Abstract
DNA replication control is a key process in maintaining genomic integrity. Monitoring DNA replication initiation is particularly important as it needs to be coordinated with other cellular events and should occur only once per cell cycle. Crucial players in the initiation of DNA replication are the ORC protein complex, marking the origin of replication, and the Cdt1 and Cdc6 proteins, that license these origins to replicate by recruiting the MCM2‐7 helicase. To accurately achieve its functions, Cdt1 is tightly regulated. Cdt1 levels are high from metaphase and during G1 and low in S/G2 phases of the cell cycle. This control is achieved, among other processes, by ubiquitination and proteasomal degradation. In an overexpression screen for Cdt1 deubiquitinating enzymes, we isolated USP37, to date the first ubiquitin hydrolase controlling Cdt1. USP37 overexpression stabilizes Cdt1, most likely a phosphorylated form of the protein. In contrast, USP37 knock down destabilizes Cdt1, predominantly during G1 and G1/S phases of the cell cycle. USP37 interacts with Cdt1 and is able to de‐ubiquitinate Cdt1 in vivo and, USP37 is able to regulate the loading of MCM complexes onto the chromatin. In addition, downregulation of USP37 reduces DNA replication fork speed. Taken together, here we show that the deubiquitinase USP37 plays an important role in the regulation of DNA replication. Whether this is achieved via Cdt1, a central protein in this process, which we have shown to be stabilized by USP37, or via additional factors, remains to be tested.
Keywords: Initiation of DNA replication, DNA damage response, Ubiquitin hydrolase, DNA damage, Protein degradation, Proteasome
Highlights
We identified a ubiquitin hydrolase that controls the DNA replication licensing protein Cdt1: USP37.
USP37 stabilizes mainly a low mobility form of Cdt1 that is unstable and likely to be phosphorylated.
USP37 and Cdt1 interact and USP37 is able to de‐ubiquitinate Cdt1 in vivo.
USP37 overexpression increases the MCM7 loading on the chromatin.
USP37 downregulation decreases replication fork speed, indicating a function in DNA replication.
Abbreviation
- DUB
deubiquitinating enzyme
1. Introduction
Replicating the genome is an essential process for living organisms. DNA replication needs to be tightly regulated and monitored in order to preserve cellular genomic stability. First, because the genome needs to be replicated only once per cell cycle to avoid differences in genome content between the mother and daughter cells and second, because while DNA synthesis occurs, the genome is particularly vulnerable to damage and errors. In eukaryotic cells, complex mechanisms control and monitor DNA replication. A critical regulated step to avoid DNA replication associated instability occurs at the initiation of DNA replication. In eukaryotes, the control of DNA initiation requires the coordination of several proteins/protein complexes (reviewed in Costa et al., 2013; Fragkos et al., 2015). The origin recognition complex (ORC), a heterohexamer with DNA‐dependent ATPase activity, directly recognizes and binds origins of replication. Subsequently, Cdc6 and Cdt1 are recruited to the origins and are able to load the minichromosome maintenance protein (MCM) complex MCM2–7, a heterohexamer that has ATPase‐dependent DNA helicase activity, onto the replication origin. The binding of the helicase to the DNA starts the licensing of replication origins and forms the so‐called prereplicative complex (pre‐RC). In this loading process, ATP hydrolysis by Cdc6 helps the stable association of MCM2‐7 with the DNA, after which the interaction with two additional factors enhance its helicase activity: the heterotetrameric GINS complex (formed by Psf1, Psf2, Psf3 and Sld5 proteins) and CDC45, forming the CMG complex. This complex is subsequently activated by S phase CDKs and CDC7–DBF4. Then, the replication protein A complex (RPA) binds to and stabilizes the single‐stranded DNA, interacts with the DNA polymerase‐α‐DNA primase complex (Pol α complex) and acts as a “fidelity‐clap” for the polymerase (Bochkareva et al., 1998; Fanning et al., 2006; Maga et al., 2001). Polymerization by the Pol α complex marks the start of DNA replication in the cell that continues with the recruitment of other DNA polymerases and the help of associated factors.
Cdt1 is a major regulatory factor during the initiation of DNA replication. Cdt1 is inhibited by Geminin, but regulation of Cdt1 protein levels also critically depends on ubiquitin‐mediated degradation by the proteasome during cell cycle progression (Saxena and Dutta, 2005). Cdt1 levels are high during mitosis and G1 phase of cell cycle and low during S and G2 phases (Nishitani et al., 2001; Rialland et al., 2002; Wohlschlegel et al., 2000). DNA origin licensing starts during metaphase (Dimitrova et al., 1999), which coincides with drop of Geminin levels that continue to be low during G1 and rise again during S/G2 phases, thereby avoiding origin licensing after G1 (Clijsters et al., 2013; McGarry and Kirschner, 1998). Moreover, Cdt1 was shown to be degraded in the presence of DNA damage as Cdt1 levels drastically drop after a genotoxic insult (Higa et al., 2003; Hu et al., 2004). Together these regulatory mechanisms help to restrict the replication licensing to only once per cell cycle and/or avoid DNA replication in the presence of damage.
Several E3 ligase complexes were described to target Cdt1 for proteasomal degradation in different conditions. Cdt1 degradation during the cell cycle was shown to depend on the SCF–Skp2, the CRL4‐Cdt2 E3 ubiquitin ligase complexes and on SCF‐FBXO31 (Chandrasekaran et al., 2011; Higa et al., 2006; Jin et al., 2006; Johansson et al., 2014; Nishitani et al., 2006; Sansam et al., 2006). Moreover, CRL4‐Cdt2 dependent degradation of Cdt1 depends on DNA‐bound PCNA, which occurs during S phase and after DNA damage (Arias and Walter, 2006; Senga et al., 2006).
Although relatively much is known about the ubiquitination of Cdt1, the reverse process or Cdt1 deubiquitination is less studied. In this article we find USP37 as a ubiquitin hydrolase for Cdt1. USP37 was first identified as key enzyme that stabilizes Cyclin A counteracting the ubiquitination by the anaphase‐promoting complex APC/CCdh1 (Huang et al., 2011). USP37 itself is a substrate of the APC/CCdh1 and its regulated during the cell cycle as USP37 levels increase at the G1/S boundary, and remain high during S and G2 phases (Huang et al., 2011). USP37 is degraded at the G2/M boundary in a SCF‐β‐TRCP and Plk1‐dependent manner and remains low in G1 phase of cell cycle (Burrows et al., 2012). Since its first description in 2011, USP37 function has been linked to a number of different proteins. These include important pro‐division regulators (Cyclin A, c‐Myc or 14‐3‐3γ) but also USP37 is able to regulate genomic stability by controlling DNA double strand break repair by homologous recombination or proper mitotic progression by controlling WAPL, a negative regulator of chromatin cohesion (Huang et al., 2011; Kim et al., 2015; Pan et al., 2014; Typas et al., 2015; Yeh et al., 2015).
In this article we describe a new role of USP37 controlling Cdt1 and DNA replication. USP37 overexpression increases Cdt1 protein levels and knock down of this deubiquitinating enzyme (DUB) leads to lower Cdt1 levels. Importantly, USP37 depletion impacts on the loading of the MCM2‐7 replication helicase and on the replication fork speed demonstrating its critical role in regulating DNA replication.
2. Materials and methods
2.1. Cell lines and plasmids
U2OS and 293T cells were grown using standard procedures.
The Addgene expression plasmid #22602 for Flag‐HA‐USP37 was obtained from JW Harper (Harvard Medical School, Boston, USA) (Sowa et al., 2009). A catalytic inactive version of USP37 was obtained by changing the Cysteine 350 to Serine into the Flag‐HA‐USP37 expressing plasmid, using the QuickChange Site‐Directed Mutagenesis Kit (Agilent Technologies). The same kit was used to generate an siRNA#1 resistant version of the Flag‐HA‐USP37 by introducing the following 4 silent mutations (shown in lower case) in the cDNA: CAGCTgtcTCAcAACATT. Cdt1 cDNA was cloned into the pEXPR‐IBA103 (Novagen) vector to obtain a Strep‐Cdt1 expressing plasmid. An expression plasmid for His‐Ubiquitin was a gift from D. Bohmann (Rochester, New York, USA) (Salghetti et al., 1999).
2.2. Cell synchronizations
Cells in G1 or mitosis were synchronized with a single thymidine block (2.5 mM thymidine for 24 h) and release. Also to enrich cells in G1 we incubated with 10 μM Lovastatin (Cayman) for 20 h. After 8 h, 0.1 μg/ml nocodazole (Sigma–Aldrich) was added for 16 h. Mitotic cells were then isolated by shake off and lysed (mitotic cells) or nocodazole was washed off and cells were replated in fresh medium for 5 h (G1 cells). For synchronization at G1/S, a single thymidine block was used. To obtain S and G2 cells, thymidine was subsequently washed off and cells were collected after 4 and 12 h, respectively.
2.3. Antibodies and reagents
Antibodies obtained from commercial sources were: anti‐β‐actin (clone AC‐15, Sigma–Aldrich), anti‐USP37 (ab190184), anti‐Cdt1 (ab70829) and anti‐Kap1 (ab190178) antibodies from Abcam, anti‐γ‐H2AX (Clone JBW301) and anti‐ubiquitinylated proteins (clone FK2, #04‐263) from Merck‐Millipore, anti‐Chk1 (G‐4), anti‐Cdt1 (H‐300), anti‐RRM2 (N‐8) and anti‐GAPDH (FL‐335) from Santa Cruz Biotechnology, anti‐pSer317‐Chk1 from R&D, anti‐pSer10 histone H3 and anti‐Flag from Genscript and anti‐RRM1 and mouse anti‐Ubiquitin from Cell Signaling. The anti‐Cdt1 antibody was raised against amino acids 1–230 of human Cdt1. Anti‐MCM7 and anti‐Rad9 were previously described (Méndez and Stillman, 2000; Toueille et al., 2004).
Thymidine and nocodazole were purchased from Sigma Aldrich and MG132 (used at 5 μM for 16 h or 6 h at 20 μM) from Calbiochem. Lambda phosphatase was purchased from New England Biolabs.
2.4. Cell transfections
Transfections of plasmids were performed using standard calcium phosphate method (Pérez‐Castro and Freire, 2012). For downregulation, siRNA oligos were transfected using Lipofectamine RNAiMAX (Thermo Fisher) according to the manufacturers instructions. The following siRNA oligos were used (Thermo Fisher and GenePharma):
| Luciferase | UCGAAGUAUUCCGCGUACGdTdT |
| USP37#1 | CAGCUAAGUCAUAACAUUAdTdT |
| USP37#2 | CCAAGGAUAUUUCAGCUAAdTdT |
| USP37#3 | GAAUAAAGUCAGCCUAGUAdTdT |
| Cdt1 | AACGUGGAUGAAGUACCCGACdTdT |
2.5. Immunoprecipitations and ubiquitin assays
Immunoprecipitations were carried out with anti‐Flag M2‐agarose (Sigma–Aldrich) or with Cdt1/control antibody crosslinked to protein A sepharose CL‐4b (GE Healthcare) as previously described (Pérez‐Castro and Freire, 2012). His‐Ubiquitin pull downs were carried out using Nickel‐NTA agarose (Qiagen) as described before (Mamely et al., 2006).
2.6. Chromatin fractionation
Biochemical fractionation of cells was performed as previously described (Méndez and Stillman, 2000; Smits et al., 2006).
2.7. Flow cytometry
Cells were collected by trypsinization and fixed in 70% ethanol at 4 °C for minimal 2 h. After fixation, cells were washed with PBS and the DNA was stained with propidium iodide. For BrdU staining, cells were incubated with BrdU 10 μM for 30 min. After fixation, cells were washed with 0.5% PBS‐T (0.5% Tween‐20 in PBS) and then incubated in denaturing solution (0.5% Triton X‐100, 2 M HCl) for 30 min at 37 °C. Then cells were neutralized with 1M Tris–HCl pH 7.5. After washing with PBS, cells were incubated with anti‐BrdU antibody (GenScript) in BSA‐T‐PBS (1% BSA, 0.5% Tween 20 in PBS) for 16 h at 4 °C. After washing with BSA‐T‐PBS, cells were incubated with Alexa 647 secondary antibody (Life Technologies) followed by staining with 25 μg/ml propidium iodide. The samples were analyzed using a MACSQuant Analyzer flow cytometer using MACSQuantify software (Miltenyi Biotec).
2.8. DNA fiber analysis
Exponentially growing cells were pulse‐labeled with 50 uM CldU (20 min) followed by 250 uM IdU (20 min). Labeled cells were collected and DNA fibers were spread in buffer containing 0.5% SDS, 200 mM Tris pH 7.4 and 50 mM EDTA. For immunodetection of labeled tracks, fibers were incubated with primary antibodies (for CldU, rat anti‐BrdU; for IdU, mouse anti‐BrdU) and developed with the corresponding secondary antibodies conjugated to Alexa dyes. Mouse anti‐ssDNA antibody was used to assess fiber integrity. Slides were examined with a Leica DM6000 B microscope, as described previously (Mourón et al., 2013). The conversion factor used was 1 μm = 2.59 kb (Jackson and Pombo, 1998). In each assay, at least 200 tracks were measured to estimate fork rate and around 300 tracks were analyzed to estimate the frequency of origin firing (first label origins – green–red–green – are shown as percentage of all red – CldU – labeled tracks) (Petermann et al., 2010).
2.9. Quantitative real‐time RT‐PCR
Total RNA was isolated from cells by the Chomczynski method (Chomczynski and Sacchi, 1987). The purity and concentration of RNA was determined by Nano‐drop 2000 (Thermo‐Fisher). The quantification of relative abundance of Cdt1 mRNA was carried out using quantitative PCR and the SYBR green detection method. Total RNA was reverse transcribed using a cDNA synthesis kit (Promega, Madison, WI), following the manufacturers instructions, and the Cdt1 cDNA was PCR amplified using gene‐specific primers. The oligonucleotides used were: 5′ TAATCTGACCTCCTGGTGCC 3′ (forward primer), and 5′ GTAGGCGTTTTGAGGAGTGC 3′ (reverse primer). The resulting increase in fluorescence during the PCR reaction was detected in the iQ5 system (Bio‐Rad, Hercules, CA) and gene expression was normalized with the reference gen GADP. The data were analyzed using 2 (‐Delta Delta C(T)) Method (Livak and Schmittgen, 2001).
3. Results and discussion
3.1. USP37 regulates Cdt1 protein levels
Cdt1 levels are regulated by ubiquitin‐dependent proteasomal degradation during the cell cycle and after DNA damage. Since so far no enzymes were described to regulate Cdt1 deubiquitination, we carried out an overexpression screen with a library of human ubiquitin and ubiquitin‐like hydrolases. We first raised an anti‐Cdt1 antibody and the specificity of this antibody was checked using extracts of Cdt1 protein‐depleted cells and cells overexpressing a Flag‐tagged version of Cdt1 (Supplemental Figure 1A and B). The antibody recognized a specific signal both in 293T and U2OS cells. The screen was then carried out with a collection of 78 different hydrolases, mainly ubiquitin hydrolases but also Sumo hydrolases in both control conditions and after DNA damage (30 min after UV light) and Cdt1 protein levels were studied by Western blot. We expected the overexpressed hydrolase enzyme targeting Cdt1 to decrease ubiquitination and therefore to increase Cdt1 protein levels. The result of the full screen is shown in Supplemental Figures 2 and 3. Surprisingly, overexpression of only one DUB in our library, USP37, was able to increase levels of Cdt1 (Supplemental Figure 3 and Figure 1A). The effect of Cdt1 upregulation after USP37 overexpression was observed under conditions of DNA damage. In unperturbed cells, Cdt1 expression levels similar to the control were observed for all deubiquitinases tested. No significant changes in Cdt1 mRNA levels were observed after USP37 overexpression, indicating that the increase of Cdt1 was due to changes at protein level only (Figure 1B). A more detailed study shows that levels of Cdt1 were upregulated after USP37 overexpression also basal conditions and at different times after DNA damage (Figure 1C). Since cell cycle progression could be affected by USP37 overexpression and Cdt1 levels change during the cell cycle, the experiment was repeated in cells synchronized in S phase. Figure 1D shows an USP37‐dependent stabilization of Cdt1 in cells with similar DNA profiles, excluding the possibility of an indirect cell cycle effect. In this experiment also the levels of the RRM1 and RRM2 subunits of the ribonucleotide reductase and phosphorylated H2AX (γH2AX) were analyzed, as changes in these proteins might be indicative of anomalous S phase progression. However, no significant differences were found after USP37 overexpression (Figure 1D). Moreover, USP37 overexpression did not affect cell cycle progression judged by BrdU/PI flow cytometry analysis (Figure 1E). If the overexpression of USP37 increases Cdt1 protein levels by affecting its ubiquitination‐ and proteasomal‐dependent degradation, lack of USP37 should have the opposite consequence. Depletion of USP37 by siRNA indeed resulted in decreased Cdt1 protein levels in basal conditions, as shown in Figure 1F. Nevertheless, the Cdt1 degradation kinetics after DNA damage is similar in Luc and USP37 siRNA treated cells, indicating that only the overexpression of the USP37 can stabilize Cdt1 after DNA damage. Also, overexpression of an siRNA resistant version of Flag‐USP37 rescued the drop in Cdt1 levels caused by USP37 depletion (Supplemental Figure 4A).
Figure 1.

USP37 controls Cdt1 levels. (A) 293T cells were transfected with an empty vector (EV) or expression plasmids for the indicated DUBs and when indicated treated with UV light (40 J/m2) and collected 30 min later. Cell lysates were analyzed by Western blot with the indicated antibodies. (B) 293T cells transfected with EV or Flag‐USP37 were lysed for analysis of mRNA levels by real‐time PCR or by Western blot with the indicated antibodies. The graph shows the average Cdt1 mRNA levels of 3 independent experiments. (C) 293T cells were transfected with EV control or Flag‐USP37. Then, cells were left untreated or treated with 40 J/m2 UV light before collecting them at the indicated times (min). Extracts were analyzed by Western blot. (D) 293T cells transfected with Flag‐USP37 were incubated with thymidine for 24 h and released in fresh medium for 4 h. Then cells were left untreated or treated with UV light (40 J/m2) and collected 30 min post‐treatment before Western blot (left) or by flow cytometry (propidium iodide) analysis. (E) 293T cells were transfected with Flag‐USP37 or empty vectors and were labeled with BrdU (30 min) for FACS analysis. The percentage of BrdU positive cells is indicated. (F) U2OS cells were transfected with the indicated siRNA oligos and left untreated or UV irradiated (40 J/m2) before collection at the indicated times and analysis by Western blot. Quantification of Cdt1 levels compared to the loading control β‐actin is shown at the bottom. In this quantification, Cdt1 levels in all samples were compared to those in siLuc cells without damage (set as 1, upper row). Also, samples of USP37 depleted cells were compared to the undamaged control (set to 1, bottom row).
Western blot analysis using the Cdt1 antibody showed two Cdt1 bands in most of the experiments, of which predominantly the upper band becomes stabilized after USP37 overexpression. We therefore pursued to characterize the nature of the upper band. Our antibody was raised against the N‐terminus of Cdt1. To explore if the upper band was a Cdt1 splice variant, two other commercial antibodies were used that recognize the Cdt1 C‐terminus. As shown in Figure 2A, the three antibodies detected both Cdt1 bands, suggesting that the upper band is a modified version of Cdt1 rather than a splice form. Since USP37 catalytic activity removes ubiquitin and Cdt1 is target for degradation by the proteasome, we reasoned that the upper band might be an unstable form of Cdt1. Indeed, addition of the proteasome inhibitor MG132 increases the level of the band that moves with lower mobility (Figure 2B) and interestingly, USP37 knock down decreased the upper band under such conditions indicating that even in the presence of MG132, the lack of USP37 destabilizes the Cdt1 upper band. The experiment was repeated with 3 different USP37 siRNA oligos with a similar outcome, indicating that the observed decrease in Cdt1 stability was due to USP37 depletion and not the result of an off target effect (Figure 2C). Next, to further investigate the nature of the unstable upper band, the possibility of an (mono‐)ubiquitin modification was tested. For that, endogenous Cdt1 was immunoprecipitated from USP37 overexpressing cells. Even though both forms of Cdt1 were efficiently immunoprecipitated (Supplemental Figure 4B and Figure 2D), no signal was observed after probing the immunoprecipitate with anti‐ubiquitin or conjugated ubiquitin antibodies (Figure 2D). This result argues against the Cdt1 slow mobility being the result of ubiquitination. Then the extracts were treated with lambda phosphatase, to examine possible phosphorylation of Cdt1. Rad9, 45 kDa in unphosphorylated form, but up to 60 kDa when phosphorylated (St Onge et al., 2001, 1999, 1999), was used as a positive control for lambda phosphatase activity (Figure 2E). Treating the extracts with phosphatase resulted in a reduction of the Cdt1 upper band that is induced by USP37 overexpression (Figure 2E). As an additional approach, Cdt1 was immunoprecipitated from USP37 overexpressing cells and the purified immunoprecipitates were treated with lambda phosphatase. Again an increase of the low mobility band was observed after USP37 overexpression. This band disappeared completely after phosphatase treatment (Figure 2F). These experiments strongly suggest that the Cdt1 mobility shift is due to phosphorylation. Instability of a low mobility, phosphorylated form of Cdt1 was also described by others and an additional study showed that this Cdt1 phosphorylation is dependent on CDKs. This phosphorylated form of Cdt1 interacts with the SCF‐Skp2 complex that targets Cdt1 for ubiquitination (Li et al., 2003; Liu et al., 2004). Therefore, is likely that USP37 counteracts this ubiquitination and degradation by binding with higher affinity.
Figure 2.

USP37 stabilizes a phosphorylated form of Cdt1. (A) 293T cells transfected with empty or Flag‐USP37 vector were lysed and loaded three times in the same gel for parallel Western blot analysis with three different Cdt1 antibodies. The anti‐Cdt1 immunoblot intensities were different and comparable exposures in each case are shown. (B) 293T cells were transfected with control or USP37 siRNA oligos and left untreated or treated with MG132 for 6 h when indicated. Cell lysates were analyzed with the indicated antibodies. (C) The same as (B), but using U2OS cells. (D) Endogenous Cdt1 was immunoprecipitated with the anti‐Cdt1 antibody from lysates of 293T cells overexpressing USP37. Input and immunoprecipitates were analyzed by Western blot with the indicated antibodies. Two different exposures of the same blot are shown. (E) 293T cells transfected with empty or Flag‐USP37 overexpression vectors were lysed. Extracts were treated or not with lambda phosphatase for 60 min prior analysis by Western blotting with the indicated antibodies. (F) 293T cells overexpressing Flag‐HA‐USP37 were lysed and anti‐Cdt1 immunoprecipitations were carried out. The indicated immunoprecipitates were incubated with lambda phosphatase for 60 min before analysis by Western blot with the indicated antibodies.
3.2. USP37 de‐ubiquitinates Cdt1
Next, to examine if USP37 was directly affecting Cdt1 levels, the interaction between the two proteins was analyzed by immunoprecipitation. Endogenous Cdt1 co‐immunoprecipitated with overexpressed Flag‐USP37 (Figure 3A). Although overexpression of USP37 stabilizes Cdt1 after DNA damage, Figure 3B shows that the interaction between the two proteins did not significantly change under these conditions, suggesting that Cdt1 stability after UV light is not due to changes in affinity towards USP37. Interestingly, Cdt1 upper band is enriched in the immunoprecipitations, indicating USP37 regulation preferentially over that isoform.
Figure 3.
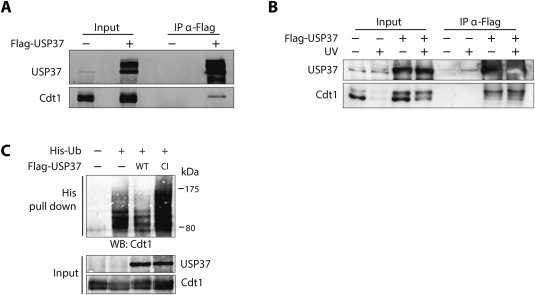
USP37 interacts with and deubiquitinates Cdt1. (A) 293T cells transfected with an empty vector or Flag‐USP37 were lysed and immunoprecipitations were carried out with anti‐Flag beads. Immunoprecipitates were analyzed by Western blot with anti‐Cdt1 and anti‐USP37 antibodies. (B) As in (A), but when indicated, cells were UV irradiated (40 J/m2, 1 h). (C) 293T cells were transfected with a plasmid expressing Strep‐Cdt1 together with control or His‐Ubiquitin expression plasmids, and wild type or catalytic inactive Flag‐USP37. 20 h after transfection, cells were incubated with MG132 for 16 h before lysis under denaturing conditions. Western blot analysis of input and His‐pull downs were carried out with the indicated antibodies.
The interaction data suggest that USP37 directly affects Cdt1 ubiquitination. We therefore tested if the ubiquitination status of Cdt1 is affected by USP37 overexpression. As a control a catalytic inactive version of USP37 (Cys350Ser) was used. After expression of His‐Ubiquitin in 293T cells and performing a pull down for His containing proteins, ubiquitinated forms of Cdt1 were detected (Figure 3C, second lane). Ubiquitinated forms of Cdt1 notably decreased upon co‐expression of wild type USP37. In contrast, Cdt1 ubiquitination increased in the presence of Cys350Ser USP37, suggesting that overexpression of the catalytic inactive protein antagonizes the endogenous wild type USP37. Altogether these data strongly indicate that Cdt1 is a target substrate for USP37.
3.3. USP37 regulates Cdt1 during G1 and G1/S
To understand the biological significance of the Cdt1 regulation by USP37, first the levels of both proteins during the cell cycle were examined. It is documented that during the cell cycle, both proteins are controlled by ubiquitin‐mediated proteasomal degradation, but in a distinct way. While Cdt1 levels are starting to raise during mitosis, continue high in G1 and decrease in S and G2 phases, USP37 levels are low in G1 and increase in S and G2 phases (Huang et al., 2011; Nishitani et al., 2000). Figure 4A confirms that USP37 levels were mainly high at the G1/S transition and in S and G2, in agreement to its described role to promote S phase entry (Huang et al., 2011). In contrast, Cdt1 protein levels were high in G1 and mitosis and low in S/G2 phases. Cdt1 damage‐specific degradation occurred during all stages of cell cycle except mitosis, similar to recently reported (Morino et al., 2015). Interestingly, after UV irradiation in S and G2 phases USP37 levels showed a slight decrease and also a mobility shift, suggesting a DNA damage‐induced posttranslational modification specifically in these phases. Importantly, USP37 protein was low but detectable during the G1 phase and likewise, Cdt1 levels were low but detectable during S/G2 (Figure 4A). We therefore questioned during what phase of cell cycle USP37 regulates Cdt1. Cells were synchronized in different stages of the cell cycle and at the same time USP37 was depleted by siRNA. Interestingly, knock down of USP37 reduced the levels of Cdt1 in G1 and G1/S but not during S phase, indicating that although USP37 levels are low in G1, this DUB still functions to stabilize Cdt1 at this moment of the cell cycle (Figure 4B). USP37 depletion clearly affected the levels of the lower Cdt1 band, whereas the upper band is not detected here. This adds proof to our hypothesis that the upper phosphorylated band of Cdt1 is an unstable form of Cdt1.
Figure 4.

USP37 controls Cdt1 levels during G1 and G1/S. (A) U2OS were left asynchronous or synchronized using different protocols as described in Materials and Methods. When indicated, 30 min before collecting, cells were treated with 40 J/m2 UV light. Then samples were collected for propidium iodide analysis by flow cytometry (right) or Western blot with the indicated antibodies (left). (B) U2OS were transfected with control or USP37 siRNA oligos and synchronized at the same time as described. A fraction of cells was lysed for Western blot analysis with the indicated antibodies (left) and another fraction of cells was collected for flow cytometry analysis after propidium iodide staining (right).
3.4. USP37 regulates DNA replication at different levels
Since the role of USP37 was linked to the transition from G1 to S phase, but not much was known about its role during DNA replication, replication fork speed was measured accurately by DNA fiber analysis. As shown in Figure 5A, replication fork progression rate was significantly delayed in USP37‐depleted cells as compared to control cells and consequently an increase of the percentage of replication firing was observed under these conditions. These data strongly suggest that USP37 plays an active role in replication fork progression. It is tempting to speculate that USP37 could stabilize one of the important proteins at the replication fork, and since there was no reduction of the percentage of replication origins firing, this effect is unlikely to be due to controlling Cdt1 directly. As shown in Figure 1D and E, this effect is also not the result of a reduced availability of nucleotides by controlling the levels of the subunits RRM1 or RRM2 of the ribonucleotide reductase. As USP37 was described to control Cyclin A, known regulator of DNA replication, this Cyclin is a candidate target for the decrease on replication fork speed after USP37 depletion (Huang et al., 2011).
Figure 5.
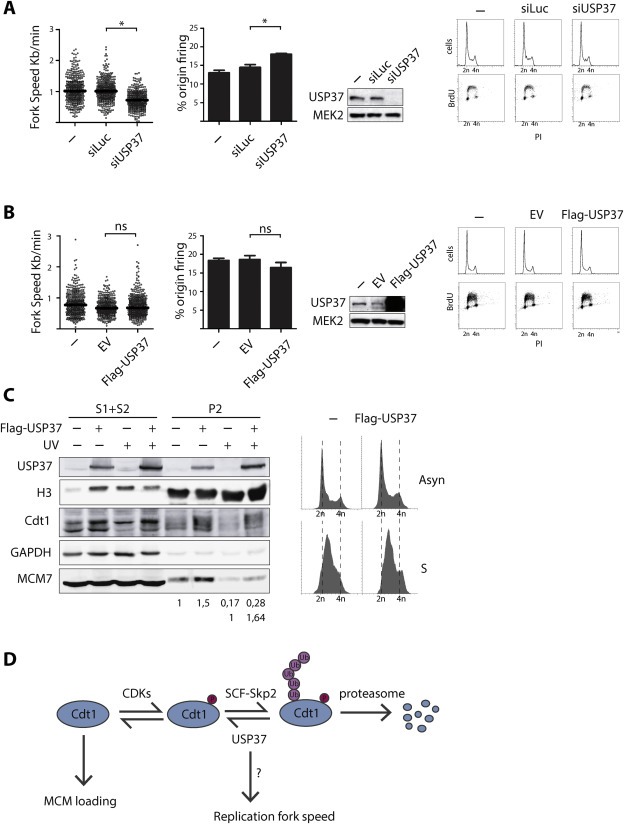
USP37 controls replication fork speed and impacts on MCM loading. U2OS cells were left untreated or transfected with control or USP37 siRNA oligos (A) or 293T were left untreated or transfected with an empty or Flag‐USP37 expressing plasmid (B). 48 h after transfections cells were labeled with nucleotide analogues as described in materials and methods. Cells were then collected for DNA fiber analysis, for Western blotting with the indicated antibodies and for flow cytometry analysis (BrdU/PI). Depicted is the replication fork speed and the percentage of origin firing from two different experiments. Horizontal lines represent the median of relative fork speed distribution. Western blots and flow cytometry graphs are shown from a representative experiment. (C) 293T cells transfected with an empty vector or Flag‐USP37 expressing plasmid were synchronized by thymidine block and collected 4 h after thymidine release for flow cytometry analysis or for biochemical fractionation and Western blot analysis with the indicated antibodies. S1 + S2 represent soluble proteins, P2 the chromatin fraction. Quantification of the MCM7 levels in the chromatin fraction compared to the levels of histone H3 is represented. In the first row the MCM7 levels are compared to EV without damage, the second row compares −/+ Flag‐USP37 expression in UV‐treated cells. (D) Model summarizing previously known data and the new data from this article. USP37 deubiquitinates a phosphorylated, unstable form of Cdt1. USP37 also delays replication fork speed by a mechanism likely independent of Cdt1.
However, USP37 overexpression did not lead to a significant change in the replicative parameters compared to control conditions (Figure 5B). Nevertheless, we continued to study the role of USP37 in modification of Cdt1 particularly, and the effect of USP37 overexpression on the loading of the MCM2‐7 complex, the major function of Cdt1 in initiation of DNA replication, was examined. To avoid any undesired cell cycle effects, cells synchronized in early S phase were used. Notably, USP37 overexpression increased MCM7 loading onto the insoluble nuclear fraction (P2; chromatin) of the chromatin fractionation (Figure 5C). This effect was also observed in UV‐irradiated S phase cells (Figure 5C), strongly suggesting that USP37 can regulate the initiation of DNA replication. Similar experiments were performed after USP37 depletion, resulting in no changes on MCM7 loading, suggesting that the there is an excess of Cdt1 to achieve the loading. Another possibility to explain the uploading of MCM7 after USP37 overexpression is a direct change in the ubiquitination status of MCM7 by USP37. However, in vivo ubiquitination experiments did not show changes in MCM7 ubiquitination status after USP37 overexpression (Supplemental Figure 4C). Interestingly, similar to others, an increase on the ubiquitination status of the USP37 catalytic inactive version comparing to the wild type UPS37 was observed, suggesting that USP37 could auto‐de‐ubiquitinate (Tanno et al., 2014).
The fact that overexpression of USP37 did not change replication parameters (Figure 5B) or cell cycle profiles (1, 5B) in spite of increasing MCM7 loading suggest that the other known mechanisms controlling Cdt1/initiation of replication are able to maintain replication levels normal after even when Cdt1 protein is upregulated. Future work will investigate this into detail. Similarly, the increase of MCM7 loading after UV treatment might not have an effect on replication parameters, as recently it was shown that a non‐degradable version of Cdt1 does not induce additional DNA synthesis after DNA damage (Tsanov et al., 2014).
Altogether, we show that USP37 is a DUB that modifies Cdt1, a central protein in DNA replication initiation, by stabilizing mainly a low mobility form that is likely a phosphorylated form of Cdt1 (Figure 5D). Our data also demonstrate that USP37 controls DNA replication fork speed (Figure 5D) and it is expected that this effect on DNA fork speed is by controlling different target proteins than Cdt1, as Cdt1 itself is not predicted to have a major role in replication fork speed regulation. This adds USP37 to USP7 and USP29, recently demonstrated by us and others, as other important DUBs in DNA replication (Jagannathan et al., 2014; Martín et al., 2015).
Further studies are needed to establish the details of regulation of USP37 itself, although our data indicate that this protein is actively controlling Cdt1 function during G1 and S phases. It is possible that the USP37 mobility change after UV light, shown in Figure 3, reflects changes in its regulation. The fact that USP37 is involved in DNA replication, and particularly in the control of the protein levels of Cdt1, a potential oncogenic protein, makes it a putative therapeutic target in cancer treatment. USP37 was shown to also stabilize other known oncogenes like c‐Myc or Cyclin A, strengthening the relevance of USP37 inhibition in cancer therapy (Huang et al., 2011; Pan et al., 2014).
3.5. Conclusions
We identified a new regulator of DNA replication: USP37. USP37 stabilizes the licensing factor Cdt1 and also plays a further role in the progression of DNA replication.
Supporting information
The following are the supplementary data related to this article:
Supplemental Figure 1 Homemade anti‐Cdt1 antibody specifically recognizes human Cdt1. (A). 293T cells were transfected with control or Cdt1 siRNA oligos or with empty vector (EV) or a vector expressing Flag‐Cdt1. Duplicate samples were ran on a gel and Western blot analysis was carried out using the anti‐Cdt1 homemade antibody and anti‐Flag as indicated. Dotted line indicates where the membrane was cut to incubate with the different antibodies. (B). 293T and U2OS cells were transfected with control or Cdt1 siRNAs and extracts were made for Western blot analysis with the indicated antibodies.
Supplemental Figure 2 No change in Cdt1 levels after overexpression of many ubiquitin or ubiquitin‐like hydrolases. 293T cells were transfected with expression plasmids for the indicated hydrolases (or empty vector) and were left untreated or UV irradiated (40 J/m2). Cells were lysed 30 min after treatment and analyzed by Western blot with anti‐Cdt1 antibodies and β‐actin as loading control. The expression plasmids for (tagged) ubiquitin or ubiquitin‐like hydrolases were kindly provided by several collaborating laboratories.
Supplemental Figure 3 Increased Cdt1 levels after USP37 overexpression. 293T cells were transfected, lysed and analyzed as in Supplemental Figure 2.
Supplemental Figure 4 Flag‐USP37 overexpression rescues Cdt1 levels in USP37 depleted cells and does not affect MCM7 ubiquitination (A) 293T cells were transfected 2 times with siRNA against Luc or USP37#1 and with siRNA resistant Flag‐USP37 and were treated with thymidine 24 h, released for 6 h and incubated with Lovastatin 10 μM for extra 20 h before being analyzed by Western blot with the indicated antibodies (B) 293T cells were transfected with Flag‐USP37, treated with MG132 for 6 h or left untreated. Immunoprecipitations with control or anti‐Cdt1 antibodies were carried out using extracts of the Flag‐USP37 expressing cells. All samples were analyzed by Western blot with the anti‐Cdt1 antibody. (B) 293T cells were transfected when indicated with control, His‐Ubiquitin, wild type or catalytic inactive Flag‐USP37 plasmids. 20 h after transfection, cells were incubated with MG132 for 16 h before lysis under denaturing conditions. Western blotting analysis of input and His pull downs were performed with the indicated antibodies.
Acknowledgments
The authors are grateful to V. Smits for careful reading of the manuscript. This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (SAF2013‐49149‐R, BFU2014‐51672‐REDC), Instituto de Salud Carlos III (BA15/00092) and Fundación CajaCanarias AP2015/008) to RF.
Supplementary data 1.
1.1.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molonc.2016.05.008.
Hernández-Pérez Santiago, Cabrera Elisa, Amoedo Hugo, Rodríguez-Acebes Sara, Koundrioukoff Stephane, Debatisse Michelle, Méndez Juan, Freire Raimundo, (2016), USP37 deubiquitinates Cdt1 and contributes to regulate DNA replication, Molecular Oncology 10, doi: 10.1016/j.molonc.2016.05.008.
References
- Arias, E.E. , Walter, J.C. , 2006. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 8, 84–90. 10.1038/ncb1346 [DOI] [PubMed] [Google Scholar]
- Bochkareva, E. , Frappier, L. , Edwards, A.M. , Bochkarev, A. , 1998. The RPA32 subunit of human replication protein A contains a single-stranded DNA-binding domain. J. Biol. Chem. 273, 3932–3936. [DOI] [PubMed] [Google Scholar]
- Burrows, A.C. , Prokop, J. , Summers, M.K. , 2012. Skp1-Cul1-F-box ubiquitin ligase (SCF(βTrCP))-mediated destruction of the ubiquitin-specific protease USP37 during G2-phase promotes mitotic entry. J. Biol. Chem. 287, 39021–39029. 10.1074/jbc.M112.390328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran, S. , Tan, T.X. , Hall, J.R. , Cook, J.G. , 2011. Stress-stimulated mitogen-activated protein kinases control the stability and activity of the Cdt1 DNA replication licensing factor. Mol. Cell. Biol. 31, 4405–4416. 10.1128/MCB.06163-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski, P. , Sacchi, N. , 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. 10.1006/abio.1987.9999 [DOI] [PubMed] [Google Scholar]
- Clijsters, L. , Ogink, J. , Wolthuis, R. , 2013. The spindle checkpoint, APC/C(Cdc20), and APC/C(Cdh1) play distinct roles in connecting mitosis to S phase. J. Cell Biol. 201, 1013–1026. 10.1083/jcb.201211019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, A. , Hood, I.V. , Berger, J.M. , 2013. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 82, 25–54. 10.1146/annurev-biochem-052610-094414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova, D.S. , Todorov, I.T. , Melendy, T. , Gilbert, D.M. , 1999. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 146, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning, E. , Klimovich, V. , Nager, A.R. , 2006. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34, 4126–4137. 10.1093/nar/gkl550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkos, M. , Ganier, O. , Coulombe, P. , Méchali, M. , 2015. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 16, 360–374. 10.1038/nrm4002 [DOI] [PubMed] [Google Scholar]
- Higa, L.A. , Banks, D. , Wu, M. , Kobayashi, R. , Sun, H. , Zhang, H. , 2006. L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 5, 1675–1680. [DOI] [PubMed] [Google Scholar]
- Higa, L.A.A. , Mihaylov, I.S. , Banks, D.P. , Zheng, J. , Zhang, H. , 2003. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5, 1008–1015. 10.1038/ncb1061 [DOI] [PubMed] [Google Scholar]
- Hu, J. , McCall, C.M. , Ohta, T. , Xiong, Y. , 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6, 1003–1009. 10.1038/ncb1172 [DOI] [PubMed] [Google Scholar]
- Huang, X. , Summers, M.K. , Pham, V. , Lill, J.R. , Liu, J. , Lee, G. , Kirkpatrick, D.S. , Jackson, P.K. , Fang, G. , Dixit, V.M. , 2011. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol. Cell 42, 511–523. 10.1016/j.molcel.2011.03.027 [DOI] [PubMed] [Google Scholar]
- Jackson, D.A. , Pombo, A. , 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan, M. , Nguyen, T. , Gallo, D. , Luthra, N. , Brown, G.W. , Saridakis, V. , Frappier, L. , 2014. A role for USP7 in DNA replication. Mol. Cell. Biol. 34, 132–145. 10.1128/MCB.00639-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, J. , Arias, E.E. , Chen, J. , Harper, J.W. , Walter, J.C. , 2006. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721. 10.1016/j.molcel.2006.08.010 [DOI] [PubMed] [Google Scholar]
- Johansson, P. , Jeffery, J. , Al-Ejeh, F. , Schulz, R.B. , Callen, D.F. , Kumar, R. , Khanna, K.K. , 2014. SCF-FBXO31 E3 ligase targets DNA replication factor Cdt1 for proteolysis in the G2 phase of cell cycle to prevent re-replication. J. Biol. Chem. 289, 18514–18525. 10.1074/jbc.M114.559930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.-O. , Kim, S.-R. , Lim, K.-H. , Kim, J.-H. , Ajjappala, B. , Lee, H.-J. , Choi, J.-I. , Baek, K.-H. , 2015. Deubiquitinating enzyme USP37 regulating oncogenic function of 14-3-3γ. Oncotarget 6, 36551–36576. 10.18632/oncotarget.5336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Zhao, Q. , Liao, R. , Sun, P. , Wu, X. , 2003. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J. Biol. Chem. 278, 30854–30858. 10.1074/jbc.C300251200 [DOI] [PubMed] [Google Scholar]
- Liu, E. , Li, X. , Yan, F. , Zhao, Q. , Wu, X. , 2004. Cyclin-dependent kinases phosphorylate human Cdt1 and induce its degradation. J. Biol. Chem. 279, 17283–17288. 10.1074/jbc.C300549200 [DOI] [PubMed] [Google Scholar]
- Livak, K.J. , Schmittgen, T.D. , 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Maga, G. , Frouin, I. , Spadari, S. , Hubscher, U. , 2001. Replication protein A as a “fidelity clamp” for DNA polymerase alpha. J. Biol. Chem. 276, 18235–18242. 10.1074/jbc.M009599200 [DOI] [PubMed] [Google Scholar]
- Mamely, I. , van Vugt, M.A. , Smits, V.A.J. , Semple, J.I. , Lemmens, B. , Perrakis, A. , Medema, R.H. , Freire, R. , 2006. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr. Biol. 16, 1950–1955. 10.1016/j.cub.2006.08.026 [DOI] [PubMed] [Google Scholar]
- Martín, Y. , Cabrera, E. , Amoedo, H. , Hernández-Pérez, S. , Domínguez-Kelly, R. , Freire, R. , 2015. USP29 controls the stability of checkpoint adaptor Claspin by deubiquitination. Oncogene 34, 1058–1063. 10.1038/onc.2014.38 [DOI] [PubMed] [Google Scholar]
- McGarry, T.J. , Kirschner, M.W. , 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Méndez, J. , Stillman, B. , 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morino, M. , Nukina, K. , Sakaguchi, H. , Maeda, T. , Takahara, M. , Shiomi, Y. , Nishitani, H. , 2015. Mitotic UV irradiation induces a DNA replication-licensing defect that potentiates G1 arrest response. PLoS One 10, e0120553 10.1371/journal.pone.0120553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourón, S. , Rodriguez-Acebes, S. , Martínez-Jiménez, M.I. , García-Gómez, S. , Chocrón, S. , Blanco, L. , Méndez, J. , 2013. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 20, 1383–1389. 10.1038/nsmb.2719 [DOI] [PubMed] [Google Scholar]
- Nishitani, H. , Lygerou, Z. , Nishimoto, T. , Nurse, P. , 2000. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404, 625–628. 10.1038/35007110 [DOI] [PubMed] [Google Scholar]
- Nishitani, H. , Sugimoto, N. , Roukos, V. , Nakanishi, Y. , Saijo, M. , Obuse, C. , Tsurimoto, T. , Nakayama, K.I. , Nakayama, K. , Fujita, M. , Lygerou, Z. , Nishimoto, T. , 2006. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 25, 1126–1136. 10.1038/sj.emboj.7601002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani, H. , Taraviras, S. , Lygerou, Z. , Nishimoto, T. , 2001. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J. Biol. Chem. 276, 44905–44911. 10.1074/jbc.M105406200 [DOI] [PubMed] [Google Scholar]
- Pan, J. , Deng, Q. , Jiang, C. , Wang, X. , Niu, T. , Li, H. , Chen, T. , Jin, J. , Pan, W. , Cai, X. , Yang, X. , Lu, M. , Xiao, J. , Wang, P. , 2014. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene 10.1038/onc.2014.327 [DOI] [PubMed] [Google Scholar]
- Petermann, E. , Woodcock, M. , Helleday, T. , 2010. Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. U.S.A. 107, 16090–16095. 10.1073/pnas.1005031107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Castro, A.J. , Freire, R. , 2012. Rad9B responds to nucleolar stress through ATR and JNK signalling, and delays the G1-S transition. J. Cell. Sci. 125, 1152–1164. 10.1242/jcs.091124 [DOI] [PubMed] [Google Scholar]
- Rialland, M. , Sola, F. , Santocanale, C. , 2002. Essential role of human CDT1 in DNA replication and chromatin licensing. J. Cell. Sci. 115, 1435–1440. [DOI] [PubMed] [Google Scholar]
- Salghetti, S.E. , Kim, S.Y. , Tansey, W.P. , 1999. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 18, 717–726. 10.1093/emboj/18.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansam, C.L. , Shepard, J.L. , Lai, K. , Ianari, A. , Danielian, P.S. , Amsterdam, A. , Hopkins, N. , Lees, J.A. , 2006. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 20, 3117–3129. 10.1101/gad.1482106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, S. , Dutta, A. , 2005. Geminin-Cdt1 balance is critical for genetic stability. Mutat. Res. 569, 111–121. 10.1016/j.mrfmmm.2004.05.026 [DOI] [PubMed] [Google Scholar]
- Senga, T. , Sivaprasad, U. , Zhu, W. , Park, J.H. , Arias, E.E. , Walter, J.C. , Dutta, A. , 2006. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 281, 6246–6252. 10.1074/jbc.M512705200 [DOI] [PubMed] [Google Scholar]
- Smits, V.A.J. , Reaper, P.M. , Jackson, S.P. , 2006. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr. Biol. 16, 150–159. 10.1016/j.cub.2005.11.066 [DOI] [PubMed] [Google Scholar]
- Sowa, M.E. , Bennett, E.J. , Gygi, S.P. , Harper, J.W. , 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403. 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge, R.P. , Besley, B.D. , Park, M. , Casselman, R. , Davey, S. , 2001. DNA damage-dependent and -independent phosphorylation of the hRad9 checkpoint protein. J. Biol. Chem. 276, 41898–41905. 10.1074/jbc.M105152200 [DOI] [PubMed] [Google Scholar]
- St Onge, R.P. , Udell, C.M. , Casselman, R. , Davey, S. , 1999. The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRAD1 and hHUS1. Mol. Biol. Cell 10, 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno, H. , Shigematsu, T. , Nishikawa, S. , Hayakawa, A. , Denda, K. , Tanaka, T. , Komada, M. , 2014. Ubiquitin-interacting motifs confer full catalytic activity, but not ubiquitin chain substrate specificity, to deubiquitinating enzyme USP37. J. Biol. Chem. 289, 2415–2423. 10.1074/jbc.M113.528372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toueille, M. , El-Andaloussi, N. , Frouin, I. , Freire, R. , Funk, D. , Shevelev, I. , Friedrich-Heineken, E. , Villani, G. , Hottiger, M.O. , Hübscher, U. , 2004. The human Rad9/Rad1/Hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: implications for DNA repair. Nucleic Acids Res. 32, 3316–3324. 10.1093/nar/gkh652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsanov, N. , Kermi, C. , Coulombe, P. , Van der Laan, S. , Hodroj, D. , Maiorano, D. , 2014. PIP degron proteins, substrates of CRL4Cdt2, and not PIP boxes, interfere with DNA polymerase η and κ focus formation on UV damage. Nucleic Acids Res. 42, 3692–3706. 10.1093/nar/gkt1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas, D. , Luijsterburg, M.S. , Wiegant, W.W. , Diakatou, M. , Helfricht, A. , Thijssen, P.E. , van de Broek, B. , Mullenders, L.H. , van Attikum, H. , 2015. The de-ubiquitylating enzymes USP26 and USP37 regulate homologous recombination by counteracting RAP80. Nucleic Acids Res. 43, 6919–6933. 10.1093/nar/gkv613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmer, E. , Karnitz, L.M. , 1999. Human homologs of Schizosaccharomyces pombe rad1, hus1, and rad9 form a DNA damage-responsive protein complex. J. Biol. Chem. 274, 567–570. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel, J.A. , Dwyer, B.T. , Dhar, S.K. , Cvetic, C. , Walter, J.C. , Dutta, A. , 2000. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290, 2309–2312. 10.1126/science.290.5500.2309 [DOI] [PubMed] [Google Scholar]
- Yeh, C. , Coyaud, É. , Bashkurov, M. , van der Lelij, P. , Cheung, S.W.T. , Peters, J.M. , Raught, B. , Pelletier, L. , 2015. The deubiquitinase USP37 regulates chromosome cohesion and mitotic progression. Curr. Biol. 25, 2290–2299. 10.1016/j.cub.2015.07.025 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The following are the supplementary data related to this article:
Supplemental Figure 1 Homemade anti‐Cdt1 antibody specifically recognizes human Cdt1. (A). 293T cells were transfected with control or Cdt1 siRNA oligos or with empty vector (EV) or a vector expressing Flag‐Cdt1. Duplicate samples were ran on a gel and Western blot analysis was carried out using the anti‐Cdt1 homemade antibody and anti‐Flag as indicated. Dotted line indicates where the membrane was cut to incubate with the different antibodies. (B). 293T and U2OS cells were transfected with control or Cdt1 siRNAs and extracts were made for Western blot analysis with the indicated antibodies.
Supplemental Figure 2 No change in Cdt1 levels after overexpression of many ubiquitin or ubiquitin‐like hydrolases. 293T cells were transfected with expression plasmids for the indicated hydrolases (or empty vector) and were left untreated or UV irradiated (40 J/m2). Cells were lysed 30 min after treatment and analyzed by Western blot with anti‐Cdt1 antibodies and β‐actin as loading control. The expression plasmids for (tagged) ubiquitin or ubiquitin‐like hydrolases were kindly provided by several collaborating laboratories.
Supplemental Figure 3 Increased Cdt1 levels after USP37 overexpression. 293T cells were transfected, lysed and analyzed as in Supplemental Figure 2.
Supplemental Figure 4 Flag‐USP37 overexpression rescues Cdt1 levels in USP37 depleted cells and does not affect MCM7 ubiquitination (A) 293T cells were transfected 2 times with siRNA against Luc or USP37#1 and with siRNA resistant Flag‐USP37 and were treated with thymidine 24 h, released for 6 h and incubated with Lovastatin 10 μM for extra 20 h before being analyzed by Western blot with the indicated antibodies (B) 293T cells were transfected with Flag‐USP37, treated with MG132 for 6 h or left untreated. Immunoprecipitations with control or anti‐Cdt1 antibodies were carried out using extracts of the Flag‐USP37 expressing cells. All samples were analyzed by Western blot with the anti‐Cdt1 antibody. (B) 293T cells were transfected when indicated with control, His‐Ubiquitin, wild type or catalytic inactive Flag‐USP37 plasmids. 20 h after transfection, cells were incubated with MG132 for 16 h before lysis under denaturing conditions. Western blotting analysis of input and His pull downs were performed with the indicated antibodies.


