Abstract
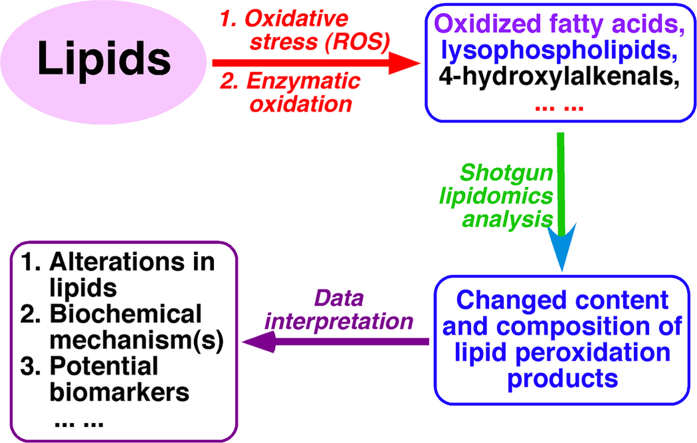
Multi-dimensional mass spectrometry-based shotgun lipidomics (MDMS-SL) has made profound advances for comprehensive analysis of cellular lipids. It represents one of the most powerful tools in analyzing lipids directly from lipid extracts of biological samples. It enables the analysis of nearly 50 lipid classes and thousands of individual lipid species with high accuracy/precision. The redox imbalance causes oxidative stress, resulting in lipid peroxidation, and alterations in lipid metabolism and homeostasis. Some lipid classes such as oxidized fatty acids, 4-hydroxyalkenal species, and plasmalogen are sensitive to oxidative stress or generated corresponding to redox imbalance. Therefore, accurate assessment of these lipid classes can provide not only the redox states, but also molecular insights into the pathogenesis of diseases. This review focuses on the advances of MDMS-SL in analysis of these lipid classes and molecular species, and summarizes their recent representative applications in biomedical/biological research. We believe that MDMS-SL can make great contributions to redox biology through substantiating the aberrant lipid metabolism, signaling, trafficking, and homeostasis under oxidative stress-related condition.
Abbreviations: AMPP, N-(4-amiomethylphenyl)pyridinium; aPC, plasmanylcholine; aPE, plasmanylethanolamine; dPC, phosphatidylcholine; dPE, phosphatidylethanolamine; DEGs, differently expressed genes; EETs, epoxyeicosatrienoic acids; 4-HDDE, 4-hydroxy-2E,6Z-dodecadienal; HDL, high-density lipoprotein; 4-HDTE, 4-hydroxy-dodecatrienal; HETE, hydroxyeicosatetraenoic acid; 4-HHE, 4-hydroxy-2E-hexenal; 4-HNDE, 4-hydroxy-nondienal; 4-HNE, 4-hydroxy-2E-nonenal; HPLC, high performance liquid chromatography; MDMS-SL, multi-dimensional MS-based shotgun lipidomics; MS, mass spectrometry; Ox-HDL, oxidized HDL; PG, phosphatidylglycerol; PLA2, phospholipase A2; pPC, plasmenylcholine; pPE, plasmenylethanolamine; PS, phosphatidylserine; PUFAs, polyunsaturated fatty acids; ROS, reactive oxygen species; SLE, systemic lupus erythematosus; TAG, triglycerides
Keywords: Shotgun lipidomics, Oxidative stress, Lipid peroxidation, Oxidized fatty acids, 4-hydroxyalkenal species, Plasmalogen
Graphical abstract
Highlights
-
•
Shotgun lipidomic analysis of lipid peroxidation products.
-
•
Determination of alternated lipid species by shotgun lipidomics.
-
•
Shotgun lipidomics revealed possible biochemical mechanisms and biomarkers.
-
•
Assessment of redox biology in SLE disease and neurotoxicity by shotgun lipidomics.
1. Introduction
1.1. Lipid, lipidomics, and multi-dimensional mass spectrometry-based shotgun lipidomics
Lipids involve numerous biological processes and play many crucial roles in cellular functions, including cellular barriers, signaling, energy storage, and growth and survival. Therefore, perturbations in lipid homeostasis are closely associated to diverse phenotypes and disease states, such as obesity, diabetes, cancer, neurodegenerative disorders, and autoimmune diseases [1], [2], [3], [4], [5]. It is clear that investigation of lipid alterations can make great contributions to elucidate disease pathogenesis and discover potential biomarkers for early diagnosis of diseases and drug efficacy.
However, cellular lipids are highly diverse and complex. These lipids consist of different polar head groups, backbones, and various aliphatic chains which connect to backbones in different linkages. The aliphatic chains are different in length (i.e., different numbers of carbon atoms), different degrees of unsaturation, different locations of double bonds, and potential branches, etc [6]. It is predicted that tens of thousands to hundreds of thousands possible lipid species exist in cellular lipidome at the levels of amol/mg to nmol/mg of protein [7], [8]. Moreover, many new lipid species are continually being discovered [9].
In addition to their diversities in chemical structures, lipids are also highly dynamic. Lipid molecular species and compositions are varied from species, cell types, cellular organelles, and subcellular membrane, leaflets of membrane bilayers, and membrane microdomains (i.e., rafts) [10]. They are dynamically changing with life cycle, environmental conditions, or pathological perturbation [11], [12], [13]. Furthermore, their metabolism is interwoven via numerous pathways and networks [14].
After genomics and proteomics, lipidomics was also coined in early 2000 as a disciplinary field to investigate all lipids in a large scale and at the levels of intact molecular species [15], [16]. It has been demonstrated that lipidomics analysis serves as a powerful tool for understanding the biochemical mechanisms underlying lipid-related disease processes through quantifying the changes of individual lipid classes, subclasses and molecular species and identifying the altered pathways and networks underlying changed lipid classes, subclasses, and molecular species [14]. In recent years, lipidomics has made great advances due to the rapid development in novel analysis strategies and approaches [17], and new instruments and techniques of MS [18].
Depending on whether LC separation is coupled to a mass spectrometer, MS-based lipidomics can be classified into two major categories, i.e., LC-MS-based and direct infusion-based lipidomics. The latter one is usually termed shotgun lipidomics. Based on the unique features and the mass spectrometers employed, at least three different approaches of shotgun lipidomics, including tandem MS-based, high mass accuracy MS-based, and multi-dimensional MS-based (MDMS-SL), have been developed and well documented in the literature [6], [19].
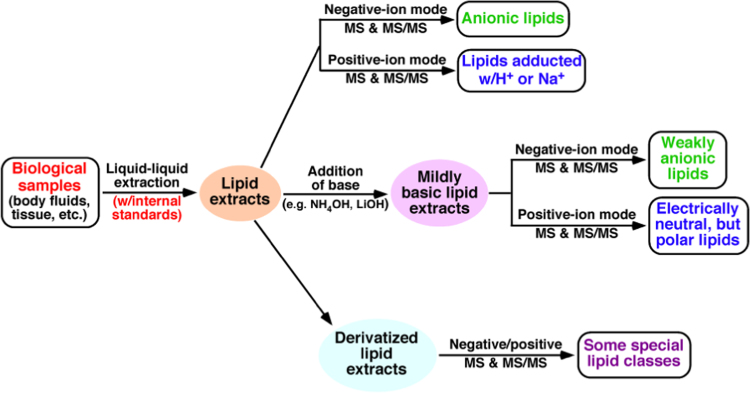
MDMS-SL, which maximally exploits the unique chemical and physical properties inherent in discrete lipid classes enabling for analysis of very low-abundance levels, overcomes the majority of the limitations of other shotgun lipidomics approaches and has many significant advantages [6], [19]. At its current development, this platform allows the researchers to analyze nearly 50 lipid classes and thousands of individual lipid species with >90% accuracy [17], including identification of fatty acyl positional isomers [20] and fatty acid isomers [21] of the species. The typical workflow of MDMS-SL analysis of biological samples is schematically illustrated in Fig. 1.
Fig. 1.
Schematic illustration of the workflow of MDMS-SL for analysis of cellular lipidomes directly from crude extracts of biological samples [20], [21].
1.2. Overview of conventional methodology for analysis of lipids indicative of oxidative stress
Oxidative stress arises due to redox imbalance between the oxidative and anti-oxidative systems of cells and/or tissues. It results in the over productions of oxidative free radical and reactive oxygen species (ROS), which could attack cellular proteins, lipids, and nucleic acids leading to cellular dysfunction [22]. A large number of studies have demonstrated that oxidative stress is tightly associated with many diseases [22], [23], [24], [25], [26]. The changed levels of a variety of lipids are indicative of oxidative stress. For example, 4-hydroxyalkenal species and eicosanoids are lipid peroxidation products generated through complex enzymatic and nonenzymatic reactions [27], [28], and plasmalogens serve as one of endogenous antioxidants [29]. Many methods have been developed to measure the levels of these classes of lipids present in biological fluid and tissue samples, including immunoassay methods [30], separation-based MS methods (i.e., GC-MS, normal phase LC-MS, and reversed phase LC-MS) [31], and shotgun lipidomics [17]. All of these methods have advantages and limitations. Enzyme-linked immunosorbent assays are popular and simple to perform and accessible to most of laboratories, but the questionable specificity when so many isomers can cross-react with the various antibodies has limited their utilities [32]. HPLC methods coupled with UV detection are only useful for analysis of relatively high abundance metabolites [33]. More attention should be paid to uniform derivatization and interfering substances when using derivatization coupled with fluorescence detection [34]. Although MS-based approaches are sometimes complex and require sample preparation involving extraction and purification, they are the “gold standard” methods for allowing researchers to measure all of the different species [35], [36], [37]. Compared with shotgun lipidomics, separation-based MS methods need more internal standards [38]. Chromatographic separation leads to the differential elusion of internal standard and analytes, whereas co-elution of an analyte with its standard is very important to compensate for matrix effects and varying ionization efficiencies during gradient elution [38]. Both 4-hydroxyalkenal species and eicosanoids are instable and would decompose during the procedure of chromatographic separation. In addition, separation-based MS methods are generally time-consuming, which is not suitable for larger sample profiling [39].
In last few years, MDMS-SL for analysis of these oxidative stress-related lipid species has been developed rapidly. Although a serial of reviews on shotgun lipidomics have been published [6], [14], [17], they have different focuses. In this review, we first introduce the principles of MDMS-SL for 4-hydroxyalkenal species, eicosanoids, and plasmalogens, and then summarize their recent representative applications under different disease states. Finally, we discuss the advantages of MDMS-SL for analysis of these oxidative-stress-related lipids and how to further explore the methods in future work.
2. Quantitative analysis of lipid molecular species associated with redox biology by shotgun lipidomics
2.1. Shotgun lipidomics of 4-hydroxyalkenal species
Among the cellular components, phospholipids, which usually contain high levels of polyunsaturated fatty acids (PUFAs), are the most sensitive to be attacked by ROS induced by oxidative stress [40]. 4-Hydroxyalkenal species, a class of α, β-unsaturated aldehyde, are considered to be one of the most reactive electrophilic end-products of lipid peroxidation generated from PUFAs. Therefore, the levels of 4-hydroxyalkenal species are the indicator of the oxidative stress of a biological system. 4-Hydroxyalkenal species are highly reactive due to the presence of three reactive functional groups in the chemical structure: a carbonyl group on C1, a conjugated double bond (alkene) between C2 and C3, and a hydroxyl group on C4 (Fig. 2). These groups make the 4-hydroxyalkenal species highly reactive toward nucleophilic thiol and amino groups, and can readily form covalent adducts with various cellular (macro)molecules (e.g., lipids, proteins, and nucleic acids). The interactions could lead to inhibition of protein and DNA synthesis, dysregulation of enzyme activities, alteration in mitochondrial coupling, etc [41], [42]. Moreover, compared with free radicals, these species are relative stable and can easily escape from initial generation sites to propagate the oxidative injury, thus to serve as “toxic second massagers”. Many investigations have demonstrated that the accumulation of those 4-hydroxyalkenal adducts is linked to the pathogenesis of diverse cardiovascular diseases, liver inflammation, renal failure, autoimmune diseases, and neurodegenerative disorders, as well as aging [43], [44], [45], [46], [47], [48].
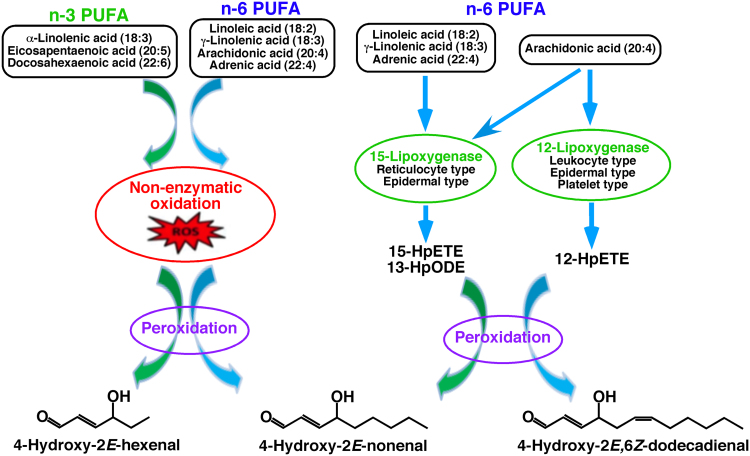
Fig. 2.
Mechanisms of 4-hydroxyalkenal species production [49]. PUFA abbreviates polyunsaturated fatty acid. 15-HpETE, 12-HpETE, and 13-HpODE stand for 15-hydroperoxyeicosatetraenoic acid, 12-hydroperoxyeicosatetraenoic acid, and 13-hydroperoxyoctadecadienoic acid.
4-Hydroxyalkenal species are generated from a variety of complex enzymatic and nonenzymatic reactions during diverse physiological and pathophysiological processes as illustrated in Fig. 2 [49]. Briefly, 4-hydroxy-2E-nonenal (4-HNE), usually the most common species among the 4-hydroxyalkenal family in most of biological systems, is generated by peroxidation of n-6 PUFAs, such as arachidonic, linoleic, and other n-6 PUFAs, as well as their 15-lipxoygenase metabolites (i.e., 15-hydroperoxyeicosatetraenoic acid, and 13-hydroperoxyoctadecadienoic acid); 4-hydroxy-2E,6Z-dodecadienal (4-HDDE) is exclusively produced from 12-lipxoygenase metabolites of arachidonic acid (e.g., 12-hydroperoxyeicosatetraenoic acid); 4-hydroxy-2E-hexenal (4-HHE) is the peroxidative product of n-3 PUFAs (e.g., linolenic, eicosapentaenoic, and docosahexaenoic acids); and other 4-hydroxyalkenal species (e.g., 4-hydroxynondienal (4-HNDE) and 4-hydroxydodecatrienal (4-HDTE)) can also be produced through peroxidation of PUFAs [50].
Although the enzymatic and nonenzymatic reactions of lipid peroxidation are very complicated, it has been demonstrated that the enzymatic pathway of lipid peroxidation is strictly consistent with the particular expression of the various types of lipoxygenases [49], [51], [52]. Therefore, the changes of different 4-hydroxyalkenal species truly reflect the activities of corresponding lipoxygenases and the extent of oxidative stress.
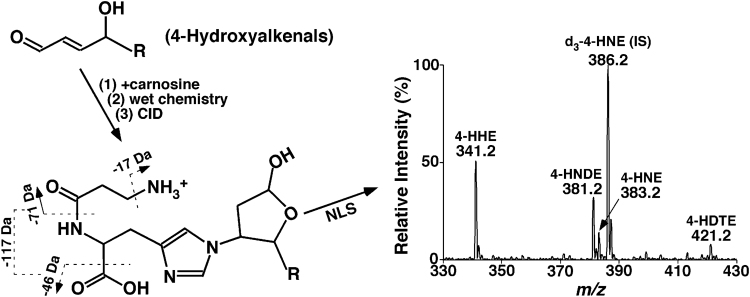
To understand the formation and biological roles of 4-hydroxyalkenals, an important aspect is to detect and measure the levels of these species in biological systems. 4-Hydroxyalkenals are present in very low abundance, very unstable, and unionizable. Varieties of methodologies have been developed and used to identify and quantify individual 4-hydroxyalkenal species and their adducts [47], [53], [54], [55], [56], [57], [58]. Our group exploited the facile Michael adduct of carnosine (β-alanyl-L-histidine) with 4-hydroxyalkenals to develop a sensitive, simple, high-throughput, and accurate MDMS-SL method for identification and quantification of these compounds [50].
Briefly, a stable isotope-labeled d3-4-HNE as an internal standard is added before lipid extraction and reaction of the carnosine with 4-hydroxyalkenals is relatively quick. After working up the reaction mixture with a modified Bligh-Dyer procedure, the aqueous phase is retained, evaporated, resuspended with a small volume of methanol/water (1:1 by volume), and directly infused into a mass spectrometer for characterization and quantitation of 4-hydroxyalkenals [50]. It has been demonstrated that (1) in ESI-MS analysis, the ionization efficiency of the generated carnosine adducts is significantly enhanced in comparison to the native form of 4-hydroxyalkenal species in the positive-ion mode; (2) product ion MS analyses of the formed adducts display many abundant, informative, and characteristic fragment ions, which can be used not only to identify and quantify 4-hydroxyalkenals, but also to discover other novel 4-hydroxyalkenals (e.g., 4-HNDE and 4-HDTE); and (3) the derivatization can stabilize 4-hydroxyalkenals for prevention of their losses during analysis. Based on the advantages of the derivatization, a sensitive and facile MDMS-SL method has been developed for quantification of 4-hydroxyalkenal species directly from chloroform extracts (Fig. 3). This method has been exploited to determine the mass levels of different 4-hydroxyalkenal species in various biological samples from clinic and animal models, such as the heart, kidney, liver, brain, skeletal muscle, and plasma/serum [23], [50], [59], [60].
Fig. 3.
Schematic illustration of the MDMS-SL method for analysis of 4-hydroxyalkenal species directly from lipid extracts of biological samples [50]. D3−4-HNE is added as an internal standard (IS). 4-HHE, 4-HNDE, 4-HNE, and 4-HDTE stand for 4-hydroxy-2E-hexenal, 4-hydroxy-2E-nondienal, 4-hydroxy-2E-nonenal, and 4-hydroxy-dodecatrienal, respectively.
2.2. Fatty acidomics of oxidized fatty acids for analysis of eicosanoids, docosanoids, nitrosylated FA, halogenated FA, and others
Fatty acids (FAs) are a large family of lipids including nonesterified (i.e., saturated and unsaturated) and modified FAs (e.g., all oxidized, halogenated, and other modified FAs). All of them contain at least one carboxylic group and a long aliphatic chain. Like 4-hydroxyalkenals, oxidized FAs are also generated by a variety of complex enzymatic and nonenzymatic oxidations of PUFAs [61]. These lipids function as cellular signaling molecules in diverse critical physiological and pathological processes, including cell growth and development, blood coagulation, kidney function, immune responses, and, most notably, inflammation [62], [63], [64], [65]. In addition, some isomers of them might have cooperative/opposite biological properties, so it would be difficult to explain the molecular mechanisms by determining only a limited number of these molecules [66]. Simultaneously measuring a wide range of them could better understand their roles in different processes and provide a set of biomarkers for disease diagnosis or prognosis.
However, identification and quantification of these lipids are a huge challenge, because they (1) are present at extremely low concentration in biological systems; (2) are not very stable; and (3) have many isomeric species. In addition to the difference in chain length, number of double bonds, and locations of these double bonds in acyl chain, the PUFA can be oxidized in different positions of its acyl chain. Thus, there exist a huge number of oxidized FAs with similar chemical structures. Although many different methods for analysis of them with different applications have been developed, only a subgroup or a few subgroups of these lipids have been analyzed at a time in each method [32], [33], [37]. Moreover, numerous internal standards should be added for accurate quantification of these molecules [61].
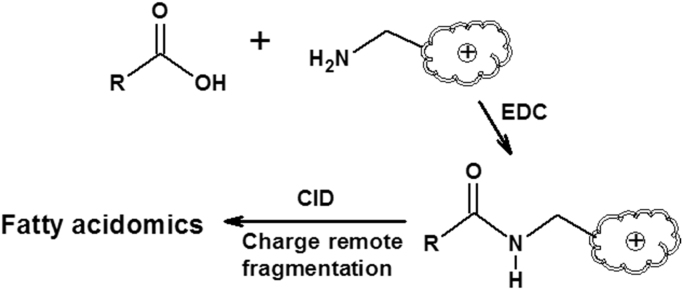
Based on the charge-fragmentation nature in mass spectrometry, we have developed a MDMS-SL based approach for global analysis of cellular lipid species containing a functional group of carboxylic acid, including identification of chain length, double bond number and locations, modified group(s) and locations, etc., and quantification of these species including individual isomeric species with only one internal standard per family (Fig. 4) [21].
Fig. 4.
Schematic illustration of an MDMS-SL approach for analysis of oxidized fatty acids which contains at least one functional group of carboxylic acid [21]. The amidation reaction is catalyzed by 1-ethyl-3-(3-dimethylaminopropyl)carbiodiimide (EDC).
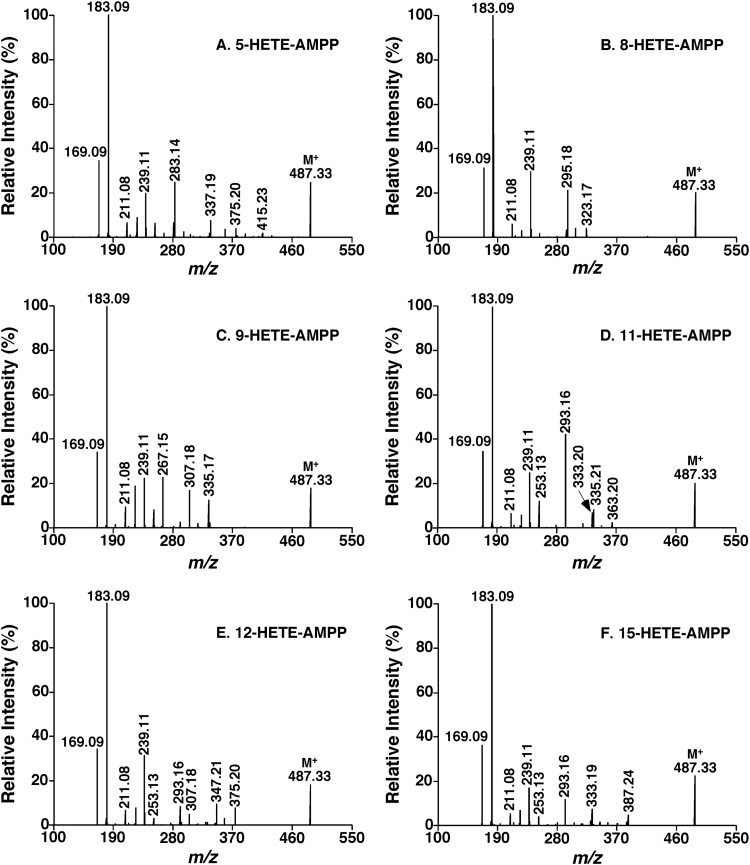
Specifically, a permanent charge site is introduced to the commonly-carried carboxylic acid of these lipids through an amidation reaction as previously reported [67]. The derivatization reaction significantly enhances the ionization efficiency and yields a lot of informative structural fragments in product ion MS analysis after collision-induced dissociation (CID). After a variety of charge-carried reagents have been tested, it has been found that all the derivatives with these reagents could yield informative charge-remote fragmentation patterns [21]. Furthermore, these fragmentation patterns could be used to identify the lipids containing a carboxylic acid including their isomers [21]. As an example, the product ion MS spectra of hydroxyeicosatetraenoic acid (HETE) isomeric derivatives with N-(4-amiomethylphenyl)pyridinium (AMPP) are shown in Fig. 5.
Fig. 5.
Product ion ESI-MS analysis of hydroxyeicosatetraenoic acid (HETE) isomers after derivatization with N-(4-amiomethylphenyl)pyridinium (AMPP) [21]. Product ion ESI-MS analysis of these derivatized HETE isomers at collision energy of 40 eV and collision gas pressure of 1 mTorr. The fragment ions at m/z 169 and 183 can be used for screening these isomers at the m/z position of molecular ions in the precursor-ion scans of m/z 169 and 189, respectively, and quantifying the total content of their mixture relative to a selected internal standard (e.g., a stable isotope labeled HETE). The fingerprints of the fragment ions between m/z 190 and 450 (i.e., m/z 239, 253, 267, 283, 293, 295, 307, 333, 335, 347, 375, 387) can be used to simulate a tandem MS spectrum of a mixture of the HETE isomers and determine their composition. Absolute amounts of individual HETE species can be derived from the total amount of the mixture and the composition.
Intriguingly, the fragmentation patterns of individual isomers are so different that the compositions of those isomers can be readily determined through simulating the tandem MS spectrum of the mixture of the isomers with the tandem MS spectra of individual isomers using multiple linear regression analysis [21]. Therefore, quantification of the identified lipid species containing carboxylic acid including isomers can be achieved by MS analysis only after derivatization without chromatographic separation. In the study, it has been demonstrated that the developed MDMS-SL approach works for analysis of almost all kinds of oxidized FAs, such as HETEs, diHETEs, epoxyeicosatrienoic acids (EETs), nitrosylated FAs, etc. Using this approach surely facilitates identification of the biochemical mechanisms underlying diverse pathological conditions [21].
2.3. Measurement of plasmalogen molecular species and metabolites for assessment of oxidative stress
Plasmalogens (1-O-alk-1′-enyl-2-acyl-glycerophospholipids) are a unique subclass of phospholipids characterized by the presence of a vinyl-ether bond at the sn-1 position, mainly including plasmenylcholine (pPC) and plasmenylethanolamine (pPE) [68]. Plasmalogens generally account approximately 10 mol% of the total phospholipid mass [29]. However, they could be very abundant in some organs [29], [69]. Plasmalogens play many crucial roles in cellular functions, including reservoirs for second messengers, working as endogenous antioxidants, as well as serving as essential structural components of the cellular membranes [29], [70]. It has been demonstrated that aberrant metabolism of plasmalogens is closely associated with insulin resistance [71], atherosclerosis [72], neurodegeneration (i.e., Alzheimer's disease, Parkinson's disease, Down syndrome, and Niemann-Pick type C) [5], [73], fatty alcohol accumulation in fibroblasts [74], aging [75], and ischemia-reperfusion injury [76], etc.
The presence of another isomeric ether-containing subclass of lipids in PE and PC often complicates the identification of plasmalogens in a cellular lipid mixture. So far, varieties of methods have been developed to identify and quantify the plasmalogens [77], [78], [79], [80]. Some of them exploit the fact that the vinyl ether bond of plasmalogens is very acid-labile. Through comparison of total molecular species profiles of both acid-treated and non-treated lipid extracts, plasmalogens can be distinguished from alkyl subclass species [68].
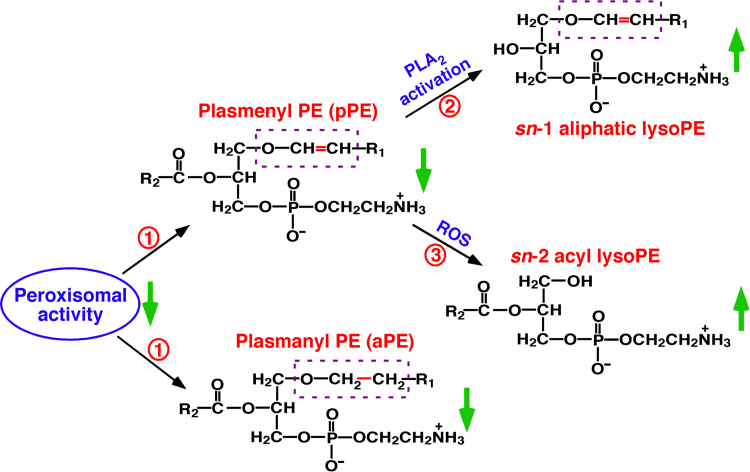
In biological systems, the total content of plasmalogens rarely increases since their biosynthesis is strictly regulated by peroxisomes [14]. Three major metabolic pathways can lead to the decreased plasmalogen content, including peroxisomal dysfunction, phospholipase A2 (PLA2) activation, and oxidative stress [14]. Interestingly, aberrant metabolism of each pathway has unique products and/or manifestations as shown in Fig. 6. For instance, reduced plasmalogen levels with elevated lysoplasmalogens suggest the activation of PLA2 [81]. Decreased plasmalogen contents with increased choline and ethanolamine lysoglycerophospholipids (particularly those containing PUFAs) indicate the increased oxidative stress [82]. Meanwhile, determining other indicators of oxidative stress, such as 4-hydroxyalkenals [50], chlorinated lipids [83], and/or eicosanoids [35], can provide further supports of this pathway. Finally, decreased plasmalogen contents accompanying the decreased levels of ether-containing plasmalogen counterparts [84] and minimal changes of choline and ethanolamine lysoglycerophospholipids suggest the dysfunction of peroxisomes. Accordingly, measuring the levels of plasmalogen species and relative metabolites by MDMS-SL can identify the corresponding pathophysiological pathways leading to the changed plasmalogen homeostasis [23].
Fig. 6.
Illustration of three major metabolic pathways responsible for plasmalogen reduction. These include peroxisomal dysfunction (Pathway 1), phospholipase A2 activation (Pathway 2), and oxidative stress (Pathway 3) [14]. Determining the levels of the plasmalogen species and relative metabolites through MDMS-SL can readily identify the corresponding aberrant pathway. PLA2 and ROS stand for phospholipase A2 and reactive oxygen species, respectively.
3. Representative applications of shotgun lipidomics for studying redox biology
As enhancing the coverage for lipid analysis, shotgun lipidomics becomes a powerful tool in determining altered lipid content in diverse states [85], such as revealing the underlying mechanism(s) for the changes of lipid homeostasis [86], discovering biomarkers for disease diagnosis or prognosis for fatty liver [87], testing the efficacy of new drugs [88], [89], supporting the exploration of new treatments [12], and further confirming the results from other “omics” (e.g., genomics, proteomics, and transcriptomics) with model systems (i.e., animals, cells, plants, bacteria, etc.) [90], [91]. A large number of reviews on the applications of shotgun lipidomics have been presented for varieties of biological studies with different emphasis [6], [14], [17], [19], [38], [68], [92]. Herein, recent representative applications of shotgun lipidomics for studying redox biology, especially those recently conducted by our group, are summarized.
3.1. Oxidative stress in systemic lupus erythematosus
Systemic lupus erythematosus (SLE) is a deadly autoimmune disease with a diversity of clinical features, mainly occurring in women at their childbearing ages [93]. The exact pathological mechanism(s) of SLE remain unknown and are usually attributed to dysfunction of immunocytes and genetic and/or environmental factors [94]. Oxidative stress is elevated in SLE patients due to mitochondrial dysfunction in lupus T cells, resulting in accumulation of ROS and depletion of reduced glutathione [95]. The increased oxidative stress is tightly associated with SLE pathogenesis [96]. However, common indicators of oxidative stress are nonspecific and non-representative of all SLE symptoms.
Oxidative stress interferes with lipid metabolism of SLE. In addition to showing a “lupus pattern” of dyslipoproteinemia characterized with high levels of very-low-density lipoprotein (VLDL) cholesterol and triglycerides (TAG), and low levels of high-density lipoprotein (HDL) cholesterol [97], SLE patients always exhibit higher serum levels of oxidized HDL (ox-HDL) [98]. The aberrant metabolism of lipids and lipoproteins may account for cardiovascular diseases pathogenesis of SLE, which is a major causal factor for the high morbidity and mortality of SLE patients [99]. Moreover, oxidative modification of self-antigens is present in SLE pathology due to oxidative stress pathology. There are more than 100 different autoantibodies in SLE, many of which are related to lipids and/or enzymes involving lipid metabolism [100]. Therefore, determining alterations in these lipids is very important for further understanding the mechanism(s) underpinning SLE pathogenesis.
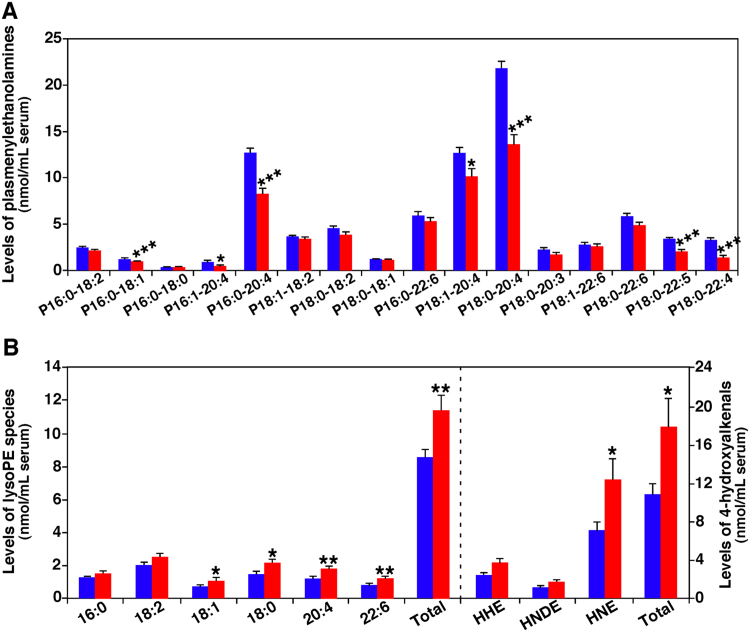
Our MDMS-SL technology has been employed to analyze the lipids of serum collected from 30 SLE patients and 30 controls, respectively [23]. It has been found that, in addition to the elevated TAG content as previously known [101], lipidomics revealed the significant decrease of all identified pPC and pPE species. Compared with control, the total losses of these lipids accounted for approximately 26 and 21 mol%, respectively (Fig. 7A shows the changes of PE species). However, other two subclasses of both PE (e.g., phosphatidylethanolamine (dPE), and plasmanylethanolamine (aPE)) and PC (i.e., phosphatidylcholine (dPC), and plasmanylcholine (aPC)) changed minimally.
Fig. 7.
Comparison of the contents of representative serum lipids between SLE patients and controls. Plasmalogen PE species (Panel A), and ethanolamine lysoglycerophospholipids (lysoPE) and 4-hydroxyalkenal species (Panel B) present in serum lipid extracts from SLE patients (n=30, red bar) and healthy controls (n=30, blue bar) were determined by MDMS-SL as previously described [23]. The data represent means±SEM from different individuals (n=30). *p<0.05, **p<0.01, and ***p<0.001 compared with those in the control group. The prefix “p” in Panel A is used to abbreviate plasmalogen PE species. HHE, HNDE, and HNE stand for 4-hydroxy-2E-hexenal, 4-hydroxy-2E-nondienal, and 4-hydroxy-2E-nonenal, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
In order to understand the mechanism(s) for the reduction of plasmalogen content, the levels of lysoPE species were also determined by MDMS-SL in the study. It has been found that significant increase of sn-2 acyl type lysoPE in SLE patients, especially those containing PUFAs (Fig. 7B). As schematically shown in Fig. 6, the decreased pPE content with breakage of sn-1 vinyl ether bond is related to lipid peroxidation [14]. To further support the identified pathway, the levels of 4-hydroxyalkenal species were also determined by MDMS-SL (Fig. 7B). As anticipated, the total level of these species substantially increased in the patients, clearly indicating the elevated oxidative stress at the SLE state. As mentioned above, HNE can propagate oxidative stress to other intracellular organelles, which may explain the varieties of clinical symptoms of SLE patients. Additionally, multivariate and multiple analysis showed significant correlations among SLE disease index, proinflammatory cytokine IL-10 level, and contents of 4-hydroxyalkenals and pPE species, suggesting that the increased mass levels of 4-hydroxyalkenals and decreased pPE content may be associated with disease severity and production of autoantibodies in SLE patients.
In summary, shotgun lipidomics analysis has not only revealed the changes of lipid species in SLE patients, but also provided insights into underlying biochemical mechanism(s) for these alterations. Moreover, the results of lipidomics are significantly associated with disease severity, autoimmunity, different clinical symptoms, and/or complications of SLE, indicating that the changed levels of those lipid species may serve as novel potential biomarkers for diagnose and/or prognosis of SLE.
3.2. The adverse effects of anesthetics on brain lipids
Considerable studies evidence the adverse effects of general anesthetics on the brain development in preclinical young animal models, including accelerated apoptosis, alterations in dendritic morphology, as well as cognitive and behavioral changes [102], [103], [104], [105]. Thus, the safety of children exposed to anesthetics receives more public concern, especially prolonged exposure (e.g., >8 h). So far, the underlying mechanism(s) responsible for the neurotoxicity and injury remain elusive.
Sevoflurane is a volatile anesthetic that is widely used in children as it has many advantages over other intravenous or inhalation anesthetics, such as more comfort, lower blood/gas solubility, less irritation to airway, pleasant smell, notably, and less adverse effects [106]. However, lots of evidence suggests that sevoflurane can also cause neuronal apoptosis and behavioral dysfunctions [107], [108], [109]. Therefore, evaluation of the relevant alterations on the developing brain after prolonged exposure of sevoflurane is of importance.
In a recent study, after infant monkeys were exposed to clinically-relevant concentration of sevoflurane for 9 h, their frontal cortical tissues were harvested for DNA microarray, shotgun lipidomics, Luminex protein, and histological assays [110]. DNA analysis revealed that sevoflurane exposure caused broad induction of differently expressed genes (DEGs) in the monkey brain. The DEGs are closely associated with nervous system development and functions, neurological diseases, and cell death/survival. Intriguingly, some of them directly involve in the networks of lipid metabolism, such as fatty acid and steroid metabolism [110]. Lipid is the most abundant component in the brain other than water, thus, its homeostasis and signaling play vital roles in cell functions and viability [111]. Therefore, lipidomics analysis could provide insights into the mechanism(s) underpinning the induced neurotoxicity and find new biomarkers.
MDMS-SL analysis has been performed on the lipid extracts of tissue samples from the brain cortices of monkeys with or without exposure to sevoflurane. It has been found that substantial alterations in content and composition of multiple lipid classes and molecular species in brain frontal cortices of infant monkeys exposed to sevoflurane for 9 h are present [110]. For example, the total mass level of PE species are significantly reduced from 131.4±1.9 nmol/mg protein in controls to 110.6±11.4 nmol/mg protein in sevoflurane-treated group (p<0.05). Moreover, several of these reduced species are plasmalogen species (Fig. 8), which act as one of the natural antioxidants and are more susceptible to ROS in the body. The markedly elevated levels of lysoPE species and 4-HNE strongly support the existence of increased oxidative stress after prolonged sevoflurane exposure.
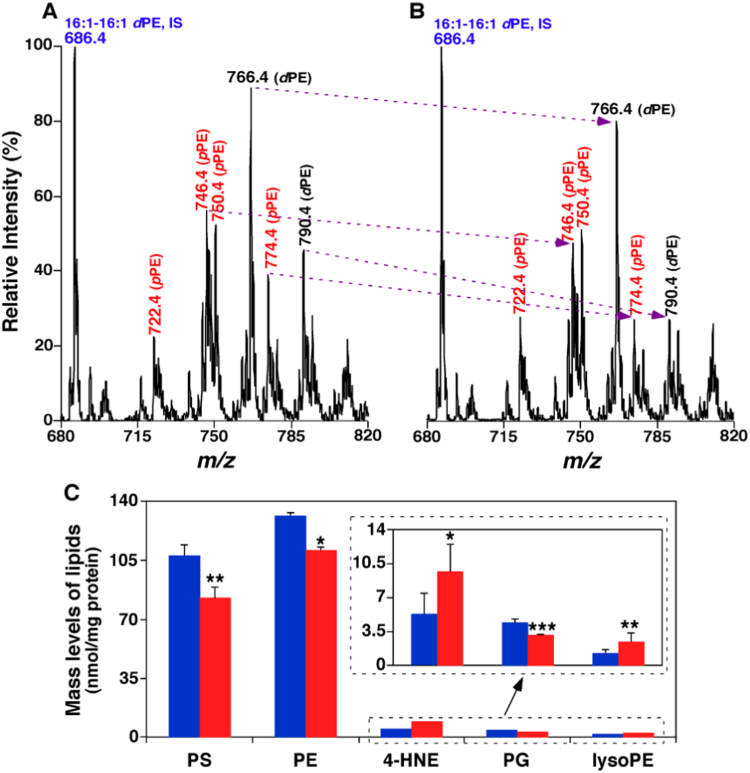
Fig. 8.
MDMS-SL analysis determines the altered levels of lipids present in brain cortices of infant monkeys exposed to sevoflurane for 9 h, indicating the existence of oxidative stress [110]. Panels A and B show the reduction of the mass levels of ethanolamine glycerophospholipid (PE) including many plasmalogen PE species (highlighted with red) by MS analysis. Panel C summarizes the total mass levels of the altered lipid classes induced by exposure to sevoflurane (Red bar) in comparison to controls (Blue bar) [110]. *p<0.05; **p<0.01; and ***p<0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Lipidomics also shows the significant reduced levels of phosphatidylserine (PS) and phosphatidylglycerol (PG) species (p<0.01 and p<0.001, respectively) in the brain of infant monkeys after sevoflurane exposure (Fig. 8). PS is located in the inner leaflet of plasma membrane, possessing unique physical and biochemical properties. PS involves in the recruitment and activation of numerous enzymes that are associated with neuronal survival, neurite growth, and synaptogenesis [112], [113], [114]. Therefore, the disruption of PS could lead to subsequent neuronal dysfunction and/or degeneration. PG species, largely present in mitochondrial membrane, play a vital role in the maintenance of mitochondrial structure and functions [115]. The reduction of PG contents in sevoflurane-treated group suggests that mitochondria might be impaired in sevoflurane exposure. As the “power house” of a cell, mitochondrion is a critical generator of ROS. The impaired mitochondria may be associated with increased oxidative stress including the elevation of HNE and lysoPE. Consistently, Fluoro-Jade C staining shows more degenerating neurons in sevoflurane-treated group with the higher levels of cytokines in comparison to the controls [110].
In summary, shotgun lipidomics reveals that mitochondria could be the major target of prolonged sevoflurane exposure, resulting in the increased oxidative stress and the alteration in lipid homeostasis. Therefore, lipidomics not only demonstrates the results supporting the findings from DNA, protein, and histological analysis, but also provides deeper insights into the mechanism underpinning the sevoflurane-induced neurotoxicity. In addition, the specific changed lipids may be sensitive biomarkers for the early detection of anesthetic-induced neuronal damage.
4. Summary and perspective
Although the lipidomics discipline has only emerged for a short period, it has been greatly advanced due to the rapid development of modern technologies and plays an important role in a vast variety of biological research. The MDMS-SL platform maximally exploits the unique chemical and physical properties inherent in diverse lipid classes and has been proved to be one of the most powerful tools for lipid analysis. The increased oxidative stress induced by redox imbalance can generate a variety of lipid peroxidation products or might involve in alterations in lipid metabolism. Usually, these oxidative stress-related lipid metabolites are present in very low abundance, instable, isomeric, and unionizable, all of which are huge challenges for their analyses. Through chemical derivatization, sensitive, simple, high-throughput, and accurate methods based on the principles of MDMS-SL for identification and quantification of these metabolites such as 4-hydroxyalkenal species and oxidized fatty acids have been developed and proved to be very powerful and useful.
Shotgun lipidomics has been used for studies on a variety of pathophysiological conditions including SLE disease and anesthetic-induced neurotoxicity. The findings from the research strongly indicate that lipidomics not only identifies the presence of lipid peroxidation in different states, which may be novel potential sensitive biomarkers for disease diagnosis and/or prognosis, but also provides insights into the underpinning biochemical mechanism(s) of the pathophysiological states. We believe that shotgun lipidomics can make great contributions to the studies of redox biology.
Conflict of interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was partially supported by National Institute of General Medical Sciences Grant R01 GM105724, Science and Technology Planning Project of Zhejiang Province (No. 2015C37025), National Natural Science Foundation of China (No. 81373633), and the National Basic Research Program “973” of China (No. 2014CB543001).
References
- 1.Wymann M.P., Schneiter R. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 2.Perry R.J., Samuel V.T., Petersen K.F., Shulman G.I. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510:84–91. doi: 10.1038/nature13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zalba S., Hagen T.L. Ten. Cell membrane modulation as adjuvant in cancer therapy. Cancer Treat. Rev. 2017;52:48–57. doi: 10.1016/j.ctrv.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mapstone M., Cheema A.K., Fiandaca M.S., Zhong X., Mhyre T.R., MacArthur L.H., Hall W.J., Fisher S.G., Peterson D.R., Haley J.M., Nazar M.D., Rich S.A., Berlau D.J., Peltz C.B., Tan M.T., Kawas C.H., Federoff H.J. Plasma phospholipids identify antecedent memory impairment in older adults. Nat. Med. 2014;20:415–418. doi: 10.1038/nm.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han X. Lipid alterations in the earliest clinically recognizable stage of Alzheimer's disease: implication of the role of lipids in the pathogenesis of Alzheimer's disease. Curr. Alzheimer Res. 2005;2:65–77. doi: 10.2174/1567205052772786. [DOI] [PubMed] [Google Scholar]
- 6.Han X., Yang K., Gross R.W. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shevchenko A., Simons K. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 8.Han X., Jiang X. A review of lipidomic technologies applicable to sphingolipidomics and their relevant applications. Eur. J. Lipid Sci. Technol. 2009;111:39–52. doi: 10.1002/ejlt.200800117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yore M.M., Syed I., Moraes-Vieira P.M., Zhang T., Herman M.A., Homan E.A., Patel R.T., Lee J., Chen S., Peroni O.D., Dhaneshwar A.S., Hammarstedt A., Smith U., McGraw T.E., Saghatelian A., Kahn B.B. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159:318–332. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira M.S., de Oliveira D.N., de Oliveira R.N., Allegretti S.M., Catharino R.R. Screening the life cycle of Schistosoma mansoni using high-resolution mass spectrometry. Anal. Chim. Acta. 2014;845:62–69. doi: 10.1016/j.aca.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Aviram R., Manella G., Kopelman N., Neufeld-Cohen A., Zwighaft Z., Elimelech M., Adamovich Y., Golik M., Wang C., Han X., Asher G. Lipidomics analyses reveal temporal and spatial lipid organization and uncover daily oscillations in intracellular organelles. Mol. Cell. 2016;62:636–648. doi: 10.1016/j.molcel.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 13.He Q., Han X. Cardiolipin remodeling in diabetic heart. Chem. Phys. Lipids. 2014;179:75–81. doi: 10.1016/j.chemphyslip.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Han X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016;12:668–679. doi: 10.1038/nrendo.2016.98. [DOI] [PubMed] [Google Scholar]
- 15.Han X., Gross R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Lagarde M., Geloen A., Record M., Vance D., Spener F. Lipidomics is emerging. Biochim. Biophys. Acta. 1634;2003:61. doi: 10.1016/j.bbalip.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 17.Wang M., Wang C., Han R.H., Han X. Novel advances in shotgun lipidomics for biology and medicine. Prog. Lipid Res. 2016;61:83–108. doi: 10.1016/j.plipres.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanksby S.J., Mitchell T.W. Advances in mass spectrometry for lipidomics. Annu. Rev. Anal. Chem. 2010;3:433–465. doi: 10.1146/annurev.anchem.111808.073705. [DOI] [PubMed] [Google Scholar]
- 19.Yang K., Han X. Lipidomics: techniques, applications, and outcomes related to biomedical sciences. Trends Biochem. Sci. 2016;41:954–969. doi: 10.1016/j.tibs.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han X., Gross R.W. Structural determination of lysophospholipid regioisomers by electrospray ionization tandem mass spectrometry. J. Am. Chem. Soc. 1996;118:451–457. [Google Scholar]
- 21.Wang M., Han R.H., Han X. Fatty acidomics: global analysis of lipid species containing a carboxyl group with a charge-remote fragmentation-assisted approach. Anal. Chem. 2013;85:9312–9320. doi: 10.1021/ac402078p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi G., Cortopassi G. Oxidative stress in inherited mitochondrial diseases. Free Radic. Biol. Med. 2015;88:10–17. doi: 10.1016/j.freeradbiomed.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu C., Zhou J., Yang S., Li H., Wang C., Fang X., Fan Y., Zhang J., Han X., Wen C. Oxidative stress leads to reduction of plasmalogen serving as a novel biomarker for systemic lupus erythematosus. Free Radic. Biol. Med. 2016;101:475–481. doi: 10.1016/j.freeradbiomed.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Li S., Tan H.Y., Wang N., Zhang Z.J., Lao L., Wong C.W., Feng Y. The role of oxidative stress and antioxidants in liver diseases. Int. J. Mol. Sci. 2015;16:26087–26124. doi: 10.3390/ijms161125942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lappas M., Hiden U., Desoye G., Froehlich J., Hauguel-deMouzon S., Jawerbaum A. The role of oxidative stress in the pathophysiology of gestational diabetes mellitus. Antioxid. Redox Signal. 2011;15:3061–3100. doi: 10.1089/ars.2010.3765. [DOI] [PubMed] [Google Scholar]
- 26.Latina A., Viticchie G., Lena A.M., Piro M.C., Annicchiarico-Petruzzelli M., Melino G., Candi E. DeltaNp63 targets cytoglobin to inhibit oxidative stress-induced apoptosis in keratinocytes and lung cancer. Oncogene. 2016;35:1493–1503. doi: 10.1038/onc.2015.222. [DOI] [PubMed] [Google Scholar]
- 27.Wang D., Dubois R.N. Eicosanoids and cancer. Nat. Rev. Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalleau S., Baradat M., Gueraud F., Huc L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013;20:1615–1630. doi: 10.1038/cdd.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braverman N.E., Moser A.B. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta. 1822;2012:1442–1452. doi: 10.1016/j.bbadis.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog. Lipid Res. 2003;42:318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 31.Harkewicz R., Dennis E.A. Applications of mass spectrometry to lipids and membranes. Annu. Rev. Biochem. 2011;80:301–325. doi: 10.1146/annurev-biochem-060409-092612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldbart A.D., Krishna J., Li R.C., Serpero L.D., Gozal D. Inflammatory mediators in exhaled breath condensate of children with obstructive sleep apnea syndrome. Chest. 2006;130:143–148. doi: 10.1378/chest.130.1.143. [DOI] [PubMed] [Google Scholar]
- 33.Maskrey B.H., Bermudez-Fajardo A., Morgan A.H., Stewart-Jones E., Dioszeghy V., Taylor G.W., Baker P.R., Coles B., Coffey M.J., Kuhn H., O'Donnell V.B. Activated platelets and monocytes generate four hydroxyphosphatidylethanolamines via lipoxygenase. J. Biol. Chem. 2007;282:20151–20163. doi: 10.1074/jbc.M611776200. [DOI] [PubMed] [Google Scholar]
- 34.Yue H., Strauss K.I., Borenstein M.R., Barbe M.F., Rossi L.J., Jansen S.A. Determination of bioactive eicosanoids in brain tissue by a sensitive reversed-phase liquid chromatographic method with fluorescence detection. J. Chromatogr. B. 2004;803:267–277. doi: 10.1016/j.jchromb.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 35.Ecker J. Profiling eicosanoids and phospholipids using LC-MS/MS: principles and recent applications. J. Sep. Sci. 2012;35:1227–1235. doi: 10.1002/jssc.201200056. [DOI] [PubMed] [Google Scholar]
- 36.Prakash C., Zhang J.Y., Falck J.R., Chauhan K., Blair I.A. 20-Hydroxyeicosatetraenoic acid is excreted as a glucuronide conjugate in human urine. Biochem. Biophys. Res. Commun. 1992;185:728–733. doi: 10.1016/0006-291x(92)91686-k. [DOI] [PubMed] [Google Scholar]
- 37.Murphy R.C., Barkley R.M., Zemski Berry K., Hankin J., Harrison K., Johnson C., Krank J., McAnoy A., Uhlson C., Zarini S. Electrospray ionization and tandem mass spectrometry of eicosanoids. Anal. Biochem. 2005;346:1–42. doi: 10.1016/j.ab.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 38.Wang M., Wang C., Han X. Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-what, how and why? Mass Spectrom. Rev. 2016 doi: 10.1002/mas.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mesaros C., Lee S.H., Blair I.A. Targeted quantitative analysis of eicosanoid lipids in biological samples using liquid chromatography-tandem mass spectrometry. J. Chromatogr. B. 2009;877:2736–2745. doi: 10.1016/j.jchromb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luczaj W., Gegotek A., Skrzydlewska E. Antioxidants and HNE in redox homeostasis. Free Radic. Biol. Med. 2016 doi: 10.1016/j.freeradbiomed.2016.11.033. [DOI] [PubMed] [Google Scholar]
- 41.Pizzimenti S., Ciamporcero E., Daga M., Pettazzoni P., Arcaro A., Cetrangolo G., Minelli R., Dianzani C., Lepore A., Gentile F., Barrera G. Interaction of aldehydes derived from lipid peroxidation and membrane proteins. Front. Physiol. 2013;4:242. doi: 10.3389/fphys.2013.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong H., Yin H. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: focusing on mitochondria. Redox Biol. 2015;4:193–199. doi: 10.1016/j.redox.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zarkovic K. 4-hydroxynonenal and neurodegenerative diseases. Mol. Asp. Med. 2003;24:293–303. doi: 10.1016/s0098-2997(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 44.Kurien B.T., Hensley K., Bachmann M., Scofield R.H. Oxidatively modified autoantigens in autoimmune diseases. Free Radic. Biol. Med. 2006;41:549–556. doi: 10.1016/j.freeradbiomed.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 45.Minko I.G., Kozekov I.D., Harris T.M., Rizzo C.J., Lloyd R.S., Stone M.P. Chemistry and biology of DNA containing 1,N(2)-deoxyguanosine adducts of the alpha,beta-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem. Res. Toxicol. 2009;22:759–778. doi: 10.1021/tx9000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castro J.P., Jung T., Grune T., Siems W. 4-Hydroxynonenal (HNE) modified proteins in metabolic diseases. Free Radic. Biol. Med. 2016 doi: 10.1016/j.freeradbiomed.2016.10.497. [DOI] [PubMed] [Google Scholar]
- 47.Andringa K.K., Udoh U.S., Landar A., Bailey S.M. Proteomic analysis of 4-hydroxynonenal (4-HNE) modified proteins in liver mitochondria from chronic ethanol-fed rats. Redox Biol. 2014;2:1038–1047. doi: 10.1016/j.redox.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ravi S., Johnson M.S., Chacko B.K., Kramer P.A., Sawada H., Locy M.L., Wilson L.S., Barnes S., Marques M.B., Darley-Usmar V.M. Modification of platelet proteins by 4-hydroxynonenal: potential mechanisms for inhibition of aggregation and metabolism. Free Radic. Biol. Med. 2016;91:143–153. doi: 10.1016/j.freeradbiomed.2015.10.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riahi Y., Cohen G., Shamni O., Sasson S. Signaling and cytotoxic functions of 4-hydroxyalkenals. Am. J. Physiol. Endocrinol. Metab. 2010;299:E879–E886. doi: 10.1152/ajpendo.00508.2010. [DOI] [PubMed] [Google Scholar]
- 50.Wang M., Fang H., Han X. Shotgun lipidomics analysis of 4-hydroxyalkenal species directly from lipid extracts after one-step in situ derivatization. Anal. Chem. 2012;84:4580–4586. doi: 10.1021/ac300695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guichardant M., Bacot S., Moliere P., Lagarde M. Hydroxy-alkenals from the peroxidation of n-3 and n-6 fatty acids and urinary metabolites. Prostaglandins Leukot. Essent. Fat. Acids. 2006;75:179–182. doi: 10.1016/j.plefa.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 52.Riahi Y., Sin-Malia Y., Cohen G., Alpert E., Gruzman A., Eckel J., Staels B., Guichardant M., Sasson S. The natural protective mechanism against hyperglycemia in vascular endothelial cells: roles of the lipid peroxidation product 4-hydroxydodecadienal and peroxisome proliferator-activated receptor delta. Diabetes. 2010;59:808–818. doi: 10.2337/db09-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Kuijk F.J., Siakotos A.N., Fong L.G., Stephens R.J., Thomas D.W. Quantitative measurement of 4-hydroxyalkenals in oxidized low-density lipoprotein by gas chromatography-mass spectrometry. Anal. Biochem. 1995;224:420–424. doi: 10.1006/abio.1995.1060. [DOI] [PubMed] [Google Scholar]
- 54.Kawai Y., Takeda S., Terao J. Lipidomic analysis for lipid peroxidation-derived aldehydes using gas chromatography-mass spectrometry. Chem. Res. Toxicol. 2007;20:99–107. doi: 10.1021/tx060199e. [DOI] [PubMed] [Google Scholar]
- 55.Williams T.I., Lovell M.A., Lynn B.C. Analysis of derivatized biogenic aldehydes by LC tandem mass spectrometry. Anal. Chem. 2005;77:3383–3389. doi: 10.1021/ac048265+. [DOI] [PubMed] [Google Scholar]
- 56.Warnke M.M., Wanigasekara E., Singhal S.S., Singhal J., Awasthi S., Armstrong D.W. The determination of glutathione-4-hydroxynonenal (GSHNE), E-4-hydroxynonenal (HNE), and E-1-hydroxynon-2-en-4-one (HNO) in mouse liver tissue by LC-ESI-MS. Anal. Bioanal. Chem. 2008;392:1325–1333. doi: 10.1007/s00216-008-2383-3. [DOI] [PubMed] [Google Scholar]
- 57.Uchida K., Szweda L.I., Chae H.Z., Stadtman E.R. Immunochemical detection of 4-hydroxynonenal protein adducts in oxidized hepatocytes. Proc. Natl. Acad. Sci. USA. 1993;90:8742–8746. doi: 10.1073/pnas.90.18.8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hartley D.P., Kolaja K.L., Reichard J., Petersen D.R. 4-Hydroxynonenal and malondialdehyde hepatic protein adducts in rats treated with carbon tetrachloride: immunochemical detection and lobular localization. Toxicol. Appl. Pharmacol. 1999;161:23–33. doi: 10.1006/taap.1999.8788. [DOI] [PubMed] [Google Scholar]
- 59.He Q., Wang M., Harris N., Han X. Tafazzin knockdown interrupts cell cycle progression in cultured neonatal ventricular fibroblasts. Am. J. Physiol. Heart Circ. Physiol. 2013;305:H1332–H1343. doi: 10.1152/ajpheart.00084.2013. [DOI] [PubMed] [Google Scholar]
- 60.Lai L., Wang M., Martin O.J., Leone T.C., Vega R.B., Han X., Kelly D.P. A role for peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1) in the regulation of cardiac mitochondrial phospholipid biosynthesis. J. Biol. Chem. 2014;289:2250–2259. doi: 10.1074/jbc.M113.523654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Massey K.A., Nicolaou A. Lipidomics of oxidized polyunsaturated fatty acids. Free Radic. Biol. Med. 2013;59:45–55. doi: 10.1016/j.freeradbiomed.2012.08.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lagarde M., Chen P., Vericel E., Guichardant M. Fatty acid-derived lipid mediators and blood platelet aggregation. Prostaglandins Leukot. Essent. Fat. Acids. 2010;82:227–230. doi: 10.1016/j.plefa.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 63.Nicolaou A., Mauro C., Urquhart P., Marelli-Berg F. Polyunsaturated fatty acid-derived lipid mediators and T cell function. Front. Immunol. 2014;5:75. doi: 10.3389/fimmu.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hao C.M., Breyer M.D. Physiologic and pathophysiologic roles of lipid mediators in the kidney. Kidney Int. 2007;71:1105–1115. doi: 10.1038/sj.ki.5002192. [DOI] [PubMed] [Google Scholar]
- 65.Smith A.N., Muffley L.A., Bell A.N., Numhom S., Hocking A.M. Unsaturated fatty acids induce mesenchymal stem cells to increase secretion of angiogenic mediators. J. Cell Physiol. 2012;227:3225–3233. doi: 10.1002/jcp.24013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobrian A.D., Lieb D.C., Cole B.K., Taylor-Fishwick D.A., Chakrabarti S.K., Nadler J.L. Functional and pathological roles of the 12- and 15-lipoxygenases. Prog. Lipid Res. 2011;50:115–131. doi: 10.1016/j.plipres.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bollinger J.G., Thompson W., Lai Y., Oslund R.C., Hallstrand T.S., Sadilek M., Turecek F., Gelb M.H. Improved sensitivity mass spectrometric detection of eicosanoids by charge reversal derivatization. Anal. Chem. 2010;82:6790–6796. doi: 10.1021/ac100720p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han X., Gross R.W. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of the cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 69.Han X., Holtzman D.M., McKeel D.W., Jr. Plasmalogen deficiency in early Alzheimer's disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J. Neurochem. 2001;77:1168–1180. doi: 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 70.Nagan N., Zoeller R.A. Plasmalogens: biosynthesis and functions. Prog. Lipid Res. 2001;40:199–229. doi: 10.1016/s0163-7827(01)00003-0. [DOI] [PubMed] [Google Scholar]
- 71.Tonks K.T., Coster A.C., Christopher M.J., Chaudhuri R., Xu A., Gagnon-Bartsch J., Chisholm D.J., James D.E., Meikle P.J., Greenfield J.R., Samocha-Bonet D. Skeletal muscle and plasma lipidomic signatures of insulin resistance and overweight/obesity in humans. Obesity. 2016;24:908–916. doi: 10.1002/oby.21448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rasmiena A.A., Barlow C.K., Stefanovic N., Huynh K., Tan R., Sharma A., Tull D., de Haan J.B., Meikle P.J. Plasmalogen modulation attenuates atherosclerosis in ApoE- and ApoE/GPx1-deficient mice. Atherosclerosis. 2015;243:598–608. doi: 10.1016/j.atherosclerosis.2015.10.096. [DOI] [PubMed] [Google Scholar]
- 73.Han X. Multi-dimensional mass spectrometry-based shotgun lipidomics and the altered lipids at the mild cognitive impairment stage of Alzheimer's disease. Biochim. Biophys. Acta. 1801;2010:774–783. doi: 10.1016/j.bbalip.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rizzo W.B., Craft D.A., Judd L.L., Moser H.W., Moser A.B. Fatty alcohol accumulation in the autosomal recessive form of rhizomelic chondrodysplasia punctata. Biochem. Med. Metab. Biol. 1993;50:93–102. doi: 10.1006/bmmb.1993.1050. [DOI] [PubMed] [Google Scholar]
- 75.Lessig J., Fuchs B. Plasmalogens in biological systems: their role in oxidative processes in biological membranes, their contribution to pathological processes and aging and plasmalogen analysis. Curr. Med. Chem. 2009;16:2021–2041. doi: 10.2174/092986709788682164. [DOI] [PubMed] [Google Scholar]
- 76.Marsche G., Heller R., Fauler G., Kovacevic A., Nuszkowski A., Graier W., Sattler W., Malle E. 2-chlorohexadecanal derived from hypochlorite-modified high-density lipoprotein-associated plasmalogen is a natural inhibitor of endothelial nitric oxide biosynthesis. Arterioscler. Thromb. Vasc. Biol. 2004;24:2302–2306. doi: 10.1161/01.ATV.0000148703.43429.25. [DOI] [PubMed] [Google Scholar]
- 77.Maeba R., Nishimukai M., Sakasegawa S., Sugimori D., Hara H. Plasma/serum plasmalogens: methods of analysis and clinical significance. Adv. Clin. Chem. 2015;70:31–94. doi: 10.1016/bs.acc.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 78.Otoki Y., Nakagawa K., Kato S., Miyazawa T. MS/MS and LC-MS/MS analysis of choline/ethanolamine plasmalogens via promotion of alkali metal adduct formation. J. Chromatogr. B. 2015;1004:85–92. doi: 10.1016/j.jchromb.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 79.Fuchs B. Analytical methods for (oxidized) plasmalogens: methodological aspects and applications. Free Radic. Res. 2015;49:599–617. doi: 10.3109/10715762.2014.999675. [DOI] [PubMed] [Google Scholar]
- 80.Fhaner C.J., Liu S., Zhou X., Reid G.E. Functional group selective derivatization and gas-phase fragmentation reactions of plasmalogen glycerophospholipids. Mass Spectrom. 2013;2:S0015. doi: 10.5702/massspectrometry.S0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ford D.A., Hazen S.L., Saffitz J.E., Gross R.W. The rapid and reversible activation of a calcium-independent plasmalogen-selective phospholipase A2 during myocardial ischemia. J. Clin. Investig. 1991;88:331–335. doi: 10.1172/JCI115296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gross R.W. High plasmalogen and arachidonic acid content of canine myocardial sarcolemma: a fast atom bombardment mass spectroscopic and gas chromatography-mass spectroscopic characterization. Biochemistry. 1984;23:158–165. doi: 10.1021/bi00296a026. [DOI] [PubMed] [Google Scholar]
- 83.Wacker B.K., Albert C.J., Ford B.A., Ford D.A. Strategies for the analysis of chlorinated lipids in biological systems. Free Radic. Biol. Med. 2013;59:92–99. doi: 10.1016/j.freeradbiomed.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van den Bosch H., Schrakamp G., Hardeman D., Zomer A.W., Wanders R.J., Schutgens R.B. Ether lipid synthesis and its deficiency in peroxisomal disorders. Biochimie. 1993;75:183–189. doi: 10.1016/0300-9084(93)90076-5. [DOI] [PubMed] [Google Scholar]
- 85.Han X., Rozen S., Boyle S., Hellegers C., Cheng H., Burke J.R., Welsh-Bohmer K.A., Doraiswamy P.M., Kaddurah-Daouk R. Metabolomics in early Alzheimer's disease: identification of altered plasma sphingolipidome using shotgun lipidomics. PLoS One. 2011;6:e21643. doi: 10.1371/journal.pone.0021643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han X. Potential mechanisms contributing to sulfatide depletion at the earliest clinically recognizable stages of Alzheimer's disease: a tale of shotgun lipidomics. J. Neurochem. 2007;103(s1):171–179. doi: 10.1111/j.1471-4159.2007.04708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sa R., Zhang W., Ge J., Wei X., Zhou Y., Landzberg D.R., Wang Z., Han X., Chen L., Yin H. Discovering a critical transition state from nonalcoholic hepatosteatosis to nonalcoholic steatohepatitis by lipidomics and dynamical network biomarkers. J. Mol. Cell Biol. 2016;8:195–206. doi: 10.1093/jmcb/mjw016. [DOI] [PubMed] [Google Scholar]
- 88.Hu C., Wang Y., Fan Y., Li H., Wang C., Zhang J., Zhang S., Han X., Wen C. Lipidomics revealed idiopathic pulmonary fibrosis-induced hepatic lipid disorders corrected with treatment of baicalin in a murine model. AAPS J. 2015;17:711–722. doi: 10.1208/s12248-014-9714-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang A., Qiu S., Sun H., Zhang T., Guan Y., Han Y., Yan G., Wang X. Scoparone affects lipid metabolism in primary hepatocytes using lipidomics. Sci. Rep. 2016;6:28031. doi: 10.1038/srep28031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang C., Wang M., Zhou Y., Dupree J.L., Han X. Alterations in mouse brain lipidome after disruption of CST gene: a lipidomics study. Mol. Neurobiol. 2014;50:88–96. doi: 10.1007/s12035-013-8626-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li M., Baughman E., Roth M.R., Han X., Welti R., Wang X. Quantitative profiling and pattern analysis of triacylglycerol species in Arabidopsis seeds by electrospray ionization mass spectrometry. Plant J. 2014;77:160–172. doi: 10.1111/tpj.12365. [DOI] [PubMed] [Google Scholar]
- 92.Dehairs J., Derua R., Rueda-Rincon N., Swinnen J.V. Lipidomics in drug development, Drug discovery today. Technologies. 2015;13:33–38. doi: 10.1016/j.ddtec.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 93.Lisnevskaia L., Murphy G., Isenberg D. Systemic lupus erythematosus. Lancet. 2014;384:1878–1888. doi: 10.1016/S0140-6736(14)60128-8. [DOI] [PubMed] [Google Scholar]
- 94.Tsokos G.C. Systemic lupus erythematosus. N. Engl. J. Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 95.Doherty E., Oaks Z., Perl A. Increased mitochondrial electron transport chain activity at complex I is regulated by N-acetylcysteine in lymphocytes of patients with systemic lupus erythematosus. Antioxid. Redox Signal. 2014;21:56–65. doi: 10.1089/ars.2013.5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Perl A. Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nat. Rev. Rheumatol. 2013;9:674–686. doi: 10.1038/nrrheum.2013.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reichlin M., Fesmire J., Quintero-Del-Rio A.I., Wolfson-Reichlin M. Autoantibodies to lipoprotein lipase and dyslipidemia in systemic lupus erythematosus. Arthritis Rheumatol. 2002;46:2957–2963. doi: 10.1002/art.10624. [DOI] [PubMed] [Google Scholar]
- 98.Frostegard J., Svenungsson E., Wu R., Gunnarsson I., Lundberg I.E., Klareskog L., Horkko S., Witztum J.L. Lipid peroxidation is enhanced in patients with systemic lupus erythematosus and is associated with arterial and renal disease manifestations. Arthritis Rheumatol. 2005;52:192–200. doi: 10.1002/art.20780. [DOI] [PubMed] [Google Scholar]
- 99.Trager J., Ward M.M. Mortality and causes of death in systemic lupus erythematosus. Curr. Opin. Rheumatol. 2001;13:345–351. doi: 10.1097/00002281-200109000-00002. [DOI] [PubMed] [Google Scholar]
- 100.Sherer Y., Gorstein A., Fritzler M.J., Shoenfeld Y. Autoantibody explosion in systemic lupus erythematosus: more than 100 different antibodies found in SLE patients. Semin. Arthritis Rheumatol. 2004;34:501–537. doi: 10.1016/j.semarthrit.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 101.Borba E.F., Bonfa E., Vinagre C.G., Ramires J.A., Maranhao R.C. Chylomicron metabolism is markedly altered in systemic lupus erythematosus. Arthritis Rheumatol. 2000;43:1033–1040. doi: 10.1002/1529-0131(200005)43:5<1033::AID-ANR11>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 102.Davidson A.J., Disma N., de Graaff J.C., Withington D.E., Dorris L., Bell G., Stargatt R., Bellinger D.C., Schuster T., Arnup S.J., Hardy P., Hunt R.W., Takagi M.J., Giribaldi G., Hartmann P.L., Salvo I., Morton N.S., von Ungern Sternberg B.S., Locatelli B.G., Wilton N., Lynn A., Thomas J.J., Polaner D., Bagshaw O., Szmuk P., Absalom A.R., Frawley G., Berde C., Ormond G.D., Marmor J., McCann M.E. G.A.S. consortium, Neurodevelopmental outcome at 2 years of age after general anaesthesia and awake-regional anaesthesia in infancy (GAS): an international multicentre, randomised controlled trial. Lancet. 2016;387:239–250. doi: 10.1016/S0140-6736(15)00608-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jevtovic-Todorovic V., Hartman R.E., Izumi Y., Benshoff N.D., Dikranian K., Zorumski C.F., Olney J.W., Wozniak D.F. Early exposure to common anesthetic agents causes widespread neurodegeneration in the developing rat brain and persistent learning deficits. J. Neurosci. 2003;23:876–882. doi: 10.1523/JNEUROSCI.23-03-00876.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sanchez V., Feinstein S.D., Lunardi N., Joksovic P.M., Boscolo A., Todorovic S.M., Jevtovic-Todorovic V. General anesthesia causes long-term impairment of mitochondrial morphogenesis and synaptic transmission in developing rat brain. Anesthesiology. 2011;115:992–1002. doi: 10.1097/ALN.0b013e3182303a63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Paule M.G., Li M., Allen R.R., Liu F., Zou X., Hotchkiss C., Hanig J.P., Patterson T.A., Slikker W., Jr., Wang C. Ketamine anesthesia during the first week of life can cause long-lasting cognitive deficits in rhesus monkeys. Neurotoxicol. Teratol. 2011;33:220–230. doi: 10.1016/j.ntt.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liang G., Ward C., Peng J., Zhao Y., Huang B., Wei H. Isoflurane causes greater neurodegeneration than an equivalent exposure of sevoflurane in the developing brain of neonatal mice. Anesthesiology. 2010;112:1325–1334. doi: 10.1097/ALN.0b013e3181d94da5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lu Y., Wu X., Dong Y., Xu Z., Zhang Y., Xie Z. Anesthetic sevoflurane causes neurotoxicity differently in neonatal naive and Alzheimer disease transgenic mice. Anesthesiology. 2010;112:1404–1416. doi: 10.1097/ALN.0b013e3181d94de1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shen X., Liu Y., Xu S., Zhao Q., Guo X., Shen R., Wang F. Early life exposure to sevoflurane impairs adulthood spatial memory in the rat. Neurotoxicology. 2013;39:45–56. doi: 10.1016/j.neuro.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 109.Takaenoki Y., Satoh Y., Araki Y., Kodama M., Yonamine R., Yufune S., Kazama T. Neonatal exposure to sevoflurane in mice causes deficits in maternal behavior later in adulthood. Anesthesiology. 2014;120:403–415. doi: 10.1097/ALN.0000435846.28299.e7. [DOI] [PubMed] [Google Scholar]
- 110.Liu F., Rainosek S.W., Frisch-Daiello J.L., Patterson T.A., Paule M.G., Slikker W., Jr., Wang C., Han X. Potential Adverse effects of prolonged sevoflurane exposure on developing monkey brain: from abnormal lipid metabolism to neuronal damage. Toxicol. Sci. 2015;147:562–572. doi: 10.1093/toxsci/kfv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.O'Brien J.S., Sampson E.L. Lipid composition of the normal human brain: gray matter, white matter, and myelin. J. Lipid Res. 1965;6:537–544. [PubMed] [Google Scholar]
- 112.Leventis P.A., Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 113.Huang B.X., Akbar M., Kevala K., Kim H.Y. Phosphatidylserine is a critical modulator for Akt activation. J. Cell Biol. 2011;192:979–992. doi: 10.1083/jcb.201005100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim H.Y., Akbar M., Lau A., Edsall L. Inhibition of neuronal apoptosis by docosahexaenoic acid (22:6n-3). role of phosphatidylserine in antiapoptotic effect. J. Biol. Chem. 2000;275:35215–35223. doi: 10.1074/jbc.M004446200. [DOI] [PubMed] [Google Scholar]
- 115.Gonzalvez F., Gottlieb E. Cardiolipin: setting the beat of apoptosis. Apoptosis. 2007;12:877–885. doi: 10.1007/s10495-007-0718-8. [DOI] [PubMed] [Google Scholar]