likelihood
- OCT
Optimal Cutting Temperature Compound
- PCNA
Proliferating Cell Nuclear Antigen
- PPL
Periplakin
- RGS20
Regulator of G‐protein Signaling 20
- RNA‐seq
RNA sequencing
- SAMD9
Sterile Alpha Motif Domain‐containing 9
- SCUBE1
Signal peptide, CUB domain, EGF‐like 1
- SLC16A3
Solute Carrier Family 16, member A3
- TCGA
The Cancer Genome Atlas
- TRC6C
Trinucleotide Repeat Containing 6C
- WFDC1
WAP Four‐Disulfide Core Domain 1
1. Introduction
Pancreatic adenocarcinoma cancer survival statistics are disheartening, with only 7% of patients surviving to 5 years post‐diagnosis, and the majority of patients succumbing to this disease within one year from diagnosis (Siegel et al., 2015). The high mortality rate associated with pancreatic cancer is due to late‐stage diagnosis and limited efficacy of the current chemotherapeutic arsenal. Due to the asymptomatic nature of pancreatic tumors, 53% of pancreatic cancer patients are not diagnosed until the tumor has metastasized. If the tumor is detected prior to metastasis, surgical resection can be combined with chemotherapy and radiation therapy, increasing the 5 year survival rate to ∼26%. However, only ∼10–15% of patients are diagnosed early enough for surgical resection to be feasible, and ∼80% of these patients relapse within 2 years of surgery because the chemotherapy does not eradicate all tumor cells (Heinemann and Boeck, 2008; Stathis and Moore, 2010). The CONKO‐001 trial recently reported a median survival time of 22.8 months for patients that undergo curative resection and are treated with adjuvant gemcitabine (Oettle et al., 2013). This indicates only marginal improvements in survival time have been achieved in the three decades since the GISTG trial, which reported a median survival time of 21 months, highlighting the need for significant progress to improve the outcomes of these early‐stage patients given the considerable risks and financial burdens imposed by surgery (Kalser and Ellenberg, 1985).
We have conducted RNA‐seq analysis on a cohort of 51 pancreatic adenocarcinoma primary tumor tissues with the goal of understanding survival differences within early‐stage pancreatic cancer patients with localized disease. Our cohort includes a significant fraction of long‐term survivors (>3 years), which provide the opportunity to understand how these patients differ from short‐term survivors. Previous studies investigating gene expression and pancreatic survival have made valuable findings (Bailey et al., 2016; Collisson et al., 2011; Donahue et al., 2012; Newhook et al., 2014; Stratford et al., 2010; Wu et al., 2011). However, there is still a need to identify distinguishing molecular phenotypes in pancreatic tumors that will aid in directing patient care. We present novel transcript expression differences in primary tumors from early‐stage pancreatic patients with vastly different survival times, and we demonstrate that these expression differences have prognostic value in multiple patient cohorts. We link a subset of these molecular alterations to intrinsic transcriptome differences that drive the response to gemcitabine treatment in vitro. Furthermore, we provide evidence that reduction of Angiopoietin‐like 4 (ANGPTL4) expression, a gene expressed higher in short‐term survivors, impairs chemoresistant pancreatic cancer cell growth.
2. Material and methods
2.1. Patient tissue
Pancreatic adenocarcinoma tissues used for this study were collected at the University of Alabama at Birmingham between November 2003 and June 2011 from patients undergoing curative surgical resection to remove pancreatic tumor tissue under an IRB‐approved protocol (Table 1). Patients were treatment‐naïve prior to tumor resection, and the majority of the patients were treated with gemcitabine/nucleoside analog therapy post‐resection (Supplemental Table 1). Pancreatic tissue was flash‐frozen in liquid nitrogen and stored at −80 °C.
Table 1.
Clinical information for 51 patients used in this study and 68 patients in the validation cohort.
| Cohort | Training cohort | Validation cohort | ||||
|---|---|---|---|---|---|---|
| Full | Short survivors | Long survivors | Full | Short survivors | Long survivors | |
| Total samples | 51 | 14 | 13 | 68 | 16 | 11 |
| T stage | ||||||
| 1 | 3 | 0 | 2 | 0 | 0 | 0 |
| 2 | 12 | 3 | 4 | 4 | 0 | 3 |
| 3 | 31 | 10 | 5 | 60 | 14 | 7 |
| 4 | 5 | 1 | 2 | 1 | 0 | 0 |
| TX | 0 | 0 | 0 | 3 | 2 | 1 |
| N stage | ||||||
| N0 | 14 | 3 | 5 | 19 | 5 | 6 |
| N1 | 37 | 11 | 8 | 46 | 9 | 4 |
| NX | 0 | 0 | 0 | 3 | 2 | 1 |
| M stage | ||||||
| M0 | 48 | 13 | 13 | 2 | 0 | 0 |
| M1 | 1 | 0 | 0 | 3 | 2 | 0 |
| MX | 2 | 1 | 0 | 63 | 14 | 11 |
| Median overall survival days (Standard error) | 572 (109.7) | 216 (24.0) | 1544 (230.2) | 413.5 (45.2) | 211.5 (23.6) | 1054 (87.9) |
| Median age (Standard error) | 66 (1.6) | 71 (2.3) | 64 (3.9) | 68 (1.3) | 70.5 (2.9) | 64 (2.6) |
| Diabetes status | ||||||
| Yes | 22 | 4 | 7 | 0 | 0 | 0 |
| No | 29 | 10 | 6 | 0 | 0 | 0 |
| N/A | 0 | 0 | 0 | 68 | 16 | 11 |
| Sex | ||||||
| Male | 32 | 9 | 8 | 33 | 7 | 5 |
| Female | 19 | 5 | 5 | 35 | 9 | 6 |
| Race | ||||||
| Caucasian | 44 | 10 | 12 | 0 | 0 | 0 |
| African‐American | 6 | 3 | 1 | 0 | 0 | 0 |
| N/A | 1 | 1 | 0 | 68 | 16 | 11 |
2.2. Macrodissection
Frozen tissues from pancreatic cancers were macrodissected to enrich the specimens in cancer. For each case, the original diagnostic slides were reviewed by the pathologist (WEG) to determine the morphology of the pancreatic cancer. The frozen tissue was embedded in optimal cutting temperature compound (OCT) and a frozen section was cut that was orientated by ink dots to the frozen tissue in the OCT block. Using the orientated frozen section as a guide, areas of uninvolved pancreas, lymphocytic infiltration, and other non‐malignant tissues were removed from the OCT block by carefully separating unwanted tissues using a single edge razor blade. The proportion of tumor and the percent tumor nuclei were estimated in the macrodissected specimens. Of note, about 30% of cases could not be macrodissected successfully, usually due to an inadequate amount of cancer in the frozen tissue or because intermixed non‐malignant tissue could not be separated adequately from the cancer cells. These were not included in the cases evaluated.
2.3. Source and culture of pancreatic cancer cell lines
AsPC‐1, Capan‐1, Capan‐2, CFPac‐1, HPAF, and PANC‐1 were obtained from ATCC. L3.6pl was obtained from Isaiah J. Fidler at The University of Texas MD Anderson Cancer Center (Bruns et al., 1999). Panc 2.03 and Panc 6.03 were obtained from Elizabeth M. Jaffee at Johns Hopkins University (Jaffee et al., 1998). Suit‐2, S2‐013, and S2‐VP10 were obtained from Michael A. Hollingsworth at The University of Nebraska Medical Center (Akisawa et al., 1999; Iwamura et al., 1997). S2‐LM7‐AA and S2‐LM7‐YB were derived from liver metastases produced from Suit‐2 (LR McNally and DJ Buchsbaum, unpublished). All cell lines were expanded upon receipt to prepare frozen cell stocks that were tested to confirm lack of mycoplasma contamination and cultured in vitro for no longer than 3 months after thawing.
2.4. Nucleic acid extraction from tissues and cell lines
To extract nucleic acid, tissues were homogenized in Qiagen RLT buffer + 1% BME using an MP FastPrep‐24 and Lysing Matrix D beads for three rounds of 45 s at 6.5 m/s (FastPrep homogenizer, Lysing Matrix D, MP Bio, Santa Ana, CA, USA). RNA was extracted from 350 μL tissue lysate (corresponding to 10 mg of tissue) using Norgen Animal RNA extraction kit according to manufacturer protocol (Norgen Animal Tissue RNA Purification Kit, Norgen Biotek Corporation, Thorold, ON, CAN).
A panel of 14 pancreatic cancer cell lines was screened for gemcitabine sensitivity by exposure to serial dilutions of the drug to calculate an IC50 value for each line. The lines were classified as sensitive or resistant based on resulting IC50 values. Subsequently, treatment naïve cells were grown in 100 mm tissue culture dishes, scraped to dissociate cells from the dish, pelleted, and frozen in liquid nitrogen. Upon thawing, cells were resuspended in PBS, and total RNA was extracted from ∼2,500,000 cells from each cell line using the Norgen Animal RNA extraction kit according to manufacturer protocol.
2.5. RNA‐seq library construction
RNA sequencing libraries were constructed using Tn‐RNA‐seq, a transposase‐mediated construction method described previously (Gertz et al., 2012). Four RNA‐seq libraries were pooled into each lane and sequenced using Illumina HiSeq 2000 instruments to generate paired‐end 50 reads (Illumina, San Diego, CA, USA). Read‐pairs (average of ∼47 million read‐pairs per library across 51 RNA‐seq libraries) were aligned to Gencode (version 9.0) using TopHat (version 1.4.1), and the relative abundance of each transcript was quantified using Cufflinks (version 1.3.0) and BEDTools (Harrow et al., 2006, 2002, 2009, 2010).
2.6. Differential expression analysis
The R package DESeq2 (version 1.8.1) was used to evaluate differential expression in pancreatic tumor tissues using a categorical variable. The two classes were: 1) patients who succumbed to the disease within 300 days from diagnosis (short‐survivors) and 2) patients who survived for at least 900 days from diagnosis (long‐survivors). DESeq2 assumes a negative binomial distribution to account for the over‐dispersion in counts in RNA‐seq data (Love et al., 2014). Transcripts with a DESeq2 FDR‐adjusted p‐value <5% were classified as significant. Transcripts expressed from X and Y‐chromosomes were removed prior to differential expression analysis. Pathway analysis was conducted using the web‐based tool LRPath using all GO term annotations, adjusting to transcript read count with RNA‐Enrich, including directionality and limiting maximum GO term size to 500 genes (Kim et al., 2012). The Cytoscape Enrichment Map plug‐in was used for visualization (Isserlin et al., 2014). The Genesetfile (.gmt) from July 24, 2015 was downloaded from http://download.baderlab.org/EM_Genesets/. Mapping parameters were: p‐value cutoff = 0.005, FDR cutoff = 0.1 and overlap coefficient = 0.5. Networks were exported as PDFs.
For the cell line analysis, DESeq2 version (1.8.1) was used to evaluate differential expression in cell lines using a categorical variable based on whether the cell line was sensitive (IC50 < 60 nM) or resistant (IC50 > 300 nM) to gemcitabine treatment after three days of treatment. Transcripts with a DESeq2 FDR‐adjusted p‐value <5% were classified as significant. Transcripts expressed from X and Y‐chromosomes were removed prior to differential expression analysis.
2.7. Random forest modeling
Random forest predictive models were trained using differentially expressed transcripts (FDR < 0.05) between short‐ and long‐survivors in the training cohort, clinical data (patient age, diabetes, gender, race and tumor stage), and 1000 randomly sampled transcript sets equivalent in size to the number of differentially expressed transcripts via the R based “randomForest” package (version 4.6–12). Models were generated as described by Griffith et al. except 501 trees were used to generate each model (Griffith et al., 2013). Model performance was assessed by out of bag error (OOB) reported by the “randomForest” function.
2.8. Hierarchical clustering
Hierarchical clustering was performed on variance‐stabilized RNA‐seq data using the hclust command in R (R version 3.2.1).
2.9. LASSO model selection
A predictive survival model was generated from transcripts differentially expressed (DESeq FDR <0.05) between short‐ and long‐survival patients in the training set using multivariate linear regression with L1 penalized log partial likelihood (LASSO) (Simon et al., 2011; Tibshirani, 1996) for feature selection. LASSO was performed with the R package “glmnet” (version 1.9–8) and the penalty parameter, λ, was selected based on three‐fold cross‐validation within the training set (Friedman et al., 2010). The resulting model was evaluated on the ICGC Australian cohort of patients as a validation. The thresholds for dichotomization (300 and 900 days) used in the validation set were identical to the training set. ICGC data was retrieved from release 18 on January 21st, 2015 at https://dcc.icgc.org/repository (Zhang et al., 2011). Model performance was evaluated based on the ability to classify test set patients as short‐survivors or long‐survivors and an area under the curve (AUC) value was generated with the R package “ROCR” (version 1.0–7) (Sing et al., 2005). Kaplan–Meier curves, Cox proportional hazards models and Concordance‐indexes (C‐Index) were generated with the R packages “survival” (version 2.38–1) and “survcomp” (version 1.16.0). C‐Index was assessed using the “concordance.index” function in the “survcomp” package with default settings. C‐Index is one of the most widely used performance measures for survival models and represents a non‐parametric ranking analysis interpreted as the proportion of all pairs of samples whose predicted survival time is correctly ordered out of all samples capable of being comparably ordered after censoring (Harrell, 2001).
2.10. Tumor subtype classification
Tumors from the training and validation cohorts were classified into the subtypes previously described by Bailey et al. (Bailey et al., 2016). Validation cohort classification was performed by simply using the labels provided by Bailey et al. in supplemental table 14. The training cohort was classified by performing k‐means clustering using the R “kmeans” function with k = 4 on the 613 subtype informative transcripts listed in Bailey et al. supplemental table 14. Assigned clusters showed nearly identical expression patterns to the validation cohort and transcript sets highly expressed in each validation cohort subtype were used to assign the proper label to each subtype in the training cohort.
2.11. Meta‐PCNA analysis
Meta‐PCNA, a previously established index of transcripts positively correlated with the proliferation marker PCNA, was used to determine if the survival transcript signature was proliferation independent (Venet et al., 2011). The meta‐PCNA value was established for each patient by using the median variance stabilized expression value for the 130 meta‐PCNA genes expressed in our patient cohort. Mann–Whitney tests were performed to demonstrate that meta‐PCNA values were not significantly different between short‐surviving and long‐surviving patients (p‐value > 0.05). Likelihood ratio tests were performed with the R package “lmtest” (version 0.9–33) (Zeileis and Hothron, 2002) to determine if our prognostic model still significantly improved classification of patients as short‐surviving or long‐surviving compared to a reduced model containing only the meta‐PCNA value in both the training cohort of patients and the validation cohort.
2.12. WGCNA analysis
The gene co‐expression network for ANGPTL4 was defined using the R package “WGCNA” (version 1.51) (Langfelder and Horvath, 2008). Clustering was performed on the 5000 protein‐coding transcripts with highest standard deviation across the training cohort using the “blockwiseModules” function with a minimum module size of 30. Pathway analysis on the genes present in co‐expression module containing ANGPTL4 was performed using DAVID (da Huang et al., 2009) and the 5000 genes used in the WGCNA clustering analysis were used as the background gene set.
2.13. ANGPTL4 knockdown
PANC‐1 cells were transfected using the appropriate Lonza (Cologne, Germany) Nucleofection kit (Kit R, Program X‐005) with either one of two siRNA directed towards ANGPTL4 (ThermoFisher, Massachusetts, USA) or a scrambled control siRNA. Transfected cells were seeded in 96 well opaque tissue culture‐treated plates (Greiner‐Bio One, North Carolina, USA) at a density of 5000 cells/well, and incubated overnight to allow for cell adherence. Twenty‐four hours post‐nucleofection, cells were dosed with gemcitabine (ThermoFisher, Massachusetts, USA) at concentrations ranging from 0 to 500 μM. Cytotoxicity levels were measured using the CellTiter‐Glo luminescent cell viability assay (Promega, Wisconsin, USA) 24, 48, and 72‐h post‐gemcitabine treatment. Knockdown efficiency of ANGPTL4 was measured via qPCR using an ANGPTL4 taqman probeset (ThermoFisher, Massachusetts, USA). CyQUANT proliferation assay (ThermoFisher, Massachusetts, USA) was used as an orthogonal method for measuring cell viability 48 and 72‐h post‐gemcitabine treatment.
To generate stable ANGPTL4 knockdown cells, three shRNAs targeting ANGPTL4 were obtained from Sigma (Sigma, Missouri, USA) and lentiviral vectors were generated for the ANGPTL4 shRNAs and a GFP control shRNA as previously described (Kutner et al., 2009). One million Panc‐1 cells were transduced with the lentiviral vectors and treated with puromycin (2 μg/ml) to generate stable cell lines. Cytotoxicity measurements and qPCR were performed as described above, with the exception that we plated 1000 cells/well for the cytotoxicity assays. To measure ANGPTL4 protein expression levels, we collected media from each of the cell lines, and performed a human ANGPTL4 enzyme‐linked immunosorbent assay (ELISA) as per manufacturer instruction (ThermoFisher, Massachusetts, USA).
To investigate if ANGPTL4 expression associated with survival in TCGA (The Cancer Genome Atlas) data, Level 3 TCGA RNA‐seq and clinical data was downloaded from the TCGA data portal (https://tcga‐data.nci.nih.gov/tcga/) on November 13th, 2015. “Days_to_death” and/or “Days_to_last_follow_up” columns in the clincal_patient_paad file were used to acquire censored survival times from the patient cohort. Kaplan–Meier analysis was performed as described above to compare survival differences between the top quartile of ANGPTL4 expressers to the bottom quartile.
3. Results
3.1. Gene expression correlates with pancreatic patient survival time
To investigate gene expression patterns with clinical utility in pancreatic cancer, we performed RNA‐seq on a cohort of 51 fresh‐frozen pancreatic tumor tissues (Table 1). Specifically, we were interested in exploring whether RNA transcript expression patterns correlated with patient survival. We employed a dichotomized approach, investigating transcriptional differences occurring in the tumors of patients with extreme survival differences, and included the top and bottom ∼25% of our total patient cohort. We observed 323 differentially expressed transcripts (FDR p‐value < 0.05) between the short‐survivors (n = 14) and the long‐survivors (n = 13). Of these 323 transcripts, 176 have higher expression in the short‐survival patients, and 147 have a higher expression in the long‐survival patients (Figure 1A; Supplemental Table 2). Expression patterns in our cohort strongly overlap with an independent cohort with 20 transcripts significant (FDR < 0.05) in both cohorts (Fisher's Exact p = 2.87e‐14). Transcripts with higher expression in the long‐survivors were enriched for immune response cellular pathways and calcium signaling pathways, whereas transcripts with higher expression levels in the tumors of short‐survivors were enriched for cell cycle regulation, EGFR signaling, cell adhesion, vesicle transport, peptidase regulation, apoptosis, protein glycosylation and keratinocyte development (Figure 1B, Supplemental Table 3). We broadly assessed the prognostic utility of these significant transcripts relative to randomly generated sets of transcripts and clinical information (including patient age, gender, race, tumor stage, nodal involvement and diabetes status) using random forest models and found that our significant transcripts far outperformed both random transcripts and clinical data (Supplemental Fig. 1).
Figure 1.
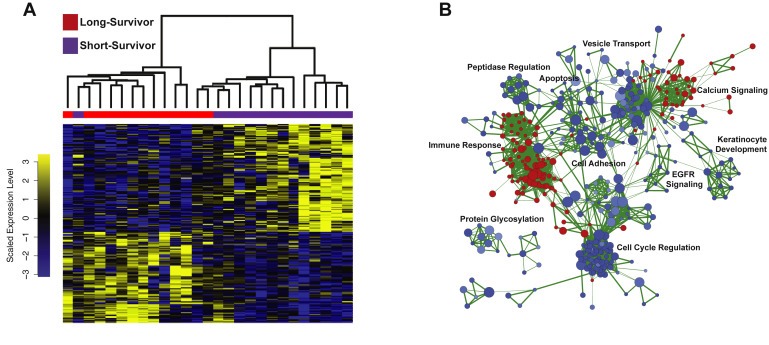
Transcripts significantly different between pancreatic short‐ survivors and long‐survivors. A) Heatmap of 323 transcripts significantly different between the short‐survivors and long‐survivors. Blue is low expression and yellow is high expression. B) Network diagram of enriched pathways. Blue is higher expression in short‐survivors and red is higher expression in long‐survivors.
3.2. A 19‐transcript signature successfully predicts patient survival
To prioritize transcripts most predictive of patient survival, we employed the multivariate linear regression with L1 penalized log partial likelihood (LASSO) for feature selection (Figure 2) (Simon et al., 2011; Tibshirani, 1996). A 19‐transcript model was identified that successfully discriminated short‐survival from long‐survival patients in the training cohort, and this model differentiated the short‐ and long‐surviving patients in an independent cohort of pancreatic RNA‐seq data from ICGC (Zhang et al., 2011) (Figure 3; Supplemental Table 4). The 19‐transcript model also demonstrated success as a continuous Cox proportional hazards model, generating a concordance index of 0.82 (p = 1.46e‐23) in the training cohort and a concordance index of 0.60 (p = 0.04) in the validation cohort (n = 68). Furthermore, Kaplan–Meier curves demonstrate that patients in the top and bottom tertiles of predicted survival have significantly different survival times in both cohorts (Figure 4). Thus, despite being optimized for distinguishing patients on the extremes of the prognosis spectrum, our model has significant predictive power when applied to full patient cohorts as a continuous model. We further investigated the performance of our survival model in the context of each of the four subtypes described by Bailey et al. (Bailey et al., 2016). Although sample sizes are insufficient to make definitive interpretations in either cohort, it appears our model successfully distinguishes the squamous subtype previously associated with poor prognosis from the other three subtypes in both the validation cohort and training cohort (Supplemental Fig. 2).
Figure 2.
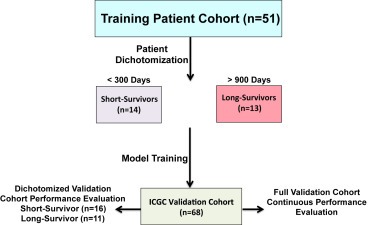
Analysis workflow for prognostic model training and validation. Patients in the training cohort were dichotomized into long‐ (survived > 900 days, n = 13) and short‐survivor (survived < 300 days, n = 14) groups. A multivariate linear regression model with LASSO for feature selection capable of classifying patients as long‐ or short‐survivors was developed on the training cohort. Next the model was applied to an independent validation cohort. We applied the model both to pre‐dichotomized long (n = 11) and short (n = 16) survivors as well as the whole cohort as a continuous model.
Figure 3.
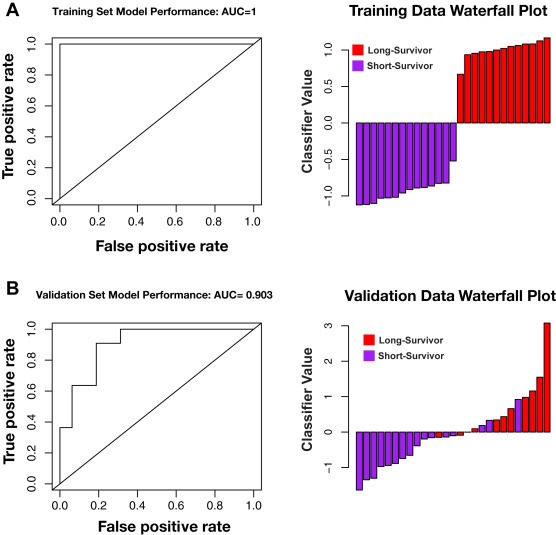
Receiver operating characteristic (ROC) curve and corresponding waterfall plots for the 19‐transcript LASSO regression model. A) Model performance on the training cohort (AUC = 1). B) Model performance on the validation cohort (AUC = 0.90). Waterfall plot data is centered at the optimal classifier threshold as defined in the training cohort.
Figure 4.
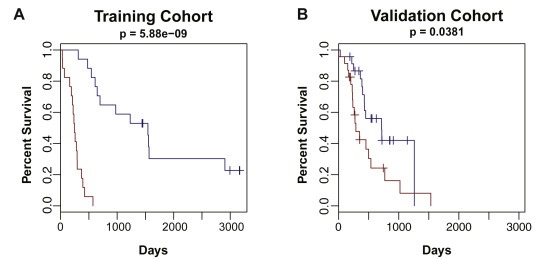
Kaplan–Meier analysis of the 19‐transcript model on the full training cohort (left) and validation cohort (right). A Cox regression model was trained on the full training cohort using the 19 transcript predictive model on short‐survival and long‐survival patients and evaluated on the full validation cohort. The short‐survival curve (red) represents patients in the lowest tertile of predicted survival time and the long‐survival curve (blue) represents patients in the top tertile of predicted survival time.
The 19‐transcript model includes genes with a variety of functions, many of which have been implicated in cancer previously (Supplemental Table 4). Solute Carrier Family 16, member A3 (SLC16A3) has a higher expression level in the short‐surviving patient tumors, and encodes a monocarboxylate transporter involved in a broad range of cellular metabolic pathways including energy metabolism of tumor cells and drug transport (Halestrap, 2013). SCL16A3 has higher expression in breast cancer metastases in comparison to breast primary tumors, and is a component of a gene signature that has been linked to poor outcome in multiple cancers, including breast, lung, and glioblastoma (Hu et al., 2009). Regulator of G‐Protein Signaling 20 (RGS20) also has higher expression in the short‐survivor patient tumors. RGS20 inhibits signal transduction of G‐protein signaling, and RGS20 has higher expression in metastatic melanoma when compared to primary melanoma (Riker et al., 2008). Trinucleotide Repeat Containing 6C (TRC6C) has higher expression in long‐surviving patient tumors and functions to promote microRNA‐mediated gene silencing (Huntzinger et al., 2010; Jinek et al., 2010). Finally, WAP Four‐Disulfide Core Domain 1 (WFDC1), which has higher expression in the long‐surviving patient tumors, is a secreted protease that is involved in inhibiting cell proliferation of tumor cells (Madar et al., 2009).
3.3. Survival is not explained by cellular proliferation differences
A recent study suggested that, in breast cancer, transcripts can be associated with patient outcome simply due to the high correlation of global gene expression patterns with proliferation, and demonstrated that 90% of randomly selected sets of 100 transcripts or more are statistically correlated with patient outcome (Venet et al., 2011). This study points to the underlying importance of cellular proliferation in overall patient outcome in breast cancer and leads to a larger question of whether the effect of cellular proliferation rates on gene expression predicts prognosis in other cancer types.
We used the established meta‐PCNA index of 131 transcripts (130 of which were expressed in our data set) presented in the Venet et al. study (Venet et al., 2011) to address whether the RNA expression pattern we present here is dependent on cellular proliferation. The meta‐PCNA index is composed of the top 1% of transcripts most highly correlated with Proliferating Cell Nuclear Antigen (PCNA), in gene expression measurements from 36 different human normal tissues (Ge et al., 2005). In both the training and validation cohorts, the meta‐PCNA values were not significantly different between the short‐survival and long‐survival patient populations (Mann–Whitney p‐value = 0.4583 and p‐value = 0.1779, respectively) (Supplemental Figure 3A).
To investigate the effects of proliferation on our datasets further, we explored whether the meta‐PCNA index affected the power of our 19‐transcript LASSO model for distinguishing short‐survival and long‐survival patients. We performed a likelihood ratio test using the meta‐PCNA index as the null model for both the training and validation cohorts, and discovered that in both patient cohorts, the 19‐transcript model added predictive information over the meta‐PCNA index alone (training p‐value = 4.443e‐05; validation p‐value = 8.124e‐04). Furthermore, Principle Component Analysis (PCA) demonstrated that our 19‐transcript model, both prior to and after normalization to the meta‐PCNA index, was able to distinguish the short‐survival and long‐survival patient populations (Supplemental Figure 3B). These data provide evidence to suggest that our gene signature is largely independent of established markers for cellular proliferation.
3.4. A subset of gemcitabine sensitivity genes is differentially expressed between the short‐survival and long‐survival patients
To explore the idea that resistance to adjuvant therapy may play a role in the survival time and contribute to our gene expression signature, we investigated whether any of our 323 significant transcripts overlapped with transcripts that are intrinsically differentially expressed in pancreatic cancer cell lines that are sensitive or resistant to gemcitabine (Supplemental Table 5A). The majority of the short‐survival and long‐survival patients for which we have complete treatment information were treated with gemcitabine (Supplemental Table 1). We performed RNA‐seq and compared gene expression patterns in seven gemcitabine‐sensitive pancreatic cancer cell lines and seven gemcitabine‐resistant pancreatic cancer cell lines, and we identified ∼1300 differentially expressed transcripts. In vitro expression differences strongly overlapped with patient data, as twenty‐two transcripts were significant in both analyses (Fishers‐exact p‐value = 2.587e‐04; Supplemental Table 3B).
A subset of the overlapping transcripts has intriguing biological links to cancer (Figure 5). One of these overlapping transcripts, UDP‐GlcNAc:betaGal beta‐1,3‐N‐acetylglucosaminyltransferase 5 (B3GNT5), is an enzyme responsible for transferring N‐acetylglucosamine (GlcNAc) to glycolipid substrates and is associated with breast cancer survival (Potapenko et al., 2015). Cathepsin H (CTSH) is a proteinase that has been implicated in angiogenic switching, vascularization, and growth of mouse models of pancreatic islet cell cancer (Gocheva et al., 2010). Sterile alpha motif domain containing 9 (SAMD9) encodes for a protein that localizes to the cell membrane and is thought to play a role in regulating cellular proliferation and apoptosis (Tanaka et al., 2010). Periplakin (PPL) is a component of desmosomes, and expression of PPL results in cisplatin resistance in endometrial carcinoma cells (Suzuki et al., 2010). Kruppel‐Like Factor 6 (KLF6) is a transcription factor with multiple splice variants, and expression of one of these splice variants in HepG2 cell lines increases gemcitabine sensitivity (Hanoun et al., 2015). Furthermore, specific splice‐isoforms of KLF6 have been previously associated with pancreatic cancer prognosis and tumor grade (Hartel et al., 2008; Stratford et al., 2010). Increased expression of Signal peptide, CUB domain, EGF‐like 1 (SCUBE1) in prostate cancer associated fibroblast cells reduced prostate tumor size in mouse models (Orr et al., 2013). Given our findings that these transcripts are associated with survival in pancreatic cancer and gemcitabine sensitivity in pancreatic cancer cell lines, these transcripts represent potential targets for sensitizing patients to adjuvant gemcitabine treatment.
Figure 5.
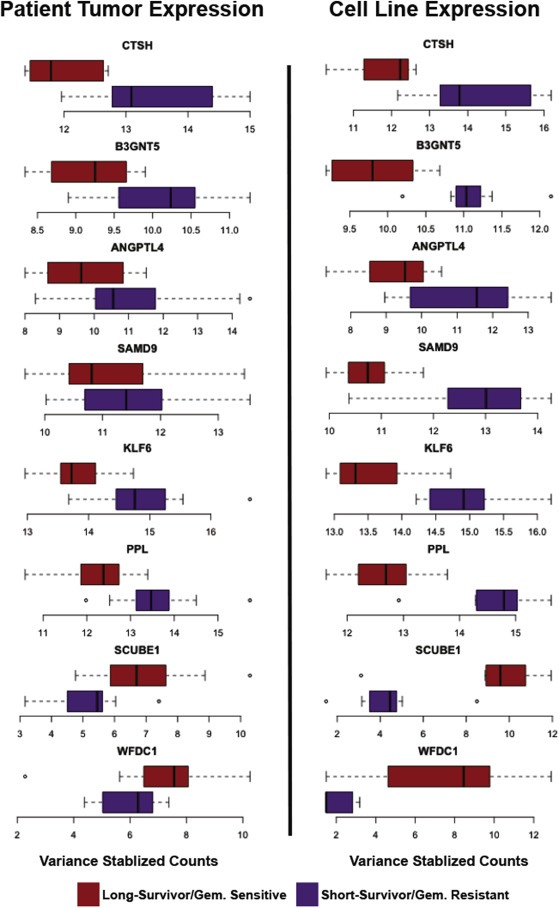
Boxplots of transcripts significant between both short‐survival and long‐survival patients and gemcitabine‐sensitive and ‐resistant cell lines. Boxplots of DESeq2 variance‐stabilized data in pancreatic cancer cell lines and patient tissues demonstrate that most of these overlapping transcripts have a higher expression in the short‐survival patients and the gemcitabine‐resistant pancreatic cell lines. Purple represents short‐survivors/resistant cancer cell lines and red represents long‐survivors/sensitive cancer cell lines.
3.5. Knockdown of ANGPTL4 expression in a gemcitabine‐resistant pancreatic cancer cell line affects cell proliferation
We were particularly interested in further investigating the functional role of Angiopoietin‐Like 4 (ANGPTL4) in pancreatic cancer cells. ANGPTL4 had higher expression in pancreatic cancer cell lines resistant to gemcitabine therapy and patients with shorter survival, and these expression patterns were confirmed via qPCR (Supplemental Figure 4A and B). Furthermore, we observed ANGPTL4 expression significantly associated with survival time in the validation and TCGA cohorts of pancreatic adenocarcinoma RNA‐seq datasets (Supplemental Figure 4C). We again separated patients into the four Bailey et al. pancreatic cancer subtypes, and observed that ANGPTL4 is expressed highest in the squamous subtype (Supplemental Figure 5A and B) and ANGPTL4 expression levels are able to prognostically stratify patients within the squamous subtype of both cohorts (Supplemental Figure 5C and D) (Bailey et al., 2016). We used Weighted Gene Co‐Expression Network Analysis (WGCNA) to determine what protein‐expressed genes are being co‐expressed with ANGPTL4, and observed enrichment for genes involved in ectoderm and epidermis differentiation (Supplemental Table 6).
PANC‐1 cells are resistant to gemcitabine treatment (IC50 > 300 nM after 3 days of treatment), so we were interested in knocking down ANGPTL4 expression in this cell line to determine effects of knockdown on gemcitabine response. We reduced ANGPTL4 expression in PANC‐1 cells using siRNA or shRNA knockdown, and we confirmed efficient knockdown through qPCR analysis of ANGPTL4 expression and ANGPLT4 ELISA of cell culture media (Figure 6A, Supplemental Figure 6A and B). We observed a striking reduction of PANC‐1 cell growth at all time points after ANGPTL4 knockdown (Figure 6B, Supplemental Figure 5C). This growth effect was independent of the effects of gemcitabine treatment as cells treated with both a siRNA targeting ANGPTL4 and gemcitabine doses ranging from 0 to 500 μM showed less proliferation than cells treated with gemcitabine alone (Figure 6C).
Figure 6.
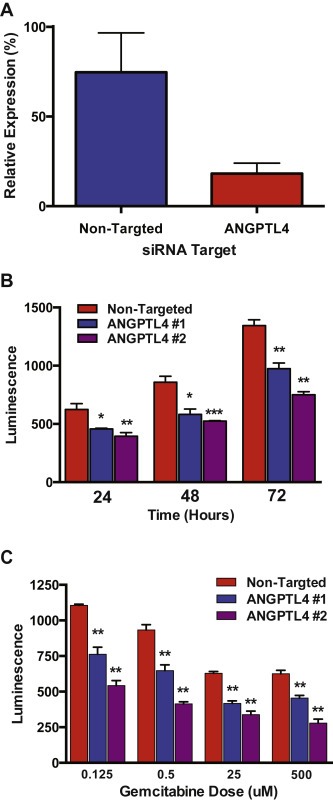
ANGPTL4 knockdown in Panc‐1 cells. A) qPCR data demonstrates efficient knockdown of ANGPTL4 at 48 h post‐nucleofection. B) ANGPTL4 knockdown in Panc‐1 cells results in a decrease of cell proliferation. C) The cell proliferation effect of ANGPTL4 knockdown is independent of the effects of gemcitabine treatment.
4. Discussion
With a 7% overall 5‐year survival rate, there is a clear and present need for improvement of the clinical care of pancreatic cancer patients, regardless of stage at diagnosis. While the survival time for patients diagnosed with operable disease is considerably longer than those diagnosed with inoperable disease, it is still a relatively short time frame given the morbidity associated with surgical resection. Among patients with resectable disease, median survival has only improved from 21 months in 1985 to 22.8 months in 2014 (Oettle et al., 2013). Recent studies in metastatic disease found combination therapy with gemcitabine‐based and non‐gemcitabine containing therapy are superior to single‐agent gemcitabine toward progression‐free and overall survival (Conroy et al., 2011; Von Hoff et al., 2011). As these treatment regimens are increasingly explored in the earlier stages of disease, our expression signature may aid in stratifying patients that may benefit from a non‐gemcitabine based therapy regimen.
While many factors contribute to a patient's overall survival, we explored biological differences that are associated with survival differences in pancreatic patients with similar clinical characteristics. Our study identified a set of transcripts differentially expressed in tumors of pancreatic cancer patients that were diagnosed early enough for curative surgery, but who had vastly differing survival times. Our confidence in these biological differences is bolstered by the ability of these expression patterns to distinguish patients with differing survival times in the validation cohort of pancreatic tumor tissues, which, to our knowledge, is the largest pancreatic cancer RNA‐seq dataset available at this time. We confirmed in the validation cohort that neither the short‐survival or long‐survival patients could be classified by mutations in pancreatic cancer driver genes and other clinically relevant genes such as KRAS, TP53, CDKN2A, SMAD4, and USP9X (Biankin et al., 2012; Jones et al., 2008; Pérez‐Mancera et al., 2012; Waddell et al., 2015). Furthermore, the short‐survival and long‐survival patients are not enriched for a particular pancreatic cancer subtype, although interestingly, our data suggest that the 19‐transcript model can distinguish the Squamous subtype, which has a significantly worse survival time than the ADEX, Immunogenic and Progenitor subtypes (Bailey et al., 2016; Collisson et al., 2011).
The biology within these 323 transcripts suggests novel therapeutic interventions, especially for those patients with the very short survival times. We are especially intrigued by the transcripts that we discovered with similar expression comparisons in short‐ vs. long‐survival patients and gemcitabine‐resistant vs. gemcitabine‐sensitive cell lines. It is perhaps not surprising that a mechanism for a shorter survival time after surgery would be resistance to adjuvant chemotherapy. A recent large retrospective study examining the clinical characteristics of pancreatic adenocarcinoma patients surviving 10 years or more highlighted adjuvant chemotherapy as the second most important predictor of extreme survivorship behind lymph node positivity ratio (Paniccia et al., 2015). The comparison of these two genomic analyses has provided us with several genes, many not previously implicated in gemcitabine resistance, which might provide insight into new pathways for therapeutic intervention.
ANGPTL4 is a secreted protein that regulates lipid and glucose metabolism; however, other complex cellular roles are emerging for this protein, including roles in cancer progression and drug response (Tan et al., 2012). ANGPTL4 expression was recently associated with chemosensitivity to cisplatin in ovarian cancer (McEvoy et al., 2015). However, overexpression of ANGPTL4 is associated with lower disease‐free survival in basal breast tumors in young women, and ANGPTL4 copy number gain within circulating tumor cells in breast cancer patients is associated with higher tumor aggressiveness (Johnson et al., 2015; Kanwar et al., 2015). Intriguingly, treatment of orthotopic liver tumors in xenograft models with an antibody to ANGPTL4 resulted in inhibition of tumor growth and metastasis (Ng et al., 2014). These conflicting roles of ANGPTL4 may be driven by differing functions of the N‐terminus and C‐terminus of ANGPTL4, which result from post‐translational cleavage of the protein. Our data suggests that ANGPTL4 functions to promote cell proliferation and is a member of a gene expression network involved in epidermal differentiation and development. Similarly, ANGPTL4 expression is highest in the tumors classified as Squamous subtype and may hold future utility as a simple marker for patients in this subtype with particularly poor prognosis. Therefore, we believe ANGPTL4 represents a novel putative therapeutic target for pancreatic cancer patients and merits further investigation in patients or tumor model systems that demonstrate resistance to gemcitabine.
5. Conclusions
Our investigation has identified gene transcripts that are differentially expressed in pancreatic adenocarcinoma tumor tissues from patients with similar clinical characteristics but vastly differing survival times. We present a 19‐transcript signature that is capable of distinguishing patients with differing survival times, and importantly, we validate the prognostic value of this signature in an independent cohort of patients. It is important to note that currently available pancreatic cancer patient cohorts, including our own, are limited in size and future studies will elucidate the value of this expression signature across larger cohorts of pancreatic adenocarcinoma patients. We discovered that a subset of the survival‐associated transcripts is also differentially expressed in pancreatic cancer cell lines with differing sensitivity to gemcitabine treatment. Through siRNA knockdown experiments, we provide evidence that ANGPTL4, a transcript with higher expression in short‐term survivors and gemcitabine‐resistant pancreatic cancer cells, has a newly discovered role in pancreatic cancer growth, and thus provides a novel therapeutic target for pancreatic cancer treatment.
Author contributions
Conception and Design: MKK, RCR, JAP, WEG, SMV, DJB, SJC, and RMM.
Acquisition of Data: MKK, RCR, JG, NSD, BEJ, PGO, KCS, EWG, JDC, MJH, JAP, WEG, SMV, DJB, SJC, and RMM.
Analysis and interpretation of the data: MKK, RCR, SJC, and RMM.
Writing, review and/or revision of the manuscript: MKK, RCR, JG, PGO, MJH, JAP, WEG, SMV, DJB, SJC, and RMM.
Administrative, technical, or material support: NSD, BEJ, PGO, KCS, EWG, JDC, MJH, JAP, and WEG.
Study Supervision: JAP, WEG, SMV, DJB, SJC, and RMM.
Conflict of interest disclosures
None reported.
Funding/Support
This work was funded by the UAB/UMN SPORE in Pancreatic Cancer (P50CA101955), the NIH‐National Institute of General Medical Sciences Medical Scientist Training Program (5T32GM008361‐21), and by The State of Alabama and the HudsonAlpha Institute.
Availability of data and materials
The datasets supporting the conclusions of this article are available in the GEO databank: GSE79670.
Supporting information
The following are the supplementary data related to this article:
Supplemental Figure 1 Random forest analysis of clinical data, random transcripts, and significant transcripts. Histogram of out of bag errors for random forest models generated on differentially expressed transcripts (red dashed line), clinical data (blue dashed line), and 1000 randomly sampled transcript sets (gray bars). Clinical data consisted of patient age, gender, race, tumor stage, nodal involvement and diabetes status. Significant transcripts were transcripts that met a 5% FDR cutoff in DESeq2 analysis of expression differences between short‐survivors and long‐survivors. Random transcripts sets were generated by randomly sampling from all transcripts measured via RNA‐seq at an equivalent number to the significant transcripts. Differentially expressed transcripts possessed an out of bag error (0.185) lower than all random transcript sets (p < 0.001) while clinical data possessed an out of bag error greater than most random transcript pulls (0.593, p = 0.979).
Supplemental Figure 2 LASSO model performance across pancreatic cancer subtypes. Boxplots of the first principle component of the 19 transcripts included in the LASSO prognostic model across all 4 pancreatic cancer subtypes in the training cohort (A) and the validation cohort (B). P‐values represent a Wilcoxon rank sum test between the squamous subtype and the subtype of interest. *** indicates a p‐value < 0.001.
Supplemental Figure 3 Meta‐PCNA analysis. A) Boxplots of meta‐PCNA index values of short‐survival and long‐survival patients in the training cohort (left) and validation cohort (right). B) Principle Component Analysis (PCA) of meta‐PCNA index normalized residual values for the 19 transcripts incorporated in the LASSO model for the UAB training cohort (left) and the ICGC validation cohort (right).
Supplemental Figure 4 ANGPTL4 expression analysis. A) qPCR results of ANGPTL4 expression in three short‐survivors and three long‐survivors. B) qPCR results of ANGPTL4 expression across pancreatic cell lines. C) Kaplan–Meier curves based on ANGPTL4 expression in three independent pancreatic tumor cohorts.
Supplemental Figure 5 ANGPTL4 expression across pancreatic cancer subtypes. Boxplots of ANGPTL4 expression across pancreatic cancer subtypes in the training cohort (A) and the validation cohort (B). p‐Values represent a Wilcoxon rank sum test between the squamous subtype and the subtype of interest. ** indicates a p‐value < 0.01 and *** indicates a p‐value < 0.001. Kaplan–Meier curves of ANGPTL4 expression within the squamous subtype show survival differences between the top half of ANGPTL4 expressers and the bottom half in both the training (C) and the validation cohort (D).
Supplemental Figure 6 Lentiviral knockdown of ANGPTL4 expression in the PANC‐1 pancreatic cancer cell line. A) qPCR results for PANC‐1 cells transduced with lentiviral vectors containing shRNA for control GFP (blue) or three separate shRNAs towards ANGPTL4 (red, green, yellow). B) ELISA measurements of ANGPTL4 protein levels in media collected from transduced PANC‐1 cells. C) Cell proliferation rates in transduced PANC‐1 cells.
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
We thank Drs. Brittany Lasseigne and Kevin Bowling for critical reading of the manuscript. We thank Dr. Nick Cochran for packaging the ANGPTL4 lentiviral vectors. We thank Dr. Shawn Levy, Braden Boone, Angela Jones, and all the members of the HudsonAlpha Genomic Services Lab for providing RNA sequencing data for this project. We acknowledge use of International Cancer Genome Consortium project Australian pancreatic RNA‐seq dataset and The Cancer Genome Atlas pancreatic RNA‐seq dataset, which were both extremely valuable in validation of our findings.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2016.05.004.
Kirby Marie K., Ramaker Ryne C., Gertz Jason, Davis Nicholas S., Johnston Bobbi E., Oliver Patsy G., Sexton Katherine C., Greeno Edward W., Christein John D., Heslin Martin J., Posey James A., Grizzle William E., Vickers Selwyn M., Buchsbaum Donald J., Cooper Sara J., Myers Richard M., (2016), RNA sequencing of pancreatic adenocarcinoma tumors yields novel expression patterns associated with long-term survival and reveals a role for ANGPTL4, Molecular Oncology 10, doi: 10.1016/j.molonc.2016.05.004.
References
- Akisawa, N. , Nishimori, I. , Iwamura, T. , Onishi, S. , Hollingsworth, M.A. , 1999. High levels of ezrin expressed by human pancreatic adenocarcinoma cell lines with high metastatic potential. Biochem. Biophys. Res. Commun. 258, 395–400. 10.1006/bbrc.1999.0653 [DOI] [PubMed] [Google Scholar]
- Bailey, P. , Chang, D.K. , Nones, K. , Johns, A.L. , Patch, A.-M. , Gingras, M.-C. , Miller, D.K. , Christ, A.N. , Bruxner, T.J.C. , Quinn, M.C. , Nourse, C. , Murtaugh, L.C. , Harliwong, I. , Idrisoglu, S. , Manning, S. , Nourbakhsh, E. , Wani, S. , Fink, L. , Holmes, O. , Chin, V. , Anderson, M.J. , Kazakoff, S. , Leonard, C. , Newell, F. , Waddell, N. , Wood, S. , Xu, Q. , Wilson, P.J. , Cloonan, N. , Kassahn, K.S. , Taylor, D. , Quek, K. , Robertson, A. , Pantano, L. , Mincarelli, L. , Sanchez, L.N. , Evers, L. , Wu, J. , Pinese, M. , Cowley, M.J. , Jones, M.D. , Colvin, E.K. , Nagrial, A.M. , Humphrey, E.S. , Chantrill, L.A. , Mawson, A. , Humphris, J. , Chou, A. , Pajic, M. , Scarlett, C.J. , Pinho, A.V. , Giry-Laterriere, M. , Rooman, I. , Samra, J.S. , Kench, J.G. , Lovell, J.A. , Merrett, N.D. , Toon, C.W. , Epari, K. , Nguyen, N.Q. , Barbour, A. , Zeps, N. , Moran-Jones, K. , Jamieson, N.B. , Graham, J.S. , Duthie, F. , Oien, K. , Hair, J. , Grützmann, R. , Maitra, A. , Iacobuzio-Donahue, C.A. , Wolfgang, C.L. , Morgan, R.A. , Lawlor, R.T. , Corbo, V. , Bassi, C. , Rusev, B. , Capelli, P. , Salvia, R. , Tortora, G. , Mukhopadhyay, D. , Petersen, G.M. , Initiative, A.P.C.G. , Munzy, D.M. , Fisher, W.E. , Karim, S.A. , Eshleman, J.R. , Hruban, R.H. , Pilarsky, C. , Morton, J.P. , Sansom, O.J. , Scarpa, A. , Musgrove, E.A. , Bailey, U.-M.H. , Hofmann, O. , Sutherland, R.L. , Wheeler, D.A. , Gill, A.J. , Gibbs, R.A. , Pearson, J.V. , Waddell, N. , Biankin, A.V. , Grimmond, S.M. , 2016. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52. [DOI] [PubMed] [Google Scholar]
- Biankin, A.V. , Waddell, N. , Kassahn, K.S. , Gingras, M.-C. , Muthuswamy, L.B. , Johns, A.L. , Miller, D.K. , Wilson, P.J. , Patch, A.-M. , Wu, J. , Chang, D.K. , Cowley, M.J. , Gardiner, B.B. , Song, S. , Harliwong, I. , Idrisoglu, S. , Nourse, C. , Nourbakhsh, E. , Manning, S. , Wani, S. , Gongora, M. , Pajic, M. , Scarlett, C.J. , Gill, A.J. , Pinho, A.V. , Rooman, I. , Anderson, M. , Holmes, O. , Leonard, C. , Taylor, D. , Wood, S. , Xu, Q. , Nones, K. , Fink, J.L. , Christ, A. , Bruxner, T. , Cloonan, N. , Kolle, G. , Newell, F. , Pinese, M. , Mead, R.S. , Humphris, J.L. , Kaplan, W. , Jones, M.D. , Colvin, E.K. , Nagrial, A.M. , Humphrey, E.S. , Chou, A. , Chin, V.T. , Chantrill, L.A. , Mawson, A. , Samra, J.S. , Kench, J.G. , Lovell, J.A. , Daly, R.J. , Merrett, N.D. , Toon, C. , Epari, K. , Nguyen, N.Q. , Barbour, A. , Zeps, N. , Initiative, A.P.C.G. , Kakkar, N. , Zhao, F. , Wu, Y.Q. , Wang, M. , Muzny, D.M. , Fisher, W.E. , Brunicardi, F.C. , Hodges, S.E. , Reid, J.G. , Drummond, J. , Chang, K. , Han, Y. , Lewis, L.R. , Dinh, H. , Buhay, C.J. , Beck, T. , Timms, L. , Sam, M. , Begley, K. , Brown, A. , Pai, D. , Panchal, A. , Buchner, N. , De Borja, R. , Denroche, R.E. , Yung, C.K. , Serra, S. , Onetto, N. , Mukhopadhyay, D. , Tsao, M.-S. , Shaw, P.A. , Petersen, G.M. , Gallinger, S. , Hruban, R.H. , Maitra, A. , Iacobuzio-Donahue, C.A. , Schulick, R.D. , Wolfgang, C.L. , Morgan, R.A. , Lawlor, R.T. , Capelli, P. , Corbo, V. , Scardoni, M. , Tortora, G. , Tempero, M.A. , Mann, K.M. , Jenkins, N.A. , Perez-Mancera, P.A. , Adams, D.J. , Largaespada, D.A. , Wessels, L.F.A. , Rust, A.G. , Stein, L.D. , Tuveson, D.A. , Copeland, N.G. , Musgrove, E.A. , Scarpa, A. , Eshleman, J.R. , Hudson, T.J. , Sutherland, R.L. , Wheeler, D.A. , Pearson, J.V. , McPherson, J.D. , Gibbs, R.A. , Grimmond, S.M. , 2012. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405. 10.1038/nature11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns, C.J. , Harbison, M.T. , Kuniyasu, H. , Eue, I. , Fidler, I.J. , 1999. In vivo selection and characterization of metastatic variants from human pancreatic adenocarcinoma by using orthotopic implantation in nude mice. Neoplasia 1, 50–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson, E.A. , Sadanandam, A. , Olson, P. , Gibb, W.J. , Truitt, M. , Gu, S. , Cooc, J. , Weinkle, J. , Kim, G.E. , Jakkula, L. , Feiler, H.S. , Ko, A.H. , Olshen, A.B. , Danenberg, K.L. , Tempero, M.A. , Spellman, P.T. , Hanahan, D. , Gray, J.W. , 2011. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503. 10.1038/nm.2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy, T. , Desseigne, F. , Ychou, M. , Bouche, O. , Guimbaud, R. , Becouarn, Y. , Adenis, A. , Raoul, J.L. , Gourgou-Bourgade, S. , de la Fouchardiere, C. , Bennouna, J. , Bachet, J.B. , Khemissa-Akouz, F. , Pere-Verge, D. , Delbaldo, C. , Assenat, E. , Chauffert, B. , Michel, P. , Montoto-Grillot, C. , Ducreux, M. , 2011. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 364, 1817–1825. 10.1056/NEJMoa1011923 [DOI] [PubMed] [Google Scholar]
- da Huang, W. , Sherman, B.T. , Lempicki, R.A. , 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Donahue, T.R. , Tran, L.M. , Hill, R. , Li, Y. , Kovochich, A. , Calvopina, J.H. , Patel, S.G. , Wu, N. , Hindoyan, A. , Farrell, J.J. , Li, X. , Dawson, D.W. , Wu, H. , 2012. Integrative survival-based molecular profiling of human pancreatic cancer. Clin. Cancer Res. 18, 1352–1363. 10.1158/1078-0432.CCR-11-1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J. , Hastie, T. , Tibshirani, R. , 2010. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22. [PMC free article] [PubMed] [Google Scholar]
- Ge, X. , Yamamoto, S. , Tsutsumi, S. , Midorikawa, Y. , Ihara, S. , Wang, S.M. , Aburatani, H. , 2005. Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 86, 127–141. [DOI] [PubMed] [Google Scholar]
- Gertz, J. , Varley, K.E. , Davis, N.S. , Baas, B.J. , Goryshin, I.Y. , Vaidyanathan, R. , Kuersten, S. , Myers, R.M. , 2012. Transposase mediated construction of RNA-seq libraries. Genome Res. 22, 134–141. 10.1101/gr.127373.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocheva, V. , Chen, X. , Peters, C. , Reinheckel, T. , Joyce, J.A. , 2010. Deletion of cathepsin H perturbs angiogenic switching, vascularization and growth of tumors in a mouse model of pancreatic islet cell cancer. Biol. Chem. 391, 937–945. 10.1515/BC.2010.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith, O.L. , Pepin, F. , Enache, O.M. , Heiser, L.M. , Collisson, E.A. , Spellman, P.T. , Gray, J.W. , 2013. A robust prognostic signature for hormone-positive node-negative breast cancer. Genome Med. 5, 1–14. 10.1186/gm496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap, A.P. , 2013. The SLC16 gene family – structure, role and regulation in health and disease. Mol. Aspects Med. 34, 337–349. [DOI] [PubMed] [Google Scholar]
- Hanoun, N. , Bureau, C. , Diab, T. , Gayet, O. , Dusetti, N. , Selves, J. , Vinel, J.-P. , Buscail, L. , Cordelier, P. , Torrisani, J. , 2015. The SV2 variant of KLF6 is down-regulated in hepatocellular carcinoma and displays anti-proliferative and pro-apoptotic functions. J. Hepatol. 53, 880–888. 10.1016/j.jhep.2010.04.038 [DOI] [PubMed] [Google Scholar]
- Harrell, F. , 2001. Regression Modeling Strategies with Applications to Linear Models, Logistic Regression, and Survival Analysis Springer-Verlag; New York: [Google Scholar]
- Harrow, J. , Denoeud, F. , Frankish, A. , Reymond, A. , Chen, C.-K. , Chrast, J. , Lagarde, J. , Gilbert, J.G.R. , Storey, R. , Swarbreck, D. , Rossier, C. , Ucla, C. , Hubbard, T. , Antonarakis, S.E. , Guigo, R. , 2006. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 7, (Suppl. 1) S4.1–S4.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartel, M. , Narla, G. , Wente, M.N. , Giese, N.A. , Martignoni, M.E. , Martignetti, J.A. , Friess, H. , Friedman, S.L. , 2008. Increased alternative splicing of the KLF6 tumour suppressor gene correlates with prognosis and tumour grade in patients with pancreatic cancer. Eur. J. Cancer 44, 1895–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann, V. , Boeck, S. , 2008. Perioperative management of pancreatic cancer. Ann. Oncol. 19, 273–278. 10.1093/annonc/mdn450 [DOI] [PubMed] [Google Scholar]
- Hu, Z. , Fan, C. , Livasy, C. , He, X. , Oh, D.S. , Ewend, M.G. , Carey, L.A. , Subramanian, S. , West, R. , Ikpatt, F. , Olopade, O.I. , van de Rijn, M. , Perou, C.M. , 2009. A compact VEGF signature associated with distant metastases and poor outcomes. BMC Med. 7, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger, E. , Braun, J.E. , Heimstädt, S. , Zekri, L. , Izaurralde, E. , 2010. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J. 29, 4146–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isserlin, R. , Merico, D. , Voisin, V. , Bader, G.D. , 2014. Enrichment Map – a Cytoscape app to visualize and explore OMICs pathway enrichment results. F1000Research 3, 141 10.12688/f1000research.4536.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamura, T. , Caffrey, T.C. , Kitamura, N. , Yamanari, H. , Setoguchi, T. , Hollingsworth, M.A. , 1997. P-selectin expression in a metastatic pancreatic tumor cell line (SUIT-2). Cancer Res. 57, 1206–1212. [PubMed] [Google Scholar]
- Jaffee, E.M. , Schutte, M. , Gossett, J. , Morsberger, L.A. , Adler, A.J. , Thomas, M. , Greten, T.F. , Hruban, R.H. , Yeo, C.J. , Griffin, C.A. , 1998. Development and characterization of a cytokine-secreting pancreatic adenocarcinoma vaccine from primary tumors for use in clinical trials. Cancer J. Sci. Am. 4, 194–203. [PubMed] [Google Scholar]
- Jinek, M. , Fabian, M.R. , Coyle, S.M. , Sonenberg, N. , Doudna, J.A. , 2010. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat. Struct. Mol. Biol. 17, 238–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.H. , Hu, P. , Fan, C. , Anders, C.K. , 2015. Gene expression in “young adult type” breast cancer: a retrospective analysis. Oncotarget 6, 13688–13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, S. , Zhang, X. , Parsons, D.W. , Lin, J.C.-H. , Leary, R.J. , Angenendt, P. , Mankoo, P. , Carter, H. , Kamiyama, H. , Jimeno, A. , Hong, S.-M. , Fu, B. , Lin, M.-T. , Calhoun, E.S. , Kamiyama, M. , Walter, K. , Nikolskaya, T. , Nikolsky, Y. , Hartigan, J. , Smith, D.R. , Hidalgo, M. , Leach, S.D. , Klein, A.P. , Jaffee, E.M. , Goggins, M. , Maitra, A. , Iacobuzio-Donahue, C. , Eshleman, J.R. , Kern, S.E. , Hruban, R.H. , Karchin, R. , Papadopoulos, N. , Parmigiani, G. , Vogelstein, B. , Velculescu, V.E. , Kinzler, K.W. , 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806. 10.1126/science.1164368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalser, M.H. , Ellenberg, S.S. , 1985. Pancreatic cancer: adjuvant combined radiation and chemotherapy following curative resection. Arch. Surg. 120, 899–903. [DOI] [PubMed] [Google Scholar]
- Kanwar, N. , Hu, P. , Bedard, P. , Clemons, M. , McCready, D. , Done, S.J. , 2015. Identification of genomic signatures in circulating tumor cells from breast cancer. Int. J. Cancer 137, 332–344. 10.1002/ijc.29399 [DOI] [PubMed] [Google Scholar]
- Kim, J.H. , Karnovsky, A. , Mahavisno, V. , Weymouth, T. , Pande, M. , Dolinoy, D.C. , Rozek, L.S. , Sartor, M.A. , 2012. LRpath analysis reveals common pathways dysregulated via DNA methylation across cancer types. BMC Genomics 13, 526 10.1186/1471-2164-13-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutner, R.H. , Zhang, X.-Y. , Reiser, J. , 2009. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 4, 495–505. [DOI] [PubMed] [Google Scholar]
- Langfelder, P. , Horvath, S. , 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 1–13. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. , Anders, S. , 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. BioRxiv 1–21. 10.1101/002832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madar, S. , Brosh, R. , Buganim, Y. , Ezra, O. , Goldstein, I. , Solomon, H. , Kogan, I. , Goldfinger, N. , Klocker, H. , Rotter, V. , 2009. Modulated expression of WFDC1 during carcinogenesis and cellular senescence. Carcinogenesis 30, 20–27. 10.1093/carcin/bgn232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy, L.M. , O'Toole, S.A. , Spillane, C.D. , Martin, C.M. , Gallagher, M.F. , Stordal, B. , Blackshields, G. , Sheils, O. , O'Leary, J.J. , 2015. Identifying novel hypoxia-associated markers of chemoresistance in ovarian cancer. BMC Cancer 15, 547 10.1186/s12885-015-1539-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newhook, T.E. , Blais, E.M. , Lindberg, J.M. , Adair, S.J. , Xin, W. , Lee, J.K. , Papin, J.A. , Parsons, J.T. , Bauer, T.W. , 2014. A thirteen-gene expression signature predicts survival of patients with pancreatic cancer and identifies new genes of interest. PLoS One 9, e105631 10.1371/journal.pone.0105631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, K.T.-P. , Xu, A. , Cheng, Q. , Guo, D.Y. , Lim, Z.X.-H. , Sun, C.K.-W. , Fung, J.H.-S. , Poon, R.T.-P. , Fan, S.T. , Lo, C.M. , Man, K. , 2014. Clinical relevance and therapeutic potential of angiopoietin-like protein 4 in hepatocellular carcinoma. Mol. Cancer 13, 196 10.1186/1476-4598-13-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettle, H. , Neuhaus, P. , Hochhaus, A. , 2013. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the conko-001 randomized trial. JAMA 310, 1473–1481. [DOI] [PubMed] [Google Scholar]
- Orr, B. , Grace, O.C. , Brown, P. , Riddick, A.C.P. , Stewart, G.D. , Franco, O.E. , Hayward, S.W. , Thomson, A.A. , 2013. Reduction of pro-tumorigenic activity of human prostate cancer-associated fibroblasts using Dlk1 or SCUBE1. Dis. Model. Mech. 6, 530–536. 10.1242/dmm.010355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniccia, A. , Hosokawa, P. , Henderson, W. , Al, E. , 2015. Characteristics of 10-year survivors of pancreatic ductal adenocarcinoma. JAMA Surg. 150, 701–710. 10.1001/jamasurg.2015.0668 [DOI] [PubMed] [Google Scholar]
- Pérez-Mancera, P.A. , Rust, A.G. , van der Weyden, L. , Kristiansen, G. , Li, A. , Sarver, A.L. , Silverstein, K.A.T. , Grützmann, R. , Aust, D. , Rümmele, P. , Knösel, T. , Herd, C. , Stemple, D.L. , Kettleborough, R. , Brosnan, J.A. , Li, A. , Morgan, R. , Knight, S. , Yu, J. , Stegeman, S. , Collier, L.S. , ten Hoeve, J.J. , de Ridder, J. , Klein, A.P. , Goggins, M. , Hruban, R.H. , Chang, D.K. , Biankin, A.V. , Grimmond, S.M. , APGI, Wessels, L.F.A. , Wood, S.A. , Iacobuzio-Donahue, C.A. , Pilarsky, C. , Largaespada, D.A. , Adams, D.J. , Tuveson, D.A. , 2012. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 486, 266–270. 10.1038/nature11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapenko, I.O. , Lüders, T. , Russnes, H.G. , Helland, Å. , Sørlie, T. , Kristensen, V.N. , Nord, S. , Lingjærde, O.C. , Børresen-Dale, A.-L. , Haakensen, V.D. , 2015. Glycan-related gene expression signatures in breast cancer subtypes; relation to survival. Mol. Oncol. 9, 861–876. 10.1016/j.molonc.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan, A.R. , 2002. BEDTools: the Swiss-army tool for genome feature analysis. Current Protocols in Bioinformatics John Wiley & Sons, Inc; 10.1002/0471250953.bi1112s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riker, A.I. , Enkemann, S.A. , Fodstad, O. , Liu, S. , Ren, S. , Morris, C. , Xi, Y. , Howell, P. , Metge, B. , Samant, R.S. , Shevde, L.A. , Li, W. , Eschrich, S. , Daud, A. , Ju, J. , Matta, J. , 2008. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genomics 1, 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R.L. , Miller, K.D. , Jemal, A. , 2015. Cancer statistics, 2015. CA Cancer J. Clin. 65, 5–29. 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- Simon, N. , Friedman, J. , Hastie, T. , Tibshirani, R. , 2011. Regularization paths for cox's proportional hazards model via coordinate descent. J. Stat. Softw. 39, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing, T. , Sander, O. , Beerenwinkel, N. , Lengauer, T. , 2005. ROCR: visualizing classifier performance in R. Bioinformatics 21, 3940–3941. 10.1093/bioinformatics/bti623 [DOI] [PubMed] [Google Scholar]
- Stathis, A. , Moore, M.J. , 2010. Advanced pancreatic carcinoma: current treatment and future challenges. Nat. Rev. Clin. Oncol. 7, 163–172. 10.1038/nrclinonc.2009.236 [DOI] [PubMed] [Google Scholar]
- Stratford, J.K. , Bentrem, D.J. , Anderson, J.M. , Fan, C. , Volmar, K.A. , Marron, J.S. , Routh, E.D. , Caskey, L.S. , Samuel, J.C. , Der, C.J. , Thorne, L.B. , Calvo, B.F. , Kim, H.J. , Talamonti, M.S. , Iacobuzio-Donahue, C.A. , Hollingsworth, M.A. , Perou, C.M. , Yeh, J.J. , 2010. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med. 7, 10.1371/journal.pmed.1000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, A. , Horiuchi, A. , Ashida, T. , Miyamoto, T. , Kashima, H. , Nikaido, T. , Konishi, I. , Shiozawa, T. , 2010. Cyclin A2 confers cisplatin resistance to endometrial carcinoma cells via up-regulation of an Akt-binding protein, periplakin. J. Cell. Mol. Med. 14, 2305–2317. 10.1111/j.1582-4934.2009.00839.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, M.J. , Teo, Z. , Sng, M.K. , Zhu, P. , Tan, N.S. , 2012. Emerging roles of angiopoietin-like 4 in human cancer. Mol. Cancer Res. 10, 677–688. 10.1158/1541-7786.MCR-11-0519 [DOI] [PubMed] [Google Scholar]
- Tanaka, M. , Shimbo, T. , Kikuchi, Y. , Matsuda, M. , Kaneda, Y. , 2010. Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int. J. Cancer 126, 1982–1991. 10.1002/ijc.24965 [DOI] [PubMed] [Google Scholar]
- Tibshirani, R. , 1996. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 58, 267–288. [Google Scholar]
- Trapnell, C. , Pachter, L. , Salzberg, S.L. , 2009. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Williams, B.A. , Pertea, G. , Mortazavi, A. , Kwan, G. , van Baren, M.J. , Salzberg, S.L. , Wold, B.J. , Pachter, L. , 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venet, D. , Dumont, J.E. , Detours, V. , 2011. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput. Biol. 7, 10.1371/journal.pcbi.1002240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff, D.D. , Ramanathan, R.K. , Borad, M.J. , Laheru, D.A. , Smith, L.S. , Wood, T.E. , Korn, R.L. , Desai, N. , Trieu, V. , Iglesias, J.L. , Zhang, H. , Soon-Shiong, P. , Shi, T. , Rajeshkumar, N.V. , Maitra, A. , Hidalgo, M. , 2011. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial. J. Clin. Oncol. 29, 4548–4554. 10.1200/JCO.2011.36.5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell, N. , Pajic, M. , Patch, A.-M. , Chang, D.K. , Kassahn, K.S. , Bailey, P. , Johns, A.L. , Miller, D. , Nones, K. , Quek, K. , Quinn, M.C.J. , Robertson, A.J. , Fadlullah, M.Z.H. , Bruxner, T.J.C. , Christ, A.N. , Harliwong, I. , Idrisoglu, S. , Manning, S. , Nourse, C. , Nourbakhsh, E. , Wani, S. , Wilson, P.J. , Markham, E. , Cloonan, N. , Anderson, M.J. , Fink, J.L. , Holmes, O. , Kazakoff, S.H. , Leonard, C. , Newell, F. , Poudel, B. , Song, S. , Taylor, D. , Waddell, N. , Wood, S. , Xu, Q. , Wu, J. , Pinese, M. , Cowley, M.J. , Lee, H.C. , Jones, M.D. , Nagrial, A.M. , Humphris, J. , Chantrill, L.A. , Chin, V. , Steinmann, A.M. , Mawson, A. , Humphrey, E.S. , Colvin, E.K. , Chou, A. , Scarlett, C.J. , Pinho, A.V. , Giry-Laterriere, M. , Rooman, I. , Samra, J.S. , Kench, J.G. , Pettitt, J.A. , Merrett, N.D. , Toon, C. , Epari, K. , Nguyen, N.Q. , Barbour, A. , Zeps, N. , Jamieson, N.B. , Graham, J.S. , Niclou, S.P. , Bjerkvig, R. , Grützmann, R. , Aust, D. , Hruban, R.H. , Maitra, A. , Iacobuzio-Donahue, C.A. , Wolfgang, C.L. , Morgan, R.A. , Lawlor, R.T. , Corbo, V. , Bassi, C. , Falconi, M. , Zamboni, G. , Tortora, G. , Tempero, M.A. , Initiative, A.P.C.G. , Gill, A.J. , Eshleman, J.R. , Pilarsky, C. , Scarpa, A. , Musgrove, E.A. , Pearson, J.V. , Biankin, A.V. , Grimmond, S.M. , 2015. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518, 495–501. 10.1038/nature14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T.T. , Gong, H. , Clarke, E.M. , 2011. A transcriptome analysis by lasso penalized cox regression for pancreatic cancer survival. J. Bioinform. Comput. Biol. 9, 63–73. [DOI] [PubMed] [Google Scholar]
- Zeileis, A. , Hothron, T. , 2002. Diagnostic checking in regression relationships. R News 2, 7–10. [Google Scholar]
- Zhang, J. , Baran, J. , Cros, A. , Guberman, J.M. , Haider, S. , Hsu, J. , Liang, Y. , Rivkin, E. , Wang, J. , Whitty, B. , Wong-Erasmus, M. , Yao, L. , Kasprzyk, A. , 2011. International Cancer Genome Consortium Data Portal—a one-stop shop for cancer genomics data. Database 2011, 10.1093/database/bar026 bar026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplemental Figure 1 Random forest analysis of clinical data, random transcripts, and significant transcripts. Histogram of out of bag errors for random forest models generated on differentially expressed transcripts (red dashed line), clinical data (blue dashed line), and 1000 randomly sampled transcript sets (gray bars). Clinical data consisted of patient age, gender, race, tumor stage, nodal involvement and diabetes status. Significant transcripts were transcripts that met a 5% FDR cutoff in DESeq2 analysis of expression differences between short‐survivors and long‐survivors. Random transcripts sets were generated by randomly sampling from all transcripts measured via RNA‐seq at an equivalent number to the significant transcripts. Differentially expressed transcripts possessed an out of bag error (0.185) lower than all random transcript sets (p < 0.001) while clinical data possessed an out of bag error greater than most random transcript pulls (0.593, p = 0.979).
Supplemental Figure 2 LASSO model performance across pancreatic cancer subtypes. Boxplots of the first principle component of the 19 transcripts included in the LASSO prognostic model across all 4 pancreatic cancer subtypes in the training cohort (A) and the validation cohort (B). P‐values represent a Wilcoxon rank sum test between the squamous subtype and the subtype of interest. *** indicates a p‐value < 0.001.
Supplemental Figure 3 Meta‐PCNA analysis. A) Boxplots of meta‐PCNA index values of short‐survival and long‐survival patients in the training cohort (left) and validation cohort (right). B) Principle Component Analysis (PCA) of meta‐PCNA index normalized residual values for the 19 transcripts incorporated in the LASSO model for the UAB training cohort (left) and the ICGC validation cohort (right).
Supplemental Figure 4 ANGPTL4 expression analysis. A) qPCR results of ANGPTL4 expression in three short‐survivors and three long‐survivors. B) qPCR results of ANGPTL4 expression across pancreatic cell lines. C) Kaplan–Meier curves based on ANGPTL4 expression in three independent pancreatic tumor cohorts.
Supplemental Figure 5 ANGPTL4 expression across pancreatic cancer subtypes. Boxplots of ANGPTL4 expression across pancreatic cancer subtypes in the training cohort (A) and the validation cohort (B). p‐Values represent a Wilcoxon rank sum test between the squamous subtype and the subtype of interest. ** indicates a p‐value < 0.01 and *** indicates a p‐value < 0.001. Kaplan–Meier curves of ANGPTL4 expression within the squamous subtype show survival differences between the top half of ANGPTL4 expressers and the bottom half in both the training (C) and the validation cohort (D).
Supplemental Figure 6 Lentiviral knockdown of ANGPTL4 expression in the PANC‐1 pancreatic cancer cell line. A) qPCR results for PANC‐1 cells transduced with lentiviral vectors containing shRNA for control GFP (blue) or three separate shRNAs towards ANGPTL4 (red, green, yellow). B) ELISA measurements of ANGPTL4 protein levels in media collected from transduced PANC‐1 cells. C) Cell proliferation rates in transduced PANC‐1 cells.
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Data Availability Statement
The datasets supporting the conclusions of this article are available in the GEO databank: GSE79670.


