Abstract
We report the main characteristics of ‘Actinomyces provencensis’ strain SN12T sp. nov., ‘Corynebacterium bouchesdurhonense’ strain SN14T sp. nov., ‘Corynebacterium provencense’ strain SN15T sp. nov. and ‘Xanthomonas massiliensis’ strain SN8T sp. nov., which were all isolated from stool samples from obese French patients.
Keywords: Culturomics, halophilic species, human gut microbiota, new species, taxonogenomics
The study of the human gut microbiota experienced a new revolution with culturomics, a new concept that enables exploring, as comprehensively as possible, the viable population of prokaryotes associated with the human gastrointestinal tract [1], [2]. In 2015, using this culturomics approach, we isolated four bacteria which were not identified by matrix-assisted desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Bremen, Germany) [1], [2]. These species were isolated by culture on Columbia agar supplemented with 5% sheep's blood (bioMérieux, Marcy l'Etoile, France) at 37°C under microaerophilic conditions using CampyGen (Thermo Scientific, Villebon-sur-Yvette, France) for 48 hours, all from stool samples from obese French patients. All subjects provided written informed consent, and the study was validated by the ethics committee of the IFR48 Federative Research Institute under number 09-022. The strains are anaerobic and facultative, but growth was better under aerobic conditions. The failure of identification by MALDI-TOF MS of these four strains led us to sequence their 16S rRNA gene using fD1-rP2 primers as described previously using a 3130-XL sequencer (Applied Biosciences, Saint-Aubin, France) [3].
Strain SN12 showed colonies bright grey and nonhaemolytic, with a diameter of 0.5 to 1 mm on 5% sheep's blood–enriched Columbia agar (bioMérieux) after 48 hours of incubation under aerobic conditions. It is a facultative anaerobic bacteria. Cells were Gram-positive, rod-shaped bacilli, spore forming and motile. The strain had positive catalase activity, but oxidase was negative. Strain SN12 had a 16S rRNA gene sequence identity of 95.04% with Actinomyces odontolyticus strain JCM 14871 (Genbank accession number NR_114395), the phylogenetically closest species with standing in nomenclature [4] (Fig. 1). This <98.65% similarity leads us to putatively classify SN12 as a member of the Actinomyces genus and belonging to Actinobacteria phylum [5]. Therefore, we propose the creation of the new species ‘Actinomyces provencensis’ (pro.ven.cen′sis, N.L. adj. neut., from Provence, the region in France, where the strain was isolated). SN12T is the type strain of the species ‘Actinomyces provencensis’.
Fig. 1.
Phylogenetic tree showing positions of ‘Actinomyces provencensis’ strain SN12T relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained with Kimura two-parameter models using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 1.000 times to generate majority consensus tree. Scale bar indicates 1% nucleotide sequence divergence.
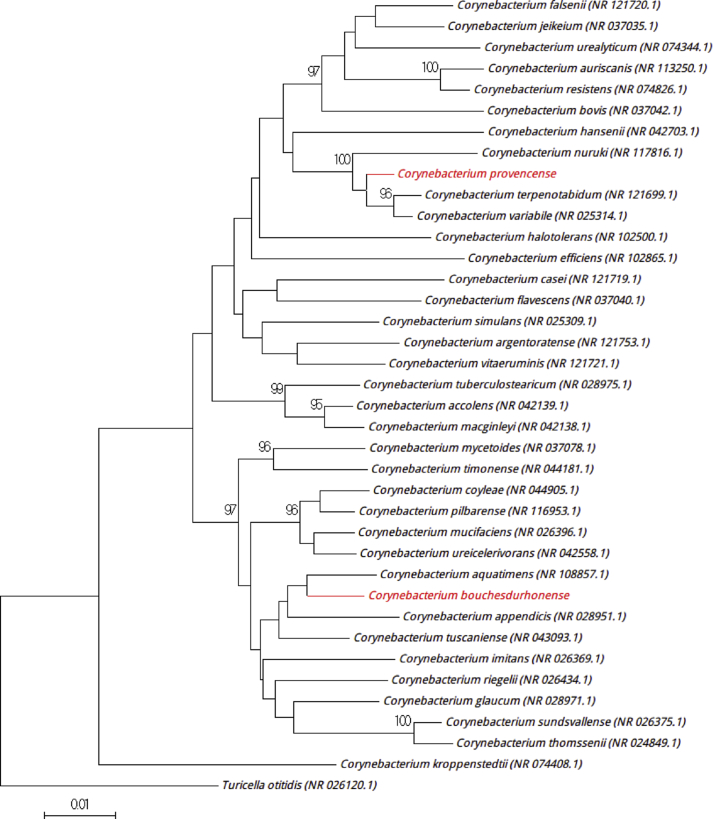
The colonies of the strain SN14 on 5% sheep's blood–enriched Columbia agar (bioMérieux) at 37°C under aerobic conditions were white, circular, nonhaemolytic and opaque, 1 mm in diameter after 48 hours of incubation. Bacterial cells were Gram positive, rod shaped, facultative anaerobic, spore forming and nonmotile. Under electron microscopy, cells had a mean length and diameter of 0.8 and 1.8–5.0 μm respectively. Catalase and oxidase activities were positive. The strain SN14 exhibited a 97.3% sequence identity with Corynebacterium tuscaniense strain ISS-5309 (NR_043093), the phylogenetically closest species with standing in nomenclature (Fig. 2). Corynebacterium tuscaniense strain ISS-5309 was isolated from culture of human blood [6]. This <98.65% similarity with its phylogenetically closest species with a validly published name leads us to putatively classify SN14 as a representative strain of a new species within the genus Corynebacterium in the Actinobacteria phylum [5]. On the basis of the 16S rRNA gene sequence results and its phylogenetical position with the others closest species, we propose the creation of the new species ‘Corynebacterium bouchesdurhonense’ (bou.ches.du.rho.nen′se, N.L. adj. neut., from Bouches du Rhône, the county in France where the strain was isolated). SN14T is the type strain of the species ‘Corynebacterium bouchesdurhonense’.
Fig. 2.
Phylogenetic tree showing position of ‘Corynebacterium bouchesdurhonense’ strain SN14T and ‘Corynebacterium provencense’ strain SN15T relative to other phylogenetically close neighbours. Alignment and phylogenetic inferences were done as described in Fig. 1.
The colonies of the strain SN15 were milky white, circular, nonhaemolytic and opaque, 1 to 1.5 mm in diameter after 48 hours of incubation on 5% sheep's blood–enriched Columbia agar (bioMérieux) under aerobic conditions. Cells were Gram positive, rod shaped (0.6 × 1.2–1.7 μm), facultative anaerobic, motile and spore forming. Strain SN15 exhibited catalase positivity and oxidase negativity. The strain SN15 had a 16S rRNA gene sequence identity of 97.3% with Corynebacterium tuscaniense strain ISS-5309 (NR_043093), the phylogenetically closest species with standing in nomenclature [6] (Fig. 2). This <98.65% similarity leads us to putatively classify SN15 as a new species in the Actinobacteria phylum [5]. From these results, we propose the creation of the new species ‘Corynebacterium provencense’ (pro.ven.cen′se, N.L. adj. neut., from Procence, the region in France where the strain was isolated). SN15T is the type strain of the species ‘Corynebacterium provencense’.
The colonies of the strain SN8 were khaki green, circular, viscous and nonhaemolytic, 1 to 2 mm in diameter on 5% sheep's blood–enriched Columbia agar (bioMérieux) under aerobic conditions after 48 hours of incubation. Bacterial cells were Gram negative, rod shaped, motile and non–spore forming. Observed under electron microscopy, bacterial cells measure 0.6 μm in diameter and 1.8–2.0 μm in length. Catalase activity test was positive. Oxidase activity test was negative. The strain SN8 had a 16S rRNA gene sequence identity of 98.08% with Xanthomonas campestris strain ATCC 33913 (NR_074936), the phylogenetically closest species with standing in nomenclature [7] (Fig. 3), which putatively classifies strain SN8 as a member of a new species within the genus Xanthomonas in the phylum Proteobacteria [5]. Thus, we propose the creation of the new species ‘Xanthomonas massiliensis’ (ma.ssi.li.en′sis, N.L. adj. neut., from Massilia, the antic name of Marseille, France, where the strain was isolated). SN8T is the type strain of the species ‘Xanthomonas massiliensis’.
Fig. 3.
Phylogenetic tree showing position of ‘Xanthomonas massiliensis’ strain SN8T relative to other phylogenetically close neighbours. Alignment and phylogenetic inferences were done as described in Fig. 1.
MALDI-TOF MS spectrum
The MALDI-TOF MS spectra of these species are available online (http://mediterranee-infection.com/article.php?laref=256&titre=urms-database).
Nucleotide sequence accession number
The 16S r RNA gene sequence was deposited in GenBank under the following accession numbers: ‘Actinomyces provencensis’ strain SN12T (LN881591), ‘Corynebacterium bouchesdurhonense’ strain SN14T (LN881599), ‘Corynebacterium provencense’ strain SN15T (LN890283) and ‘Xanthomonas massiliensis’ strain SN8T (LN881611).
Deposit in a culture collection
The strains were deposited in the Collection de Souches de l'Unité des Rickettsies (CSUR, WDCM 875) under numbers P2166 (‘Actinomyces provencensis’ strain SN12T), P2067 (‘Corynebacterium bouchesdurhonense’ strain SN14T), P2161 (‘Corynebacterium provencense’ strain SN15T) and P2129 (‘Xanthomonas massiliensis’ strain SN8T).
Acknowledgement
This study was funded by the Fondation Méditerranée Infection.
Conflict of Interest
None declared.
References
- 1.Lagier J.C., Hugon P., Khelaifia S., Fournier P.E., La Scola B., Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skerman V.B.D., McGowan V., Sneath P.H.A. Approved lists of bacterial names. Int J Syst Evol Microbiol. 1980;30:225–420. [Google Scholar]
- 5.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 6.Riegel P., Creti R., Mattei R., Nieri A., Hunolstein C.V. Isolation of Corynebacterium tuscaniae sp. nov. from blood cultures of a patient with endocarditis. J Clin Microbiol. 2006;44:307–312. doi: 10.1128/JCM.44.2.307-312.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vauterin L., Hoste B., Kersters K., Swings J. Reclassification of Xanthomonas. Int J Syst Bacteriol. 1995;45:472–489. [Google Scholar]